Abstract
Spinal Muscular Atrophy (SMA), an autosomal recessive neuromuscular disorder, is a leading genetic cause of infant mortality. SMA is caused by the homozygous loss of Survival Motor Neuron-1 (SMN1). However, low, but essential, levels of SMN protein are produced by a nearly identical copy gene called SMN2. Detailed analysis of neuromuscular junctions in SMA mice has revealed a selective vulnerability in a subset of muscle targets, suggesting that while SMN is reduced uniformly, the functional deficits manifest sporadically. Additionally, in severe SMA models, it is becoming increasing apparent that SMA is not restricted solely to motor neurons. Rather, additional tissues including the heart, vasculature, and the pancreas contribute to the complete SMA-associated pathology. Recently, transgenic models have been utilized to examine the tissue-specific requirements of SMN, including selective depletion and restoration of SMN in motor neurons. To determine whether the cortical neuronal populations expressing the Emx-1 promoter are involved in SMA pathology, we generated a novel SMA mouse model in which SMN expression was specifically induced in Emx-1 expressing cortical neurons utilizing an Emx-1-Cre transgene. While SMN expression was robust in the central nervous system as expected, SMA mice did not live longer. Weight and time-to-right motor function were not significantly improved.
Keywords: Survival Motor Neuron (SMN), Spinal Muscular Atrophy (SMA), EMX-1, neurodegeneration, animal models of disease, central nervous system
Introduction
Spinal Muscular Atrophy (SMA) is caused by the homozygous loss of Survival Motor Neuron-1 (SMN1) (Crawford and Pardo 1996; Oskoui et al. 2007). Importantly, however, SMA is not due to complete absence of SMN, rather, it is due to dramatically reduced SMN levels. A nearly identical copy gene present in humans, SMN2, is a critical disease modifier that produces low amounts of SMN protein (Lefebvre et al. 1995; Rochette et al. 2001). The majority of SMN2-derived transcripts are alternatively spliced and produce a truncated and unstable isoform; therefore, SMN2 cannot fully compensate for the loss of SMN1 (Hofmann et al. 2000; Cartegni and Krainer 2002). Despite ubiquitous SMN expression, low levels of full-length SMN result in the preferential loss of lower motor neurons, leading to SMA development.
Recently, SMA animal models have been developed to examine the temporal and spatial role of SMN in SMA development. The human skeletal actin (HSA) and prion promoters (PrP) were used to drive SMN expression in severe SMA mice that typically die within the first week of life (Le et al. 2011). High levels of muscle expression from the HSA promoter failed to alter the life span of the animals; however, SMN expression driven by the pan-neuronal PrP cassette significantly extended survival beyond 150 days. The PrP cassette is widely expressed in the central nervous system including a broad range of neurons as well as astrocytes. Olig2-Cre mediated depletion of SMN within motor neurons and oligodendrocytes in an otherwise wildtype background led to an SMA phenotype that significantly impacted skeletal muscle and motor neuron pathology as well as motor neuron function (Park et al. 2010). Interestingly, however, Olig2-Cre mediated depletion of SMN did not lead to an early death similar to several SMA mouse models that express uniformly low SMN levels systemically. Hb9-driven restoration of SMN in motor neurons significantly improved the neuromuscular junction phenotypes in severe SMA mice; however, survival in Hb9-rescued mice was only extended ~ 5 days, to a total lifespan of~ 2 weeks (Gogliotti et al. 2012). Similarly, Nestin-mediated restoration of SMN in neuronal populations (including motor neurons) does mitigate central and peripheral synaptic defects, however, the average life span is only extended from ~2.5 days to ~10 days (Lee et al. 2012) These results suggest that while motor neurons are clearly important, additional cellular populations could contribute to the complex SMA pathology. Consistent with this, in severe SMA mice, a severe and progressive defect is present in proprioceptive reflexes that correlate with decreased function and number of synapses on motor neuron somata and proximal dendrites. (Mentis et al. 2011). These results and the identification of peripheral defects in SMA mice (Bevan et al. 2010; Heier et al. 2010; Shababi et al. 2010; Bowerman et al. 2012b; Somers et al. 2012) demonstrate that the comprehensive pathological picture within severe SMA contexts is remarkably complex and will likely encompass not only motor neurons, but additional cells within the central nervous system as well as the periphery.
