Abstract
The sirtuins are a family of highly conserved NAD+-dependent deacetylases that act as cellular sensors to detect energy availability and modulate metabolic processes. Two sirtuins that are central to the control of metabolic processes are mammalian sirtuin 1 (SIRT1) and sirtuin 3 (SIRT3), which are localized to the nucleus and mitochondria, respectively. Both are activated by high NAD+ levels, a condition caused by low cellular energy status. By deacetylating a variety of proteins that induce catabolic processes while inhibiting anabolic processes, SIRT1 and SIRT3 coordinately increase cellular energy stores and ultimately maintain cellular energy homeostasis. Defects in the pathways controlled by SIRT1 and SIRT3 are known to result in various metabolic disorders. Consequently, activation of sirtuins by genetic or pharmacological means can elicit multiple metabolic benefits that protect mice from diet-induced obesity, type 2 diabetes, and nonalcoholic fatty liver disease.
I. INTRODUCTION
From budding yeast to humans, the fundamental principles of cellular energy metabolism are nearly identical. In all living organisms, cellular energy is produced and expended using highly homologous pathways, and energy is stored and transferred using universal “energy currencies” such as ATP and NADH. The tight balance between such anabolic and catabolic pathways ensures that cells do not deplete essential energy stores, which would ultimately cause cellular damage or death. Accordingly, evolutionarily conserved mechanisms have been established to protect cells from low cellular energy availability and to store excess energy for future use. Such mechanisms include the mTOR signaling pathway to detect branched-chain amino acids like leucine, carbohydrate responsive element binding protein (ChREBP) to respond to elevated glucose levels, AMP-activated protein kinase (AMPK) to sense low cellular ATP levels, ghrelin O-acetyltransferase (GOAT) to detect medium-chain fatty acids, and sirtuins to detect NAD+/NADH levels. This review focuses on SIRT1 and SIRT3, the two most prominent sirtuins involved in mammalian energy homeostasis, and gives an overview on the manyfold metabolic benefits elicited by both fuel sensors.
A. Historical Background
Almost a century ago, the Nobel laureate Francis Peyton Rous reported that chronic calorie restriction elicits beneficial metabolic effects on the spontaneous occurrence of tumors in rats (279). At the same time, Osborne et al. (238) showed that calorie restriction of young rats (1.5–6 mo of age) restores fertility at a later age and prolongs life span (238). While initially these findings were widely ignored, a growing number of studies in the following decades corroborated these benefits of calorie restriction, and accumulated evidence demonstrated that calorie restriction also protects from other age-related diseases, such as chronic kidney failure. In 1960, Berg and Simms (38) proposed that the reduction in body fat plays a decisive role in mediating the beneficial effects of calorie restriction on fertility, age-related pathologies, and longevity in rats. Subsequent studies, however, suggested that the benefits of calorie restriction may be due to a decrease in the absolute amount of calories consumed (40). More recently, this concept has been challenged (211), and evidence points to the effect of specific amino acids in determining the effects of long-term calorie restriction on life span and fertility (118, 219). In the past two decades, new techniques made it possible to focus also on the molecular underpinnings of metabolic benefits through calorie restriction. Several mechanisms have been identified that were shown to play a role, such as the attenuation of oxidative damage, decreases in insulin and glucose levels, an impairment of the growth hormone-insulin-like growth factor I (IGF-I) axis, and an activation of the deacetylase family of sirtuins.
In parallel with the work on calorie restriction, a genetic screen for long-lived mutants of Saccharomyces cerevisiae led to the discovery that the silent information regulation-2 (Sir2) gene could slow aging in this species (164, 306). Sir2 was later identified as NAD+-dependent deacetylase for hi-stone proteins that is required for calorie restriction to extend yeast life span (150, 203). Further studies identified structural and functional homologs in numerous other organisms (217). This review summarizes current research on sirtuin physiology and function with particular emphasis on metabolic effects elicited by pharmacological, genetic, or physiological manipulation of sirtuins.
B. Overview
The mammalian sirtuins are a family of NAD+-dependent enzymes with homology to the Saccharomyces cerevisiae gene silent information regulator 2 (Sir2). Humans have seven sirtuins, SIRT1-SIRT7. SIRT1, the most studied member of this family, plays an important role in several processes ranging from cell cycle regulation to energy homeostasis. SIRT3 has recently emerged as a sirtuin with considerable impact on mitochondrial energy metabolism and function. Numerous studies have also demonstrated that both SIRT1 and SIRT3 play an important role in different types of cancer. SIRT1 has tumor suppression activity in ageing- and metabolic syndrome-associated cancer (123, 133, 134, 283), and SIRT3 has also been identified as a tumor suppressor (35, 100, 171, 245).
The basic sirtuin structure and function have remained highly conserved across species, from bacteria to humans. In humans, sirtuins exist throughout the body; for example, SIRT1 is expressed in the brain, liver, pancreas, adipose tissue, muscle, and heart. Sirtuins may function as deacetylases and/or ADP-ribosyltransferases. Sirtuins with deacetylase activity remove the acetyl groups from acetylated lysine residues of numerous target proteins, including histones and transcription factors. Section II details the structure, function, and localization of sirtuins.
Sirtuins are cellular energy sensors that require NAD+ for their enzymatic activity. As a result, their activity is directly linked with metabolism. Certain cellular stressors or a low energy state in the cell increases the NAD+/NADH ratio, decreases nicotinamide levels, and activates sirtuins (13, 203). Section III describes the relationship of NAD+, calorie restriction, and the sirtuin family, as well as the mechanisms for sirtuin-catalyzed NAD+-dependent reactions.
Mammalian SIRT1 deacetylates a host of target proteins that are important for apoptosis, the cell cycle, circadian rhythms, mitochondrial function, and metabolism. In particular, much current research focuses on the impact of SIRT1 in glucose homeostasis, lipid metabolism, and energy balance. While SIRT1 plays an important role in metabolic function, sirtuins 3–5 are localized in mitochondria and may regulate mitochondrial energy metabolism. SIRT3, the most studied of the mitochondrial sirtuins, deacetylates a number of mitochondrial proteins and might also play a role in regulating ATP production. Section IV focuses on the molecular targets of SIRT1 and SIRT3.
Mammalian sirtuins are not only regulated by NAD+/NADH ratio or cellular stressors, but also by endogenous proteins involved in signal transduction and transcription, as well as by a number of microRNAs. Section V will describe the complex regulation of SIRT1 and SIRT3 by endogenous factors.
Sirtuins 1 and 3 are expressed in a wide variety of tissues and target numerous proteins. Section VI depicts how the activation of SIRT1 influences metabolically active tissues, such as liver, skeletal muscle, pancreas, adipose tissue, or brain, by inducing a wide range of physiological processes. Section VII focuses on the specific roles of SIRT3 in diverse, metabolically active tissues.
Several mouse models have been used to characterize the metabolic functions of sirtuins. Inbred whole body SIRT1 knockout mice are born underweight and do not live past the early postnatal stage. SIRT1 knockout mice on an outbred background exhibit developmental and metabolic abnormalities including cardiac defects and decreased locomotor activity, but they also exhibit improved glucose homeostasis. Several studies focus on liver-specific ablation of SIRT1 and the role of hepatic SIRT1; however, many of these findings are contradictory. In addition to SIRT1 deficiency, SIRT1 overexpression is also under investigation. Mice with global SIRT1 overexpression show resistance to metabolic dysfunction as a result of high-fat diet exposure. Gain- and loss-of-function studies on sirtuins other than SIRT1 have also been performed. Of particular importance to metabolic regulation are the mitochondrial sirtuins. Section VIII outlines the metabolic phenotypes of global or tissue-specific SIRT1 and SIRT3 loss- and gain-of-function models and depicts specific roles of SIRT1 for circadian rhythms and diabetes-induced cardiac function.
Section IX describes the current knowledge on genetic polymorphisms in SIRT1 and SIRT3 and their implication in metabolic diseases.
It has been suggested that pharmacological SIRT1 activation may alleviate metabolic dysfunction associated with obesity or other metabolic disorders. Resveratrol is a proposed SIRT1 activator, but its metabolic benefits, when administered pharmacologically in humans, are still a matter of controversy. Several small-molecule SIRT1 activators with improved pharmacological activity are under investigation for their potential therapeutic benefits in humans. Section X covers the metabolic consequences of SIRT1 activation by resveratrol and other natural or synthetic ligands.
Finally, section XI gives a general view on future directions and perspectives for sirtuin research.
II. THE STRUCTURE, FUNCTION, AND DIVERSITY OF SIRTUINS
A. Sirtuins Are Highly Conserved in Evolution
The Saccharomyces cerevisiae gene silent information regulator 2 (Sir2) was identified as a NAD+-dependent histone deacetylase (150, 309) involved in life span extension associated with calorie restriction (13, 201). Sir2 homologs, known as sirtuins, have been identified in numerous higher organisms including Drosophila melanogaster (31, 231), Caenorhabditis elegans (327), mice (366), and humans (51, 107, 108, 298). Seven sirtuins (SIRT1–SIRT7) comprise the family of Sir2 homologs in humans (107, 108). Mammalian sirtuins share the conserved sirtuin domain, but vary in subcellular localization and function. Of note, sirtuins have been lost in many species including insects, nematodes, and plants (121). Thereby, it seems that the loss of individual sirtuins might be compensated for by redundant functions in remaining sirtuin family members (121).
In yeast, Sir2 deacetylates the acetyl-lysine residues of histones (150) by catalyzing a unique chemical reaction that requires NAD+ and generates the novel product O-acetyl-ADP-ribose (O-AADPR) (48, 72, 321, 324). As discussed below, this deacetylase activity is conserved in mammalian sirtuins. In addition to histone deacetylation, mammalian sirtuins are also able to catalyze reactions for a number of protein substrates (107). Furthermore, certain mammalian sirtuins possess ADP-ribosyltransferase activity (107).
Consistent with the evolutionarily conserved activity of Sir2 and its homologs, increased protein levels of Sir2 not only confer longevity benefits to S. cerevisiae (164, 173), but also to C. elegans (327) and D. melanogaster (277). However, other lines of evidence suggest that Sir2 does not affect life span in flies (231). Elevated expression of sirtuins in normal human cells does not extend replicative life span (218), and there is no evidence that a sirtuin can extend the life span of a mammal, unless it is under metabolic stress (34, 246).
Today, we know that biochemical features are highly conserved in sirtuins. Their physiological roles, however, differ between species. While Sir2 in yeast is ostensibly confined to deacetylation of histones, but also controls segregation of protein carbonylation, mammalian sirtuins target multiple proteins, regulating many diverse processes ranging from cell cycle progression and mitochondrial function to metabolism and energy homeostasis.
B. Structural Properties of SIRT1 and SIRT3
Among the yeast proteins that comprise the Sir silencing complex (Sir1–4), Sir2 is unique (reviewed in Ref. 117), as it is the only homolog that has been evolutionarily conserved from bacteria to humans (3, 51, 107, 298). Accordingly, all Sir2-like proteins possess a sirtuin core domain containing a series of sequence motifs that is conserved across organisms (51). Molecular phylogenetic analyses of a diverse array of organisms (including archaea bacteria, yeasts, plants, protozoans, and metazoans) have shown that the conserved sirtuin core domain sequences of eukaryotic organisms can be grouped into four main classes: SIRT1, SIRT2, and SIRT3 compose class I, SIRT4 composes class II, SIRT5 composes class III, and SIRT6 and SIRT7 compose class IV (108). Sir2 protein is also unique among the silencing factors in Saccharomyces cerevisiae because it silences the rDNA as well as the silent mating-type loci and telomeres (303). Silencing is a universal form of transcriptional regulation in which regions of the genome are reversibly inactivated by a myriad of mechanisms. Sir2 is also required to segregate damaged proteins into the mother cell, thus sparing the daughter cell (4). However, the molecular target of this action and how this relates to mammalian aging is not yet known.
In budding yeast, the sirtuins also include a set of genes known as homologs of Sir2 (HST1–4), which play roles in gene silencing, DNA repair, and life span extension by calorie restriction (51, 77, 86, 187, 330, 357). The proteins encoded by the HSTs are ~30–65% identical to Sir2 and are characterized by a conserved core domain, which is up to 84% identical to Sir2 and essential for Sir2 silencing (298).
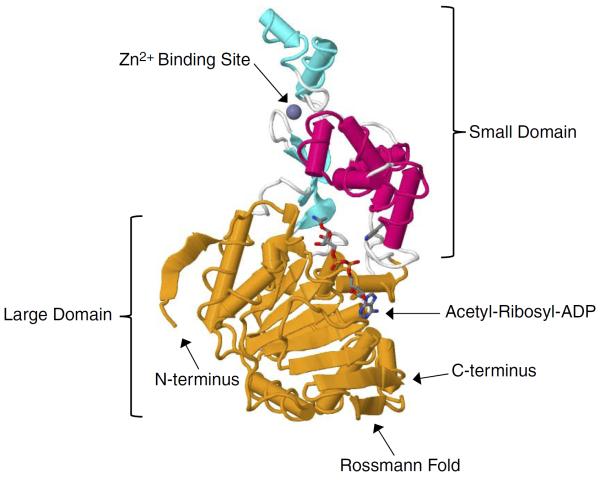
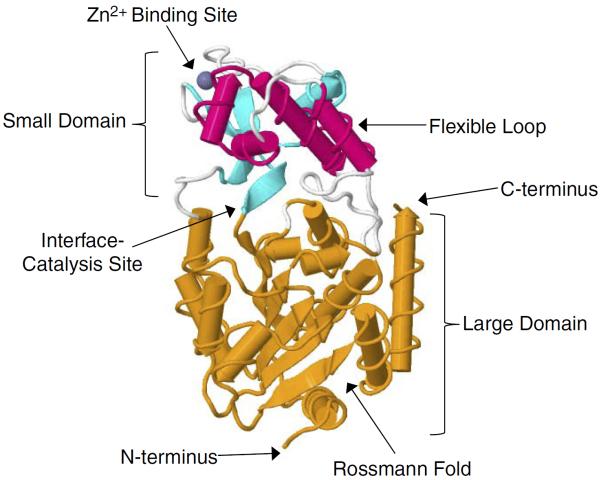
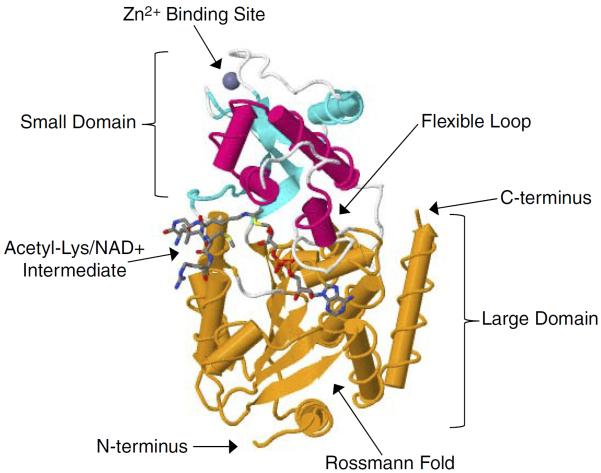
To understand the structural basis of the enzymatic mechanism of the sirtuins, a number of crystal structures have been determined. In Archaeoglobus fulgidus, two crystal structures of Sir2-Af1 complexed with NAD were solved at 2.1 and 2.4 Å resolution (101, 221). The structure revealed that the protein consists of a large domain having a Rossmann fold and a small domain containing a three-stranded zinc ribbon motif. NAD is bound in a pocket between the two domains (221) (FIGURE 1). On the other hand, the 1.7 Å crystal structure of the 323-amino acid catalytic core of human SIRT2 reveals an NAD-binding domain, which is a variant of the Rossmann fold, and a smaller domain composed of a helical module and a zinc-binding module. Mutagenesis studies suggest that a conserved large groove at the interface of the two domains is the likely site of catalysis (101). More recently, several crystal structures for human SIRT3 have been also reported (160), including the apo-structure without substrate, a structure with a peptide containing acetyl lysine of its natural substrate trapped by a thioacetyl peptide, and a structure with the dethioacetylated peptide bound (160) (FIGURES 2 and 3). Finally, the structure of human SIRT6 has revealed unique features such as a splayed zinc-binding domain and the absence of a helix bundle that in other sirtuin structures connects the zinc-binding motif and Rossmann fold domain (243). SIRT6 also lacks the conserved, highly flexible, NAD+-binding loop and instead contains a stable single helix (243).
FIGURE 1.
Structure of yeast Sir2 in complex with an acetyl-ribosyl-ADP intermediate. Yeast Sir2 is depicted in the cartoon representation with the zinc binding site highlighted in cyan, the helical module highlighted in magenta, and the Rossmann foldlike NAD+ binding site highlighted in orange. The acetyl-ADP intermediate is depicted as a stick representation with carbon atoms in gray, oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in yellow, and phosphate atoms in orange. The structure was assembled from Protein Data Bank code 2HJH (Protein Data Bank: Hall BE, Buchberger JR, Gerber SA, Ambrosio ALB, Gygi SP, Filman D, Moazed D, Ellenberger T.).
FIGURE 2.
Structure of apo SIRT3. SIRT3 is depicted in the cartoon representation with the zinc binding site highlighted in cyan, the helical module highlighted in magenta, the Rossmann foldlike NAD+ binding site highlighted in orange, and the domain interface loops highlighted in white. The structure was assembled from Protein Data Bank code 3GLS (Jin et al., Ref. 160).
FIGURE 3.
Structure of SIRT3 in complex with AceCS2-Ks-acetyl-ADPR. SIRT3 is depicted in the cartoon representation with the zinc binding site highlighted in cyan, the helical module highlighted in magenta, and the Rossmann foldlike NAD+ binding site highlighted in orange. The acetyl-lysine substrate, which here is the trapped reaction intermediate AceCS2-Ks-acetyl-ADPR peptidal compound, inserts into the domain interface populated by loops that are highlighted in white where catalysis of the deacetylation occurs. The AceCS2-Ks-acetyl-ADPR is depicted as a stick representation with carbon atoms in gray, oxygen atoms in red, nitrogen atoms in blue, sulfur atoms in yellow, and phosphate atoms in orange. The structure was assembled from PDB code 3GLT (Jin et al., Ref. 160).
C. Tissue Distribution of SIRT1
SIRT1 is expressed in a wide range of tissues and organs and has been detected in the liver, pancreas, heart, muscle, and adipose tissue of mice (151, 191, 374). Other reports, utilizing a mCherry reporter gene in C. elegans, identified Sir2 mainly in muscle and intestinal cells (27). In humans and mice, SIRT1 is ubiquitously expressed in the brain, especially in areas related to neurodegenerative diseases, such as the prefrontal cortex, hippocampus, and basal ganglia (379). SIRT1 is also expressed in important metabolic centers of the brain (268), including the hypothalamic arcuate, ventromedial, dorsomedial, and paraventricular nuclei, and the area postrema and the nucleus of the solitary tract in the hindbrain. During aging, SIRT1 expression is decreased in specific nuclei of the hypothalamus of mice, but not in all brain regions (185). Other studies focusing on the function of SIRT1 in development found that SIRT1 is expressed at high levels in most of the tissues of embryos, where expression was found in the heart and the brain, and to a lesser extent in liver, spleen, kidney, lung, thymus, testis, and ovary (280). Overall, these studies suggest that SIRT1 is expressed in a wide variety of tissues.
D. Subcellular Distribution of SIRT1
The subcellular localization of SIRT1 likely depends on cell type, stress status, and molecular interactions. SIRT1 has been observed in both the nucleus and the cytoplasm, where it interacts with both nuclear and cytosolic proteins (62, 218, 322). For example, SIRT1 is ubiquitously present in the prefrontal cortex, hippocampus, and basal ganglia. Throughout the rodent brain and spinal cord, both cytosolic and nuclear localization can be detected, although immunocytochemical and Western blot analyses indicate that SIRT1 is predominantly nuclear (379). The primarily nuclear localization of SIRT1 can be attributed to two nuclear localization signals and two nuclear export signals (322). Thus the extent of cytosolic versus nuclear localization of SIRT1 may be dictated by the relative strengths of the nuclear localization and export signals in each cell type or environmental signal (123).
E. Tissue Distribution of SIRT3
Like SIRT1, mammalian SIRT3 is expressed in a variety of tissues. Studies carried out with quantitative RT-PCR in different mouse tissues show the highest expression in kidney, brain, and heart, followed by liver and testes, with lower expression in lung, ovary, spleen, and thymus (159). Similar expression patterns are seen in human tissues (314, 354).
