Abstract
Cellular senescence is an intrinsic defense mechanism to various cellular stresses: while still metabolically active, senescent cells stop dividing and enter a proliferation arrest. Here, we identify DPY30, a member of all mammalian histone H3K4 histone methyltransferases (HMTases), as a key regulator of the proliferation potential of human primary cells. Following depletion of DPY30, cells show a severe proliferation defect and display a senescent phenotype, including a flattened and enlarged morphology, elevated level of reactive oxygen species (ROS), increased SA-β-galactosidase activity, and formation of senescence-associated heterochromatin foci (SAHFs). While DPY30 depletion leads to a reduced level of H3K4me3-marked active chromatin, we observed a concomitant activation of CDK inhibitors, including p16INK4a, independent of H3K4me3. ChIP experiments show that key regulators of cell-cycle progression, including ID proteins, are under direct control of DPY30. Because ID proteins are negative regulators of the transcription factors ETS1/2, depletion of DPY30 leads to the transcriptional activation of p16INK4a by ETS1/2 and thus to a senescent-like phenotype. Ectoptic re-introduction of ID protein expression can partially rescue the senescence-like phenotype induced by DPY30 depletion. Thus, our data indicate that DPY30 controls proliferation by regulating ID proteins expression, which in turn lead to senescence bypass.
Keywords: chromatin, DPY30, ID proteins, MLL, senescence
Introduction
Transcriptional regulation involves the action of chromatin remodelling complexes as well as histone modifying enzymes. Post-translational modifications (PTMs) of histone proteins, such as acetylation, methylation, phosphorylation, and ubiquitination, affect numerous molecular processes, including gene expression. Histone lysine methylation can either activate or repress genes, depending on the lysine residue that is modified. In general, trimethylation of lysine 27 on histone H3 (H3K27me3) is a ‘repressive’ mark, while methylation on lysine 4 of the same histone (H3K4me3) is an ‘active’ mark. These two histone modifications are synthesized by the antagonistic action of the Polycomb group (PcG) complexes and the Trithorax group (TrxG) complexes, respectively, and are involved in many biological readouts, including cell fate determination, differentiation, cellular memory, cell-cycle control and when deregulated in tumorigenesis (Richly et al, 2010; Schuettengruber et al, 2011).
Only two histone methyltransferases (HMTases) within the PcG complex family have been described in mammals, but TrxG complexes are known to be more diverse. In yeast, SET domain-containing 1 (Set1) of the COMPASS complex is the only H3K4 HMTase that specifically catalyses the mono-, di-, and trimethylation of H3K4 (Miller et al, 2001; Roguev et al, 2001). In contrast, at least six COMPASS-like complexes with non-redundant functions have been described in mammals. The complexes, named after their respective HMTase (hSet1a, hSet1b, MLL1 (mixed linage leukaemia), MLL2, MLL3, and MLL4) are structurally different but contain at least one evolutionary conserved SET domain specific for H3K4 deposition (Schuettengruber et al, 2011). All Set1 and MLL complexes also share four common structural components (ASH2L, RBBP5, WDR5, and DPY30) that are conserved in yeast (Bre2, Swd1, Swd3, and Sdc1, respectively) and are essential for stability and proper HMTase activity of the complex (Tenney and Shilatifard, 2005; Steward et al, 2006; van Nuland et al, 2013).
While specific functions of ASH2L, RBBP5, and WDR5 have been described, DPY30 remains fairly uncharacterized. It was shown that DPY30 is a common subunit of all Set1 and MLL complexes and that it directly interacts with ASH2L (Cho et al, 2007; Wang et al, 2009; van Nuland et al, 2013). Originally, Dpy-30 was described in C. elegans, where it is implicated in embryogenesis and dosage compensation (Hsu et al, 1995; Dong et al, 2005). The yeast DPY30 homologue Sdc1 is part of the COMPASS complex. Its deletion affects global H3K4 methylation and growth; however, the phenotype is less severe than those observed after deletion of other subunits of the complex (Dehe et al, 2006). In mammals, it was recently shown that DPY30 is implicated in the differentiation potential of mouse embryonic stem cells (mESCs), but not in their self-renewal (Jiang et al, 2011).
Genome-wide expression studies combined with ChIP-on-chip and ChIP sequencing experiments in human embryonic fibroblasts and mESCs suggest that PcG and their antagonist TrxG proteins play a major role in developmental and differentiation processes (Bracken et al, 2006; Ang et al, 2011; Jiang et al, 2011). In addition, PcG and TrxG proteins play a crucial role in cell cycle and senescence (Simboeck et al, 2011). Cellular senescence is caused by various cellular stresses and leads to an irreversible proliferation arrest. Senescent cells share several common features, such as flattened and enlarged morphology, nuclei that can display the appearance of senescence-associated heterochromatin foci (SAHFs), an enlarged lysosomal compartment and consequently more SA-β-galactosidase activity, activation of DNA damage response (DDR) pathway, elevated levels of ROS, and the expression of several cell-cycle inhibitors (Dimri et al, 2000; Narita et al, 2003; Campisi and d'Adda di Fagagna, 2007). Recently, it was reported that senescent cells secrete pro-inflammatory factors, a phenomenon called senescence-associated secretory phenotype (SASP) (Coppe et al, 2008). Replicative senescence results from shortened chromosome telomeres after repeated rounds of DNA replication coupled to cell division (Hayflick, 1965; Campisi and d'Adda di Fagagna, 2007). Oncogene-induced senescence (OIS) is induced by various cancer-causing cellular stresses, including oxidative stress, activation of oncogenes, and inactivation of tumour suppressor genes, and is therefore considered to be an important barrier to cellular transformation (Krimpenfort et al, 2001; Sharpless et al, 2001). Two major pathways involved in senescence are the pRb and p53 tumour suppressor pathways, which control the expression of the INK4/ARF locus and the cyclin-dependent kinase inhibitors (CDKIs) p21CIP1WAF1 and p27KIP1. p16INK4a and p15INK4b are also CDKIs that inhibit cyclinD/CDK4 (CDK6) complexes and, concomitantly, pRb phosphorylation in early G1 phase, leading to a block in cell-cycle progression. p14ARF prevents p53 degradation, which consequently can lead to apoptosis or cell-cycle arrest by activating p21CIP1WAF1 and p27KIP1. These CDKIs specifically inhibit cyclinE/CDK2 complexes in late G1 phase and maintain a stable growth arrest in senescence (Adams, 2009; Simboeck et al, 2011).
The INK4/ARF locus, a critical regulator of senescence, is tightly regulated by the PcG and TrxG proteins. In proliferating cells, the locus is silenced by PcG complexes (PRC2 and PRC1) and the chromatin is enriched in H3K27me3 (Bracken et al, 2007). When senescence is triggered, the PRC2 complex is displaced from the locus and H3K27me3 is actively removed by JMJD3 (Agger et al, 2009). Co-activator complexes are then recruited to the locus, leading to H3K4me3 deposition; of these, the MLL1 complex is the best-studied one to date (Kia et al, 2008). In addition, remodelling complexes, such as SWI/SNF (which contains the TrxG protein SNF5), alter the chromatin structure and thereby enable the binding of ETS family transcription factors. The p16INK4a promoter has ETS binding sites, and p16INK4a expression after exposure to senescence triggers has been shown to be due especially to the binding of ETS1/2 (Ohtani et al, 2001; Huot et al, 2002; von Brandenstein et al, 2012). The action of ETS1/2 can be suppressed through interactions with a member of the ID protein family (Ohtani et al, 2001).
In line with this simplified model, overexpression of PcG proteins (e.g., BMI1, CBX7, and CBX8) delays the onset for replicative senescence by repressing the INK4/ARF locus (Gil et al, 2004; Dietrich et al, 2007). On the other hand, MLL1 directly activates the p16INK4a promoter after oncogene activation and deletion of MLL1 bypasses OIS induced by ectopic expression of the oncogene H-Ras (Kia et al, 2008; Kotake et al, 2009).
Here, we analysed the function of DPY30, a relatively uncharacterized subcomponent of all mammalian hSet1 and MLL complexes. Unexpectedly, depletion of DPY30 led to a dramatic proliferation arrest. The phenotype resembled cellular senescence, with increased SA-β-galactosidase and ROS activity, increased DDR, an induction of the INK4/ARF locus independent of H3K4me3, and formation of SAHFs. Genome-wide expression and ChIP sequencing studies revealed that DPY30 directly controls cell-cycle regulators, including ID proteins that secondarily contribute to a stable cell-cycle arrest. These findings reveal a novel mechanism of INK4/ARF regulation in senescence and show for the first time, to our knowledge, that key regulators of cell-cycle progression are under the control of DPY30 and, therefore, of hSet1 and MLL complexes.