To further examine the role of SMN within the central nervous system, we generated a novel SMA mouse model in which SMN expression was specifically restored in cortical neurons utilizing an Emx-1-Cre transgene (Guo et al. 2000; Chan et al. 2001). While SMN was expressed highly as anticipated, the overall pathology and survival of the SMA-Emx1-Cre mice was not significantly altered. While Emx-1 mediated induction of SMN does not alter the SMA phenotype, this work further delineates tissues which are and are not responsible for the complex SMA pathology.
Materials and Methods
Genotyping and Mouse Handling
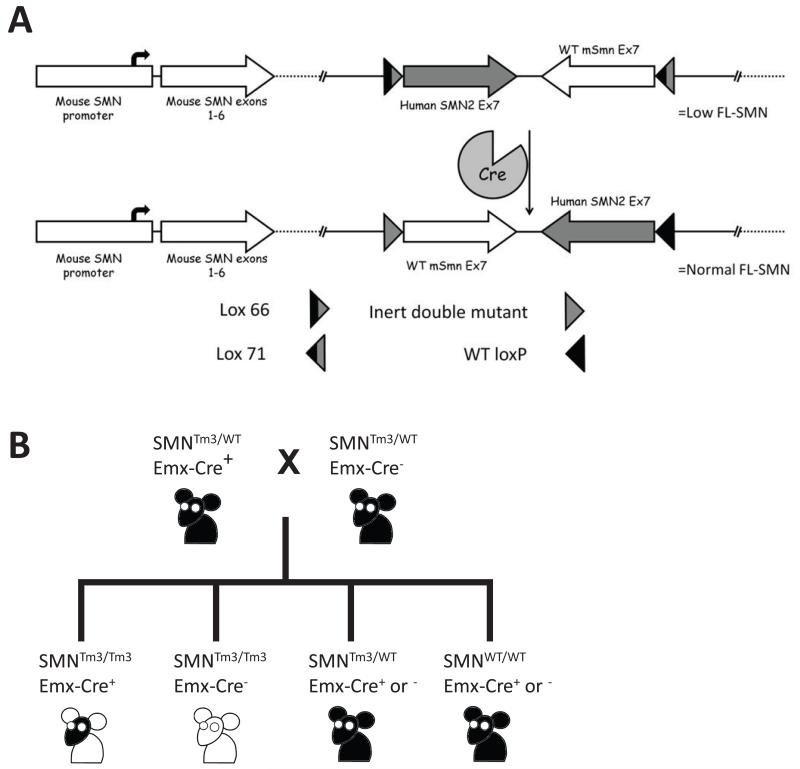
Animals were handled according to the University of Missouri Animal Care and Use Committee approved protocols. Mice were bred according to the schematic presented in Figure 1b. The day of birth was counted as PND1 with neonates being genotyped within 24 hours. Animals were genotyped using PCR conditions as previously described (Coady et al. 2008). SMA mice were raised with 2 heterozygous siblings. Additional heterozygous and wild-type animals were culled at the time of injection in experimental cages to control for litter size.
Figure 1.
Tissue Collection for PCR
In order to harvest tissue for DNA purification, the vertebral column was separated from the torso and the spinal cord was removed. The brain was dissected from the skull and divided into four equal sections. Total DNA was purified from the tissues using the Qiagen DNeasy Blood and Tissue Kit.
Western Blot Analysis
Tissues were harvested at indicated times and analysis was performed as previously described (Mattis et al. 2006; Shababi et al. 2011). Mouse monoclonal anti-SMN (BDBiolabs), 1:2,000, was used for SMN detection. Equal protein amounts were loaded as analyzed by Bradford protein assay (Bradford 1976).
Motor Function Analysis
Time to right was measured from P5 to death. Mice were placed on their backs and given a maximum of 30 seconds to right themselves on to their paws. Inability to right within 30 seconds was considered failure.