F. Subcellular Distribution of SIRT3
SIRT3 was the first mammalian sirtuin shown to be localized to mitochondria (218, 236, 294). It is localized to the mitochondrial matrix, and cleavage of its signal sequence is necessary for enzymatic activity (294). The major isoform of mouse SIRT3 is a 257-amino acid protein that aligns with the COOH-terminal portion of human SIRT3 (residues 143–399) (366). However, subsequent reports have questioned the exclusivity of the mitochondrial distribution of SIRT3. This ambiguity likely arises from the two different isoforms (short and full length) of SIRT3, which are expressed in both humans and mice and appear to be differentially distributed in a tissue-specific manner (124, 253, 319). Additionally several splice variants (M1, M2, and M3) have been reported (64). The variants M1 and M2 appear to be localized to the mitochondria, whereas the M3 splice variant displays nuclear localization (64). While full-length mouse SIRT3 has been reported to be exclusively localized to the mitochondria (159), other groups report that in the heart the long form of mSIRT3 can be detected in mitochondria, the cytoplasm, and the nucleus (319). However, after overexpression of short and long forms of mSIRT3 in mouse embryonic fibroblasts and H9C2 cells, both forms were located in the nucleus as well as the cytoplasm (29).
As with the mouse SIRT3 protein, there are also two forms of the human SIRT3 protein. hSIRT3 is found in both long and short isoforms (65), and initially it was thought that the full-length form is localized exclusively to the mitochondria, whereas the short form, which loses the NH2-terminal 142 residues, is present in the cytoplasm and nucleus (65). However, recent findings have questioned whether the localization of human SIRT3 is exclusive to mitochondria (65, 236, 289). One study now suggests that SIRT3 is present in the nucleus under basal conditions, but translocates to the mitochondria during cellular stress (289). Indeed, the location of SIRT3 in the nucleus is a matter of controversy, and studies using various hSIRT3 expression constructs and subcellular fractionation failed to find hSIRT3 in the nucleus, suggesting it is exclusively mitochondrial (65). The distinct methodologies and antibodies used by the different laboratories might explain these differences (65).
III. CELLULAR FUEL SENSING BY SIRT1 AND SIRT3
Sirtuins function as cellular energy sensors that are either activated or inhibited by the level of metabolic cofactors and intermediates in the cell, including NAD+, NADH, and nicotinamide (42, 51, 150, 188, 287, 309). The main organelle that produces energy for use in cellular metabolism is the mitochondrion. Mitochondria are equipped with machinery to facilitate the oxidation of substrate (e.g., carbohydrates and fatty acids) to produce ATP. This oxidative reaction generates energy, which is stored in reduced carrier molecules (as FADH2 and NADH) that are utilized in the mitochondrial membrane to generate ATP through the oxidative phosphorylation cascade. Thus the generation of energy (i.e., ATP) in the cells is linked to the production of FADH2 and NADH. When these carriers are oxidized, they become FAD and NAD+, and all four molecules are important regulators of numerous biochemical reactions inside the cell.
A. Cellular Energy: NAD+/NADH Ratios and the NAD+ Salvage Pathway
An absolute requirement of the sirtuin reaction is the cosubstrate NAD+, which places the sirtuins at the nexus of cellular energy metabolic regulation and provides a possible link between cytosolic energy status and nuclear signaling. Catabolic reactions such as β-oxidation, glycolysis, protein degradation, and citric acid cycling reduce NAD+ to NADH. When energy is plentiful, intracellular NADH levels rise while NAD+ levels drop, although the ratio always favors NAD+ over NADH (232). NADH from cytosolic metabolism feeds into the mitochondria through the malate-aspartate shuttle or by reducing pyruvate to lactate to join NADH produced via the citric acid cycle (25). Regardless of its origin, NADH is oxidized in the mitochondria by the electron transport chain (ETC). Specifically, NADH feeds into the ETC through its oxidation to NAD+ at complex I (137). Subsequent reactions of the ETC produce energy that is used to pump protons across the mitochondrial matrix. This generates the proton gradient that drives ATP synthesis via oxidative phosphorylation.
Cellular NAD+ is derived from two main sources: de novo synthesis from the amino acid tryptophan (215) and via the salvage of nicotinamide back to NAD+. The product of the sirtuin reaction, nicotinamide, acts as a feedback inhibitor by binding in a conserved regulatory pocket on the enzymes called the C-pocket (20, 42).
In yeast, the removal of nicotinamide, the rate-limiting step of the NAD salvage pathway, is catalyzed by pyrazinamidase/nicotinamidase 1 (PNC1), a nicotinamide deaminase that is upregulated in response to caloric restriction and cellular stress (13). Overexpression of NAD salvage genes extends life span in a Sir2-dependent manner (12), while deletion of the PNC1 gene blocks life span extension by calorie restriction (13). These findings are consistent with the hypothesis that the ability of dietary manipulations to extend life span in mammals is mediated, in part, by the induction of enzymes that increase the rate at which nicotinamide is recycled back to NAD+ (361).
In humans, the rate-limiting step of the NAD+ salvage pathway is catalyzed, not by PNC1, but by a nicotinamide phosphoribosyltransferase (named as NAMPT in bacteria, but also known as PBEF), which converts nicotinamide to nicotinamide mononucleotide (209, 273). Like the yeast PNC1 gene, the expression of this essential enzyme appears to be regulated by stress (154, 209, 361), and its activity is specifically elevated in times of prolonged fasting (362). Additional findings suggest that NAMPT could directly influence the NAD+/NADH ratio in the cell (273, 335). Furthermore, findings by Revollo et al. (273) suggest that overexpression of NAMPT directly increases SIRT1 activity, implicating NAD+ concentration in the regulation of SIRT activity.
Studies in yeast bolstered the hypothesis that the NAD+/NADH and the NAD+/nicotinamide ratios regulate sirtuin activity. Calorie restriction induces a decrease in both NADH and nicotinamide concentrations and is associated with increased life span and Sir2 activity (13, 201–203, 286). Despite strong genetic evidence, a subsequent biochemical study questioned whether NADH is in sufficient concentrations in vivo to inhibit sirtuins. This study suggested that the ratio of NAD+ to nicotinamide is the primary mechanism of sirtuin regulation (291).
Studies in mice suggest that prolonged fasting can induce a 50% increase in hepatic NAD+, which was associated with increased SIRT1 activity (275). Similarly, Yang et al. (362) demonstrated that food deprivation increased the levels of NAMPT and mitochondrial NAD+ levels, and that the ensuing stress protection required both SIRT3 and SIRT4. Indeed, the dependency of mitochondrial SIRT3 on NAD+ has been clearly demonstrated in multiple studies (236, 289, 294).
B. Mechanism of NAD+-Dependent Deacetylation
The Sir2 family of class III deacetylases utilizes a distinctive chemical reaction to deacetylate lysine residues. This reaction consumes NAD+ to produce nicotinamide, the deacetylated substrate, and O-AADPR (48, 72, 321, 324). The NAD+-dependent deacetylation reaction is a six-step reaction eloquently reviewed by Sauve (284). Briefly, the reaction begins with the binding of NAD+ to the sirtuin catalytic site, a process mediated by highly conserved catalytic phenylalanine (129, 138) and histidine residues (221, 285, 307). The first step in the sirtuin-catalyzed deacetylation utilizes amide cleavage of NAD+ to produce nicotinamide and a peptidylimidate intermediate (285). This is followed by formation of a 1′,2′-acyloxonium via nucleophilic attack of the 2′-OH group of the imidate intermediate. This collapse is facilitated by the conserved catalytic histidine (221, 307), which acts as an activating base to the 2′-OH group. The nucleophilic attack is followed by hydrolysis to 2′-AADPR via attack by water, releasing the free amino group of the previously acetylated lysine residue, AADPR, and nicotinamide. A caveat of this efficient reaction is its inhibition by the reaction product nicotinamide (42). This product can bind to the active site, reacting with peptidyl imidate to form the reverse reaction yielding NAD+. In addition to the sirtuin-mediated deacetylation, a spontaneous isomeric transition occurs in the solvent to maintain equilibrium of 2′- and 3-AADPR (285).
C. Mechanism of NAD+-Dependent ADP-Ribosylation
In addition to the more widely studied deacetylation reaction, sirtuin enzymes were first identified by their ability to catalyze ADP-ribosylation of target proteins (107, 323). While the mechanisms of ADP-riboslytransfer are still in debate, several reports have proposed a possible mechanism for this reaction (98, 129, 130, 284). A leading hypothesis by Hawse and Wolberger (130) suggests that the targets of sirtuin-mediated ADP-riboslytransfer contain a nucleophilic residue +2 positions from the acetylated lysine. In this mechanism, NAD+ reacts with acetyllysine to form an O-alkyamidate and release nicotinamide. The +2 nucleophilic residue next attacks the O-alkylamidate intermediate to produce ADP-ribosylation and acetyllysine (130). A recent study on SIRT6 indicates that this sirtuin physically associates with poly-ADP-ribose polymerase 1 (PARP1), mono-ADP-ribosylates, and PARP1 on lysine residue 521, thereby stimulating PARP1 poly-ADP-ribosylase activity and enhancing double-strand break repair under oxidative stress (212).
D. Regulation of Sirtuin Activity by Calorie Restriction and Metabolic Stressors
Although some exceptions exist, studies in the area of aging have shown repeatedly that caloric restriction (CR) improves glucose metabolism, increases mitochondrial activity, and extends life span in a broad range of species from yeast to mammals (66, 127, 132, 144). In yeast, a reduction of glucose from 2 to 0.5% in the media extends life span ~30%. Furthermore, Sir2 and respiration are required for this effect (13, 187, 203).
However, the benefits of CR in yeast are not solely dependent on Sir2. Severe CR (0.05% glucose) extends yeast replicative life span but does not require Sir2 nor mitochondrial respiration (161, 162). Moreover, Sir2 has no effect on yeast survival under starvation (97) or in worms under CR (41).
It is important to point out that the amount of yeast Sir2 protein does not increase during CR (13), and there are alternative hypotheses concerning the mechanism for CR-mediated longevity. Surprisingly, Sir2 levels and NAD+ concentrations are not elevated following CR in yeast (12). Instead, enzymatic activity is believed to increase in response to changes in the concentrations of nicotinamide and NADH, where PNC1 plays an important role as described below.
While there are seven known mammalian homologs of Sir2, SIRT1 appears to be the one that most closely resembles the yeast Sir2 enzyme (108). CR studies in rodent models have shown that, unlike its effect in yeast, CR increases SIRT1 in several metabolically relevant tissues including brain, kidney, liver, white adipose tissue, and skeletal muscle (63). The CR-stimulated increase in SIRT1 protein is accompanied by increased NAD+, suggesting elevated activity in addition to the elevated enzyme levels (227). Supporting the link between CR and SIRT1, analyses of genetically elevated SIRT1 suggest phenotypes that are highly reminiscent of those displayed during treatment with CR and SIRT1 agonists (34, 186, 220). Nevertheless, the hypothesis that SIRT1 activity is increased in all tissues after CR has been called into question recently by a report showing SIRT1 activity in the liver is decreased by CR, and that this reduction by CR is correlated with the reduced role of this organ in fat synthesis (74). However, the time of day, species, and type of CR are all variables that may explain differences between these studies.
Overexpression of SIRT1 in mice is associated with positive metabolic outcomes. Specifically, these mice display a decrease in adiposity, serum cholesterol, and insulin, while displaying increased resistance to obesity-generated glucose intolerance and insulin resistance (28, 46). On the other hand, studies utilizing mice deficient for the SIRT1 gene have also provided insight into its role in the physiology of CR (5). Specifically, the global SIRT1-deficient mice in a mixed background elicit a shortened life span, which is resistant to CR (75, 196). Moreover, SIRT1-deficient mice in a clean background are perinatally lethal, and therefore, it is difficult to predict in this case the full effect of the mixed background.
In addition to its activation during CR, SIRT1 is also activated in the presence of oxidative stress (54, 177, 311). This activity is important in the context of tissue preservation. For example, the SIRT1-mediated deacetylation of FOXO appears to protect β-cells from oxidative stress (174, 175). Similarly, while HFD exposure induces oxidative stress via NF-κB (58), SIRT1 overexpression has been shown to increase antioxidant defense enzymes and inhibit NF-κB activation, thereby providing powerful protection from hepatic inflammation, glucose intolerance, and NAFLD (249). SIRT1 has been also proposed as a link between caloric intake and mood. Specifically, in conditions of CR, when SIRT1 is activated, anxious behavior is increased (93, 198).
Likewise, the effect of SIRT1 in neuronal tissues appears to be protective in the context of oxidative stress. Recent studies have suggested that SIRT1 is responsible for protection from neuronal, and specifically axonal, degeneration (16, 90, 169, 336), suggesting that the activation of SIRT1 might be important in the treatment of Alzheimer's disease or other neurodegenerative diseases such as Huntington's disease (156, 158). Although SIRT1 is known to protect tissues from oxidative damage, work by several groups suggests that SIRT1 may also have a role in providing genetic stability during times of oxidative damage. Specifically, SIRT1 deacetylates several genes important for DNA repair (377, 378) and prevents immediate cell death in favor of senescent growth arrest (295). Additionally, SIRT1 is directly recruited to sites of broken DNA and helps recruit key DNA repair proteins such as Rad51 and Nbs1 (235). Further supporting a role in genetic stability, SIRT1−/− embryos exhibit impaired DNA repair and increased chromosomal abnormalities (348), while overexpression of SIRT1 reduces aneuploidy and delays lymphoma (235).
E. Regulation of SIRT3 Activity by Nutrient Intake and Metabolic Stressors
During CR, expression of SIRT3 is increased in the mitochondria of murine skeletal and cardiac muscle, as well as in white and brown adipose tissues (241, 300, 319). This is of interest as SIRT3 deacetylates, and therefore activates, the enzyme responsible for formation of acetyl-CoA from acetate [acetyl-CoA-synthase (AceCS)] (293). AceCS is critical during times of prolonged fasting, where the acetate must first be converted to acetyl-CoA before it can be utilized as a metabolite in the citric acid cycle. Conversely, in genetically obese mice, SIRT3 and genes important for mitochondrial function show decreased expression in brown adipose tissue (300).
IV. MOLECULAR TARGETS OF SIRT1 AND SIRT3
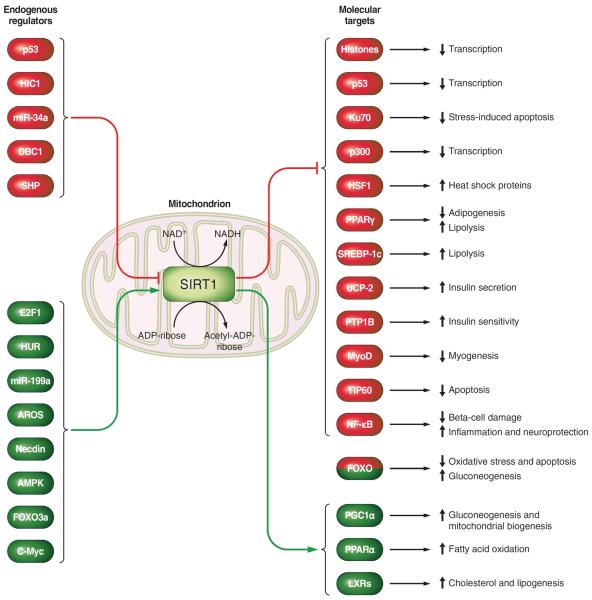
SIRT1 interacts with and regulates a number of histone and non-histone protein substrates (FIGURE 4). The wide variety of endogenous targets is correlated with the different biological functions modulated by this deacetylase. It has roles in developmental and aging regulation, energy metabolism, inflammation, and the repair of DNA double-strand break, amongst others.
FIGURE 4.
Schematic overview of the endogenous regulators and molecular targets of SIRT1.
A. Cytosolic and Nuclear Targets of SIRT1
1. Histones
Histones are proteins that are essential for the tight packaging of DNA into chromatin. The level of histone acetylation has been shown to regulate gene transcription. Specifically, increased acetylation prevents histones from binding and condensing DNA, thus enabling more transcription, whereas deacetylation promotes histone binding and decreases transcription. Each year, different reports identify new histones deacetylated by SIRT1, including histone 1 on lysine 27 (H1K27) (184); histone 3 on lysines 9, 14, 18, and 56 (H3K9, H3K14, H3K18, H3K56) (150, 337, 376); and histone 4 on lysines 12 (H4K12) and 6 (H4K6) (109, 150, 338). The molecular mechansims that target SIRT1 to its substrates are not well understood. This lack of significant substrate specificity has increased the difficulty in identifying bona fide targets for deacetylation (43). Hence, credence is given to studies wherein gain and loss of SIRT1 function have reciprocal effects on substrate acetylation.
2. p53
SIRT1 has been shown to target and deacetylate the lysine residues of not only histones but numerous protein substrates as well. This diversity of function has important implications for mammalian cell survival and senescence. The substrates of SIRT1 that regulate apoptosis and cell cycle regulation include transcription factors and DNA repair factors. p53 is an important tumor-suppressing protein that regulates many cellular activities, such as cell cycle regulation, DNA repair, and programmed cell death. As SIRT1 binds and deacetylates p53 to decrease its transcriptional activity, p53 is suggested to play a central role in SIRT1-mediated functions in tumorigenesis and senescence (78, 189, 208, 339). For this reason, SIRT1 was originally considered to be a potential tumor promoter. However, new evidence suggests that SIRT1 acts as a tumor suppressor based on its role in negatively regulating β-catenin and survivin (370). Therefore, even though the interaction between SIRT1 and p53 is clear, the current role of the SIRT1/p53 pathway on tumorigenesis is controversial. Recent evidence suggests that the interaction between SIRT1 and p53 is regulated by the methyltransferase Set7/9, since the presence of the methyltransferase Set7/9 suppresses this interaction (204). Set7/9 is a SET domain-containing histone 3 lysine 4 (H3-K4) methyltransferase (234, 344). Set7 is known to stimulate activator-induced transcription in vivo (234), and one of these transcription factors might be p53.
3. DNA damage proteins: Ku70, FOXL2, NBS1, WRN, and XPC
Ku70 is one of the two subunits (Ku70 and Ku80) of the Ku protein. The Ku protein plays a key role in multiple nuclear processes, including DNA repair, chromosome maintenance, transcription regulation, and recombination (272). It has been reported that SIRT1 prevents apoptosis by interactions with Ku70 (15, 49, 63, 157, 197). In response to CR, SIRT1 deacetylates Ku70, which in turn reduces stress-induced apoptosis by sequestering Bax, a pro-apoptotic factor, from mitochondria (63). SIRT1 can also bind and deacetylate Ku70 to increase DNA repair activity in cells subjected to radiation exposure (157).
When the transcription factor FOXL2 is upregulated, it promotes cell accumulation in G1 phase and protects cells from oxidative damage (254). SIRT1 inhibition increases both amount and activity of FOXL2, thereby limiting proliferation (37).
The Nijmegen breakage syndrome (NBS1) protein is a DNA repair factor, and its phosphorylation delays cell cycle progression (116). SIRT1 deacetylates NBS1 and thereby it is one of the suggested pathways to regulate cell survival (377).
Werner Syndrome (WRN) is a gene with essential functions in maintaining genome stability. After DNA damage, SIRT1 deacetylates WRN, decreasing its helicase and exonuclease activities (193). Thus the interaction between SIRT1 and WRN is another mechanism to regulate DNA damage.
Nucleotide excision repair (NER) is a particularly important mechanism by which the cell can prevent unwanted mutations. The xeroderma pigmentosum C (XPC) protein is essential for the initiation of the NER. SIRT1 enhances XPC expression acting as a tumor suppressor through its role in DNA repair (222).
4. FOXO1, -3, and -4
The Forkhead box, group O (FOXO) subfamily of fork-head transcription factors is able to sense nutrient availability and regulates various cellular processes, including apoptosis and the cell cycle (2, 83). SIRT1 deacetylates the FOXO proteins increasing their nuclear retention and transcriptional activity (82, 175, 260). Activation of certain members of the FOXO family can increase resistance to oxidative stress (181), and SIRT1 is responsible for potentiating this function (334). Specifically, SIRT1 regulates FOXO3 by both inhibiting FOXO3-induced apoptosis and potentiating the ability of FOXO3 to induce cell cycle arrest and resist oxidative stress (54, 334). Other studies suggest that SIRT1 modulation of FOXO1 is important for proper glucose homeostasis, angiogenesis, and feeding behaviors (96, 206, 281).
5. p300
p300 is an acetyltransferase that regulates numerous signaling pathways by facilitating transcriptional activity of a broad array of transcription factors through modular subdomains. It was demonstrated that SIRT1 interacts with and represses p300 transactivation in a NAD-dependent manner (50). SIRT1 repression involves a transcriptional repression domain of p300 named CRD1, which has two residues (Lys-1020 and Lys-1024) that are essential for SIRT1 repression and serve as substrates for SIRT1 deacetylation (50). These residues also serve as acceptor lysines for modification by the small ubiquitin-like modifier (SUMO) protein. The SUMO-specific protease SSP3 relieves SIRT1 repression of p300, indicating that p300 serves as a deacetylase substrate for SIRT1 through a conserved SUMO consensus motif (50).