Results
Loss of DPY30 leads to a senescence-like phenotype
DPY30 depletion significantly alters the differentiation potential of mESCs, in particular along the neural linage (Jiang et al, 2011). We initially investigated the role of DPY30 in differentiation of human teratocarcinoma NT2 cells, which differentiate into a neuron-like lineage upon retinoic acid stimulation (Buschbeck et al, 2009). We stably depleted DPY30 in NT2 cells using three different short-hairpin RNAs (shRNA) against the human DPY30 mRNA, to avoid off-target effects, and one unspecific control shRNA (referred to as shDPY30 and shCtrl, respectively). We monitored knockdown efficiencies on DPY30 mRNA and protein levels (Supplementary Figure 1A) and their effects on the H3K4 methylation levels by western blot and immunofluorescence (Supplementary Figure 1B and C). Reflecting the role of DPY30 in MLL-mediated H3K4 trimethylation, we observed strong reductions in global H3K4me3 upon depletion of DPY30. In addition, we observed that NT2 cells, in which DPY30 was stably interfered, stopped proliferating (Supplementary Figure 1F and G) and underwent a change in their morphology that resembled senescence (Supplementary Figure 1D). shDPY30 cells had γH2A.X stained foci, a marker for DNA double-strand breaks and activated DDR (Supplementary Figure 1C). In addition, staining for SA-β-galactosidase activity revealed that >80% of the shDPY30 cells were indeed positive for this senescence marker, whereas control cells exhibited a much lower level (12%) (Supplementary Figure 1E). Thus, the loss of DPY30 in NT2 cells severely affects global H3K4me3 levels and leads to a senescence-like phenotype. Indeed, global H3K4me3 levels are known to decrease in senescent cells, in accordance with their reduced transcriptional output.
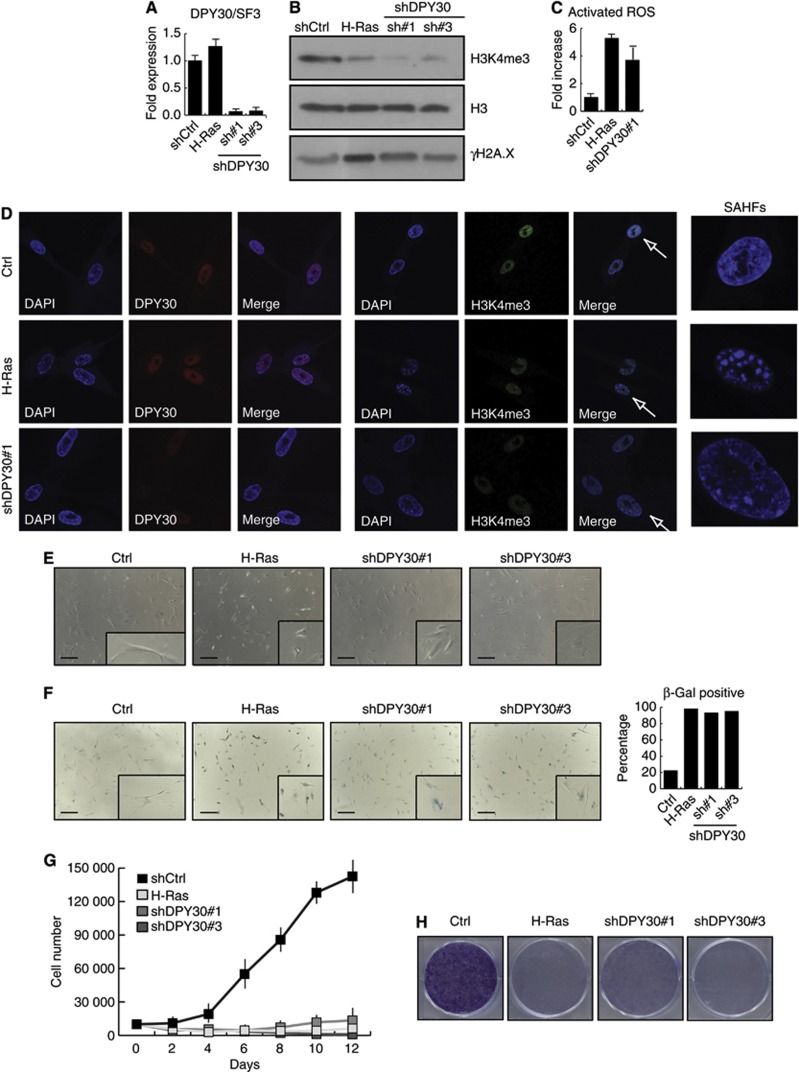
The senescent state of human cells has been best characterized in primary fibroblasts that undergo senescence in response to extensive passaging (replicative senescence or ageing) or upon exogenous expression of oncogenic Ras (H-Ras) (OIS) (Hayflick, 1965; Serrano et al, 1997). We therefore infected early passage IMR90 human fibroblasts to stably knockdown the expression of DPY30 using two different shRNAs. As a positive control for senescence, we also infected IMR90 cells to ectopically express H-RasV12 (H-Ras). Depletion of DPY30 in IMR90 cells was almost as efficient as that in NT2 cells, while ectopic expression of H-Ras did not alter DPY30 expression levels (Figure 1A and D). Global H3K4me3 levels were significantly reduced in both, shDPY30 and H-Ras expressing cells (Figure 1B and D). DPY30 knockdown cells presented increased DNA double-strand breaks (γH2A.X, Figure 1B and Supplementary Figure 2; TP53, Supplementary Figure 2) and elevated levels of activated ROS (Figure 1C). shDPY30 IMR90 cells also contained DAPI-stained SAHFs, another characteristic for senescent human fibroblasts (Narita et al, 2003) (Figure 1D). Additionally, the overall morphology of the DPY30 knockdown IMR90 cells changed, towards a more rounded and flattened phenotype with enlarged nuclei (Figure 1E). SA-β-galactosidase assays showed that >90% of the shDPY30 IMR90 cells expressed active SA-β-galactosidase (Figure 1F). Further, in proliferation assays, growth curves, and colony formation assays, shDPY30 IMR90 cells arrested and stopped proliferating (Figure 1G and H). We conclude that the loss of DPY30 results in a senescence-like phenotype in human fibroblasts.
Figure 1.
Loss of DPY30 leads to senescence-like phenotype. (A) Stable depletion of DPY30 in IMR90 human fibroblasts. mRNA expression levels of DPY30 were monitored by RT–PCR 6 days after selection in IMR90 control cells (shCtrl), those expressing oncogenic H-Ras, and those interfered for DPY30 using two different short-hairpin constructs (DPY30 sh#1 and sh#3). Expression of DPY30 was normalized to the housekeeping gene SF3B2 (SF3) and is shown relative to the IMR90 control cells as fold-reduction. These cell lines were used for the assays in this figure 6 days after selection. (B) H3K4me3 levels are reduced, while γH2A.X levels are increased, in IMR90 cells interfered for DPY30. Acid-extracted histones from control (shCtrl), senescent (H-Ras), and DPY30-depleted (DPY30 sh#1 and sh#3) IMR90 cells (6 days after selection) were separated by SDS–PAGE and analysed by western blot for H3K4me3 and γH2A.X. An antibody against total histone H3 was used to ensure equal loading. (C) Total ROS activity is increased in shDPY30 IMR90 cells. Total ROS activity was determined in control (shCtrl), H-Ras expressing and DPY30-depleted (shDPY30 sh#1) IMR90 by flow cytometry and is illustrated as fold-increase relative to control cells (n=3). (D) Immunofluorescence for DPY30 and H3K4me3 in control (Ctrl), senescent (H-Ras), and DPY30-depleted (DPY30 sh#1) cells. SAHFs in senescent and DPY30-depleted cells were stained by DAPI. (E) Morphological changes upon interference of DPY30. Pictures were taken by optical microscopy. Scale bar corresponds to 325 μm. A flattened morphology typical for senescence is observed for the IMR90 cells expressing H-Ras (as expected) as well as for the DPY30-depleted cells (DPY30 sh#1 and sh#3). (F) SA-β-galactosidase assay performed with control (Ctrl), IMR90 expressing H-Ras or DPY30-depleted (shDPY30#1 and #3) IMR90 cells. For quantification, 600 cells of each cell line were counted. The scale bar corresponds to 325 μm. (G) DPY30-depleted fibroblasts stop proliferating. Growth curves of control IMR90 (Ctrl), IMR90 expressing H-Ras, and IMR90 interfered for DPY30 (shDPY30#1 and #3) were generated by seeding 10 000 cells on day 0 after selection; the cell number was counted every second day (n=3). (H) Colony formation assay of control IMR90 (Ctrl), IMR90 expressing H-Ras, and IMR90 interfered for DPY30 (shDPY30#1 and #3) was performed by seeding 20 000 cells on day 0 after selection. After 7 days, the colonies were stained with crystal violet.
Source data for this figure is available on the online supplementary information page.