Results
To determine if restoration of SMN levels in a subset of neurons, specifically Emx-1 expressing cortical neurons, impacted disease development in a relatively severe model of SMA we utilized a previously developed mouse strain that expresses a targeted mutation within the Smn gene (JaxStock#7951). The Smn gene has been modified such that it consists of Smn genomic sequence fused to the 3′ end of the human SMN2 gene, allowing for a Cre-mediated recombination event that replaces the human SMN2 exon 7 sequence with the murine Smn exon 7 sequence (Lutz et al. 2011). The recombined allele effectively restored full-length SMN expression in a tissue-specific manner and does not generate the typical SMN2 alternative splicing event. In the current model, the Emx-1 promoter was used to drive expression of Cre recombinase (JaxStock#5628) (Guo et al. 2000; Gorski et al. 2002; Kummer et al. 2012), resulting in a restoration of full-length SMN protein in cortical neurons. In all other tissues, in the absence of Cre recombinase, the hybrid allele results in an mSmn gene analogous to human SMN2 and produces low levels of full length SMN protein. This promoter drives the expression of Cre within the cortical neurons of the motor cortex, but not in the lower motor neurons of the spinal cord (Fig. 1a). Mice homozygous for the non-recombined targeted mutation display a phenotype similar to that of the well-characterized SMNΔ7 mouse model (JaxStock#5025) (Le et al. 2005). Thus, the experimental mice express wild type levels of SMN only in the cortical neurons of the brain in the presence of Cre recombinase (Fig. 1b).
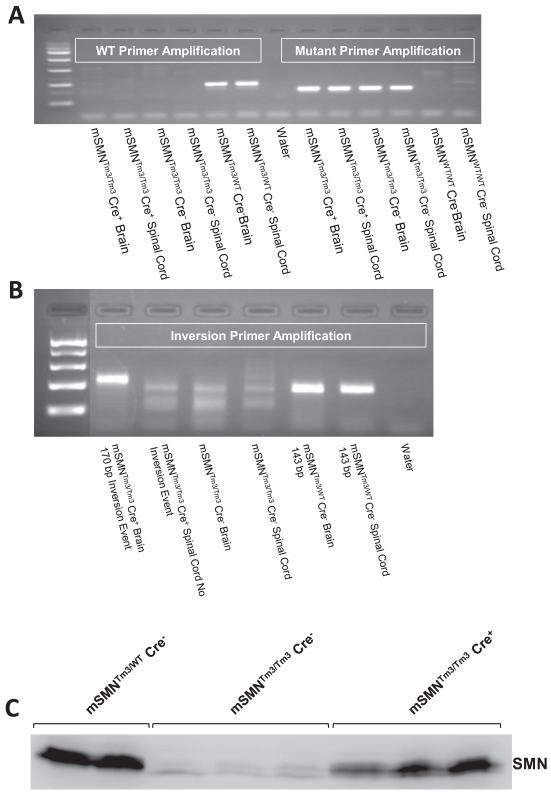
To ensure that the Cre-mediated recombination event has in fact occurred in the experimental mice, PCR was used to identify the unique recombined genomic element (Fig. 2a and b). As anticipated, the recombination event was easily detected in brain tissue in mice expressing Cre recombinase. Conversely, recombination was not detected in spinal cord tissue even in the presences of Cre recombinase, indicating that Emx-1 Cre promoter was not expressing in lower motor neurons. To examine SMN levels following the recombination event, Western blot analysis was performed on brain extract from animals using an anti-SMN monoclonal antibody (Fig. 2c). Smn levels were expectedly low in the SMA animals (mSMNTm3/Tm3,Cre−) and high in the unaffected control extracts (mSMNTm3/WT, Cre−), while SMN was increased in the experimental animals expressing Cre (mSMNTm3/Tm3,Cre+), demonstrating that the recombination event was efficient and resulted in increased SMN protein levels.
Figure 2.