6. HSF1
Heat shock factor 1 (HSF1) increases transcription of heat shock proteins in response to cell stress (142). These proteins act to stabilize other proteins, thereby reducing protein misfolding. A recent study shows that SIRT1 deacetylates HSF1, increasing its binding activity with heat shock promoter Hsp70 (350).
7. PPARγ
The peroxisome proliferator-activated receptor (PPAR) γ belongs to the nuclear hormone receptor superfamily and regulates gene expression upon heterodimerization with the retinoid X receptor by ligating to peroxisome proliferator response elements (PPREs) in the promoter region of target genes. PPARγ is considered to be one of the master regulators of adipocyte differentiation (329). It has been shown that upon food withdrawal, SIRT1 stimulates fat mobilization in white adipose tissue by repressing PPARγ (250). The mechanism by which SIRT1 represses PPARγ involves the cofactors NCoR (nuclear receptor corepressor) and SMRT (silencing mediator of retinoid and thyroid hormone receptors) (250). These effects are also observed in 3T3-L1 adipocytes, where activation of SIRT1 inhibits adipogenesis, while the inhibition of SIRT1 increases adipogenesis (250).
8. PGC-1α
PGC-1α is the inducible coactivator-1α of PPARγ, which is expressed at low levels in the liver during the fed state, but at high levels after food deprivation (135). PGC-1α plays a critical role in the regulation of hepatic gluconeogenesis and fatty acid oxidation. Consistent with this role, increased expression of PGC-1α in the liver results in enhanced hepatic glucose production (135). As discussed in more detail below, SIRT1 deacetylates PGC-1α (275), and this appears to be a crucial signal for the actions of SIRT1 at hepatic level.
9. PPARα
PPARα is a ligand-activated transcription factor that belongs to the steroid hormone receptor superfamily. PPARα is expressed predominantly in tissues that have a high level of fatty acid catabolism, such as liver, heart, and muscle, where it regulates the expression of a number of genes critical for lipid and lipoprotein metabolism. Hepatic deletion of SIRT1 leads to impaired PPARα signaling and decreases fatty acid beta-oxidation, whereas overexpression of SIRT1 activates PPARα (259). Importantly, the actions of SIRT1 on PPARα require the deacetylation of PGC-1α (259).
10. LXRs
The liver X receptors (LXRs) alpha and beta (LXRα and LXRβ) are nuclear receptors that act as cholesterol sensors. SIRT1 deacetylates a single conserved lysine on each receptor (K432 in LXRα and K433 in LXRβ), inducing receptor activity, which exerts important regulatory action on cholesterol homeostasis (194). Consistent with this SIRT1-mediated regulation, SIRT1 knockout (KO) mice have lower levels of HDL cholesterol, while their low-density lipoprotein (LDL) cholesterol levels are unaffected (194). Moreover, SIRT1 KO mice show a reduced cholesterol transport in macrophages and the liver, and this leads to an accumulation of hepatic cholesterol (194).
11. SREBP-1c
Sterol regulatory element-binding protein (SREBP) 1c is a transcriptional regulator of fatty acid synthesis enzymes. It has been demonstrated that SIRT1 deacetylates and inhibits SREBP1c activity in the liver (256, 342, 373), and it appears that the lipolytic action of SIRT1 in the liver is dependent on SREBP1c (256). The inhibition of hepatic SIRT1 activity was associated with an increase in the acetylated active nuclear form of SREBP-1c in the livers of ethanol-fed mice, suggesting that the effect of ethanol on SREBP-1 is mediated, at least in part, through SIRT1 inhibition (373).
12. UCP2
Uncoupling protein 2 (UCP2) belongs to the family of mitochondrial carriers. UCP2 promotes longevity by shifting a given cell towards fatty acid fuel utilization (14). SIRT1 represses UCP2 transcription by binding directly to its promoter (47). Consistent with this regulatory role, it was observed that transgenic mice overexpressing SIRT1 in pancreatic β-cells have lower levels of UCP2 and enhanced insulin secretion (225). Additionally, the effects of SIRT1 inhibition in the brain decreasing food intake and promoting synaptic changes are abrogated in mice deficient for UCP2 (87).
13. PTP1B
PTP1B is a member of the protein tyrosine phosphatase family. PTP1B plays a crucial role as an insulin receptor phosphatase, and PTP1B-deficient mice are more sensitive to insulin, have improved glucose metabolism, and are resistant to diet-induced obesity (94). It has been shown that the beneficial effects of SIRT1 activation on insulin sensitivity are, at least partially, mediated by the repression of PTP1B transcription at the chromatin level (317). Resveratrol, which may be an activator of SIRT1, also suppresses PTP1B, which might be a key step for enhancing insulin sensitivity (317).
14. MyoD
The transcription factors of the MyoD family have essential functions in myogenic lineage determination and skeletal muscle development. These myogenic regulatory factors activate muscle-specific transcription of numerous genes. SIRT1 has been shown to retard muscle differentiation through its interaction with MyoD (111). However, SIRT1 does not interact directly with MyoD, but forms a complex with the acetyltransferase p300/CBP-associated factor (PCAF), and then deacetylates both PCAF and MyoD (111).
15. TIP60
The histone acetyltransferase TIP60 can acetylate many substrates, including histones and p53, promoting apoptosis. SIRT1 deacetylates and inactivates TIP60 activity in vivo, suggesting that SIRT1 plays an important role in the control of histone acetyltransferase activity and function in response to DNA damage (346).
16. NF-κB
The nuclear factor kappa enhancer binding protein (NF-κB) regulates diverse biological processes including immunity, inflammation, and apoptosis. SIRT1 physically interacts with the RelA/p65 subunit of NF-κB and inhibits transcription by deacetylating RelA/p65 at lysine 310 (369). The interaction between SIRT1 and the NF-κB signaling pathway is also involved in the cigarette smoke-mediated proinflammatory cytokine release (364). This interaction was observed in a monocyte-macrophage cell line and in inflammatory cells of rat lungs. However, one important role of NF-κB is to mediate pancreatic β-cell damage. Studies in isolated rat islets or RINm5F cells have demonstrated that overexpression of SIRT1 completely blocked cytokine-induced cell damage (192), indicating that the SIRT1/NF-κB pathway plays an important role mediating β-cell damage. Overexpression of SIRT1 was further shown to attenuate hepatic NF-κB activation in vitro and in vivo (249).
17. IRS
Studies performed in vitro have shown that resveratrol inhibited the phosphorylation of insulin receptor substrate 1 (IRS1) Ser-307 and IRS2 Thr-348, which are markers of insulin resistance (92), indicating that resveratrol improves insulin signaling (106). In agreement with this, SIRT1 overexpression increased the phosphorylation of PKB, a downstream target of insulin signaling, in muscle cells and HEK293 cells, whereas the inhibition of SIRT1 induced the opposite response (106). Furthermore, suppression of SIRT1 activity inhibited the tyrosine phosphorylation of insulin receptor substrate 2 (IRS2) in HEK293 cells (380). In neurons, the inhibition of SIRT1 increased the acetylation and reduced the phosphorylation of IRS2 (196). The reduced activity of SIRT1 also reduced Ras activation and ERK1/2 phosphorylation (both markers of oxidized proteins and lipids) (196). Thus this article suggests that SIRT1 can protect neurons from oxidative damage.
18. E2F1/p73
E2F1 is known to induce the transcription of several apoptotic genes and can induce apoptosis after DNA damage events through both p53-dependent and p53-independent mechanisms. SIRT1 inhibits E2F1 (343), and this inhibition suppresses p73 transcriptional activity (81, 247). It seems that SIRT1 interacts with PCAF and E2F1 on the P1p73 promotor (247), and thereby can modulate tumor proliferation.
19. MEF2
MEF2 is a family of transcription factors with important functions in muscle cell differentiation and apoptosis (216). SIRT1 can potently induce MEF2 deacetylation during muscle cell differentiation (384), indicating that this mechanism might be responsible of the negative regulation of myogenesis by SIRT1.
20. TORC2
The CREB regulated transcription coactivator 2 (CRTC2 or TORC2) plays an essential role in gluconeogenesis. For instance, the stimulation of the gluconeogenic pathway induced by glucagon requires the dephosphorylation and nuclear translocation of TORC2. SIRT1 reduces TORC2 activity in the liver, while the inhibition of SIRT1 signaling induces TORC2 activity (206), suggesting that this pathway is an important modulator of the gluconeogenesis (discussed below).
21. PARP1
Sirtuins and PARPs act as survival and death inducing factors. These two protein families are both dependent on NAD+ for their activities. In response to DNA damage, SIRT1-dependent deacetylation blocks PARP1 activity, and it protects cells from PARP1-mediated cell death (178, 263).
22. HIF1α
Hypoxia-inducible factor 1α (HIF1α) is a transcription factor that under hypoxic conditions regulates the transcription of hundreds of genes in a cell type-specific manner (297). SIRT1 binds and deacetylates HIF1α, and therefore inactivates HIF1α by blocking p300 recruitment (199). In hypoxic conditions, SIRT1 is downregulated due to decreased NAD+ levels, which allows the acetylation and activation of HIF1α (199).
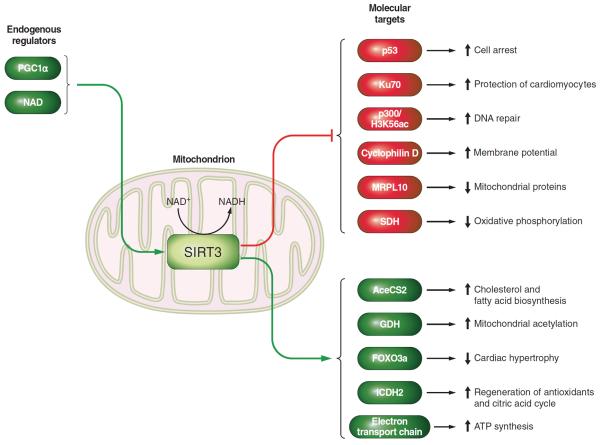
B. Mitochondrial Targets of SIRT3
The NAD+-dependent deacetylase SIRT3 is localized in mitochondria (207, 218, 236, 290, 294, 300) and plays an important role in mitochondrial metabolism (300). To exert these metabolic actions, SIRT3 regulates the expression of many mitochondrial proteins through a reversible deacetylation process (125, 293). Some of the SIRT3 substrates discussed below are nuclear, and given the fact that nuclear SIRT3 is a matter of controversy, further studies are necessary to clarify the interaction between SIRT3 and nuclear proteins (FIGURE 5).
FIGURE 5.
Schematic overview of the endogenous regulators and molecular targets of SIRT3.
1. AceCS2
Acetyl-CoA synthase 2 (AceCS2) was the first SIRT3 mitochondrial protein substrate to be identified (125, 293) and is an enzyme that catalyzes the production of acetyl-CoA from acetate (110). AceCS2 undergoes regulation by reversible acetylation; SIRT3 deacetylates and activates AceCS2, whereas acetylation of AceCS2 inhibits its activity (125, 293). Since Acetyl-CoA is an important regulator of several metabolic pathways, including cholesterol and fatty acid biosynthesis, it is possible that SIRT3 modulates those biological actions.
2. HMGCS2
Hydroxyl methylglutaryl CoA synthase 2 (HMGCS2), the rate-limiting step in β-hydroxybutyrate synthesis, was shown to be deacetylated and activated by SIRT3 in the mitochondria (301). Upon fasting, SIRT3-mediated HMGCS2 activation was required to induce the production of ketone bodies (301).
3. GDH
Glutamate dehydrogenase (GDH) is a key metabolic enzyme that is colocalized with SIRT3 in the mitochondrial matrix (290). SIRT3 deacetylates and activates GDH in vitro (290) and in vivo (207).
4. p53
SIRT3 suppresses p53 activity leading to growth arrest and senescence (363). This was demonstrated in the EJ-p53 cell line, where SIRT3 colocalizes with p53 in the mitochondria before inducing cell arrest (363). A close interaction between SIRT3 and p53 also plays an important function in mouse preimplantation in vitro (167), even though mice lacking SIRT3 are fertile.
5. Ku70
SIRT3 protects cardiomyocytes from stress-induced cell death by deacetylating Ku70, leading to the interaction between Ku70 and the proapoptotic protein Bax. This pathway is associated with a protective function of SIRT3 on cardiomyocytes under stress conditions (318, 319).
6. FOXO3a
SIRT3 activates FOXO3a in cardiomyocytes leading to the activation of a molecular pathway involving ROS/Ras/MAPK/ERK and PI3K/Akt, which ultimately inhibits cardiac hypertrophy (318). The actions of SIRT3 on cardiomyocytes have been also demonstrated in vivo, as mice lacking SIRT3 shows signs of cardiac hypertrophy at 8 wk of age (318). Similar results were obtained in C. elegans, where daf-16, the homolog of the human FOXO family, was also deacetylated by SIRT3 (155).
7. p300/H3-K56
Acetylation of histone H3 core domain lysine 56 (H3-K56) is essential for the compaction of DNA into chromatin, and the histone acetyltransferase p300 acetylates H3-K56 in vivo. Human SIRT3 deacetylates H3K-56 (341), indicating that SIRT3 regulates the DNA damage response pathway. As previously mentioned, the nonmitochondrial localization of SIRT3 remains to be fully confirmed. As such, the validity of p300 as a substrate for SIRT3 will require further investigation.
8. Cyclophilin D
Increasing evidence implicates a multi-protein complex called the mitochondrial permeability transition pore (mPTP) in the decline in mitochondrial function with age. Acute triggering of the mPTP can lead to apoptosis, whereas low-level, chronic triggering results in mitochondrial swelling, membrane depolarization, and the destruction of defective mitochondria by autophagy. SIRT3 deacetylates the regulatory component of the mPTP, cyclophilin D (CypD) on lysine 166, adjacent to the binding site of cyclosporine A, a CypD inhibitor (122, 304). Consistent with this, cardiac myocytes from mice lacking SIRT3 exhibit an age-dependent increase in mitochondrial swelling due to increased mPTP opening (122). SIRT3−/− mice show accelerated signs of aging in the heart including cardiac hypertrophy and fibrosis at 13 mo of age and are hypersensitive to heart stress. Deacetylation of cyclophilin D also induces dissociation of hexokinase II from the mitochondria and prevents cell death (304), which produces a transition from a reliance on glycolysis to oxidative phosphorylation (304).
9. MRPL10
The mitochondrial ribosomal protein L10 (MRPL10) plays an important function in the acetylation in the mitochondrial chromosome. SIRT3 overexpression in C2C12 cells induces the deacetylation of MRPL10 and thereby diminishes the synthesis of proteins in the mitochondria (367). Consistent with these findings, the inhibition of SIRT3 in those cells leads to an increased synthesis of mitochondrial proteins (367).
10. SDH
Succinate dehydrogenase (SDH) is a unique enzyme, as it participates in both the tricarboxylic acid cycle and oxidative phosphorylation in mitochondria. Therefore, it is essential for mitochondrial metabolism, and its deletion is lethal. One of its subunits, SDhA, is a substrate for SIRT3, allowing this sirtuin to regulate oxidative phosphorylation (59).
11. OTC
SIRT3 was shown to deacetylate and thereby activate ornithine transcarbamoylase (OTC), a rate-limiting enzyme in the urea cycle (126). Calorie restriction increased both SIRT3 and OTC activity; in SIRT3 null mice, low OTC activity during fasting was connected with increased orotic acid levels, indicating a disturbed nitrogen balance. Hallow et al. (126) further conducted LC/MS analyses of mitochondrial proteins in SIRT3 null and wild-type mice and revealed a number of novel SIRT3 targets involved in β-oxidation and amino acid catabolism.
12. ICDH2
The isocitrate dehydrogenase 2 (ICDH2) is another key metabolic regulator in the mitochondrial matrix that is also colocalized with SIRT3 in the mitochondrial matrix. ICDH2 is also deacetylated and activated by SIRT3 (290), an effect that was abolished by a sirtuin inhibitor. The precise acetylation sites for ICDH2 (K75 and K241) were found to be deacetylated by SIRT3 (290). The increased activity of ICDH2 promotes regeneration of antioxidants and catalyzes the citric acid cycle (290). Very recently, the deacetylation of ICDH2 by SIRT3 was shown to augment the mitochondrial glutathione antioxidant defense system in mice via increasing NADPH levels and the ratio of reduced-to-oxidized glutathione in mitochondria (310). Importantly, calorie restriction-induced SIRT3 activation was thereby linked to the prevention from extensive age-related oxidative damage (310).
13. MnSOD
Recently, SIRT3 was shown to deacetylate manganese superoxide dismutase (MnSOD), thereby increasing MnSOD activity (318, 325). Such SIRT3-mediated MnSOD activation consequently decreased mitochondrial superoxide. Conversely, SIRT3 ablation in mice decreased hepatic MnSOD activity, which might contribute to the tumor-permissive environment seen in SIRT3−/− animals. Accordingly, SIRT3−/− mice displayed higher susceptibility to radiation-induced liver damage (325). On the other hand, SIRT3 blocked cardiac hypertrophy via MnSOD activation, suggesting that SIRT3 is an important negative regulator of cardiac hypertrophy (318).
14. ATP and the electron transport chain
A recent study shows that SIRT3 deacetylates one or more proteins of the electron transport chain complex I, including NDUFA9 (5). Complex I activity is inhibited in SIRT3−/− mice and potentiated in mitochondria that have been exposed to increased levels of SIRT3 (5). These results suggest that SIRT3 plays an important role in regulating ATP synthesis in mitochondria and thus is a potential regulator of mitochondrial energy metabolism (5).
V. ENDOGENOUS REGULATORS OF SIRT1 AND SIRT3
Besides the classical activation of sirtuins by increased NAD+/NADH ratios, a number of other endogenous processes regulate SIRT1 and SIRT3 activity. Transcription of sirtuins can be controlled by a number of transcription factors and transcriptional coactivators or repressors. Furthermore, mRNA stability of sirtuins depends on the presence of specific microRNAs. Last, sirtuin activity can be regulated by posttranslational modifications through protein-protein interaction, stimulating or repressing sirtuin activity (FIGURES 4 and 5).
A. Endogenous Regulators of SIRT1
1. E2F1
E2F1 induces SIRT1 expression at the transcriptional level (53, 343). E2F1 is also a substrate of SIRT1, and deacetylation of E2F1 inhibits its activity as a transcriptional activator. This action is modulated by the interaction of SIRT1 with PCAF, which controls the E2F1/p73 apoptotic pathway (247). Accordingly, the downregulation of SIRT1 increases E2F1 transcriptional and apoptotic functions (53, 343). Therefore, this SIRT1-E2F1 negative-feedback loop might act as a regulatory switch that can determine the apoptotic fate of a cell.
2. p53
In addition to being a target for SIRT1 deacetylation, p53 can also repress SIRT1 transcription through binding to two response elements within the SIRT1 promoter. p53-null mice have increased levels of SIRT1 in different tissues, and several p53-null tumor cell lines display SIRT1 overexpression (104, 229). Therefore, SIRT1 and p53 also form a regulatory feedback loop since SIRT1 is known to deacetylate p53.
3. HIC1
The growth regulator and tumor repressor gene Hypermethylated In Cancer 1 (HIC1) also suppresses SIRT1 transcription. HIC1, COOH-terminal binding protein 1 (CTBP1), and SIRT1 form a corepressor complex (76) that binds enhancer elements upstream of the SIRT1 promoter and inhibits SIRT1 expression. In both mouse and human prostate cancer cells, as well as Hic−/− mouse embryonic fibroblasts, reduction or ablation of HIC1 is associated with an increase in SIRT1 expression levels (143), indicating one possible explanation of the increased levels of SIRT1 during tumorigenesis.
4. HuR
HuR is a ubiquitously expressed RNA-binding protein that stabilizes mRNAs and regulates translation to protein. The tumor suppressor HuR (also known as ELAVL1) is an mRNA binding protein that binds the 3′ UTR of SIRT1 mRNA, stabilizes the SIRT1 mRNA, and increases SIRT1 expression levels (1). Moreover, HUR gene expression correlates with the reduced levels of SIRT1 expression in senescent cells (1).
5. miR-34a
MicroRNAs are posttranscriptional gene regulators that are differentially expressed under several physiological conditions. The microRNA miR-34a also binds the 3′ UTR of SIRT1 mRNA (358, 359). In contrast to HUR, miR-34a prevents translation of SIRT1 and so inhibits SIRT1 deacetylase activity, subsequently inducing the accumulation of acetylated p53.