Senescence induced by loss of DPY30 correlates with activation of established senescence signalling pathways
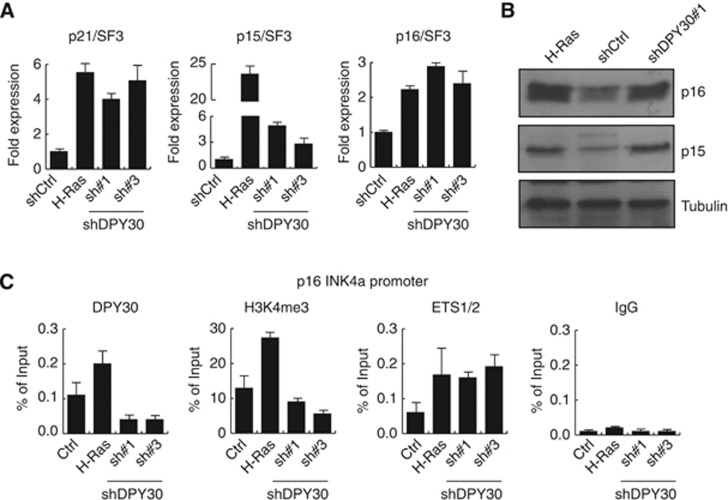
Since the shDPY30 phenotype was similar to OIS, we wondered whether it is established through activation of the two major pathways (p53 and pRB) involved in senescence (Adams, 2009; Simboeck et al, 2011). We thus monitored expression of mRNA levels of p21CIP1WAF1, p16INK4a, and p15INK4b, which are downstream targets of the p53 and pRB pathways, in shCtrl IMR90 cells, shDPY30 cells, and H-Ras expressing cells (Figure 2A). Expression of p21CIP1WAF1 and p16INK4a was strongly upregulated in shDPY30 cells, similar to in H-Ras-induced senescent cells. p15INK4b expression levels were also upregulated in all senescent cells, although much more in H-Ras-induced cells (Figure 2A). However, we observed comparable expression of the p16INK4a and p15INK4b protein levels in shDPY30 and H-Ras expressing IMR90 cells (Figure 2B). Consistent with the effect of cell division on the senescent phenotype, mRNA expression levels of the INK4 locus and the p21CIP1WAF1 gene varied depending on which day after infection the sample was taken (Supplementary Figure 3). Additionally, we confirmed that the loss of DPY30 in NT2 cells also resulted in activation of these CDKIs (Supplementary Figure 4A and B).
Figure 2.
Senescence induced by loss of DPY30 correlates with activation of tumour suppressors of the INK4/ARF and CIP/KIP family. (A) mRNA expression levels of p21CIP1WAF1 (p21), p15INK4b (p15), and p16INK4a (p16) were monitored in control (shCtrl), H-Ras-expressing, or DPY30-depleted (shDPY30#1 and #3) IMR90 cells. Gene expression was quantified 6 days after selection by RT–PCR. Expression values were normalized to the housekeeping gene SF3B2 (SF3) and are illustrated relative to IMR90 control cells as fold expression (n=3). (B) p16INK4a (p16) and p15INK4b (p15) upregulation was measured by western blot using nuclear extracts of cells as in (A). Western blots were probed with a tubulin antibody to ensure equal loading. (C) Chromatin immunoprecipitation (ChIP) in the same cell types as in (A) 6 days after selection. DPY30, H3K4me3, and ETS1/2 were monitored at the ETS1/2 binding site within the promoter region of the p16INK4a gene. An unspecific IgG antibody was included as a negative control. H3K4me3 was normalized to histone H3. Values are presented as percentage of input (n=3).
Source data for this figure is available on the online supplementary information page.
Upon onset of senescence, the p16INK4a promoter is activated by MLL1 complexes with concomitant H3K4me3 deposition (Kia et al, 2008; Kotake et al, 2009). As DPY30 is a common member of all mammalian H3K4 HMTase complexes (including the MLL1, MLL2, MLL3, MLL4, and Set1 complexes), we wondered if loss of DPY30 affects the levels of H3K4me3 at the p16INK4a promoter. We therefore performed ChIP analyses of shCtrl, shDPY30, and H-Ras-induced senescent IMR90 cells (Figure 2C). As expected, DPY30 was recruited to the p16INK4a promoter upon induction of senescence by ectopic expression of H-Ras. This correlated with an increase in H3K4me3 and with binding of the ETS1/2 transcription factor, which was shown to activate p16INK4a transcription (Ohtani et al, 2001). In contrast, in shDPY30 cells, DPY30 binding was clearly diminished with reduced levels of H3K4me3. However, in correlation with the transcriptional activation of the p16INK4a gene, the ETS1/2 transcription factor was still bound to the p16INK4a promoter.
Similarly, ChIP experiments revealed a loss of DPY30 binding and reduced H3K4me3 levels within the p16INK4a promoter in shCtrl and shDPY30 NT2 cells (Supplementary Figure 4C). In accordance with transcriptional activation of p16INK4a, binding of the polycomb component EZH2 was reduced as well as the repressive H3K27me3 mark. Additionally, monitoring pan-acetylation of histone H3 and H4 showed that the p16INK4a promoter was hyperacetylated. Overall, these data suggest that, contrary to the common model of p16INK4a regulation, activation of p16INK4a can occur independently of DPY30/MLL and even independently of increased H3K4me3 at its promoter.
Identification of deregulated genes upon depletion of DPY30
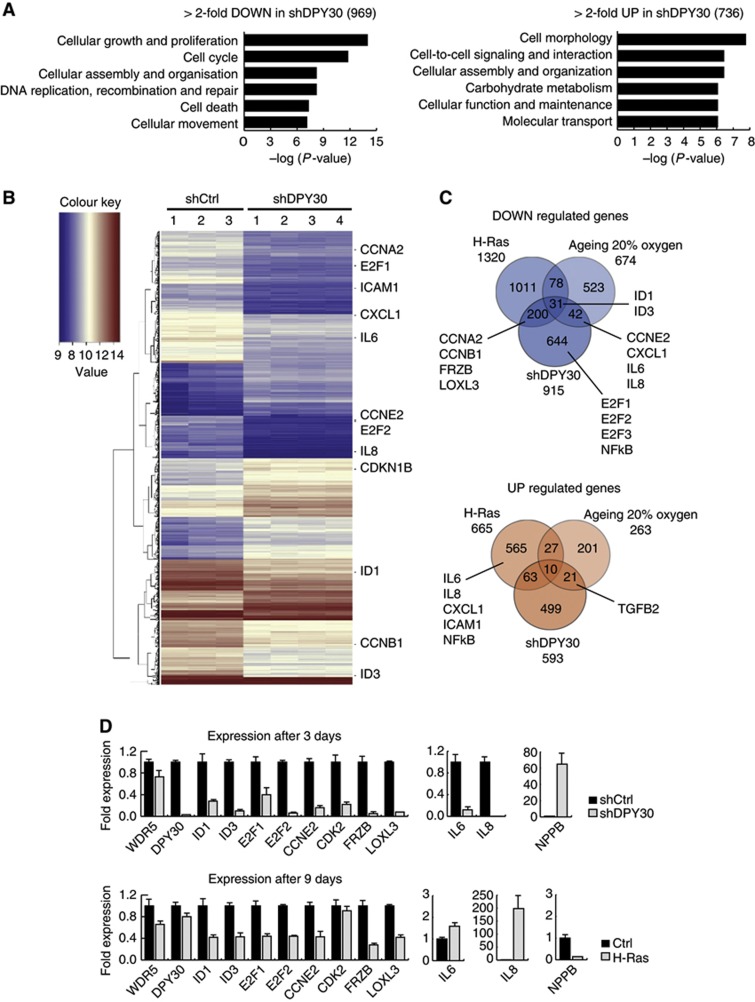
To elucidate how the loss of DPY30 can lead to a senescence-like phenotype yet also activate p16INK4a expression independently of H3K4me3, we monitored which additional genes are deregulated by performing a global expression analysis in shCtrl and shDPY30 IMR90 cells. Using stringent criteria, we observed a total of 1705 genes that were deregulated following shDPY30, with 969 genes less expressed and 736 genes high expressed (Figure 3A). GO analyses of the deregulated genes showed that downregulated genes were mainly involved in proliferation and cell cycle, while upregulated genes functioned in cell morphology and metabolism; this is in line with the observed senescence-like phenotype upon loss of DPY30. Note that we chose an early time point (of 3 days after selection) to enrich for direct DPY30 targets; however, at this time point, senescence is not fully established (Supplementary Figure 3), and the upregulation for p21CIP1WAF1 (CDKN1A), p16INK4a (CDKN2A), and p15INK4b (CDKN2B) was mild (1.5- to 2.0-fold; Supplementary Figure 5A).
Figure 3.