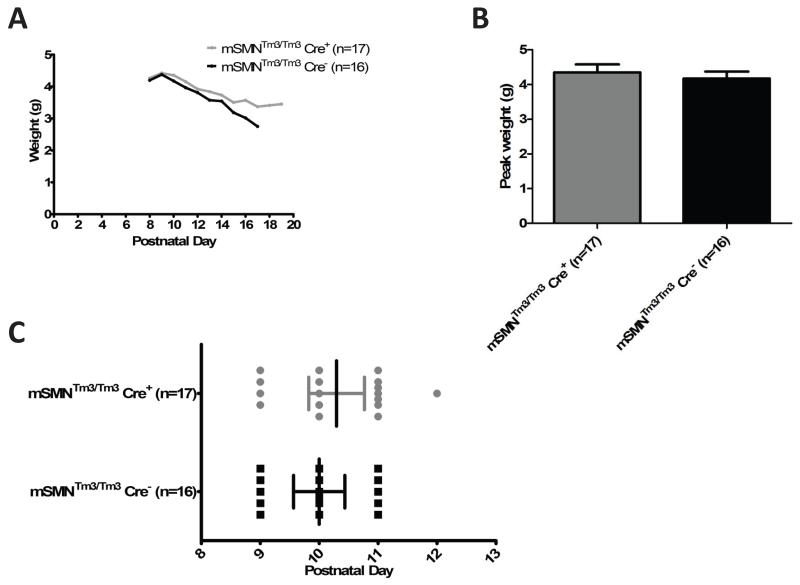
To determine whether an increase in SMN in Emx-1 expressing cortical neurons decreased disease severity in the SMA mice, well characterized components of the disease phenotype were examined. In the SMA mice, disease progression is very severe and animals rarely gain much weight, typically no more than 4 grams total body weight. To determine whether SMA-Emx1-Cre; Tm3+/+animals exhibited increased weight gain, animals were weighed daily from birth until death. At no time point was there a statistically significant difference in the average weight between the SMA control and Emx1-SMN transgenic animals (Fig. 3a). To examine these data more closely, two additional factors were analyzed: the peak weight (Fig. 3b) and the onset of weight loss (Fig. 3c). In each instance, SMA control and experimental SMA-Emx1-Cre; Tm3+/+animals initiated weight loss at approximately the same time point, P10, and both groups achieved the same peak weight of approximately 4 g. Collectively, these results demonstrate that while weight is an important component of the SMA phenotype, there was no statistically significant difference between SMA and SMA-Emx1-Cre; Tm3+/+animals, even though high levels of SMN were present in cortical neurons.
Figure 3.
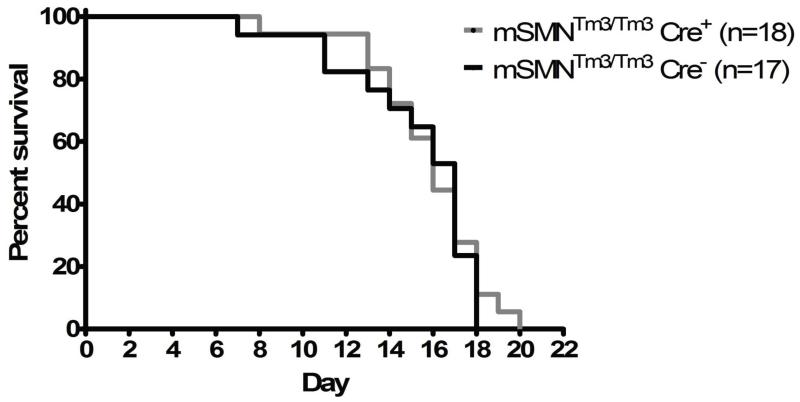
In addition to poor weight gain, the control SMA mice have a severely shortened life span, averaging approximately 14 days (Fig. 4). To determine whether increasing SMN in cortical neurons driven by Emx1 extended survival, the lifespans for SMA control and SMA-Emx1-Cre; Tm3+/+animals were determined. While there were modest differences in the Kaplan-Meir curves, the average lifespans for the two groups were nearly identical and were statistically insignificant (P = 0.66) (Fig. 4). Furthermore, the SMA-Emx1-Cre; Tm3+/+ animals did not exhibit increased motor function as measured by time-to-right assays compared to their untreated SMA littermates (data not shown). These results demonstrate that while a growing list a tissues and cell types have been implicated as contributing factors to the complex SMA phenotype, repletion of SMN specifically within cortical neurons mediated by the Emx-1 promoter, is not sufficient to modify the SMA phenotype.
Figure 4.