6. miR-199a
In addition to miR-34a, miR-199a has also been associated with SIRT1 regulation. Knockdown of miR-199a results in the upregulation of SIRT1 and HIF1α (271). As this report found that miR-199a levels were decreased in cardiac myocytes upon hypoxia, the data indicate that SIRT1 is important for hypoxic damage (271).
7. DBC1
Deleted in breast cancer 1 (DBC1, also known as KIAA1967) is an inhibitor of SIRT1 activity (170, 172, 382). Moreover, reduction of DBC1 inhibits p53-mediated apoptosis after induction of double-stranded DNA breaks owing to SIRT1-mediated p53 deacetylation. Although little is known about the normal function of DBC1, its loss in several cancer cell lines and its inhibition of SIRT1 suggest it may have an important role in tumorigenesis.
8. AROS
Active regulator of SIRT1 (AROS, also known as RPS19BP1) is a 142-amino acid nuclear protein, which directly activates SIRT1 activity and attenuates p53-dependent transcriptional activation (170). Consistent with this role, reduction of AROS increased cell susceptibility to apoptosis after DNA damage (170).
9. Necdin
Necdin is a member of the melanoma-associated antigen (MAGE) family of proteins. These proteins play multiple functions in cellular regulation, including tumorigenesis or neuronal differentiation and survival. Necdin has been shown to negatively regulate p53 by potentiating SIRT1-mediated deacetylation of p53 (128). Inhibition of p53 acetylation through a necdin-SIRT1-p53 complex prevents p53-mediated apoptosis in response to DNA damage.
10. AMPK
AMPK controls the expression of genes involved in energy metabolism in mouse skeletal muscle by acting in coordination with SIRT1. AMPK increases SIRT1 activity by increasing cellular NAD+ levels (57). Interestingly, this interaction between SIRT1 and AMPK seems to be reciprocal, as SIRT1 activation stimulates fatty acid oxidation and indirectly activates AMPK (99).
11. FOXO3a
Forkhead box O-class (FOXO) transcription factors function as tumor-suppressor proteins by inhibiting cell proliferation, promoting apoptotic cell death, and protecting cells from DNA damage and oxidative stress. FOXO3a stimulates SIRT1 transcription through two p53 binding sites present in the SIRT1 promoter (229). Consistently, knockdown of FOXO3a expression inhibits the starvation-induced increase in SIRT1 expression (229).
12. c-Myc
c-Myc regulates processes involved in many, if not all, aspects of cell fate. c-Myc and SIRT1 form a negative-feedback loop that inhibits c-Myc-induced cellular transformation. On one hand, c-Myc binds to the SIRT1 promoter and induces SIRT1 expression (375). However, SIRT1 interacts with and deacetylates c-Myc, resulting in decreased c-Myc stability and subsequent suppression of SIRT1 expression (375).
13. SHP
Orphan nuclear receptor small heterodimer partner (SHP) is a transcriptional corepressor of a wide variety of nuclear receptors. It has been reported that SHP interacts with and colocalizes with SIRT1, inhibiting the transcriptional activity of SIRT1 (71).
14. PARP1 and PARP2
The deletion of PARP1, a gene encoding a major NAD+-consuming enzyme, increases SIRT1 activity in brown adipose tissue and muscle. Also, the pharmacological inhibition of PARP in vitro and in vivo increased SIRT1 activity (24). Similar to PARP1, PARP2 deficiency also stimulated SIRT1 activity in cultured myotubes (23). However, unlike PARP1, PARP2 acts as a negative regulator of the SIRT1 promoter, and this regulation is independent of NAD+ levels.
B. Endogenous Regulators of SIRT3
Little is known about endogenous regulators of SIRT3. One report suggested that PGC-1α stimulates mouse SIRT3 activity in both muscle cells and hepatocytes (180). In agreement with this, SIRT3 knockdown suppressed the actions of PGC-1α on mitochondrial biogenesis in myotubes (180).
VI. METABOLIC TISSUES TARGETED BY SIRT1
A. Roles of SIRT1 in Liver Metabolism
1. SIRT1 and hepatic glucose metabolism
In liver, SIRT1 is upregulated during negative energy balance, as occurs during fasting and calorie restriction (7, 61, 229, 233, 275). In 2005, research led by Puigserver and collaborators (275) revealed that during fasting SIRT1 stimulates gluconeogenesis and inhibits glycolysis by deacetylating the transcriptional coactivator PGC-1α. The authors showed that SIRT1 interacts with and deacetylates PGC-1α in a NAD+-dependent manner (230, 275). Importantly, the regulation of PGC-1α by SIRT1 was found to be essential for fasting- or pyruvate-mediated increases of gluconeogenic genes (PEPCK and G6Pase) and glucose output in cultured hepatocytes. Furthermore, this deacetylation also acts to decrease the expression of glycolytic genes (glucokinase and LPK) (275). This effect of SIRT1 stimulating hepatic glucose output during fasting was later confirmed in mice (96, 274).
SIRT1 has been shown to regulate hepatic glucose metabolism (FIGURE 6) by interacting with the FOXO family of transcription factors (166, 177). FOXO proteins are inactivated by the PI3k-Akt/PKB pathway in response to hormonal signaling (32, 120). When phosphorylated, these transcription factors are inactive and are localized in the cytosol; upon dephosphorylation, they have nuclear localization and are active (383). Several groups have shown the close and intricate interaction between SIRT1 and FOXO. Motta et al. (224) found that SIRT1 binds to FOXO3a and promotes its deacetylation with consequent inhibition of FOXO3a transcriptional activity. On the other hand, FOXO3a induced by starvation promotes SIRT1 expression in a p53-dependent manner (229). Additionally, SIRT1 was also shown to interact and deacetylate FOXO1 (365) and FOXO4 (177). The FOXO family of transcription factors has been shown to stimulate the expression of genes involved in gluconeogenesis (pepck and g6pase) (353, 368), while decreasing expression of glucose kinase (gk) (21, 292, 381). During prolonged fasting, FOXO1 activity supported by SIRT1 seems to be a key element in the maintenance of glucose production (205). SIRT1 activation has been shown to stimulate FOXO1 translocation to hepatocyte nuclei (105). Under conditions of SIRT1 activation or oxidative stress, FOXO1 is confined to a nuclear subdomain where it boosts transcription of genes that promote gluconeogenesis and hepatic glucose production (105). Importantly, mutations of the FOXO1 coactivator-interacting LXXLL motif eliminated FOXO1 interaction with SIRT1, indicating that this motif is essential for SIRT1 actions mediated by FOXO1 (226).
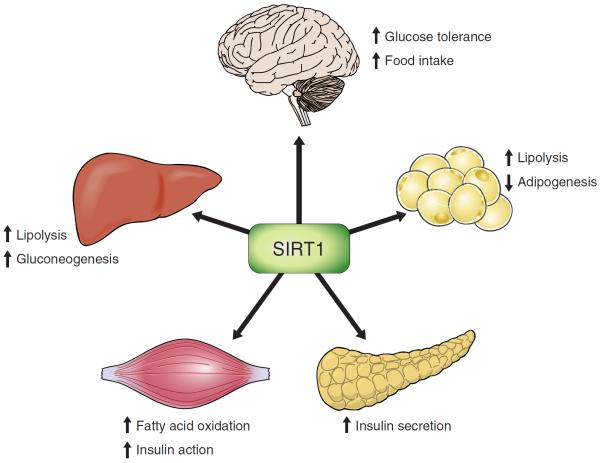
FIGURE 6.
Metabolic roles of SIRT1 in peripheral tissues and the central nervous system.
SIRT1 is also known to regulate gluconeogenesis via its interaction with STAT3 (233). STAT3 suppresses gluconeogenesis by inhibiting the transcription of PEPCK1 and g6pase (152, 153), and its activity is dependent on the phosphorylation of amino acid residues. STAT3 contains several lysine sites that are acetylated. This acetylation has been shown to regulate the activity and phosphorylation of this protein. SIRT1 deacetylases STAT3, thus decreasing STAT3 activity and its inhibitory effect on gluconeogenesis. Therefore, activation of SIRT1 during fasting stimulates gluconeogenesis by inhibiting STAT3 while activating PGC-1α and FOXO1.
In contrast, a number of studies suggest that the activation of SIRT1 inhibits insulin-induced hepatic glucose production in obese rats (220). Moreover, a recently identified fasting-inducible switch, consisting of SIRT1 and p300, causes a transfer between two key regulators of glucose production (206). The switch occurs during nutrient deprivation, when regulation of the gluconeogenic program is shifted from CREB-regulated transcription coactivator 2 (CRTC2, also TORC2) to FOXO1 (206). During early fasting, glucagon induces p300/CBP-mediated acetylation of CRTC2, which briefly increases gluconeogenic activity. During prolonged fasting, SIRT1 deacetylates and downregulates CRTC2 and promotes a FOXO1-mediated gluconeogenic program (206). Therefore, SIRT1 may also act to suppress hepatic glucose production during prolonged fasting (206).
2. SIRT1 and hepatic fatty acid metabolism
SIRT1 plays a prominent role in the regulation of hepatic fatty acid metabolism (FIGURE 6). Specifically, SIRT1 regulated hepatic lipid metabolism by activating the AMPK/LKB1 signaling pathway (140). Notably, LKB1 was essential to produce downstream effects of SIRT1 on fatty acid oxidation and lipogenesis (140). In a hyperglycemic environment, SIRT1-mediated activation of AMPK prevents lipid accumulation and FAS induction (140). Consistent with this regulation, mice fed a high-fat diet treated with resveratrol or the SIRT1 agonist SRT1720 exhibited improved liver physiology and metabolic function (34, 220, 249).
A subsequent study suggests that although SIRT1 improves insulin sensitivity, transgenic mice overexpressing SIRT1 on an atherogenic diet display elevated hepatic lipid accumulation and secretion, in spite of enhanced glucose homeostasis (261).
3. SIRT1 and hepatic cholesterol metabolism
SIRT1 also plays an important role in cholesterol homeostasis. The cholesterol-sensing LXR proteins are nuclear receptors involved in maintaining cholesterol homeostasis. LXRs act to enhance the reverse transport of cholesterol from peripheral tissues by stimulating the expression of the ATP-binding cassette transporter A1 (ABCA1). These transport proteins direct cholesterol to apolipoprotein AI to form high-density lipoproteins (194). SIRT1 interacts with and deacetylates the LXRs, influencing several targets including the ABCA1 (194), and therefore, regulating HDL production (19). SIRT1 also modulates cholesterol metabolism through the critical regulator SREBP. In cultured cells, the activation of SIRT1 increases SREBP ubiquitination, decreases SREBP nuclear levels, and diminishes the pool of active SREBP, thereby decreasing SREBP gene expression (342). SIRT1 seems to have a conserved role in metazoans in the repression of SREBP during fasting, resulting in inhibition of lipid synthesis and fat storage (342). In vivo, the administration of the synthetic SIRT1 activator SRT1720 to leptin-deficient mice fed a high-fat diet inhibited the expression of SREBP and improved the hepatosteatosis (342).
B. Roles of SIRT1 in Pancreatic β-Cells
1. SIRT1 and insulin secretion
In addition to the regulation of hepatic glucose metabolism, SIRT1 has also been linked to glucose-stimulated insulin secretion in pancreatic β-cells (47, 225) (FIGURE 6). A selective increase in the dosage of SIRT1 in pancreatic β-cells improves glucose tolerance and insulin release in response to glucose and KCl (225). Conversely, SIRT1 knockdown in β-cells had the opposite effect on insulin secretion (47). This effect appears to be partially induced via SIRT1-mediated repression of ucp2 (47). Intriguingly, this improved insulin sensitivity is not exclusive to the β-cell. SIRT1 was shown to improve insulin sensitivity by decreasing the transcription of ptp1b in vitro in myotubes (317). PTP1B acts as a negative regulator of insulin receptor signaling, and its downregulation improves insulin sensitivity (94). Transgenic mice with mild overexpression of SIRT1 also showed improved glucose tolerance in several models of insulin resistance, an effect that was mediated by decreased hepatic production of glucose and increased adiponectin levels, and not by differences in β-cell sensing of glucose (28). Alternatively, SIRT1 may act as a mediator for other signals. For instance, wallerian degeneration slow, a fusion protein that is highly expressed in the pancreas and improves glucose homeostasis, increases SIRT1 activity to downregulate UCP2 expression, leading to an improvement in glucose homeostasis (355). On the other hand, glucagon-like peptide 1 (GLP-1) inhibits SIRT1. This inhibition is essential for the GLP-1 stimulation of β-cell mass expansion (33).
2. SIRT1 and β-cell protection
SIRT1 also plays a role in pancreatic beta cell protection. In the hyperglycemic state, oxidative stress can induce pancreatic beta cell failure (175). SIRT1, FOXO1, and Pml (promyelocytic leukemia protein) form a complex that upregulates transcription factors (NeuroD and MafA) to protect β-cells from hyperglycemia-induced damage (175). Also, the SIRT1-mediated regulation of NF-κB may prevent pancreatic β-cell toxicity (192). SIRT1 has been shown to regulate NF-κB by deacetylating lysine 310 on its relA/p65 subunit (369).
C. Roles of SIRT1 in Skeletal Muscle
1. SIRT1 and skeletal muscle fatty acid metabolism
When skeletal muscle is nutrient-deprived, glucose oxidation shifts to fatty acid oxidation. SIRT1 is activated during low energy states, and therefore, it makes sense that SIRT1 would induce fatty acid oxidation as the cell shifts from glucose consumption to fatty acid oxidation (114) (FIGURE 6). In skeletal muscle, SIRT1 deacetylates PGC-1α to induce mitochondrial fatty acid oxidation in a glucose-scarce environment (114). SIRT1 deacetylates and activates acetyl-CoA synthetase (AceCS) 1, which can induce substantial fatty acid synthesis (125). AceCS1 is regulated by reversible acetylation, a process in which acetylation inactivates AceCS1 and SIRT1-induced deacetylation reactivates it. In vivo studies have shown that mice treated with the potential SIRT1 activator resveratrol (the specificity of resveratrol for SIRT1 activation is critically discussed in section X) significantly increased their aerobic capacity (186). The effects of resveratrol were also associated with an induction of genes for oxidative phosphorylation and mitochondrial biogenesis. Furthermore, these effects were largely explained by the deacetylation of PGC-1α and subsequent increases in PGC-1α activity (186). Importantly, resveratrol protected these mice against diet-induced obesity and insulin resistance (186). SIRT1 may also act on uncoupling protein 3 (UCP3), a mitochondrial protein expressed in skeletal muscle that lowers membrane potential and prevents fatty acid accumulation (11). This regulation occurs via SIRT1-mediated deacetylation of histones near the UCP3 promoter, thereby preventing UCP3 transcription and subsequently the accumulation of fatty acids (11).
2. SIRT1 and skeletal muscle glucose metabolism
As discussed in section IVA13, in vitro studies demonstrated that SIRT1 improves insulin sensitivity by repressing the protein tyrosine phosphatase 1B (PTP1B) gene in skeletal myotube cells (317). PTP1B is a key insulin receptor phosphatase, and PTP1B-deficient mice have been shown to be more insulin sensitive and more resistant to diet-induced obesity compared with controls (94). In vivo studies have also shown that resveratrol was able to increase glucose uptake, an effect independent of insulin and dependent on AMPK, through the increase of the intrinsic activity of the glucose transporter GLUT4 (52). Further in vivo study has demonstrated an increased activity of SIRT1 in muscle when mice were under caloric restriction, an effect that was completely abrogated in mice lacking SIRT1 (288). The mechanisms for this activation likely involve the Stat3/PI3K pathway as SIRT1 is known to inactivate the transcription factor Stat3 during caloric restriction, resulting in more efficient PI3K signaling during insulin stimulation (288).
3. SIRT1 and skeletal muscle differentiation
SIRT1 was shown to negatively regulate muscle gene expression and differentiation. To exert these actions, SIRT1 does not directly interact with muscle transcriptional regulators, but rather associates with the complex PCAF/GCN5. In the presence of PCAF/GCN5, SIRT1 associates with and deacetylates both transcriptional factors PCAF and myogenic determining factor (MyoD) (111). Moreover, in C2C12 myotubes, when MyoD is present with SIRT1 on the PGC-1α promoter, PGC-1α overexpression increases due to positive feedback of PGC-1α on its own promoter (10). Therefore, MyoD enhances the overexpression of PGC-1α in a SIRT1-dependent manner (10).
D. Roles of SIRT1 in White Adipose Tissue Metabolism
1. SIRT1 and adipose tissue fatty acid metabolism
SIRT1 favors lipolysis and fatty acid mobilization in response to fasting by repressing PPARγ (250), which is essential for adipogenesis (FIGURE 6). SIRT1 interacts with PPARγ DNA binding sites, but it is unclear whether SIRT1 deacetylates PPARγ directly (250). Another pathway modulating the lipolytic activity of SIRT1 in adipocytes involves the deacetylation of FOXO1 and stimulation of adipose triglyceride lipase (ATGL) gene transcription (69). Activation of SIRT1 inhibits differentiation and proliferation in pig preadipocytes, whereas inhibition of SIRT1 activates adipocyte differentiation substantially (22). In pig preadipocytes, SIRT1 may inhibit FOXO1 or other genes associated with pig adipocyte development (22). SIRT1 has been shown to regulate adipose tissue inflammation by suppressing induction of proinflammatory transcription in response to fatty acids, hypoxia, and endoplasmic reticulum stress (79, 115). For instance, the reduction of SIRT1 in vivo stimulates macrophage recruitment to adipose tissue, whereas overexpression of SIRT1 prevents adipose tissue macrophage accumulation during chronic high-fat feeding (115).
2. SIRT1 and adipose tissue glucose metabolism
SIRT1 also regulates the production and/or the secretion of insulin-sensitizing factors such as adiponectin and FGF21 through the regulation of FOXO1 and PPARγ (28, 262, 345). These studies suggest that in adipocytes, PPARγ selectively represses the expression of a group of genes containing FGF21. However, in vivo studies have yet to corroborate these findings. Moreover, the adipocytes of mice lacking the leptin receptor (db/db mice), which develop obesity and type 2 diabetes, show lower levels of FOXO1 and SIRT1 than their littermates (315), whereas the activation of SIRT1 by resveratrol rescued FOXO1 levels in the adipocytes of db/db mice (315). The beneficial role of SIRT1 on glucose metabolism might be at least partially explained by the activation of adiponectin. Adiponectin is an important regulator of energy homeostasis, and its expression is reduced in obesity and type II diabetes. SIRT1 activates FOXO1, which forms a transcription complex with CCAAT/enhancer-binding protein α (C/EBPα), thus promoting adiponectin gene transcription (262). SIRT1 may also play a role in the attenuation of ROS. In adipocytes exposed to high levels of free fatty acids, activation of SIRT1 leads to nuclear translocation of FOXO1 and is accompanied by a parallel decrease in ROS production (315). SIRT1-mediated activation of FOXO1 may therefore protect from obesity-induced inflammation and subsequently insulin resistance.
E. Central Nervous System SIRT1 Regulates Metabolic Control
SIRT1 is present in metabolically important areas of the brain, including the arcuate, ventromedial, dorsomedial, and paraventricular nuclei of the hypothalamus, as well as the area postrema and the nucleus of the solitary tract in the hindbrain (268) (FIGURE 6). SIRT1 protein levels are modulated by nutrient status, since fasting increased the hypothalamic levels of SIRT1, and this change was blunted in the hypothalamus of leptin-deficient mice (268). Specifically, SIRT1 mRNA is expressed in pro-opiomelanocortin neurons, which are essential integrators of proper glucose homeostasis and body weight regulation (268). Several studies suggest that SIRT1 regulates food intake and body weight through the central melanocortin system of the brain (55, 87, 281). For instance, a decrease in hypothalamic SIRT1 signaling by pharmacological inhibition or by siRNA prevents FOXO1 deacetylation and downregulates food intake and body weight gain (55, 87, 340). Central administration of an orexigenic melanocortin receptor antagonist reversed the effects of SIRT1 inhibition (55, 87), supporting the hypothesis that hypothalamic SIRT1 interacts with the central melanocortin system in a FOXO1-dependent manner. Pharmacological inhibition of brain SIRT1 decreased the activity of AgRP neurons and the inhibitory tone of POMC neurons (87). This inhibition of SIRT1 decreased food intake through the central melanocortin receptors in a UCP2-dependent manner (87). Moreover, pharmacological blockade of SIRT1 in the central nervous system prevents the orexigenic action of ghrelin (87, 340). Consistent with these pharmacological results, the selective disruption of SIRT1 in AgRP neurons in mice using the cre-lox approach blunted electric responses of AgRP neurons to ghrelin and decreased food intake (87). Also, the AgRP-SIRT1 KO mice displayed decreased fat mass and impaired metabolic response to fast (87). During fasting, increased SIRT1 expression represses FOXO1 activity through increased deacetylation (55). Increased SIRT1 expression also appears to activate S6K signaling (55). Consistently, the overexpression of SIRT1 in the mediobasal hypothalamus rescues hyperphagia and body weight gain induced by constitutive nuclear FOXO1 expression and thereby suppresses the expression of the orexigenic neuropeptide AgRP (281).