Microarray expression analyses of control and DPY30-depleted IMR90 cells, as compared to oncogene-induced and replicative senescence. (A) Gene ontology (GO) of >2-fold deregulated genes with a P-value of <0.05 in IMR90 cells depleted of DPY30 (shDPY30) 3 days after selection. In total, 969 genes were downregulated and 736 were upregulated. (B) Heatmap of genes deregulated >2-fold with a P-value of <0.05. Three biological replicates of IMR90 control cells (shCtrl), and four biological replicates of IMR90 DPY30 knockdown cells (shDPY30), were used for RNA extraction and array hybridization. Several genes involved in cell cycle and cytokine signalling are highlighted within the heatmap. (C) Venn diagram of overlapping deregulated genes in H-Ras-induced senescent IMR90 cells (H-Ras; Affymetrix), replicative senescent IMR90 cells grown under atmospheric conditions (ageing 20% oxygen; Affymetrix), and senescent IMR90 cells induced by knockdown of DPY30 (shDPY30; Agilent). Only genes common on both arrays were taken into account. Several genes involved in cell cycle and cytokine signalling are highlighted. (D) Verification of microarray expression analysis by RT–PCR. Samples were taken 3 days after selection for DPY30 knockdown, and 9 days after selection for H-Ras-expressing, IMR90 cells. Changes in gene expression were verified for several genes, which were mainly downregulated in DPY30 knockdown cells. Absolute expression values were normalized to the housekeeping gene SF3B2 (SF3) and are shown relative to the expression level in control IMR90 (shCtrl) cells, which was set to 1 (n=2).
Genes that were significantly downregulated include the activating E2F transcription factors (E2F1, E2F2, and E2F3) and several of their downstream targets, such as cyclins (CCNE2, CCNA2, and CCNB1) (Figure 3B; Supplementary Figure 5A). E2F transcription factors are crucial regulators of cell-cycle progression, regulating transcription of genes required for DNA synthesis, and E2F1 has been shown to be downregulated in senescent fibroblasts (Dimri et al, 1994). Additionally, all four of the ID proteins were transcriptionally downregulated in shDPY30 cells (ID1 and ID3 are highlighted in Figure 3B). ID proteins are linked to cellular senescence as they can directly interact with and repress the actions of pRb and ETS1/2 (Tournay and Benezra, 1996; Alani et al, 1999; Nickoloff et al, 2000).
Several cytokines and chemokines are known to be highly expressed and secreted in OIS and lead to the so-called SASP (Coppe et al, 2008). Surprisingly, several of those, including the chemokines IL6, IL8, CXCL1, and ICAM1, were significantly downregulated in shDPY30 cells (Figure 3B; Supplementary Figure 5B). This can reflect differences between the senescence phenotype induced by DPY30 and OIS; indeed, SASP is not an essential characteristic of senescent fibroblasts and has been shown to be regulated independently of p16INK4a (Coppe et al, 2011).
Gene set enrichment analysis (GSEA) confirmed the observation of the GO analysis. The ‘cell cycle’ and the ‘chemokine receptor binding’ gene sets were highly enriched and predominantly downregulated (Supplementary Figure 5A and B).
Comparison of deregulated genes in OIS, ageing, and shDPY30 cells
To investigate how DPY30 loss can induce a senescence-like phenotype that however does not correlate with the transcriptional output of classical senescence, we compared the transcriptome of shDPY30 cells with those of OIS and replicative senescent cells. For this, we used a published data set of growing and H-Ras-induced senescent IMR90 cells (Chicas et al, 2010; GEO data set: GSE19864) and a data set of young and aged (replicative senescence) IMR90 cells grown under atmospheric (20% oxygen) or physiologic (3% oxygen) conditions (GEO data set: GSE19018) (Supplementary Figure 6A–C). We identified the overlap of deregulated genes from all four arrays with each other, with a focus on replicative senescent cells grown under 20% oxygen, as shDPY30 cells had been grown under these conditions (Figure 3C; Supplementary Figure 7A). By and large, there was very little overlap of common downregulated genes between replicative senescence (20% oxygen) and either shDYP30-induced senescence or OIS (42 and 78 genes, respectively). ID1 and ID3 were among the few genes commonly downregulated. Interestingly, while DPY30 expression levels were unchanged in OIS cells, it was significantly reduced in replicative senescent IMR90 (20% oxygen) (Supplementary Figures 7C and 12D). Furthermore, factors upregulated in OIS SASP were usually downregulated in replicative senescence (20% oxygen) and shDPY30 senescent cells, indicating a genetic similarity between both phenotypes with respect to SASP. Interestingly, the overlap of deregulated genes in replicative senescent cells grown under different conditions (20% versus 3% oxygen) was poor, with a few SASP factors (IL8 and IL1B) actually upregulated under physiologic conditions (3% oxygen) (Supplementary Figure 7A).
Even fewer genes were commonly upregulated in the four senescent data sets, with no obvious common function. Interestingly, TGFB2, a member of the transforming growth factor beta family of cytokines, was upregulated in shDPY30 and ageing IMR90 cells (20% oxygen), but not in OIS, suggesting that the SASP profile of different types of senescence is composed of different cytokines, and that shDPY30 and aged senescence cells could share similarities in this aspect.
Deregulated genes upon DPY30 depletion were confirmed by qPCR (Figure 3D), with a focus on downregulated genes with a known role in cell cycle and senescence (ID1, ID3, E2F1, E2F2, CCNE2, and CDK2). We included the SASP-associated genes IL6 and IL8, two genes with the strongest decrease in expression (FRZB and LOXL3), and one upregulated gene (NPPB). WDR5 expression, a member of all H3K4 HMTase complexes, remained unchanged (as did all members of MLL complexes; Supplementary Figure 7B). In parallel, expression was also monitored in IMR90 cells that ectopically expressed H-Ras. After 3 days of selection, the expression of ID1 and ID3 remained unchanged in H-Ras-expressing cells, while E2F1, E2F2, and CCNE2 were repressed and IL6 and IL8 were strongly induced (Supplementary Figure 7C). However, 9 days after selection (the time point for the microarray analysis), ID1 and ID3 were downregulated also in OIS (Figure 3D). While loss of DPY30 likely affected ID protein expression immediately and probably directly, H-Ras-induced senescence led to a delayed ID repression that seemed to occur after the onset of senescence. This was also reflected at the protein level by immunoblot analysis against ID1 3 and 9 days after selection (Supplementary Figure 7D).
Identification of direct DPY30 target genes
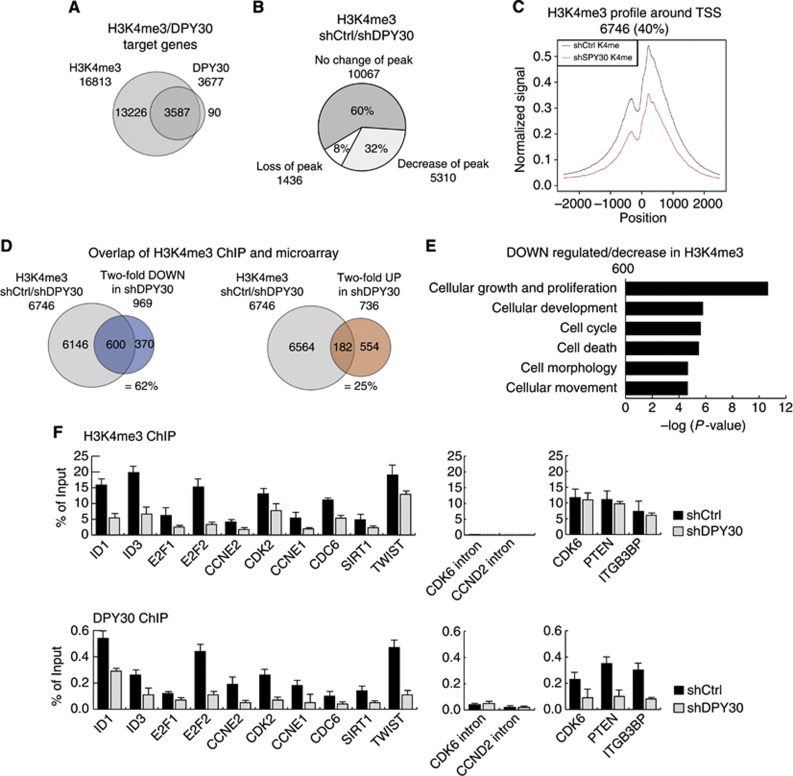
To elucidate which of the downregulated genes are direct DPY30 targets, we performed genome-wide high-throughput ChIP sequencing of DPY30 and H3K4me3 in control IMR90 fibroblasts. This revealed that DPY30 and the H3K4me3 mark are highly enriched at promoters and 5′ UTR regions of genes but are almost absent at 3′ ends of genes and in intergenic regions (Supplementary Figure 8A). H3K4me3 profiles were highly enriched within 2 kb around the transcriptional start sites (TSSs) of target genes, with a major dip slightly upstream of the TSS that coincides with sites poor in nucleosomes. In contrast, DPY30 profiles were centered over the TSS of target genes (Supplementary Figure 8A).