Discussion
Only in this type of controlled genetic experiment was it possible to ask whether SMN restoration in Emx-1 expressing cortical neurons is sufficient to decrease disease severity. While our results indicate that SMN restoration in Emx1-positive neurons does not reduce disease severity, they are not wholly unexpected as the phenotype of this novel animal model is consistent with the clinical descriptions of SMA regarding the role of cortical neurons (Pioro et al. 1994; Farrar et al. 2012). In this study, we restored SMN expression in Emx-1 expressing cortical neurons on a constitutively low full-length SMN background, closely resembling the well-characterized severe model of SMA referred to as SMNΔ7. It is possible, however, that by restoring SMN expression to Emx-1 expressing cortical neurons in a less severe model, this could result in phenotypic improvement as the SMNΔ7 model likely represents a relatively severe subpopulation of SMA patients analogous to severe Type I individuals. Less severe models have recently been described that appear to be more amenable to SMN-independent therapeutic strategies, such as the ROCK inhibitor Y-27632 (Hammond et al. 2010; Bowerman et al. 2012a). Alternatively, it is possible that other subpopulations of cortical neurons and/or glia that do not express Emx-1 could contribute to disease pathology.
Recent publications confirm the need to continue to investigate the role of other neuronal and glial cell types, especially those present in brain regions that are particularly vulnerable to the low SMN levels present in SMA such as the motor cortex, hippocampus, and dentate gyrus (Wishart et al. 2010). Somewhat surprisingly, the specific restoration of SMN in motor neurons in SMA mice, or the specific depletion of SMN in motor neurons in wildtype mice, results in relatively mild changes in pathology (Park et al. 2010; Gogliotti et al. 2012). These results suggest that either that the drivers are insufficiently restoring/depleting SMN, or, that other cells contribute to SMA pathology. It is also necessary to look for overall transmission defects in the motor circuitry composed of these cells. Perhaps, the cells populations of the corticospinal tract between the brain and spinal cord, such as cortical neurons, are more resistant to low SMN levels that those cells involved in the reflex arc between the muscle and spinal cord where massive and progressive transmission failure has been reported (Mentis et al. 2011). Additionally, recent reports have shown a loss of the central synapses of the spinal cord, highlighting the need for the examination of the glial cell populations, such as astrocytes and microglia, that have key supporting roles at these and other synapses (Ling et al. 2010). Continuing to look at the role of different neuronal and glial cell populations will help to further unravel the mechanism behind SMA development in addition to delineating tissue targets for potential therapeutics.
Acknowledgements
We thank John R. Marston for expert technical assistance and the SMA Foundation for their support of this project in its early stages. This work was supported by grants from the National Institutes of Health [R01 HD054413], and a training fellowship for JJG [NIGMS T32 5T32GM008396].
References
- Bevan AK, Hutchinson KR, Foust KD, Braun L, McGovern VL, Schmelzer L, Ward JG, Petruska JC, Lucchesi PA, Burghes AH, Kaspar BK. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum Mol Genet. 2010;19:3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman M, Murray LM, Boyer JG, Anderson CL, Kothary R. Fasudil improves survival and promotes skeletal muscle development in a mouse model of spinal muscular atrophy. BMC Med. 2012a;10:24. doi: 10.1186/1741-7015-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman M, Swoboda KJ, Michalski JP, Wang GS, Reeks C, Beauvais A, Murphy K, Woulfe J, Screaton RA, Scott FW, Kothary R. Glucose metabolism and pancreatic defects in spinal muscular atrophy. Ann Neurol. 2012b;72:256–268. doi: 10.1002/ana.23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Chan CH, Godinho LN, Thomaidou D, Tan SS, Gulisano M, Parnavelas JG. Emx1 is a marker for pyramidal neurons of the cerebral cortex. Cerebral Cortex. 2001;11:1191–1198. doi: 10.1093/cercor/11.12.1191. [DOI] [PubMed] [Google Scholar]
- Coady TH, Baughan TD, Shababi M, Passini MA, Lorson CL. Development of a single vector system that enhances trans-splicing of SMN2 transcripts. PLoS ONE. 2008;3:e3468. doi: 10.1371/journal.pone.0003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Farrar MA, Vucic S, Johnston HM, Kiernan MC. Corticomotoneuronal integrity and adaptation in spinal muscular atrophy. Arch Neurol. 2012;69:467–473. doi: 10.1001/archneurol.2011.1697. [DOI] [PubMed] [Google Scholar]