There are controversial data surrounding intracerebroventricular (ICV) infusion of the proposed SIRT1 activator resveratrol. Chronic ICV infusion of resveratrol significantly decreased insulin resistance and normalized blood glucose levels in diet-induced obese and diabetic mice (267). Interestingly, despite diminished Iκ-Bα mRNA levels after ICV resveratrol infusion, hypothalamic NF-κB signaling was augmented via deacetylation of RelA/p65 and total RelA/p65 protein (267). Another study using resveratrol has shown that the acute central injections of this compound in the mediobasal hypothalamus during basal pancreatic insulin clamp studies improved glucose homeostasis through a central SIRT1-dependent pathway (176). The mechanism(s) behind these controversial effects has yet to be determined, but it is important to note that several studies suggest that resveratrol is not an activator of SIRT1, and therefore, the effects observed after central administration of resveratrol are not exerted through hypothalamic SIRT1.
Additional studies have reported the effect of the lack of SIRT1 in the entire brain as well as in POMC neurons. Mice lacking SIRT1 in the brain showed defects in somatotropic signaling and in responding to caloric restriction (60). More specifically, brain SIRT1 specific KO mice are dwarfed and old mice are glucose intolerant and display altered caloric restriction responses (60). Another report investigating the lack of SIRT1 in POMC neurons found that these mice were heavier than their littermates and had more perigonadal fat due to lower energy expenditure (266). The deletion of SIRT1 in POMC neurons also abolished the capacity of leptin to activate phosphoinositol 3-kinase (PI3K) signaling in POMC neurons and altered brown adipose tissue and perigonadal white adipose tissue metabolism (266).
In addition to genetic disruption of SIRT1 signaling in the central nervous system, the effect of overexpression of SIRT1 in the brain has been also studied. Elevated levels of SIRT1 in hypothalamic dorsomedial and lateral nucleus increased body temperature and physical activity in response to caloric restriction (282). In summary, central SIRT1 signaling seems to be a key mediator of energy metabolism and the physiological response to calorie restriction. However, the exact mechanisms of central SIRT1 action remain to be elucidated.
F. Roles of SIRT1 in Other Tissues Involved in Metabolic Control
SIRT1 is expressed in a number of additional tissues that are important for metabolic control. For instance, SIRT1 is abundantly expressed in the mouse kidney, particularly in renal medullary interstitial cells (131). Studies in cultured renal medullary interstitial cells have shown that SIRT1 protects from oxidative injury (131) and hypoxia (182), via deacetylation and activation of HIF2α (88). In keeping with this observation, SIRT1 is downregulated during conditions of hypoxia, and therefore activates HIF1α (199). SIRT1 is further expressed in a number of myeloid cells, where it can exert metabolic control. For instance, macrophage SIRT1 was shown to improve insulin sensitivity by attenuating inflammatory pathways (372). To date, nothing is known on the role of SIRT1 in metabolically relevant tissues such as the thyroid or adrenal glands. In the intestine, SIRT1 is known to suppress intestinal tumorigenesis and colon cancer growth (102). However, nothing is known yet on potential roles of intestinal SIRT1 in metabolic control.
VII. METABOLIC TISSUES TARGETED BY SIRT3
A. Roles of SIRT3 in Liver Metabolism
Gene and protein expression of SIRT3 in the liver were activated by fasting (136) and decreased when the animals were fed a high-fat diet (HFD) (30, 168). HFD feeding in SIRT3 KO mice induced an increase in the acetylation of several hepatic proteins, suggesting that SIRT3 is a crucial regulator of hepatic mitochondrial function (168). Mechanistic findings have shown that palmitate modulates oxygen consumption and ROS levels in hepatocytes in a SIRT3-dependent manner (30). Palmitate increases oxygen consumption in hepatocytes, and the inhibition of SIRT3 abolished those actions (30). Similar results were obtained in mice lacking SIRT3, as a palmitate test in these mice induced lipotoxicity in hepatocytes, whereas this effect was abrogated when SIRT3 levels were restored (30) (FIGURE 7). The relevance of SIRT3 in hepatic metabolism was also confirmed in another study showing that SIRT3 overexpression in hepatocytes decreases the accumulation of lipids (299). This effect appeared to be mediated by the activation of AMPK and ACC (299); when AMPK was inhibited by compound C, SIRT3 failed to decrease lipid accumulation (299). In this line, another study demonstrated that SIRT3 induced fatty acid oxidation through the deacetylation of long-chain acyl coenzyme A dehydrogenase (LCAD) and mice lacking SIRT3 showed lower fatty acid oxidation (136). Very recently, it was suggested that hepatic ketone body production is regulated by SIRT3; HMGCS2, the rate limiting enzyme in the synthesis of β-hydroxybutyrate, needs deacetylation to induce the physiological response to prolonged fasting (301).
FIGURE 7.
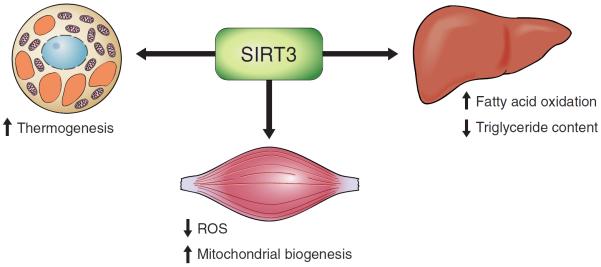
Metabolic roles of SIRT3 in peripheral tissues and the central nervous system.
B. Roles of SIRT3 in Skeletal Muscle Metabolism
Skeletal muscle plays an important role in the regulation of lipid metabolism, as lipid catabolism provides a high percentage of the energy usage for resting muscle. Similar to its regulation in the liver, SIRT3 levels are decreased in the muscle of mice fed HFD, whereas they are increased after fasting and caloric restriction (242). In addition to its regulation by nutritional status, muscle SIRT3 is also modulated by exercise, as trained mice showed higher SIRT3 levels compared with non-exercised mice (139, 242) (FIGURE 7). Furthermore, the lack of SIRT3 inactivates AMPK and pCREB, leading to the inhibition of PGC-1α activity in muscle (242).
C. Roles of SIRT3 in Adipose Tissue Function
SIRT3 has been detected at high levels in brown adipose tissue, while lower levels were found in white adipose tissue (300). In brown adipose tissue, SIRT3 is activated by caloric restriction and cold exposure, and increased temperatures reverse the higher levels of SIRT3 (136, 300) (FIGURE 7). Furthermore, constitutive expression of SIRT3 in brown adipocytes increases the expression of PGC-1α and UCP1, leading to higher thermogenesis and oxygen consumption (300).
VIII. GENETIC MODELS OF SIRT1 AND SIRT3 UNCOVER NEW ROLES IN METABOLIC CONTROL AND CIRCADIAN RHYTHMICITY
A. SIRT1 Gain- and Loss-of-Function Models and Metabolic Dysfunction
1. Global SIRT1 deficiency affects metabolic control
Early studies of lower organisms lacking Sir2 demonstrated that Sir2 deficiency eliminates the life-extending effects of calorie restriction in yeast (200) and decreases life span of flies (18). In SIRT1-deficient mice, the results are somewhat controversial, and different phenotypes have been found. Inbred SIRT1-null mice were born underweight and showed early postnatal lethality (214). Specifically, SIRT1 knockouts died between E9.5 and E14.5 and exhibit problems with proper DNA repair and histone modulation (348). Accordingly, the lack of SIRT1 also produced important developmental defects of the retina and heart (78). Male (inbred) SIRT1-null mice were shown to be sterile, potentially due to reduced expression (and thus signaling) of hypothalamic gonadotropin-releasing hormone and luteinizing hormone (179) and thus attenuated spermatogenesis (67). Nevertheless, approximately half the SIRT1-null mice on an outbred background survived past adulthood (214). Outbred SIRT1-null mice were visibly smaller (25%) than their littermates at 2–4 mo, and this difference peaked (40%) when the mice were 13–20 mo old (45). The surviving mice have a characteristic defect in eyelid development rendering them sightless (214). This body weight difference was not due to food intake, which was similar to that of wild-type mice; in fact, SIRT1-null mice were hyperphagic when food intake was normalized to body weight. However, these mice were hypermetabolic, contained inefficient liver mitochondria, and had elevated rates of lipid oxidation (45).
When the caloric intake was restricted 40%, SIRT1 knockout mice did not show the increased physical activity seen in wild-type mice, and their metabolic rate was lower (45). These findings are similar to those observed after 9 mo of CR, in which SIRT1 null mice failed to increase their physical activity, even though they performed better than wild-type mice on an accelerating treadmill or rotaroad (75). Importantly, CR did not extend the life span of SIRT1-null mice (45). Notably, although mice lacking SIRT1 had reduced levels of markers of oxidative damage in the brain, the life span of these mice was shorter under both normal and caloric restricted conditions (196). In mice, life span is often determined by the individuals and strain-specific susceptibility to develop tumors, and SIRT1 might increase life span by decreasing tumor rates. However, controversy also surrounds the functions of SIRT1 on tumorigenesis. While one study suggested that SIRT1-null mice developed tumors at an increased rate relative to wild-type mice (348), subsequent studies refuted that claim; SIRT1-null mice developed tumors at normal rates, and resveratrol treatment did not have a protective effect against tumorigenesis (44).
The complete lack of SIRT1 has differential metabolic effects on glucose homeostasis and lipid metabolism. Studies by Bordone et al. (47) showed that SIRT1 deficiency seems to improve glucose homeostasis. Global SIRT1-knockout mice demonstrated improved glucose tolerance with decreased levels of insulin and blood glucose (47). SIRT1 suppressed UCP2, which inhibited insulin secretion. Accordingly, SIRT1 knockout mice showed high UCP2 expression (47). It is possible, though, that an improvement in glucose homeostasis could be due simply to the reduced body weight, fat mass, and size of the SIRT1-knockout mice. On the other hand, partial lack of SIRT1 (SIRT1 +/− mice) led to liver steatosis when mice were fed a medium-fat diet (356). The steatosis was accompanied by an elevated liver-to-body ratio, liver size, and hepatic lipid deposition (356). Therefore, it seems that the disruption of SIRT1 activity may cause beneficial effects on glucose, but it favors lipid deposition in the liver.
Li et al. (194) determined that global SIRT1 deficiency also affects cholesterol regulation. The LXRs are important for cholesterol and triglyceride homeostasis (194). SIRT1-null mice demonstrated abnormal cholesterol homeostasis due to the decreased expression of LXR targets and, as a consequence, a reduction of reverse cholesterol transport as well as high-density lipoproteins (194), but low-density lipoprotein cholesterol was unchanged compared with wild-type mice (194).
2. Liver-specific SIRT1 knockdown affects metabolic control
The metabolic consequences of SIRT1 deficiency in liver-specific SIRT1-KO mice are even more variable and disputed than those of global SIRT1-knockout mice. In 2007, Rodgers and Puigserver (276) observed that hepatic SIRT1 knockdown in mice led to mild hypoglycemia, decreased glucose production, increased insulin sensitivity, decreased blood cholesterol levels, and increased levels of cholesterol and free fatty acids in the liver during fasting. These results were consistent with SIRT1 regulation of gluconeogenesis, cholesterol homeostasis, and free fatty acid oxidation in the liver (276). Most of these results were observed only under fasting conditions (276), and the overexpression of SIRT1 reversed many of these changes. However, another study suggests that SIRT1 knockdown in the liver may be beneficial in a rat model of type 2 diabetes (96). The knockdown of SIRT1 in those rats improved insulin sensitivity, lowered fasting glucose levels, decreased hepatic glucose production, and decreased plasma total cholesterol levels (96).
Studies in liver-specific SIRT1-knockout mice on a high-calorie “Western” diet suggested that these mice had at least some protection from accumulating fat, compared with wild-type mice. In contrast, while under calorie restriction, the liver-specific KO mice displayed the same phenotype as wild-type mice. These observations suggested that in normal mice hepatic SIRT1 may be inactivated as a result of a calorie-restricted diet and activated as a result of a high-calorie diet (74). As discussed before, these results are in direct contrast to the dogma that calorie restriction universally increases SIRT1 activity, and new questions are brought to light concerning the unique role of hepatic SIRT1 (74). Contradicting the above findings, a 2009 study by Purushotham et al. (259) suggested that hepatic SIRT1 knockout was harmful for mice that were challenged with a high-fat diet. These mice developed many metabolic problems including liver steatosis, hepatic inflammation, and endoplasmic reticulum stress (259). Accordingly, later research indicated that the specific deletion of hepatic SIRT1 in mice impaired the mTorc2/Akt signaling and resulted in hyperglycemia, oxidative damage, and insulin resistance (347). In this sense, the hepatic overexpression of SIRT1 in mice ameliorated insulin resistance and restored glucose homeostasis (195).
Overall, based on the existing evidence collected in SIRT1-null mice, it is difficult to establish the precise role of hepatic SIRT1 in regulating glucose, cholesterol, and lipid metabolism. Species differences, methodological differences (viral knockdown vs. germline mutation), the overall problem of genetic background (inbred vs. outbred), and the compromised health of SIRT1-null mice are factors that make it hard to evaluate the role of hepatic SIRT1.
3. Beneficial effects of global SIRT1 overexpression on systemic metabolism
As happened with global SIRT1 deletion, the overexpression of SIRT1 in mice also showed different results with respect to body weight and feeding behavior. However, most studies show compelling evidence that SIRT1 overexpression offers substantial benefits on serum cholesterol and insulin levels and increased resistance to high-fat diet induced glucose intolerance and insulin resistance (28, 46, 249). Mice overexpressing SIRT1 under the control of a β-actin promoter were leaner and more metabolically active than their standard-diet fed littermates. In addition, they had lower cholesterol, insulin, and fasting glucose levels as well as improved glucose tolerance (46). Furthermore, these transgenic mice performed better on a rotarod assay (46). All these characteristics resembled the phenotype showed by mice under CR. However, subsequent studies in mice that overexpressed SIRT1 under its own promotor showed slightly different results (28, 249). Banks et al. (28) showed that on a standard diet SIRT1 transgenic mice had normal insulin sensitivity but decreased food intake and locomotor activity, resulting in decreased energy expenditure but normal body weight. Concomitant overexpression of SIRT1 in environmental and genetic models of insulin resistance and diabetes improved glucose tolerance due to decreased hepatic glucose production and increased adiponectin levels; however, body weight and body composition remained unchanged (28). Accordingly, SIRT1 overexpression did not protect mice from high-fat diet-induced obesity (28, 249). However, contrary to previous studies, SIRT1 transgenic mice displayed higher energy expenditure, which was compensated for by a parallel increase in food intake (249). Nevertheless, in agreement with previous studies, high-fat diet fed SIRT1 overexpressing mice displayed lower lipid-induced inflammation and better glucose tolerance and were protected from hepatic steatosis (249). The mechanisms modulating these actions involved the induction of antioxidant proteins MnSOD and Nrf1, possibly via stimulation of PGC-1α, and a lower activation of proinflammatory cytokines, such as tumor necrosis factor-α and interleukin-6, via downmodulation of NF-κB activity (249). The overexpression of SIRT1 in mice also potentiated the stimulation of fatty acid oxidation and energy expenditure through its NAD+-independent actions on the cAMP/PKA pathway (113).
4. Pancreatic β-cell-specific SIRT1 overexpression improves glucose homeostasis
In RINm5F cells or isolated rat islets, the pharmacological stimulation and ectopic expression of SIRT1 completely prevented cytokine-mediated cytotoxicity, indicating that SIRT1 participates in the maintenance of normal insulin-secreting responses to glucose in isolated rat islets (192). In addition, SIRT1 positively regulated insulin secretion in pancreatic β-cells by repressing UCP2 (47). These results were later corroborated in mice with genetic manipulation of SIRT1 in the pancreas. For instance, the Imai laboratory has studied beta cell-specific SIRT1-overexpressing (BESTO) transgenic mice over the past few years (225, 269). In these mice, the higher dosage of SIRT1 potentiated glucose-stimulated insulin secretion and improved glucose tolerance (225), potentially via SIRT1-mediated inhibition of UCP2 (225). However, when BESTO mice were ageing to 18–24 mo, this beneficial glucose-stimulated insulin secretion was lost (269).
B. SIRT3 Gain- and Loss-of-Function Models and Mitochondrial Function
Mice deficient in SIRT3 were viable and did not show any metabolic disorder. They had normal body weight, body composition, oxygen consumption, respiratory quotient, and physical activity (207). They also responded normally to food deprivation, and adaptive thermogenesis was not altered (207). However, the lack of SIRT3 induced global mitochondrial lysine acetylation through the maintenance of basal ATP levels (5, 171, 207). Another important effect of the global deficiency of SIRT3 was that these mice had increased stress-induced superoxide levels and genomic instability, leading to a tumor-permissive phenotype (171). Prolonged SIRT3 expression in the mitochondria of murine adipocytes potentiated cellular respiration, lowered membrane potential, and attenuated reactive oxygen species production (300).
In addition to modulating metabolism, SIRT3 controls mitochondrial membrane potential by deacetylating and inhibiting the regulatory component of the mPTP cyclophilin D (122). Cardiac myocytes from mice lacking SIRT3 experience an age-dependent increase in mPTP opening and accelerated signs of aging in the heart, known phenotypes of excessive mPTP opening. Knockout mice experience cardiac hypertrophy and fibrosis at 13 mo of age and are also sensitive to transverse aortic constriction, as evidenced by cardiac hypertrophy, fibrosis, and increased mortality. Together, these findings in the mouse KO show that SIRT3 is critical for delaying mitochondrial dysfunction during aging.
C. SIRT1 and the Molecular Clock
Animals are capable of maintaining an autonomous rhythm that closely resembles the 24-h daily cycle, even in constant light conditions (320). This autonomous circadian oscillation is closely connected to the body's metabolism, and alterations in the circadian cycle can have an enormous impact on metabolism. For instance, earlier reports had shown that dysregulation of circadian rhythms leads to metabolic dysfunction and correlates with obesity as well as other metabolic problems (331). Notably, recent reports also showed that global absence of SIRT1 led to dysregulation of circadian rhythms (352). Thus, in the following section, the relationship between SIRT1, circadian rhythms, NAD+, and metabolic dysfunction are discussed in more detail.
1. Molecular underpinnings for a circadian clock
In 1994, Takahashi and colleagues (62) identified the protein CLOCK as main regulator of the autonomous circadian rhythm in cells. CLOCK is a transcription factor from the group of bHLH (basic helix-loop-helix) proteins that dimerizes with BMAL1 to regulate the transcription of several circadian genes (e.g., Cryptochrome and Period genes) (17, 26, 51). Accumulation of the transcripts (Cry and Per) facilitates complex formation with CLOCK-BMAL1, silencing the transcription of circadian genes (31, 61, 69). Interestingly, CLOCK is a histone acetyltransferase (13) that directly acetylates histone H3 and its partner BMAL1. Moreover, when Per2 binds to the CLOCK-BMAL1 complex, it is also acetylated (2). These acetylation reactions are essential for the binding and regulation of the transcription of the circadian genes (39).
2. Circadian fluctuations in NAD+ levels fine-tune the circadian oscillation of the CLOCK-BMAL1 complex via Sirt1
Recently, studies in mammalian cells suggest that NAD+ levels are regulated in a circadian rhythm. This rhythm is due to the salvage pathway, a two-step reaction that converts nicotinamidetonicotinamide mononucleotide (NMN) to NAD+ (210, 228, 270). The two enzymes responsible for catalyzing these reactions are nicotinamidephophoribosyltransferase (NAMPT) and NMN adenylytransferases (NMNAT) (95, 273). What makes this scenario interesting for the understanding of sirtuins' physiology is that SIRT1 utilizes NAD+ in its biochemical reaction to generate nicotinamide. Thus NAMPT, NMNAT, and SIRT1 close a loop in a biochemical cascade from nicotinamide down to NMN, NAD+, and finally nicotinamide (228, 270).