We found a very strong overlap (>98%) of genes enriched for DPY30 binding with genes enriched for H3K4me3 around their TSS, indicating that virtually all DPY30 target sites simultaneously carry an H3K4me3-modified histone mark signature (Figure 4A). These genes play a role in various molecular processes (Supplementary Figure 8B). To identify DPY30-regulated genes that could be involved in establishing the observed senescent-like phenotype, we performed ChIP sequencing for H3K4me3 in shDPY30 IMR90 fibroblasts (3 days after selection). Only a minor fraction of H3K4me3-positive genes (1436 genes; 8%) lost the mark completely in DPY30 knockdown cells. However, a larger fraction of target genes (5310 genes; 32%) had strongly reduced H3K4me3 peaks at their TSS (Figure 4B). We hypothesize that even more target genes would display reduced H3K4me3 levels at a time point later than 3 days after selection. To visualize the decrease in the H3K4me3 mark at these target genes in the shDPY30 sample, we plotted the averaged and normalized H3K4me3 peaks of these target genes (Figure 4C). While GO analysis for target genes with stable H3K4me3 revealed various molecular functions (Supplementary Figure 8C), target genes with decreased H3K4me3 peaks were predominately involved in cellular growth, proliferation, and cell cycle, which reflects the senescence-like phenotype of shDPY30 IMR90 cells (Supplementary Figure 8D).
Figure 4.
Global ChIP sequencing analysis of DPY30 and H3K4me3. (A) ChIPseq was performed for H3K4me3 and DPY30 in IMR90 control cells (shCtrl) 3 days after selection. The Venn diagram shows the overlap of genes that are trimethylated on H3K4 (H3K4me3) and bound by DPY30. (B) ChIPseq for H3K4me3 performed in DPY30-depleted IMR90 cells, 3 days after selection (shDPY30). H3K4me3 peak intensities were quantitatively compared for target genes in shCtrl and shDPY30 cells. The Pie chart represents a group of genes with trimethylated H3K4 (H3K4me3) in both, shCtrl and shDPY30 cells (60%), as well as genes with either loss (8%) or strong reduction (32%) in H3K4me3 peaks upon knockdown of DPY30. (C) Composite profiling of H3K4me3 peaks 2.5 kb upstream and downstream of the TSS of target genes that showed loss or a significant reduction in H3K4me3 levels in DPY30 knockdown IMR90 cells (red line). (D) Overlap of ChIPseq and expression array. The Venn diagrams show the overlap of genes with decreased H3K4me3 and those with >2-fold downregulation (=62%) or upregulation (=25%) in DPY30 knockdown IMR90 cells. (E) Gene ontology (GO) of genes with decreased H3K4me3 that were downregulated by >2-fold in DPY30 knockdown IMR90 cells. (F) Verification of ChIPseq. H3K4me3 levels and DPY30 binding were monitored on several candidate genes by RT–PCR. Three genes that revealed conserved H3K4me3 levels as well as intronic regions were included as negative controls. Values are represented as percentage of input (n=2).
We next compared genes deregulated at the transcriptome level with direct DPY30 and H3K4me3 target genes identified in ChIP sequencing. Given the presence of DPY30 in the activating MLL complexes, we were especially interested whether genes with decreased H3K4me3 were indeed transcriptionally downregulated in shDPY30 cells. We found that there was a large overlap (600 genes; 62%) of downregulated genes among the genes with reduced or lost H3K4me3 levels (Figure 4D). Of these 600 genes, about 17% (95 genes) were direct targets of DPY30 (Supplementary Figure 8E). A much smaller overlap (182 genes; 25%) was observed when upregulated genes and genes with reduced H3K4me3 levels in the shDPY30 cells were compared; this suggests that this subset of genes are not under the direct control of DPY30 and H3K4me3 but rather are activated via other mechanisms (Figure 4D). GO analysis of downregulated genes with less H3K4me3 in shDPY30 cells revealed enrichment in the molecular functions of cell cycle, cellular growth, and proliferation (Figure 4E). GSEA revealed the gene set for ‘cell cycle’ to be significantly enriched (Supplementary Figure 8F).
Interestingly, ChIP sequencing results revealed that p16INK4a (CDKN2A), p15INK4b (CDKN2B), and p21CIPKIP (CDKN1A) are also direct targets of DPY30 and H3K4me3. However, even though H3K4me3 levels were decreased at their promoters upon DPY30 knockdown, these genes were still transcriptionally active (Supplementary Figure 8F; Figure 2).
Next, to confirm the ChIP sequencing results, we tested H3K4me3 levels and DPY30 binding by RT–PCR following ChIP in several downregulated DPY30 target genes that play a role in cell-cycle control, as these could be involved in the senescence-like phenotype in shDPY30 cells (Figure 4F). We found that DPY30 binding and H3K4me3 were clearly diminished at the promoters of ID1, ID3, E2F1, E2F2, CCNE2, and CDK2. We also included four genes in the ChIP validation that have also been described to play a role in the senescence onset, namely, CCNE1, CDC6, SIRT1, and TWIST; we found that these also showed decreased H3K4me3 marks and a loss of DPY30 binding in shDPY30 cells. Finally, two regions negative for H3K4me3 were also negative in RT–PCR (CDK6 and CCND2 intronic regions), and three genes (CDK6, PTEN, and ITGB3BP) with unchanged H3K4me3 levels in shDPY30 cells were also unchanged in RT–PCR. Interestingly, we observed that these latter promoters were actually bound by DPY30, indicating that the active H3K4me3 mark was either maintained or possible changes appear only later.
ID proteins are key regulators in bypassing the senescence phenotype
To understand how the loss of DPY30 in IMR90 cells could lead to a senescence-like phenotype, we identified direct DPY30 target genes that become deregulated with the loss of DPY30. Importantly, several of those are key regulators of cell-cycle progression, including E2F family transcription factors, cyclins, CDKs, and ID proteins (for E2F transcription factors and ID proteins, see Supplementary Figure 9A and B). The role of E2Fs in senescence is still unclear: for instance, E2F1 has been reported to have a role in bypassing senescence (Park et al, 2006) but also to be involved in senescence-associated growth arrest (Dimri et al, 2000). In contrast, ID1 and ID3, which are repressed in all three expression arrays (OIS, replicative, and shDPY30-induced senescence), have already been linked to senescence in several cellular models (Tournay and Benezra, 1996; Alani et al, 1999; Nickoloff et al, 2000).
Given that ID proteins and/or E2F transcription decrease their expression in shDPY30 cells, their overexpression could prevent, or delay, senescence onset. To test for a direct involvement of these factors in establishing senescence after DPY30 knockdown, we ectopically overexpressed E2F2, ID1, or ID3 alone, or E2F2 with either ID1 or ID3, in shCtrl and shDPY30 IMR90 cells. Overexpressing these proteins did not affect the proliferation potential of these cells. In the DPY30-depleted cells, ectopic overexpression of E2F2 alone or in combination with one of the ID proteins did not rescue the senescence-like phenotype (Supplementary Figure 10).
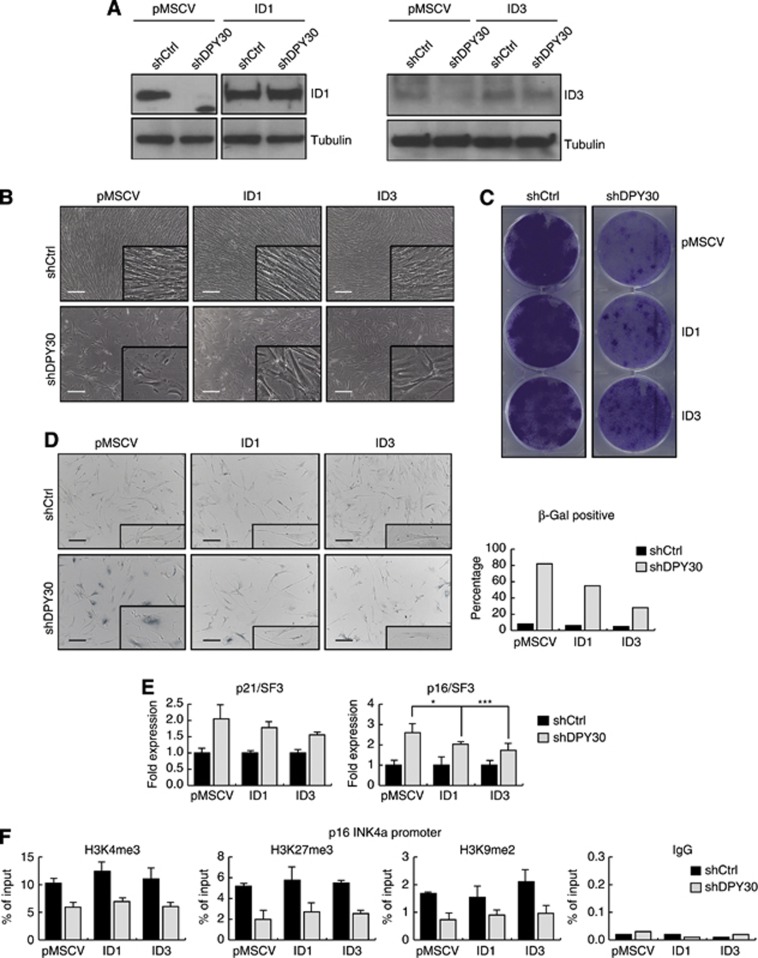
In contrast, overexpression of either ID1 or ID3 led to delayed, partial recovery of the senescence-like phenotype upon depletion of DPY30. Note that, in ID1 and ID3 overexpressing cells, DPY30 knockdown reduced mRNA levels, but not proteins levels, of ID1 and ID3 (Figure 5A; Supplementary Figure 11). All cell lines (e.g., pMSCV control and ID1 or ID3 overexpressing cells) initially had a proliferation arrest following DPY30 depletion, with high expression levels of the senescence markers p21CIP1WAF1 and p16INK4a, positive staining for SA-β-galactosidase activity, and senescence-like morphology and growth potential. However, after 2–3 weeks, the shDPY30 cells with ID1 or ID3 overexpression regained their fibroblast features and began to proliferate again, albeit not to the same degree as the IMR90 control cells, while the shDPY30 cells remained in a senescence-like state (Figure 5B). This partial rescue of proliferation is also illustrated by their ability to form colonies when plated at low density (Figure 5C). A reduced SA-β-galactosidase staining was observed in shDPY30 cells overexpressing either ID1 or ID3 (from 82 to 55 or 28%, respectively) (Figure 5D). Furthermore, the expression of p16INK4a was also significantly reduced, while that of p21CIP1WAF1 was only slightly affected (Figure 5E). The analysis of several repressive histone marks (e.g., H3K27me3 and H3K9me2) suggested that the re-silencing of p16INK4a promoter is mainly controlled by the reduced occupancy of ETS1/2 transcription factor due to overexpression of ID proteins, rather than to an epigenetic mechanism (Figure 5F).