- Gogliotti RG, Quinlan KA, Barlow CB, Heier CR, Heckman CJ, Didonato CJ. Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J Neurosci. 2012;32:3818–3829. doi: 10.1523/JNEUROSCI.5775-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Hong S, Jin XL, Chen RS, Avasthi PP, Tu YT, Ivanco TL, Li Y. Specificity and efficiency of Cre-mediated recombination in Emx1-Cre knock-in mice. Biochem Biophys Res Commun. 2000;273:661–665. doi: 10.1006/bbrc.2000.2870. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Gogliotti RG, Rao V, Beauvais A, Kothary R, DiDonato CJ. Mouse survival motor neuron alleles that mimic SMN2 splicing and are inducible rescue embryonic lethality early in development but not late. PLoS ONE. 2010;5:e15887. doi: 10.1371/journal.pone.0015887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heier CR, Satta R, Lutz C, DiDonato CJ. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum Mol Genet. 2010;19:3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc Natl Acad Sci U S A. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer M, Kirmse K, Witte OW, Holthoff K. Reliable in vivo identification of both GABAergic and glutamatergic neurons using Emx1-Cre driven fluorescent reporter expression. Cell Calcium. 2012;52:182–189. doi: 10.1016/j.ceca.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Le TT, McGovern VL, Alwine IE, Wang X, Massoni-Laporte A, Rich MM, Burghes AH. Temporal requirement for high SMN expression in SMA mice. Hum Mol Genet. 2011;20:3578–3591. doi: 10.1093/hmg/ddr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Lee AJ, Awano T, Park GH, Monani UR. Limited Phenotypic Effects of Selectively Augmenting the SMN Protein in the Neurons of a Mouse Model of Severe Spinal Muscular Atrophy. PLoS ONE. 2012;7:e46353. doi: 10.1371/journal.pone.0046353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Ling KK, Lin MY, Zingg B, Feng Z, Ko CP. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS ONE. 2010;5:e15457. doi: 10.1371/journal.pone.0015457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CM, Kariya S, Patruni S, Osborne MA, Liu D, Henderson CE, Li DK, Pellizzoni L, Rojas J, Valenzuela DM, Murphy AJ, Winberg ML, Monani UR. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J Clin Invest. 2011;121:3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis VB, Rai R, Wang J, Chang CW, Coady T, Lorson CL. Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum Genet. 2006;120:589–601. doi: 10.1007/s00439-006-0245-7. [DOI] [PubMed] [Google Scholar]
- Mentis GZ, Blivis D, Liu W, Drobac E, Crowder ME, Kong L, Alvarez FJ, Sumner CJ, O’Donovan MJ. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskoui M, Levy G, Garland CJ, Gray JM, O’Hagen J, De Vivo DC, Kaufmann P. The changing natural history of spinal muscular atrophy type 1. Neurology. 2007;69:1931–1936. doi: 10.1212/01.wnl.0000290830.40544.b9. [DOI] [PubMed] [Google Scholar]
- Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci. 2010;30:12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioro EP, Antel JP, Cashman NR, Arnold DL. Detection of cortical neuron loss in motor neuron disease by proton magnetic resonance spectroscopic imaging in vivo. Neurology. 1994;44:1933–1938. doi: 10.1212/wnl.44.10.1933. [DOI] [PubMed] [Google Scholar]
- Rochette CF, Gilbert N, Simard LR. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- Shababi M, Glascock J, Lorson CL. Combination of SMN trans-splicing and a neurotrophic factor increases the life span and body mass in a severe model of spinal muscular atrophy. Hum Gene Ther. 2011;22:135–144. doi: 10.1089/hum.2010.114. [DOI] [PubMed] [Google Scholar]
- Shababi M, Habibi J, Yang HT, Vale SM, Sewell WA, Lorson CL. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum Mol Genet. 2010;19:4059–4071. doi: 10.1093/hmg/ddq329. [DOI] [PubMed] [Google Scholar]
- Somers E, Stencel Z, Wishart TM, Gillingwater TH, Parson SH. Density, calibre and ramification of muscle capillaries are altered in a mouse model of severe spinal muscular atrophy. Neuromuscul Disord. 2012;22:435–442. doi: 10.1016/j.nmd.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Wishart TM, Huang JP, Murray LM, Lamont DJ, Mutsaers CA, Ross J, Geldsetzer P, Ansorge O, Talbot K, Parson SH, Gillingwater TH. SMN deficiency disrupts brain development in a mouse model of severe spinal muscular atrophy. Hum Mol Genet. 2010;19:4216–4228. doi: 10.1093/hmg/ddq340. [DOI] [PMC free article] [PubMed] [Google Scholar]