It is intuitive to hypothesize that if NAD+ fluctuates according to the circadian cycle, then SIRT1 activity should vary as well, because it is linked to NAD+ levels. Several reports shed light on these mechanisms, showing that SIRT1 interacts with CLOCK to generate a protein complex (SIRT1-CLOCK). SIRT1 was shown to bind and deacetylate the CLOCK complex, leading to degradation of the complex and inactivation of circadian gene transcription (2, 39). Thus SIRT1 deacetylase activity affects the CLOCK complex by regulating acetyl residues, driving degradation of the complex. Additionally, the CLOCK-BMAL1 complex was shown to bind to the NAMPT promoter where it upregulates NAMPT expression (40, 47). Increases in NAMPT levels contribute to enhance the levels of NAD+, which in turn activates SIRT1. SIRT1 subsequently deacetylates the CLOCK complex and releases it from the DNA, decreasing the transcription of NAMPT. With less NAMPT, levels of NAD+ decrease and, consequently, so does SIRT1 activity. This NAMPT-SIRT1 loop acts to fine-tune the circadian oscillation of the CLOCK-BMAL1 complex, linking metabolism variances to the circadian rhythm (40, 47).
3. Circadian rhythm control by SIRT1 may be mediated by the central nervous system
The master clock of mammals is located in the suprachiasmatic nucleus (SCN) of the basal hypothalamus (349). Lesion of the SCN abolished the cyclic nature of locomotor activity in rodents (147–149, 223, 312, 313). Transplantation of SCN grafts to the lesioned SCN could restore the oscillatory behavior in lesioned animals (264, 265, 305). The nature of the oscillatory behavior of the transplanted graft was located intracellularly, as evidenced by the fact that when neurons from the SCN are dissociated in cell culture their oscillatory (circadian) activity is maintained for many days. Thus it is well accepted that the SCN represents the dominant molecular clock that orchestrates the body's circadian rhythm based on the Zeitgeber light.
SIRT1 is expressed in the SCN, next to the ubiquitously expressed CLOCK, BMAL1, and all other components of the circadian machinery (119). Thus influence of Sirt1 on the main control of SCN clock seems likely. However, to date, NAMPT has not been identified in brain tissue (48). Nevertheless, specific groups of neurons could express minute amounts of NAMPT, which could be sufficient to regulate SCN clock rhythmicity. Thus the heterogeneity of the brain tissue, and the open question of NAMPT expression in brain or the SCN, warrants further research. Ultimately, despite the dominant role of the SCN in circadian rhythm control, the exact roles of metabolic oscillations, NAD+, and SIRT1 in the CNS and specifically in the SCN remain unclear.
4. SIRT1 and the molecular clock in peripheral tissues
Even though the SCN maintains the body's circadian rhythm, several other tissues maintain an independent, cell-autonomous oscillatory rhythm, which in most cases resembles the 24-h light-dark cycle. However, in contrast to the SCN, where the light is the main circadian regulatory input, in peripheral tissues cellular energy metabolism (feeding) serves as a potent synchronizer of subsidiary oscillations. Since cellular energy status directly affects NAD levels and thus SIRT1 activity, cell-autonomous molecular interactions of SIRT1 with CLOCK in peripheral organs may play a major role in circadian gene oscillation. Indeed, although most findings on the role of SIRT1-CLOCK were derived from studies in mammalian cells, most have also been confirmed in peripheral tissues such as liver (17, 227). In conclusion, there is compelling evidence that SIRT1 acts as a metabolic rheostat for oscillatory rhythms in peripheral organs.
D. SIRT1 and Diabetes-Induced Cardiac Dysfunction
SIRT1 conferred cardioprotection in the heart (8, 9, 252) during oxidative stress (251). Resveratrol, a proposed SIRT1 activator (reviewed in section X), prevented hypoxia-induced apoptosis in cardiomyocytes through SIRT1-mediated FOXO1 regulation (73). SIRT1-mediated cardiac benefits occurred when SIRT1 expression levels were low to moderate; in contrast, high doses of SIRT1 exacerbated oxidative stress and induced cardiomyopathy (8).
Hyperglycemia during diabetes can cause significant cardiac deterioration and dysfunction. Recent studies have demonstrated that diabetes-induced cardiac dysfunction may be alleviated through activation of the SIRT1 pathway (89, 316, 333). SIRT1 regulated angiogenic activity in endothelial cells by deacetylating and inhibiting FOXO1 (257), which is a negative regulator of angiogenesis (112, 258). In the diabetic state, repression of FOXO1 activity causes endothelial dysfunction and diminished angiogenesis. A study by Balestrieri et al. (26) proposed that SIRT1 is an important regulator of endothelial progenitor cell dysfunction in a high-glucose environment. When endothelial progenitor cells were exposed to a high-glucose environment, downregulation of EPC activity correlated with reduced SIRT1 expression and elevated acetyl-FOXO1 expression (26). In addition to these effects on endothelial progenitor cells, SIRT1 has been implicated in cardiac dys-function. Cardiac dysfunction in diabetic cardiomyopathy reduced sarcoplasmic calcium ATPase (SERCA2a) expression levels. Sulaiman et al. (316) showed that activation of SIRT1 by resveratrol increased levels of SERCA2a mRNA and improved cardiac function in mice injected with streptozotocin, a chemical used to model type 1 diabetes in animals.
According to Orimo et al. (237), the mechanism of SIRT1's beneficial action was through p53 regulation. The authors describe that hyperglycemia induced vascular senescence unless SIRT1 was introduced or p53 was removed; activation of SIRT1 by resveratrol lessened the extent of vascular dysfunction in diabetic mice, specifically through the SIRT1-p53 pathway (237).
IX. GENETIC POLYMORPHISMS OF SIRT1 AND SIRT3 IN HUMANS
A. Genetic Polymorphisms of SIRT1
Very little is known about the genetic variation of SIRT1 and its effects on energy homeostasis in humans, and the data are somewhat confusing. A recent report has examined the relationship between variants in SIRT1 and obesity in 1,068 obese patients and 313 normal-weight control subjects (248). The authors found that males but not females with a SIRT1 single nucleotide polymorphism (SNP) were associated with increased visceral adiposity (248). A similar study using a large sample-size population of 6,251 elderly subjects (population-based Rotterdam Study) found that there were two variants in SIRT1 that were associated with lower body mass index, decreased risk of obesity, and lower weight gain (385). In a smaller study using 389 patients with metabolic syndrome and 547 controls, a significant association between one SIRT1 SNP and metabolic syndrome was found (68). However, another study performed in 917 overweight individuals investigating the associations of SIRT1 SNPs with metabolic response did not find an association of SIRT1 SNPs with baseline BMI or with BMI change after a 9 mo follow-up (351).
The genetic variation of SIRT1 and its effects on human longevity are also poorly studied. One report using DNA from 1,573 long-lived individuals (centenarians and nonagenarians) found no evidence between the five SNPs analyzed and longevity (103). Similar findings were observed in another population-based Leiden85-plus Study of 1,245 subjects (183). Therefore, it seems that SNPs for SIRT1 are not related with human longevity, but further studies using larger-size populations are needed to elucidate this issue.
B. Genetic Polymorphisms of SIRT3
The knowledge about the potential role and genetic alterations of SIRT3 in humans is even lower than for SIRT1. SIRT3 is located at the telomeric terminal on 11p15.5 chromosome, and studies performed in people over 100 years have demonstrated that there is a correlation between longevity and polymorphism of four genes located in this region (85). The researchers found a SNP marker in exon 3 of the SIRT3 gene, and there was a male-specific relationship between this marker and longevity (278). The same laboratory discovered a VNTR polymorphism in intron 5 of the SIRT3 gene (36). Moreover, by analyzing allele frequencies as a function of age, the researchers found that the allele without enhancer activity was absent in males older than 90 years (36). Thus this study suggests that low expression of SIRT3 is detrimental for longevity in humans. Another study using 640 individuals found two human SIRT3 SNPs and suggested that SIRT3 increased cellular respiration (91). Another work assessed the association between SIRT3 and age and between SIRT3 and endurance exercise (190). The results showed that the protein levels of SIRT3 in muscle were lower in old sedentary subjects compared with young sedentary subjects (190). Although not statistically significant, this study suggests that endurance-trained individuals might be protected against diminishing levels of SIRT3 (190).
X. METABOLIC CONSEQUENCES OF PHARMACOLOGICAL SIRT1 ACTIVATION
A. Resveratrol: A Specific SIRT1 Activator?
Early studies showing the role of Sir2/SIRT1 in the mechanisms responsible for the effects of calorie restriction in prolonging life span (165, 200, 327) raised the possibility that SIRT1 could be implicated in glucose metabolism and insulin sensitivity, key factors impaired during aging. Initial screening studies identified the polyphenol resveratrol as a potent activator of SIRT1 (141). Utilizing this chemical tool to stimulate SIRT1 activity in vivo, several groups reported benefits of resveratrol in models of metabolic disorders (34, 186). Resveratrol administration was shown to protect against detrimental effects of high-fat diet exposure, such as glucose intolerance, insulin resistance, or life span reduction, potentially through activation of SIRT1, although no beneficial effects on body weight could be observed (6, 34, 186). However, other studies failed to show activation of SIRT1 by resveratrol (163, 239) and suggested that previous reports on the activation of SIRT1 by resveratrol could be due to an interaction between resveratrol and the fluorophore used in the assay (163, 239). This suggestion was challenged by a subsequent report showing that the fluorophore is dispensible for SIRT1 activation to occur (80). Instead, the authors suggest that the fluorophore mimics a bulky-hydrophobic amino acid in natural substrates (80). Resveratrol has been shown to be an indirect activator of several proteins such as AMPK, an important enzyme for energy and glucose homeostasis (34, 70, 84, 146, 244, 296, 332). AMPK senses the levels of ATP in the cell milieu. Metformin, a well-known activator of AMPK, is widely used to treat type 2 diabetes. Several reports showed that AMPK and SIRT1 share similar molecular pathways, and activation of SIRT1 by resveratrol could be a consequence of AMPK activation (332). Indeed, it was recently reported that the effects of resveratrol improving metabolic parameters in models of metabolic disorders are dependent on the expression of the AMPK subunit α (332). In this report, the authors tested the effects of resveratrol in AMPKα1 and AMPKα2 KO mice; resveratrol failed to improve insulin sensitivy, glucose tolerance, and mitochondria biogenesis in an AMPK-dependent manner, and failed to decrease fat mass. The authors also showed that resveratrol increased the NAD+/NADH ratio, which is known to activate SIRT1 activity (332). This effect was also dependent on the expression of AMPK, thus shedding light on a possible indirect mechanism by which SIRT1 is activated by resveratrol through modulation of NAD+/NADH levels via AMPK (57).
On the other hand, SIRT1 regulates LKB1, an upstream regulator of AMPK (140, 386). Thus the effects of resveratrol may be mediated by inhibition of LKB1. Indeed, knockdown of LKB1 reduces the ability of resveratrol to protect cells from mitochondrial dysfunction (302). Clearly, the interplay between SIRT1 and AMPK is complex, and it is not possible to conclude whether resveratrol acts on AMPK independently of SIRT1. The testing of resveratrol in a SIRT1 KO mouse would likely clarify this debate (for further review, see Refs. 56, 123).
Interestingly, considerable insight can be derived from a randomized double-blind crossover study in healthy, obese men treated with resveratrol (326). In this study resveratrol significantly reduced sleeping and resting metabolic rate. Resveratrol elevated intramyocellular lipid levels; decreased intrahepatic lipid content, circulating glucose, triglycerides, alanine-aminotransferase, and inflammation markers; improved systolic blood pressure; and decreased the HOMA index (326). Therefore, this study suggests that the treatment with resveratrol induces metabolic changes in obese humans mimicking the effects of calorie restriction.
B. Natural SIRT1 Activators
It is well known that nicotinamide adenine dinucleotides (NAD+ and NADH) are essential mediators of energy homeostasis (39). Consequently, increased intracellular levels of NAD+ activate sirtuin-dependent metabolic control. Thus compounds that modulate NAD+/NADH ratio are likely to also exert effects on SIRT1-medulated metabolic control. Indeed, β-lapachone, an o-naphthoquinone extracted from bark of the lapacho tree (Tabebuiaavellanedae), has been shown to stimulate NADH oxidation through interaction with NADH:quinoneoxidoreductase 1 (NQO1); treatment of diet-induced obese and leptin-deficient ob/ob mice with β-lapachone markedly increased fatty acid oxidation and ameliorated the symptoms broadly summarized as metabolic syndrome, such as increased adiposity, glucose intolerance, dyslipidemia, and fatty liver (145). Therefore, the pharmacological administration of β-lapachone mimics effects of SIRT1 activation/overexpression, and as a matter of fact, the treatment with β-lapachone increased SIRT1 mRNA expression in muscle and white adipose tissue (145).
Kaempferol, a flavonoid with anti- and pro-oxidant activity present in various natural sources, was shown to activate SIRT3 in myelogenous leukemia cell line K562 and promyelocitic human leukemia U937 (213). Consistently, downregulation of SIRT3 in these cell lines abolished the actions of Kaempferol (213). Since Kaempferol induced apoptosis, it seems possible that SIRT3 might play an important function in the mitochondrially mediated apoptosis.
C. Novel Synthetic SIRT1 Activators Improve Metabolic Control
After the discovery of resveratrol as a SIRT1 activator, several synthetic small molecule activators of SIRT1 were reported (220). These molecules, 1,000 times more potent than resveratrol but structurally distinct (220), were shown to act through the same enzymatic mechanism, i.e., binding to an allosteric site exposed in the enzyme-substrate complex and lowering the Km of SIRT1 for its acetylated peptide substrates. Importantly, these molecules, unlike resveratrol, exhibited good oral bioavailability in rodents, and their administration substantially improved both glucose and insulin homeostasis in ob/ob mice, diet-induced obese mice, and Zucker fa/fa rats (220). Consistent with these findings, the same researchers have shown that these activators recapitulate many of the molecular events downstream of CR in vivo, such as enhancing mitochondrial biogenesis, improving metabolic signaling pathways, and blunting proinflammatory pathways in mice fed a high-fat, high-calorie diet (308). Similarly, an independent group treated mice with nonalcoholic fatty liver disease (NAFLD) with one of these activators (SRT1720) and found that the compound reduced the expression of lipogenic enzymes, serum lipid profiles, expressions of marker genes for oxidative stress, and inflammatory cytokines in the liver of these mice (360). SRT1720 administration was further shown to enhance endurance and protect from diet-induced obesity and insulin resistance, potentially by enhancing oxidative metabolism in skeletal muscle, liver, and brown adipose tissue (99). In vitro studies using SRT1720 have shown that this drug increases insulin-stimulated glucose uptake in adipocytes (371).
However, a recent study attributed activation of SIRT1 to drug binding to the fluorphore, casting doubts on the specificity of these molecules (239). In contrast to the two independent studies, in vivo administration of SRT1720 was lethal to mice and failed to decrease plasma glucose or improve mitochondrial capacity in mice fed a high-fat diet, although insulin levels were decreased (239). This report concludes that these synthetic activators, as well as resveratrol, exhibit multiple off-target activities against receptors, enzymes, transporters, and ion channels and, therefore, are not direct activators of SIRT1 (239). Since this report, activation of SIRT1 has been shown to occur on natural peptides, and the enzyme kinetics for activation by SRT1720 and other small molecules are consistent with an allosteric activation mechanism (80).
XI. FUTURE PERSPECTIVES
Decades of biomedical progress and vast improvements in our living conditions allow us to lead productive lives into old age. At the same time, we have to face unprecedented negative impacts of our Western life-style, resulting in the rapid rise in obesity and its comorbidities, diabetes and fatty liver disease. Although major efforts are being made to attenuate such negative impacts of our Western life-style on obesity and its sequelae, to date there are no efficient pharmacological treatment options for weight control available, and surgical options such as bariatric surgeries, albeit being highly efficient, may not (yet) be feasible for large proportions of the affected population. In addition, although changes in life-style may hold the greatest promise to avoid obesity and metabolic disorders for a majority of us, dietary regimens such as calorie restriction and exercise have repeatedly been shown not to lead to adequate weight loss if not continued with highest perseverance and diligence. Furthermore, drugs that can mimic caloric restriction hold the promise of being able to prevent and treat numerous other diseases such as cancer and heart disease.
Although pharmacological or genetic activation of sirtuins, and particularly SIRT1, resembles the beneficial effects of caloric restriction, making them attractive drug targets, we should not forget that sirtuins act on many different transcription factors, which are involved in numerous biological activities. Clinical trials will be necessary to address if SIRT1 analogs have beneficial effects in obese and/or diabetic patients.
SIRT3 is still a largely unexplored drug target. Although there are promising in vitro studies overexpressing SIRT3, to our knowledge, there are no specific SIRT3 activators available, and its potential pharmacological actions in vivo remain completely unknown.
It is certain that new targets modulated by SIRT1 and SIRT3 will appear in the near future, and some of those targets will be important for several metabolic actions. To precisely dissect the molecular pathways modulating sirtuins is a crucial step in elucidating the real potential of these deacetylases, and will require years of investigation. With the information obtained on sirtuins to date, it seems that this big challenge deserves to be weighed out, and we will certainly enjoy more exciting studies on this topic during the next years.
ACKNOWLEDGMENTS
Address for reprint requests and other correspondence: M. H. Tschöp, Institute for Diabetes and Obesity, German Center for Diabetes Research, Helmholtz Center Munich, German Research Center for Environmental Health (GmbH), Ingolstaedter Landstr. 1, 85764 Neuherberg/Munich, Germany (tschoep@helmholtz-muenchen.de).