Figure 5.
Ectopic expression of ID proteins partially rescues the senescence-like phenotype induced by loss of DPY30. (A) Ectopic overexpression of ID1 and ID3 in IMR90. IMR90 cells were infected with plasmids expressing empty pMSCV or pMSCV-ID1 (ID1) or pMSCV-ID3 (ID3). After selection, these cells were infected with a control hairpin (shCtrl) or with a hairpin to stably knockdown DPY30 (shDPY30). After the second selection, the expression levels of ID proteins were monitored by western blot using whole cell extracts. Tubulin was used to ensure equal loading. (B) Ectopic expression of ID1 and ID3 partially rescues the growth defect and morphological changes induced by loss of DPY30 (18 days after the second infection). Images were taken by optical microscopy. Scale bar corresponds to 325 μm. (C) Colony formation assay of control (shCtrl) and DPY30 knockdown (shDPY30) IMR90 cells, either not expressing (pMSCV) or expressing ID1 or ID3. About 20 000 cells were seeded on day 0 after the second selection. After 24 days, the colonies were stained with crystal violet. (D) SA-β-galactosidase assay was performed with control (shCtrl) and DPY30 knockdown (shDPY30) IMR90 cells that expressed ID1 or ID3, as indicated. Cells were seeded 18 days after the second selection and fixed for the assay 2 days later. For quantification, 400 cells of each cell line were counted. (E) Ectopic expression of ID1 and ID3 partially rescues the induced expression of p16INK4a by loss of DPY30. Eighteen days after selection, expression of p21CIP1WAF1 (p21) and p16INK4a (p16) was monitored by RT–PCR. Overexpression of the ID proteins resulted in a reduced induction of p21, which was not significant (ID1, P-value=0.3; ID3, P-value=0.1; Student’s t-test) and p16, which was significant in both cases (ID1, *P-value=0.06; ID3, ***P-value=0.01; Student’s t-test). Absolute expression values were normalized to the housekeeping gene SF3B2 (SF3) and are shown relative to the expression level in control IMR90 (pMSCV/shCtrl) cells as fold expression (n=5). (F) Chromatin immunoprecipitation (ChIP) in control (shCtrl) and DPY30 knockdown (shDPY30) cells, either not expressing (pMSCV) or expressing ID1 or ID3 18 days after the second infection. H3K4me3, H3K27me3, and H3K9me2 were monitored at the p16INK4a promoter region. The histone modifications were normalized to histone H3. An unspecific IgG antibody was included as a negative control. Values are presented as percentage of input (n=3).
Source data for this figure is available on the online supplementary information page.
These data suggest that the expression of ID1 or ID3 is crucial for cell-cycle progression. Their expression is dependent on DPY30 and H3K4me3 and is almost entirely lost as a consequence of DPY30 knockdown. Loss of DPY30 and, concomitantly, of the ID proteins results in a senescence-like proliferation arrest, partially due to enhanced ETS1/2 transcription factor binding to p16INK4a. Ectopic overexpression of ID1 and (to a greater degree) ID3 rescues the observed senescence-like phenotype, albeit with a delay and resulting in a slower growth pattern, suggesting that ID proteins are key regulators in cell-cycle progression and bypassing senescence. Furthermore, data obtained in young (3 months) versus old (18 months) mice revealed decreased DPY30 as well as ID protein expression in skin, suggesting that DPY30 regulation of ID protein expression could indeed be involved in ageing (Supplementary Figure 12).
Discussion
Knockdown of DPY30 results in a senescence-like phenotype
PcG and TrxG proteins are well-studied regulators of cell fate determination and differentiation. DPY30 is a member of all mammalian Set1 and MLL complexes. Recently, DPY30 has been shown to be implicated during cell fate specification in mESCs along the neuronal linage (Jiang et al, 2011). Depletion of DPY30 changed the differentiation potential of mESCs, accompanied by reduced global H3K4me3 levels and impaired plasticity in transcriptional reprogramming, while mESC self-renewal was not affected.
Interestingly, when interfering for DPY30 in the human teratocarcinoma cell line NT2, we observed a severe defect in the proliferation potential of these cells. NT2 cells are considered stem-like, due to their capacity to differentiate into a neuron lineage upon retinoic acid signalling and are therefore an established in vitro differentiation cell system (Buschbeck et al, 2009). However, in contrast to fully pluripotent mESCs, NT2 cells are already committed for neuronal differentiation, which could explain a different readout upon loss of DPY30 in mESCs and NT2 cells.
Loss of DPY30 in differentiated human fibroblasts (IMR90) resulted in a proliferation arrest with features of cellular senescence, including active SA-β-galactosidase staining, expression of cell-cycle inhibitors (e.g., p16INK4a), activation of DDR pathways, elevated levels of ROS, and formation of SAHFs. These heterochromatin structures occur in senescent but not in quiescent cells. Other heterochromatin markers, like macroH2A, were also present in SAHFs of DPY30 knockdown cells (unpublished observation). Global expression analysis of DPY30 knockdown cells showed that mainly genes involved in metabolism, cellular function, and maintenance were induced. Thus, although the loss of DPY30 dramatically affects H3K4me3 levels, which has been suggested to be problematic for cell viability, cells compensate for this by entering a senescence-like state while remaining metabolically active.
H3K4me3-independent activation of p16INK4a
In human NT2 as well as IMR90 cells, loss of DPY30 caused a severe proliferation stop that correlates with upregulated expression of p21CIP1WAF1, p15INK4b, and p16INK4a. While this is in agreement with the observed senescence-like phenotype, it was nonetheless surprising that p16INK4a could be activated without an active DPY30–MLL complex, since it has been previously demonstrated that the MLL1 complex is required for p16INK4a activation, and that depletion of MLL1 bypasses OIS (Kotake et al, 2009). We therefore wondered whether an H3K4-specific HTMase complex might still be active in the absence of DPY30, which might explain the activation of the p16INK4a gene in shDPY30 cells. However, in both cell systems (NT2 and IMR90) we observed reduced H3K4me3 levels on the p16INK4a promoter upon DPY30 knockdown, suggesting that DPY30 is required for an active MLL complex at this locus. Therefore, an active MLL complex and concomitant H3K4me3 are not strictly required to activate p16INK4a expression. Several studies have shown that transcriptional activation of p16INK4a and p21CIP1WAF1 by HDAC inhibitors leads to hyperactetylation of histones H3 and H4 at their promoters (Zhou and Zhu, 2009; Zupkovitz et al, 2010; Wang et al, 2012), and we indeed found that the p16INK4a promoter was hyperacteylated in DPY30 knockdown cells. We additionally observed increased ETS1/2 transcription factor binding in DPY30 knockdown cells, which is required for proper p16INK4a gene activation. The activity of the ETS1/2 transcription factors is inhibited when they are bound to ID proteins (Ohtani et al, 2001). By genome-wide expression and ChIPseq analyses, we identified ID proteins as direct DPY30 target genes, whose expression was repressed following DPY30 knockdown. Therefore, we propose that, in DPY30 knockdown cells, p16INK4a expression is independent of the H3K4me3 mark and is instead activated by increased ETS1/2 transcription factor binding, which is due to a decrease in ID protein expression.