Footnotes
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
REFERENCES
- 1.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 3.Afshar G, Murnane JP. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene. 1999;234:161–168. doi: 10.1016/s0378-1119(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 4.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 5.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ajmo JM, Liang X, Rogers CQ, Pennock B, You M. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295:G833–G842. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Regaiey KA, Masternak MM, Bonkowski M, Sun L, Bartke A. Long-lived growth hormone receptor knockout mice: interaction of reduced insulin-like growth factor I/insulin signaling and caloric restriction. Endocrinology. 2005;146:851–860. doi: 10.1210/en.2004-1120. [DOI] [PubMed] [Google Scholar]
- 8.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 9.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 10.Amat R, Planavila A, Chen SL, Iglesias R, Giralt M, Villarroya F. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma Co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J Biol Chem. 2009;284:21872–21880. doi: 10.1074/jbc.M109.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amat R, Solanes G, Giralt M, Villarroya F. SIRT1 is involved in glucocorticoid-mediated control of uncoupling protein-3 gene transcription. J Biol Chem. 2007;282:34066–34076. doi: 10.1074/jbc.M707114200. [DOI] [PubMed] [Google Scholar]
- 12.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 13.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews ZB. Uncoupling protein-2 and the potential link between metabolism and longevity. Curr Aging Sci. 2010;3:102–112. doi: 10.2174/1874609811003020102. [DOI] [PubMed] [Google Scholar]
- 15.Anekonda TS, Adamus G. Resveratrol prevents antibody-induced apoptotic death of retinal cells through upregulation of Sirt1 and Ku70. BMC Res Notes. 2008;1:122. doi: 10.1186/1756-0500-1-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 17.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 18.Astrom SU, Cline TW, Rine J. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163:931–937. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Attie AD. ABCA1: at the nexus of cholesterol, HDL and atherosclerosis. Trends Biochem Sci. 2007;32:172–179. doi: 10.1016/j.tibs.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 21.Ayala JE, Streeper RS, Desgrosellier JS, Durham SK, Suwanichkul A, Svitek CA, Goldman JK, Barr FG, Powell DR, O'Brien RM. Conservation of an insulin response unit between mouse and human glucose-6-phosphatase catalytic subunit gene promoters: transcription factor FKHR binds the insulin response sequence. Diabetes. 1999;48:1885–1889. doi: 10.2337/diabetes.48.9.1885. [DOI] [PubMed] [Google Scholar]
- 22.Bai L, Pang WJ, Yang YJ, Yang GS. Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. Mol Cell Biochem. 2008;307:129–140. doi: 10.1007/s11010-007-9592-5. [DOI] [PubMed] [Google Scholar]
- 23.Bai P, Canto C, Brunyanszki A, Huber A, Szanto M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, Gergely P, Menissier-de Murcia J, Schreiber V, Sauve AA, Auwerx J. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakker BM, Overkamp KM, van Maris AJ, Kotter P, Luttik MA, van Dijken JP, Pronk JT. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 26.Balestrieri ML, Rienzo M, Felice F, Rossiello R, Grimaldi V, Milone L, Casamassimi A, Servillo L, Farzati B, Giovane A, Napoli C. High glucose downregulates endothelial progenitor cell number via SIRT1. Biochim Biophys Acta. 2008;1784:936–945. doi: 10.1016/j.bbapap.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Bamps S, Wirtz J, Savory FR, Lake D, Hope IA. The Caenorhabditis elegans sirtuin gene, sir-2.1, is widely expressed and induced upon caloric restriction. Mech Ageing Dev. 2009;130:762–770. doi: 10.1016/j.mad.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Banks AS, Kon N, Knight C, Matsumoto M, Gutierrez-Juarez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, Samsel L, McCoy JP, Jr, Leclerc J, Nguyen P, Gius D, Sack MN. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J Cell Biochem. 2010;110:238–247. doi: 10.1002/jcb.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao J, Scott I, Lu Z, Pang L, Dimond CC, Gius D, Sack MN. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med. 2010;49:1230–1237. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barlow AL, van Drunen CM, Johnson CA, Tweedie S, Bird A, Turner BM. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp Cell Res. 2001;265:90–103. doi: 10.1006/excr.2001.5162. [DOI] [PubMed] [Google Scholar]
- 32.Barthel A, Schmoll D, Unterman TG. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Bastien-Dionne PO, Valenti L, Kon N, Gu W, Buteau J. Glucagon-like peptide 1 inhibits the sirtuin deacetylase SirT1 to stimulate pancreatic beta-cell mass expansion. Diabetes. 2011;60:3217–3222. doi: 10.2337/db11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Benayoun BA, Georges AB, L'Hote D, Andersson N, Dipietromaria A, Todeschini AL, Caburet S, Bazin C, Anttonen M, Veitia RA. Transcription factor FOXL2 protects granulosa cells from stress and delays cell cycle: role of its regulation by the SIRT1 deacetylase. Hum Mol Genet. 2011;20:1673–1686. doi: 10.1093/hmg/ddr042. [DOI] [PubMed] [Google Scholar]
- 38.Berg BN, Simms HS. Nutrition and longevity in the rat. II. Longevity and onset of disease with different levels of food intake. J Nutr. 1960;71:255–263. [PubMed] [Google Scholar]
- 39.Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Bertrand HA, Lynd FT, Masoro EJ, Yu BP. Changes in adipose mass and cellularity through the adult life of rats fed ad libitum or a life-prolonging restricted diet. J Gerontol. 1980;35:827–835. doi: 10.1093/geronj/35.6.827. [DOI] [PubMed] [Google Scholar]
- 41.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 43.Blander G, Olejnik J, Krzymanska-Olejnik E, McDonagh T, Haigis M, Yaffe MB, Guarente L. SIRT1 shows no substrate specificity in vitro. J Biol Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- 44.Boily G, He XH, Pearce B, Jardine K, McBurney MW. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009;28:2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- 45.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, Moffat C, Crawford S, Saliba S, Jardine K, Xuan J, Evans M, Harper ME, McBurney MW. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 47.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borra MT, Langer MR, Slama JT, Denu JM. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 49.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 51.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 52.Breen DM, Sanli T, Giacca A, Tsiani E. Stimulation of muscle cell glucose uptake by resveratrol through sirtuins and AMPK. Biochem Biophys Res Commun. 2008;374:117–122. doi: 10.1016/j.bbrc.2008.06.104. [DOI] [PubMed] [Google Scholar]
- 53.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 55.Cakir I, Perello M, Lansari O, Messier NJ, Vaslet CA, Nillni EA. Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS One. 2009;4:e8322. doi: 10.1371/journal.pone.0008322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carlsen H, Haugen F, Zadelaar S, Kleemann R, Kooistra T, Drevon CA, Blomhoff R. Diet-induced obesity increases NF-kappaB signaling in reporter mice. Genes Nutr. 2009;4:215–222. doi: 10.1007/s12263-009-0133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen H, Miller C, Bitterman K, Wall N, Hekking B, Kessler B, Howitz K, Gorospe M, de Cabo R, Sinclair D. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 62.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 63.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 64.Cooper HM, Huang JY, Verdin E, Spelbrink JN. A new splice variant of the mouse SIRT3 gene encodes the mitochondrial precursor protein. PLoS One. 2009;4:e4986. doi: 10.1371/journal.pone.0004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411:279–285. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 66.Cooper TM, Mockett RJ, Sohal BH, Sohal RS, Orr WC. Effect of caloric restriction on life span of the housefly, Musca domestica. FASEB J. 2004;18:1591–1593. doi: 10.1096/fj.03-1464fje. [DOI] [PubMed] [Google Scholar]
- 67.Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One. 2008;3:e1571. doi: 10.1371/journal.pone.0001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cruz M, Valladares-Salgado A, Garcia-Mena J, Ross K, Edwards M, Angeles-Martinez J, Ortega-Camarillo C, de la Pena JE, Burguete-Garcia AI, Wacher-Rodarte N, Ambriz R, Rivera R, D'Artote AL, Peralta J, Parra EJ, Kumate J. Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metab Res Rev. 2010;26:261–270. doi: 10.1002/dmrr.1082. [DOI] [PubMed] [Google Scholar]
- 69.Chakrabarti P, English T, Karki S, Qiang L, Tao R, Kim J, Luo Z, Farmer SR, Kandror KV. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. J Lipid Res. 2011;52:1693–1701. doi: 10.1194/jlr.M014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan AY, Dolinsky VW, Soltys CL, Viollet B, Baksh S, Light PE, Dyck JR. Resveratrol inhibits cardiac hypertrophy via AMP-activated protein kinase and Akt. J Biol Chem. 2008;283:24194–24201. doi: 10.1074/jbc.M802869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chanda D, Xie YB, Choi HS. Transcriptional corepressor SHP recruits SIRT1 histone deacetylase to inhibit LRH-1 transactivation. Nucleic Acids Res. 2010;38:4607–4619. doi: 10.1093/nar/gkq227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang JH, Kim HC, Hwang KY, Lee JW, Jackson SP, Bell SD, Cho Y. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J Biol Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- 73.Chen CJ, Yu W, Fu YC, Wang X, Li JL, Wang W. Resveratrol protects cardiomyocytes from hypoxia-induced apoptosis through the SIRT1-FoxO1 pathway. Biochem Biophys Res Commun. 2009;378:389–393. doi: 10.1016/j.bbrc.2008.11.110. [DOI] [PubMed] [Google Scholar]
- 74.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 76.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 77.Chen XJ, Clark-Walker GD. sir2 mutants of Kluyveromyces lactis are hypersensitive to DNA-targeting drugs. Mol Cell Biol. 1994;14:4501–4508. doi: 10.1128/mcb.14.7.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cho KW, Lumeng CN. SirT1: a guardian at the gates of adipose tissue inflammation. Diabetes. 2011;60:3100–3102. doi: 10.2337/db11-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Perni RB, Riera TV, Szczepankiewicz B, Vlasuk GP, Stein RL. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010;285:32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai JM, Wang ZY, Sun DC, Lin RX, Wang SQ. SIRT1 interacts with p73 and suppresses p73-dependent transcriptional activity. J Cell Physiol. 2007;210:161–166. doi: 10.1002/jcp.20831. [DOI] [PubMed] [Google Scholar]
- 82.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dansen TB, Burgering BM. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 2008;18:421–429. doi: 10.1016/j.tcb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 84.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci USA. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Luca M, Rose G, Bonafe M, Garasto S, Greco V, Weir BS, Franceschi C, De Benedictis G. Sex-specific longevity associations defined by tyrosine hydroxylase-insulin-insulin growth factor 2 haplotypes on the 11p15.5 chromosomal region. Exp Gerontol. 2001;36:1663–1671. doi: 10.1016/s0531-5565(01)00146-2. [DOI] [PubMed] [Google Scholar]
- 86.Derbyshire MK, Weinstock KG, Strathern JN. HST1, a new member of the SIR2 family of genes. Yeast. 1996;12:631–640. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C631::AID-YEA960%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 87.Dietrich MO, Antunes C, Geliang G, Liu ZW, Borok E, Nie Y, Xu AW, Souza DO, Gao Q, Diano S, Gao XB, Horvath TL. Agrp neurons mediate Sirt1's action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J Neurosci. 2010;30:11815–11825. doi: 10.1523/JNEUROSCI.2234-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. 2010. [DOI] [PubMed] [Google Scholar]
- 89.Dong F, Ren J. Fidarestat improves cardiomyocyte contractile function in db/db diabetic obese mice through a histone deacetylase Sir2-dependent mechanism. J Hyper-tens. 2007;25:2138–2147. doi: 10.1097/HJH.0b013e32828626d1. [DOI] [PubMed] [Google Scholar]
- 90.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Dransfeld CL, Alborzinia H, Wolfl S, Mahlknecht U. SIRT3 SNPs validation in 640 individuals, functional analyses and new insights into SIRT3 stability. Int J Oncol. 2010;36:955–960. doi: 10.3892/ijo_00000574. [DOI] [PubMed] [Google Scholar]
- 92.Draznin B. Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- 93.Drew LJ, Hen R. Food for thought: linking caloric intake to behavior via sirtuin activity. Cell. 2011;147:1436–1437. doi: 10.1016/j.cell.2011.11.052. [DOI] [PubMed] [Google Scholar]
- 94.Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, Ramachandran C, Gresser MJ, Tremblay ML, Kennedy BP. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 95.Emanuelli M, Carnevali F, Saccucci F, Pierella F, Amici A, Raffaelli N, Magni G. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J Biol Chem. 2001;276:406–412. doi: 10.1074/jbc.M008700200. [DOI] [PubMed] [Google Scholar]
- 96.Erion DM, Yonemitsu S, Nie Y, Nagai Y, Gillum MP, Hsiao JJ, Iwasaki T, Stark R, Weismann D, Yu XX, Murray SF, Bhanot S, Monia BP, Horvath TL, Gao Q, Samuel VT, Shulman GI. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc Natl Acad Sci USA. 2009;106:11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 98.Fahie K, Hu P, Swatkoski S, Cotter RJ, Zhang Y, Wolberger C. Side chain specificity of ADP-ribosylation by a sirtuin. FEBS J. 2009;276:7159–7176. doi: 10.1111/j.1742-4658.2009.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 100.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 102.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Flachsbart F, Croucher PJ, Nikolaus S, Hampe J, Cordes C, Schreiber S, Nebel A. Sirtuin 1 (SIRT1) sequence variation is not associated with exceptional human longevity. Exp Gerontol. 2006;41:98–102. doi: 10.1016/j.exger.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 104.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 105.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 106.Frojdo S, Durand C, Molin L, Carey AL, El-Osta A, Kingwell BA, Febbraio MA, Solari F, Vidal H, Pirola L. Phosphoinositide 3-kinase as a novel functional target for the regulation of the insulin signaling pathway by SIRT1. Mol Cell Endocrinol. 2011;335:166–176. doi: 10.1016/j.mce.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 107.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 108.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 109.Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG. Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol. 2006;26:8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fujino T, Kondo J, Ishikawa M, Morikawa K, Yamamoto TT. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 111.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 112.Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, Hisatsune H, Nishikawa S, Nakayama K, Ikeda K, Motoyama N, Mori N. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 113.Gerhart-Hines Z, Dominy JE, Jr, Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gillum MP, Kotas ME, Erion DM, Kursawe R, Chatterjee P, Nead KT, Muise ES, Hsiao JJ, Frederick DW, Yonemitsu S, Banks AS, Qiang L, Bhanot S, Olefsky JM, Sears DD, Caprio S, Shulman GI. SirT1 regulates adipose tissue inflammation. Diabetes. 2011;60:3235–3245. doi: 10.2337/db11-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gorospe M, de Cabo R. AsSIRTing the DNA damage response. Trends Cell Biol. 2008;18:77–83. doi: 10.1016/j.tcb.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gottschling DE. Gene silencing: two faces of SIR2. Curr Biol. 2000;10:R708–711. doi: 10.1016/s0960-9822(00)00714-4. [DOI] [PubMed] [Google Scholar]
- 118.Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 121.Greiss S, Gartner A. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cell. 2009;28:407–415. doi: 10.1007/s10059-009-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, Sinclair DA. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging. 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3? Functional localization of an NAD+-dependent protein deacetylase. Biochem J. 2008;411:e11–13. doi: 10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hallows WC, Yu W, Smith BC, Devires MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Harper JM, Leathers CW, Austad SN. Does caloric restriction extend life in wild mice? Aging Cell. 2006;5:441–449. doi: 10.1111/j.1474-9726.2006.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008;28:8772–8784. doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, Zheng W, Wolberger C. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure. 2008;16:1368–1377. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hawse WF, Wolberger C. Structure-based mechanism of ADP-ribosylation by sirtuins. J Biol Chem. 2009;284:33654–33661. doi: 10.1074/jbc.M109.024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.He W, Wang Y, Zhang MZ, You L, Davis LS, Fan H, Yang HC, Fogo AB, Zent R, Harris RC, Breyer MD, Hao CM. Sirt1 activation protects the mouse renal medulla from oxidative injury. J Clin Invest. 2010;120:1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heilbronn LK, Ravussin E. Calorie restriction and aging: review of the literature and implications for studies in humans. Am J Clin Nutr. 2003;78:361–369. doi: 10.1093/ajcn/78.3.361. [DOI] [PubMed] [Google Scholar]
- 133.Herranz D, Munoz-Martin M, Canamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, Serrano M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 136.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hirst J. Towards the molecular mechanism of respiratory complex I. Biochem J. 2010;425:327–339. doi: 10.1042/BJ20091382. [DOI] [PubMed] [Google Scholar]
- 138.Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 139.Hokari F, Kawasaki E, Sakai A, Koshinaka K, Sakuma K, Kawanaka K. Muscle contrac-tile activity regulates Sirt3 protein expression in rat skeletal muscles. J Appl Physiol. 2010;109:332–340. doi: 10.1152/japplphysiol.00335.2009. [DOI] [PubMed] [Google Scholar]
- 140.Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, Cohen RA, Zang M. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 142.Hu Y, Mivechi NF. HSF-1 interacts with Ral-binding protein 1 in a stress-responsive, multiprotein complex with HSP90 in vivo. J Biol Chem. 2003;278:17299–17306. doi: 10.1074/jbc.M300788200. [DOI] [PubMed] [Google Scholar]
- 143.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 144.Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu Rev Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- 145.Hwang JH, Kim DW, Jo EJ, Kim YK, Jo YS, Park JH, Yoo SK, Park MK, Kwak TH, Kho YL, Han J, Choi HS, Lee SH, Kim JM, Lee I, Kyung T, Jang C, Chung J, Kweon GR, Shong M. Pharmacological stimulation of NADH oxidation ameliorates obesity and related phenotypes in mice. Diabetes. 2009;58:965–974. doi: 10.2337/db08-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hwang JT, Kwon DY, Park OJ, Kim MS. Resveratrol protects ROS-induced cell death by activating AMPK in H9c2 cardiac muscle cells. Genes Nutr. 2008;2:323–326. doi: 10.1007/s12263-007-0069-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ibuka N, Inouye SI, Kawamura H. Analysis of sleep-wakefulness rhythms in male rats after suprachiasmatic nucleus lesions and ocular enucleation. Brain Res. 1977;122:33–47. doi: 10.1016/0006-8993(77)90660-6. [DOI] [PubMed] [Google Scholar]
- 148.Ibuka N, Kawamura H. Loss of circadian rhythm in sleep-wakefulness cycle in the rat by suprachiasmatic nucleus lesions. Brain Res. 1975;96:76–81. doi: 10.1016/0006-8993(75)90574-0. [DOI] [PubMed] [Google Scholar]
- 149.Ibuka N, Nihonmatsu I, Sekiguchi S. Sleep-wakefulness rhythms in mice after suprachiasmatic nucleus lesions. Waking Sleeping. 1980;4:167–173. [PubMed] [Google Scholar]
- 150.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 151.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Inoue H, Ogawa W, Asakawa A, Okamoto Y, Nishizawa A, Matsumoto M, Teshigawara K, Matsuki Y, Watanabe E, Hiramatsu R, Notohara K, Katayose K, Okamura H, Kahn CR, Noda T, Takeda K, Akira S, Inui A, Kasuga M. Role of hepatic STAT3 in brain-insulin action on hepatic glucose production. Cell Metab. 2006;3:267–275. doi: 10.1016/j.cmet.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 153.Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, Hashimoto N, Kido Y, Mori T, Sakaue H, Teshigawara K, Jin S, Iguchi H, Hiramatsu R, LeRoith D, Takeda K, Akira S, Kasuga M. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 154.Iqbal J, Zaidi M. TNF regulates cellular NAD+ metabolism in primary macrophages. Biochem Biophys Res Commun. 2006;342:1312–1318. doi: 10.1016/j.bbrc.2006.02.109. [DOI] [PubMed] [Google Scholar]
- 155.Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, Deng CX, Spitz DR, Gius D. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int J Biol Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jeong H, Cohen DE, Cui L, Supinski A, Savas JN, Mazzulli JR, Yates JR, 3rd, Bordone L, Guarente L, Krainc D. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2011;18:159–165. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 158.Jiang M, Wang J, Fu J, Du L, Jeong H, West T, Xiang L, Peng Q, Hou Z, Cai H, Seredenina T, Arbez N, Zhu S, Sommers K, Qian J, Zhang J, Mori S, Yang XW, Tamashiro KL, Aja S, Moran TH, Luthi-Carter R, Martin B, Maudsley S, Mattson MP, Cichewicz RH, Ross CA, Holtzman DM, Krainc D, Duan W. Neuroprotective role of Sirt1 in mammalian models of Huntington's disease through activation of multiple Sirt1 targets. Nat Med. 2011;18:153–158. doi: 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, Hoffmann E, Carney DP, Perni RB, Jirousek MR, Bemis JE, Milne JC, Sinclair DA, Westphal CH. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci. 2009;18:514–525. doi: 10.1002/pro.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, Milne JC, Westphal CH, Perni RB. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem. 2009;284:24394–24405. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 164.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 167.Kawamura Y, Uchijima Y, Horike N, Tonami K, Nishiyama K, Amano T, Asano T, Kurihara Y, Kurihara H. Sirt3 protects in vitro-fertilized mouse preimplantation embryos against oxidative stress-induced p53-mediated developmental arrest. J Clin Invest. 2010;120:2817–2828. doi: 10.1172/JCI42020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Vanhove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 171.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 173.Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kitamura T, Ido Kitamura Y. Role of FoxO proteins in pancreatic beta cells. Endocr J. 2007;54:507–515. doi: 10.1507/endocrj.kr-109. [DOI] [PubMed] [Google Scholar]
- 175.Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 176.Knight CM, Gutierrez-Juarez R, Lam TK, Arrieta-Cruz I, Huang L, Schwartz G, Barzilai N, Rossetti L. Mediobasal hypothalamic SIRT1 is essential for resveratrol's effects on insulin action in rats. Diabetes. 2011;60:2691–2700. doi: 10.2337/db10-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kobayashi Y, Furukawa-Hibi Y, Chen C, Horio Y, Isobe K, Ikeda K, Motoyama N. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16:237–243. [PubMed] [Google Scholar]
- 178.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G. Sassone-Corsi P Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 179.Kolthur-Seetharam U, Teerds K, de Rooij DG, Wendling O, McBurney M, Sassone-Corsi P, Davidson I. The histone deacetylase SIRT1 controls male fertility in mice through regulation of hypothalamic-pituitary gonadotropin signaling. Biol Reprod. 2009;80:384–391. doi: 10.1095/biolreprod.108.070193. [DOI] [PubMed] [Google Scholar]