Loss of DPY30 causes a ‘senescence’ readout through the E2F and ID pathways
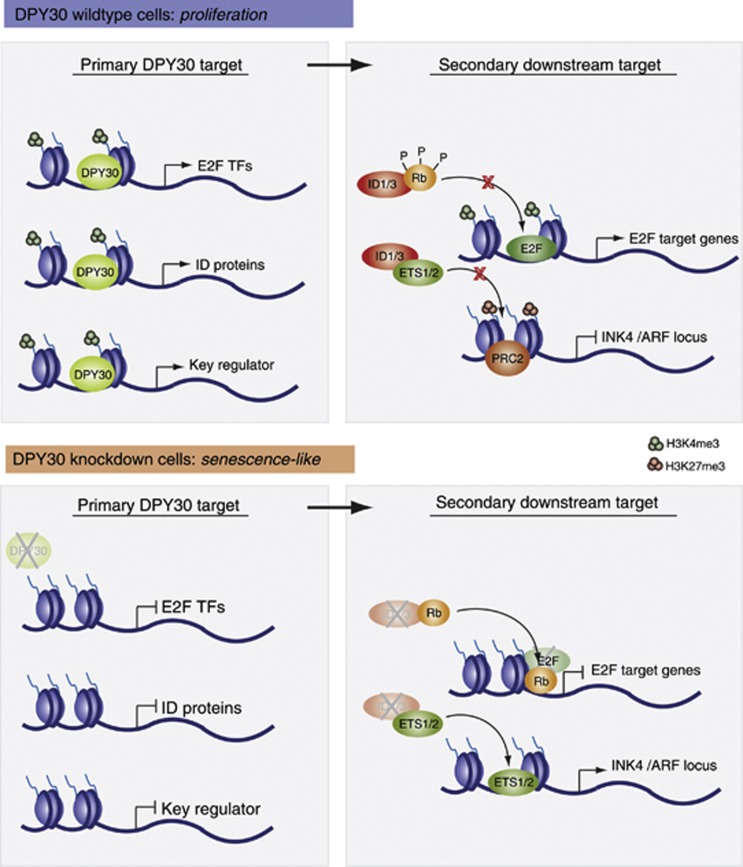
Using a genome-wide approach that combined ChIP sequencing for DPY30 and H3K4me3 with expression arrays in IMR90 control and DPY30 knockdown cells, we identified at least 600 genes that are directly targeted and regulated by DPY30. These DPY30 target genes are mainly involved in regulating proliferation and cell cycle, which correlates well with the observed phenotype of a senescence-like state in the absence of DPY30. We speculate that loss of DPY30 represses regulators of these processes, leading to secondary effects that result in a senescence-like proliferation stop (Figure 6). Interestingly, we found that the E2F transcription factors and the ID proteins are direct DPY30 targets.
Figure 6.
Potential molecular mechanisms involved in the senescence-like phenotype induced by the loss of DPY30. In DPY30 wild-type cells, expression of various target genes is under control of a functionally active DPY30-Set1/MLL complex that ensures proper H3K4me3 on these genes. This leads to the expression of the E2F transcription factors and ID proteins (among others), which are involved in regulating secondary downstream target genes: E2F binds and activates transcription of genes involved in cell-cycle progression, while the ID proteins negatively influence the action of other transcription factors, such as Rb and ETS1/2. As a consequence, Rb cannot act as a repressor at E2F target genes, and ETS1/2 cannot activate the expression of the INK4/ARF locus, which is required for the cell to proliferate. In DPY30 knockdown cells, expression of various target genes is repressed (including E2F transcription factors and ID proteins), which correlates with impaired H3K4me3 levels. Expression of E2F target genes is thus shut down since, first, E2F transcription factors are absent, and second, hypophosphorylated Rb is no longer blocked by ID proteins and can repress E2F target genes. Additionally, ETS1/2 transcription factors are free to activate the INK4/ARF locus, which contributes to a senescence-like phenotype. In addition to the E2F transcription factors and ID proteins, other key regulators could be under the control of DPY30. Upon depletion of DPY30, these key regulators would be deregulated, leading to secondary downstream effects that could also play a role in senescence pathways.
The ‘activating E2Fs’ (E2F1, E2F2, and E2F3) positively regulate progression of cell cycle by activating transcription of genes required for DNA synthesis (Attwooll et al, 2004). Several studies suggest a role of E2Fs in senescence; for instance, E2F1 knockdown in tumour cells resulted in a senescence-like cell state, while ectopic expression of E2F1 in these cells bypassed cellular senescence (Park et al, 2006). We therefore considered that deregulation of E2F1, E2F2, and E2F3 in shDPY30 cells could be responsible for the induced senescence-like proliferation arrest. However, ectopically overexpressing E2F2 in DPY30 knockdown cells did not rescue the senescence phenotype (Supplementary Figure 10). This could be explained by other E2F1 activities: ectopic expression of E2F1 was reported to induce cellular senescence by directly activating the p14ARF tumour suppressor gene (Dimri et al, 2000). Even though the senescence-like phenotype was not rescued by E2F2 overexpression in our experimental set-up, we suggest that the deregulation of these transcription factors, and the subsequent deregulation of their target genes, plays a critical role in the proliferation arrest induced by loss of DPY30 (Figure 6).
In contrast, we found that overexpression of ID1 and ID3 could partially rescue the senescence-like phenotype induced by loss of DPY30. ID proteins are thought to affect the balance between cell growth and differentiation by negatively regulating the function of basic-helix-loop-helix (bHLH) transcription factors (Zebedee and Hara, 2001). It has been shown that ectopic expression of ID1, ID2, and ID3 extends the life span of primary human keratinocytes, and that it correlates with hyper-phosphorylation of Rb and repression of p16INK4a expression, suggesting that ID proteins regulate Rb activity in general (Alani et al, 1999; Nickoloff et al, 2000). In addition to Rb, ID1 was shown to negatively regulate the action of ETS1/2 transcription factors, which activate the expression of p16INK4a (Ohtani et al, 2001). ETS1/2 are phosphorylated and activated by MAP kinases during the cell cycle. ID proteins counter balance ETS1/2 activity and therefore tightly regulate p16INK4a expression (Tournay and Benezra, 1996). In OIS, however, ETS1/2 are constitutively phosphorylated by oncogenic Ras/MAPK signalling, thus overwriting the steady state controlled by ID proteins. As a consequence, p16INK4a is highly expressed. In replicative senescence, p16INK4a expression appears to occur as a consequence of increased expression levels of ETS1/2 and decreased expression levels of ID proteins (Zebedee and Hara, 2001). How these changes in ETS1/2 and ID protein expression are regulated during ageing has not yet been addressed. However, studies in mouse epidermal stem cell ageing revealed a significant decrease in DPY30 expression, while expression of other Set1 and MLL members remained unchanged, leading to the speculation that DPY30 could be a crucial regulator of ID proteins in young epidermal stem cells (Doles et al, 2012). In accordance, we found that DPY30 and ID protein expression was decreased in old mouse keratinocytes as well as in replicative senescent human fibroblasts, suggesting a physiological relevance of DPY30-regulated ID protein expression in vivo (Supplementary Figure 12).
Interestingly, silencing of p16INK4a upon ID protein overexpression appeared to occur independently of Polycomb activity and H3K27me3 deposition. We identified the PRC2 component EZH2 as a direct DPY30 target gene whose expression is indeed downregulated in shDPY30 cells. Consequently, deposition of the H3K27me3 mark at the p16INK4a promoter was not fully restored. Similarly, the level of another repressive chromatin mark (H3K9me2) also remained low, suggesting that in this cellular system, the initial re-silencing is likely the result of reduced ETS1/2 binding due to overexpression of ID proteins.
While ectopic ID protein expression resulted in a partial rescue of the senescent-like phenotype in shDPY30 IMR90 cells, simultaneous co-expression of E2F2 reverted this effect (Supplementary Figure 10). This is not surprising given the role of E2F transcription factors on cell cycle. As major regulators of cell-cycle progression, expression of several E2F family members could lead to a senescence bypass. On the other hand, E2F1, similarly to Ras or Raf, is potentially oncogenic when highly expressed, and thus OIS is triggered to prevent tumorigenesis.
Interestingly, ID proteins often have elevated levels of expression in diverse human tumours. In some cases, high ID protein levels are associated with disease severity and poor prognosis (Perk et al, 2005). Even though ID proteins are not considered to be classical oncogenes, their expression is under the control of well-established oncoproteins, such as Myc and RAS (Tournay and Benezra, 1996; Bain et al, 2001). Preliminary results suggest that depleting DPY30 correlated with a proliferation defect in four out of five tested cancer cell lines. Interestingly, the absolute ID protein levels were high in three cancer cell lines, and we observed a robust decrease in expression when DPY30 was depleted (unpublished observation).
In summary, we suggest that DPY30, a common and fairly uncharacterized member of all Set1 and MLL complexes, is involved in the regulation of proliferation and cell-cycle progression. Binding of DPY30 to and concomitant H3K4me3 of key regulator genes in cell cycle ensure their proper expression and activity. When perturbed, cells enter a senescence-like state and p16INK4a, a senescence marker, is induced independently of H3K4me3. Its activation relies most likely on increased ETS1/2 transcription factor recruitment due to decreased expression of ID proteins. Our results contribute to better understand the role of DPY30 and suggest a detailed alternative mechanism of p16INK4a regulation in cellular senescence.