- 180.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 182.Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kuningas M, Putters M, Westendorp RG, Slagboom PE, van Heemst D. SIRT1 gene, age-related diseases, and mortality: the Leiden 85-plus study. J Gerontol A Biol Sci Med Sci. 2007;62:960–965. doi: 10.1093/gerona/62.9.960. [DOI] [PubMed] [Google Scholar]
- 184.Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lafontaine-Lacasse M, Richard D, Picard F. Effects of age and gender on Sirt 1 mRNA expressions in the hypothalamus of the mouse. Neurosci Lett. 2010;480:1–3. doi: 10.1016/j.neulet.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 186.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 187.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 188.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouza-rides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–2942. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins-novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 192.Lee JH, Song MY, Song EK, Kim EK, Moon WS, Han MK, Park JW, Kwon KB, Park BH. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-kappaB signaling pathway. Diabetes. 2009;58:344–351. doi: 10.2337/db07-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, Ge Q, Gu W, Orren D, Luo J. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 194.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 195.Li Y, Xu S, Giles A, Nakamura K, Lee JW, Hou X, Donmez G, Li J, Luo Z, Walsh K, Guarente L, Zang M. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. 2011;25:1664–1679. doi: 10.1096/fj.10-173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li Y, Yokota T, Gama V, Yoshida T, Gomez JA, Ishikawa K, Sasaguri H, Cohen HY, Sinclair DA, Mizusawa H, Matsuyama S. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ. 2007;14:2058–2067. doi: 10.1038/sj.cdd.4402219. [DOI] [PubMed] [Google Scholar]
- 198.Libert S, Pointer K, Bell EL, Das A, Cohen DE, Asara JM, Kapur K, Bergmann S, Preisig M, Otowa T, Kendler KS, Chen X, Hettema JM, van den Oord EJ, Rubio JP, Guarente L. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell. 2011;147:1459–1472. doi: 10.1016/j.cell.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 200.Lin S, Defossez P, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 201.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 202.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 204.Liu X, Wang D, Zhao Y, Tu B, Zheng Z, Wang L, Wang H, Gu W, Roeder RG, Zhu WG. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proc Natl Acad Sci USA. 2011;108:1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers D, Cole P, Yates Olefsky J, Jr, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, Olefsky J, 3rd, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 209.Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S. Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci. 2004;61:19–34. doi: 10.1007/s00018-003-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Magni G, Amici A, Emanuelli M, Raffaelli N, Ruggieri S. Enzymology of NAD+ synthesis. Adv Enzymol Relat Areas Mol Biol. 1999;73:135–182. doi: 10.1002/9780470123195.ch5. [DOI] [PubMed] [Google Scholar]
- 211.Mair W, Piper MD, Partridge L. Calories do not explain extension of life span by dietary restriction in Drosophila. PLoS Biol. 2005;3:e223. doi: 10.1371/journal.pbio.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, Fini M, Russo MA. Kaempferol induces apoptosis in two different cell lines via Akt inactivation, Bax and SIRT3 activation, and mitochondrial dysfunction. J Cell Biochem. 2009;106:643–650. doi: 10.1002/jcb.22044. [DOI] [PubMed] [Google Scholar]
- 214.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.McCreanor GM, Bender DA. The metabolism of high intakes of tryptophan, nicotinamide and nicotinic acid in the rat. Br J Nutr. 1986;56:577–586. doi: 10.1079/bjn19860138. [DOI] [PubMed] [Google Scholar]
- 216.McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- 217.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 222.Ming M, Shea CR, Guo X, Li X, Soltani K, Han W, He YY. Regulation of global genome nucleotide excision repair by SIRT1 through xeroderma pigmentosum C. Proc Natl Acad Sci USA. 2010;107:22623–22628. doi: 10.1073/pnas.1010377108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 224.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 225.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 226.Nakae J, Cao Y, Daitoku H, Fukamizu A, Ogawa W, Yano Y, Hayashi Y. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Invest. 2006;116:2473–2483. doi: 10.1172/JCI25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 230.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 231.Newman BL, Lundblad JR, Chen Y, Smolik SM. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics. 2002;162:1675–1685. doi: 10.1093/genetics/162.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nicholls DG, Ferguson SJ. Bioenergetics. Vol. 3. Academic; New York: 2002. [Google Scholar]
- 233.Nie Y, Erion D, Yuan Z, Dietrich M, Shulman G, Horvath T, Gao Q. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Orimo M, Minamino T, Miyauchi H, Tateno K, Okada S, Moriya J, Komuro I. Protective role of SIRT1 in diabetic vascular dysfunction. Arterioscler Thromb Vasc Biol. 2009;29:889–894. doi: 10.1161/ATVBAHA.109.185694. [DOI] [PubMed] [Google Scholar]
- 238.Osborne TB, Mendel LB, Ferry EL. The effect of retardation of growth upon the breeding period and duration of life of rats. Science. 1917;45:294–295. doi: 10.1126/science.45.1160.294. [DOI] [PubMed] [Google Scholar]
- 239.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Iii JL, Goodyear LJ, Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging. 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, 3rd, Goodyear LJ, Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging. 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Park CE, Kim MJ, Lee JH, Min BI, Bae H, Choe W, Kim SS, Ha J. Resveratrol stimulates glucose transport in C2C12 myotubes by activating AMP-activated protein kinase. Exp Mol Med. 2007;39:222–229. doi: 10.1038/emm.2007.25. [DOI] [PubMed] [Google Scholar]
- 245.Park SH, Ozden O, Jiang H, Cha YI, Pennington JD, Aykin-Burns N, Spitz DR, Gius D, Kim HS. Sirt3, mitochondrial ROS, ageing, and carcinogenesis. Int J Mol Sci. 2011;12:6226–6239. doi: 10.3390/ijms12096226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Pediconi N, Guerrieri F, Vossio S, Bruno T, Belloni L, Schinzari V, Scisciani C, Fanciulli M, Levrero M. hSirT1-dependent regulation of the PCAF-E2F1-p73 apoptotic pathway in response to DNA damage. Mol Cell Biol. 2009;29:1989–1998. doi: 10.1128/MCB.00552-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Peeters AV, Beckers S, Verrijken A, Mertens I, Roevens P, Peeters PJ, Van Hul W, Van Gaal LF. Association of SIRT1 gene variation with visceral obesity. Hum Genet. 2008;124:431–436. doi: 10.1007/s00439-008-0567-8. [DOI] [PubMed] [Google Scholar]
- 249.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pillai JB, Gupta M, Rajamohan SB, Lang R, Raman J, Gupta MP. Poly(ADP-ribose) polymerase-1-deficient mice are protected from angiotensin II-induced cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2006;291:H1545–H1553. doi: 10.1152/ajpheart.01124.2005. [DOI] [PubMed] [Google Scholar]
- 252.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 253.Pillai VB, Sundaresan NR, Jeevanandam V, Gupta MP. Mitochondrial SIRT3 and heart disease. Cardiovasc Res. 2010;88:250–256. doi: 10.1093/cvr/cvq250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Pisarska MD, Barlow G, Kuo FT. Minireview: roles of the forkhead transcription factor FOXL2 in granulosa cell biology and pathology. Endocrinology. 2011;152:1199–1208. doi: 10.1210/en.2010-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, Zeiher AM, Dimmeler S. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, Aicher A, Kollipara R, DePinho RA, Zeiher AM, Dimmeler S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Qiang L, Banks AS, Accili D. Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem. 2010;285:27396–27401. doi: 10.1074/jbc.M110.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Qiang L, Lin HV, Kim-Muller JY, Welch CL, Gu W, Accili D. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell Metab. 2011;14:758–767. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J Biol Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 263.Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 265.Ralph MR, Hurd MW. Pacemaker interactions in the mammalian circadian system. Braz J Med Biol Res. 1996;29:77–85. [PubMed] [Google Scholar]
- 266.Ramadori G, Fujikawa T, Fukuda M, Anderson J, Morgan DA, Mostoslavsky R, Stuart RC, Perello M, Vianna CR, Nillni EA, Rahmouni K, Coppari R. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12:78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Ramadori G, Gautron L, Fujikawa T, Vianna CR, Elmquist JK, Coppari R. Central administration of resveratrol improves diet-induced diabetes. Endocrinology. 2009;150:5326–5333. doi: 10.1210/en.2009-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ramadori G, Lee CE, Bookout AL, Lee S, Williams KW, Anderson J, Elmquist JK, Coppari R. Brain SIRT1: anatomical distribution and regulation by energy availability. J Neurosci. 2008;28:9989–9996. doi: 10.1523/JNEUROSCI.3257-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Rathaus M, Lerrer B, Cohen HY. DeubiKuitylation: a novel DUB enzymatic activity for the DNA repair protein, Ku70. Cell Cycle. 2009;8:1843–1852. doi: 10.4161/cc.8.12.8864. [DOI] [PubMed] [Google Scholar]
- 273.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotin-amide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 274.Rodgers J, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 276.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 279.Rous P. The influence of diet on transplanted and spontaneous mouse tumors. J Exp Med. 1914;20:433–451. doi: 10.1084/jem.20.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Sakamoto J, Miura T, Shimamoto K, Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004;556:281–286. doi: 10.1016/s0014-5793(03)01444-3. [DOI] [PubMed] [Google Scholar]
- 281.Sasaki T, Kim HJ, Kobayashi M, Kitamura YI, Yokota-Hashimoto H, Shiuchi T, Minokoshi Y, Kitamura T. Induction of hypothalamic Sirt1 leads to cessation of feeding via agouti-related peptide. Endocrinology. 2010;151:2556–2566. doi: 10.1210/en.2009-1319. [DOI] [PubMed] [Google Scholar]
- 282.Satoh A, Brace CS, Ben-Josef G, West T, Wozniak DF, Holtzman DM, Herzog ED, Imai S. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 284.Sauve AA. Sirtuin chemical mechanisms. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 286.Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 287.Sauve AA, Schramm VL. SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Curr Med Chem. 2004;11:807–826. doi: 10.2174/0929867043455675. [DOI] [PubMed] [Google Scholar]
- 288.Schenk S, McCurdy CE, Philp A, Chen MZ, Holliday MJ, Bandyopadhyay GK, Osborn O, Baar K, Olefsky JM. Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J Clin Invest. 2011;121:4281–4288. doi: 10.1172/JCI58554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 291.Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- 292.Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 293.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 296.Shang J, Chen LL, Xiao FX, Sun H, Ding HC, Xiao H. Resveratrol improves non-alcoholic fatty liver disease by activating AMP-activated protein kinase. Acta Pharmacol Sin. 2008;29:698–706. doi: 10.1111/j.1745-7254.2008.00807.x. [DOI] [PubMed] [Google Scholar]
- 297.Sharp FR, Bernaudin M. HIF1 and oxygen sensing in the brain. Nature Rev Neurosci. 2004;5:437–448. doi: 10.1038/nrn1408. [DOI] [PubMed] [Google Scholar]
- 298.Sherman JM, Stone EM, Freeman-Cook LL, Brachmann CB, Boeke JD, Pillus L. The conserved core of a human SIR2 homologue functions in yeast silencing. Mol Biol Cell. 1999;10:3045–3059. doi: 10.1091/mbc.10.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Shi T, Fan GQ, Xiao SD. SIRT3 reduces lipid accumulation via AMPK activation in human hepatic cells. J Dig Dis. 2010;11:55–62. doi: 10.1111/j.1751-2980.2009.00416.x. [DOI] [PubMed] [Google Scholar]
- 300.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 301.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Shin SM, Cho IJ, Kim SG. Resveratrol protects mitochondria against oxidative stress through AMP-activated protein kinase-mediated glycogen synthase kinase-3beta inhibition downstream of poly(ADP-ribose)polymerase-LKB1 pathway. Mol Pharmacol. 2009;76:884–895. doi: 10.1124/mol.109.058479. [DOI] [PubMed] [Google Scholar]
- 303.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 304.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 305.Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 306.Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 307.Smith BC, Denu JM. Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Smith JJ, Kenney RD, Gagne DJ, Frushour BP, Ladd W, Galonek HL, Israelian K, Song J, Razvadauskaite G, Lynch AV, Carney DP, Johnson RJ, Lavu S, Iffland A, Elliott PJ, Lambert PD, Elliston KO, Jirousek MR, Milne JC, Boss O. Small molecule activators of SIRT1 replicate signaling pathways triggered by calorie restriction in vivo. BMC Syst Biol. 2009;3:31. doi: 10.1186/1752-0509-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 312.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Stetson MH, Watson-Whitmyre M. Nucleus suprachiasmaticus: the biological clock in the hamster? Science. 1976;191:197–199. doi: 10.1126/science.942799. [DOI] [PubMed] [Google Scholar]
- 314.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Subauste AR, Burant CF. Role of FoxO1 in FFA-induced oxidative stress in adipocytes. Am J Physiol Endocrinol Metab. 2007;293:E159–E164. doi: 10.1152/ajpendo.00629.2006. [DOI] [PubMed] [Google Scholar]
- 316.Sulaiman M, Matta MJ, Sundaresan NR, Gupta MP, Periasamy M, Gupta M. Resveratrol, an activator of SIRT1, upregulates sarcoplasmic calcium ATPase and improves cardiac function in diabetic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2010;298:H833–H843. doi: 10.1152/ajpheart.00418.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 318.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Szymanski J. Versuche uber die Fahigkeit der Hunde zur Bildung von optischen Association. Pflügers Arch. 1918;171 [Google Scholar]
- 321.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 323.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 324.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Timmers S, Konings E, Bilet L, Houtkooper RH, van de Weijer T, Goossens GH, Hoeks J, van der Krieken S, Ryu D, Kersten S, Moonen-Kornips E, Hesselink MK, Kunz I, Schrauwen-Hinderling VB, Blaak EE, Auwerx J, Schrauwen P. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14:612–622. doi: 10.1016/j.cmet.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Tissenbaum H, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 329.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 330.Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 331.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, Kim MK, Viollet B, Chung JH. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59:554–563. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Vahtola E, Louhelainen M, Merasto S, Martonen E, Penttinen S, Aahos I, Kyto V, Virtanen I, Mervaala E. Forkhead class O transcription factor 3a activation and Sirtuin1 overexpression in the hypertrophied myocardium of the diabetic Goto-Kakizaki rat. J Hypertens. 2008;26:334–344. doi: 10.1097/HJH.0b013e3282f293c8. [DOI] [PubMed] [Google Scholar]
- 334.Van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2 (SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 335.Van der Veer E, Nong Z, O'Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97:25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- 336.Van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA. C. elegans model identifies genetic modifiers of alpha-synuclein inclusion formation during aging. PLoS Genet. 2008;4:e1000027. doi: 10.1371/journal.pgen.1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 338.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 339.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 340.Velasquez DA, Martinez G, Romero A, Vazquez MJ, Boit KD, Dopeso-Reyes IG, Lopez M, Vidal A, Nogueiras R, Dieguez C. The central Sirtuin 1/p53 pathway is essential for the orexigenic action of ghrelin. Diabetes. 2011;60:1177–1185. doi: 10.2337/db10-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, Whetstine JR, Mostoslavsky R, Puigserver P, Li X, Dyson NJ, Hart AC, Naar AM. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, Nemoto S, Finkel T, Gu W, Cress WD, Chen J. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 344.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 345.Wang H, Qiang L, Farmer SR. Identification of a domain within peroxisome proliferator-activated receptor gamma regulating expression of a group of genes containing fibroblast growth factor 21 that are selectively repressed by SIRT1 in adipocytes. Mol Cell Biol. 2008;28:188–200. doi: 10.1128/MCB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Wang J, Chen J. SIRT1 regulates autoacetylation and histone acetyltransferase activity of TIP60. J Biol Chem. 2010;285:11458–11464. doi: 10.1074/jbc.M109.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wang RH, Kim HS, Xiao C, Xu X, Gavrilova O, Deng CX. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J Clin Invest. 2011;121:4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, Kim S, Xu X, Zheng Y, Chilton B, Jia R, Zheng ZM, Appella E, Wang XW, Ried T, Deng CX. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Weaver DR. The suprachiasmatic nucleus: a 25-year retrospective. J Biol Rhythms. 1998;13:100–112. doi: 10.1177/074873098128999952. [DOI] [PubMed] [Google Scholar]
- 350.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Weyrich P, Machicao F, Reinhardt J, Machann J, Schick F, Tschritter O, Stefan N, Fritsche A, Haring HU. SIRT1 genetic variants associate with the metabolic response of Caucasians to a controlled lifestyle intervention–the TULIP Study. BMC Med Genet. 2008;9:100. doi: 10.1186/1471-2350-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wijnen H. Circadian rhythms. A circadian loop asSIRTs itself. Science. 2009;324:598–599. doi: 10.1126/science.1174132. [DOI] [PubMed] [Google Scholar]
- 353.Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–1032. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- 354.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Wu J, Zhang F, Yan M, Wu D, Yu Q, Zhang Y, Zhou B, McBurney MW, Zhai Q. WldS enhances insulin transcription and secretion via a SIRT1-dependent pathway and improves glucose homeostasis. Diabetes. 2011;60:3197–3207. doi: 10.2337/db11-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Xu F, Gao Z, Zhang J, Rivera CA, Yin J, Weng J, Ye J. Lack of SIRT1 (Mammalian Sirtuin 1) activity leads to liver steatosis in the SIRT1+/− mice: a role of lipid mobilization and inflammation. Endocrinology. 2010;151:2504–2514. doi: 10.1210/en.2009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Yahiaoui B, Taibi A, Ouaissi A. A Leishmania major protein with extensive homology to silent information regulator 2 of Saccharomyces cerevisiae. Gene. 1996;169:115–118. doi: 10.1016/0378-1119(95)00785-7. [DOI] [PubMed] [Google Scholar]
- 358.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 360.Yamazaki Y, Usui I, Kanatani Y, Matsuya Y, Tsuneyama K, Fujisaka S, Bukhari A, Suzuki H, Senda S, Imanishi S, Hirata K, Ishiki M, Hayashi R, Urakaze M, Nemoto H, Kobayashi M, Tobe K. Treatment with SRT1720, a SIRT1 activator, ameliorates fatty liver with reduced expression of lipogenic enzymes in MSG mice. Am J Physiol Endocrinol Metab. 2009;297:E1179–E1186. doi: 10.1152/ajpendo.90997.2008. [DOI] [PubMed] [Google Scholar]
- 361.Yang H, Lavu S, Sinclair DA. Nampt/PBEF/Visfatin: a regulator of mammalian health and longevity? Exp Gerontol. 2006;41:718–726. doi: 10.1016/j.exger.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Yang J, Kong X, Martins-Santos ME, Aleman G, Chaco E, Liu GE, Wu SY, Samols D, Hakimi P, Chiang CM, Hanson RW. Activation of SIRT1 by resveratrol represses transcription of the gene for the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) by deacetylating hepatic nuclear factor 4alpha. J Biol Chem. 2009;284:27042–27053. doi: 10.1074/jbc.M109.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-kappaB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. 2007;292:L567–L576. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 365.Yang Y, Hou H, Haller EM, Nicosia SV, Bai W. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 2005;24:1021–1032. doi: 10.1038/sj.emboj.7600570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Yang YH, Chen YH, Zhang CY, Nimmakayalu MA, Ward DC, Weissman S. Cloning and characterization of two mouse genes with homology to the yeast Sir2 gene. Genomics. 2000;69:355–369. doi: 10.1006/geno.2000.6360. [DOI] [PubMed] [Google Scholar]
- 367.Yang Z, Kahn BB, Shi H, Xue BZ. Macrophage alpha1 AMP-activated protein kinase (alpha1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J Biol Chem. 2010;285:19051–19059. doi: 10.1074/jbc.M110.123620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Yeagley D, Guo S, Unterman T, Quinn PG. Gene- and activation-specific mechanisms for insulin inhibition of basal and glucocorticoid-induced insulin-like growth factor binding protein-1 and phosphoenolpyruvate carboxykinase transcription. Roles of forkhead and insulin response sequences. J Biol Chem. 2001;276:33705–33710. doi: 10.1074/jbc.M101215200. [DOI] [PubMed] [Google Scholar]
- 369.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Yi J, Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim Biophys Acta. 2010;1804:1684–1689. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, Lu JC, Smith JJ, Jirousek MR, Olefsky JM. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. 2009;29:1363–1374. doi: 10.1128/MCB.00705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh DY, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298:E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.You M, Liang X, Ajmo JM, Ness GC. Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am J Physiol Gastrointest Liver Physiol. 2008;294:G892–G898. doi: 10.1152/ajpgi.00575.2007. [DOI] [PubMed] [Google Scholar]
- 374.Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann NY Acad Sci. 2009;1173(Suppl 1):E10–19. doi: 10.1111/j.1749-6632.2009.04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Yuan Z, Seto E. A functional link between SIRT1 deacetylase and NBS1 in DNA damage response. Cell Cycle. 2007;6:2869–2871. doi: 10.4161/cc.6.23.5026. [DOI] [PubMed] [Google Scholar]
- 378.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Zakhary SM, Ayubcha D, Dileo JN, Jose R, Leheste JR, Horowitz JM, Torres G. Distribution analysis of deacetylase SIRT1 in rodent and human nervous systems. Anat Rec. 2010;293:1024–1032. doi: 10.1002/ar.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Zhang J. The direct involvement of SirT1 in insulin-induced insulin receptor substrate-2 tyrosine phosphorylation. J Biol Chem. 2007;282:34356–34364. doi: 10.1074/jbc.M706644200. [DOI] [PubMed] [Google Scholar]
- 381.Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, Heidenreich KA, Sajan MP, Farese RV, Stolz DB, Tso P, Koo SH, Montminy M, Unterman TG. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 382.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Zhao X, Gan L, Pan H, Kan D, Majeski M, Adam SA, Unterman TG. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14–3-3-dependent and -independent mechanisms. Biochem J. 2004;378:839–849. doi: 10.1042/BJ20031450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Zillikens MC, van Meurs JB, Rivadeneira F, Amin N, Hofman A, Oostra BA, Sijbrands EJ, Witteman JC, Pols HA, van Duijn CM, Uitterlinden AG. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009;58:2828–2834. doi: 10.2337/db09-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Zu Y, Liu L, Lee MY, Xu C, Liang Y, Man RY, Vanhoutte PM, Wang Y. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circ Res. 2010;106:1384–1393. doi: 10.1161/CIRCRESAHA.109.215483. [DOI] [PubMed] [Google Scholar]