Materials and methods
Cell culture
IMR90 human fibroblasts were grown in minimum essential medium (MEM) supplemented with 10% fetal calf serum, 1% glutamine, and 1% antibiotics. NT2 teratocarcinoma cells were grown in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% FCS, 1% glutamine, and 1% antibiotics.
Infections, plasmids, and RNA interference
pLKO-puro-shDPY30 and pLKO-puro-shCtrl plasmids were purchased from Sigma-Aldrich (#1: NM_032574.1-263s1c1; #2: NM_032574.1-403s1c1; #3: NM_032574.1-436s1c1). Lentiviral particles were produced in 293T cells following the recommended protocol from Addgene. Puromycin selection was started 1 day after infection using a 4-μg/ml final concentration. Unless otherwise indicated, samples were taken 3 days after selection, which corresponded to 5 days following infection.
pBabe-puro-H-RasV12, pMSCV-hygro-E2F2, pMSCV-neo-ID1, pMSCV-neo-ID3, and the corresponding pMSCV empty control plasmids were used to produce retroviral particles in PhoenixA cells following standard protocols. Selection was performed using final concentrations of 500 μg/ml for hygromycin and 500 μg/ml for G418, and samples were harvested as described for DPY30-interfered cells.
Antibodies
Antibodies used in this study are listed in Supplementary data
Immunofluorescence
Cells were fixed in 4% paraformaldehyde (PFA) in PBS for 30 min, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and then blocked with 0.2% gelatin in PBS for 30 min at room temperature. DPY30, H3K4me3, γH2A.X, and TP53PB1 were labelled using the corresponding primary antibodies diluted 1:200, followed by appropriate fluorescent-coupled secondary antibodies diluted 1:500 in 0.2% gelatin in PBS for 30 min at room temperature. Nuclei were stained with DAPI (1:1000) for 10 min, followed by several washes with PBS, and mounted in Mowiol 4.88.
ROS activity
ROS activity was determined by flow cytometry using the ‘total ROS detection kit’ from Enzo Life Sciences (ENZ-51011) following the instructor’s guidelines.
SA-β galactosidase assay
Cell were fixed with 0.5% gluteraldehyde solution for 15 min at room temperature, washed twice with PBS/MgCl2, pH 6, and incubated in X-gal staining (1 mg/ml X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside), 5 mM K4Fe(CN)6-3H2O, and 5 mM K3Fe(CN)6 in PBS/MgCl2 pH 6) solution at 37°C overnight. Cells were washed three times with H2O, and brightfield pictures were taken at various magnifications. Approximately 400–600 cells were counted to quantify SA-β galactosidase-positive cells.
Proliferation assays, growth curves, and colony formation assay
For growth curves, 10 000 cells were seeded on day 0 after selection in 12-well plates, and the cell number was counted every 2 days.
For colony formation assays, 20 000 cells were seeded on day 0 after selection in 6-well plates. After 7–21 days, colonies were fixed with 10% formalin and stained with 0.05% crystal violet.
Histone isolation, preparation of whole-cell extracts, nuclear extracts, and western blot
For detailed information, see Supplementary data.
RNA extraction, RT–PCR and gene expression microarray analysis
mRNA was extracted with the RNA extraction kit (RNAeasy Quiagen). cDNA was generated from 1 μg of total mRNA using First Strand cDNA Synthesis Kit (Fermentas). cDNA was diluted 1:10, and 2 μl was used in RT–PCR with SYBR green (Roche). Primers are listed in Supplementary Table 1. For detailed information on gene expression microarray analysis, see Supplementary data.
ChIP
ChIP analyses were performed according to standard protocols. For detailed information, see Supplementary data.
ChIPseq analysis
Reads produced by ChIPseq experiments of each of the samples were aligned with the human genome (version GRCh37) using Bowtie tool (version 0.12.7), allowing two mismatches within the seed alignment (Langmead et al, 2009). Sequence tags aligned to the genome were subsequently analysed with MACS software (version 1.3.7.1) to detect genomic regions enriched for multiple overlapping DNA fragments (peaks) that we considered to be putative binding sites (Zhang et al, 2008). Model for peaks detection was created for shCtrl K4me3 and shDPY30 K4me3 by looking for enrichment ratio against background of 32, while for shCtrl DPY30, this ratio was limited to 24.
MACS estimated the false discovery rate (FDR) by comparing the peaks obtained from the samples with those from the control samples, using the same P-value cutoff (1e−5). For this study, all the peaks for shCtrl DPY30 but only the ones below 10% FDR for shCtrl K4me3 and shDPY30 K4me3 were considered.
The Ensembl annotation (version 61) was used to detect genes overlapping with peaks and the closest TSS. ChIPseq signal around TSSs was calculated by using BEDtools over TSS overlapping peaks and normalizing the coverage for the total number of mapped reads × 1 000 000 (Quinlan and Hall, 2010). The plots were drawn with R scripts.
Supplementary Material
Acknowledgments
We are indebted to VA Raker for help in preparing the manuscript, to K Helin and members of the Di Croce laboratory for discussions, and to the CRG Genomic Unit. This work was supported by grants from the Spanish ‘Ministerio de Educación y Ciencia’ (BFU2010-18692), from AGAUR, and from by European Commission’s 7th Framework Program 4DCellFate grant number 277899 to LDC.
Author Contributions: ES performed all experiments except the ones mentioned below. AG performed experiments presented in Figure 1C and Supplementary Figure 12A and B. LC performed computational analysis presented in Figure 4A–C and Supplementary Figures 8A and 9A. MB and LC performed experiments presented in Supplementary Figure 12. LDC and ES designed experiments. WMK, ES, and LDC analysed experiments and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD (2009) Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell 36: 2–14 [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K (2009) The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev 23: 1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani RM, Hasskarl J, Grace M, Hernandez MC, Israel MA, Munger K (1999) Immortalization of primary human keratinocytes by the helix-loop-helix protein, Id-1. Proc Natl Acad Sci USA 96: 9637–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR (2011) Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwooll C, Lazzerini Denchi E, Helin K (2004) The E2F family: specific functions and overlapping interests. EMBO J 23: 4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C (2001) Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol 2: 165–171 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K (2006) Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, Hansen KH, Helin K (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A, Morey L, Guigo R, Lopez-Schier H, Di Croce L (2009) The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 16: 1074–1079 [DOI] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F (2007) Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW (2010) Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 17: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Scuoppo C, Wang X, Fang X, Balgley B, Bolden JE, Premsrirut P, Luo W, Chicas A, Lee CS, Kogan SC, Lowe SW (2011) Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev 25: 2125–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K (2007) PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: 2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Rodier F, Patil CK, Freund A, Desprez PY, Campisi J (2011) Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J Biol Chem 286: 36396–36403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehe PM, Dichtl B, Schaft D, Roguev A, Pamblanco M, Lebrun R, Rodriguez-Gil A, Mkandawire M, Landsberg K, Shevchenko A, Rosaleny LE, Tordera V, Chavez S, Stewart AF, Geli V (2006) Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 281: 35404–35412 [DOI] [PubMed] [Google Scholar]
- Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH (2007) Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J 26: 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Hara E, Campisi J (1994) Regulation of two E2F-related genes in presenescent and senescent human fibroblasts. J Biol Chem 269: 16180–16186 [PubMed] [Google Scholar]
- Dimri GP, Itahana K, Acosta M, Campisi J (2000) Regulation of a senescence checkpoint response by the E2F1 transcription factor and p14(ARF) tumor suppressor. Mol Cell Biol 20: 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doles J, Storer M, Cozzuto L, Roma G, Keyes WM (2012) Age-associated inflammation inhibits epidermal stem cell function. Genes Dev 26: 2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Peng Y, Xu F, He X, Wang F, Peng X, Qiang B, Yuan J, Rao Z (2005) Characterization and crystallization of human DPY-30-like protein, an essential component of dosage compensation complex. Biochim Biophys Acta 1753: 257–262 [DOI] [PubMed] [Google Scholar]
- Freund A, Patil CK, Campisi J (2011) p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J 30: 1536–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D (2004) Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol 6: 67–72 [DOI] [PubMed] [Google Scholar]
- Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37: 614–636 [DOI] [PubMed] [Google Scholar]
- Hsu DR, Chuang PT, Meyer BJ (1995) DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development 121: 3323–3334 [DOI] [PubMed] [Google Scholar]
- Huot TJ, Rowe J, Harland M, Drayton S, Brookes S, Gooptu C, Purkis P, Fried M, Bataille V, Hara E, Newton-Bishop J, Peters G (2002) Biallelic mutations in p16(INK4a) confer resistance to Ras- and Ets-induced senescence in human diploid fibroblasts. Mol Cell Biol 22: 8135–8143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG (2011) Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP (2008) SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol 28: 3457–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Zeng Y, Xiong Y (2009) DDB1-CUL4 and MLL1 mediate oncogene-induced p16INK4a activation. Cancer Res 69: 1809–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A (2001) Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413: 83–86 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ (1984) A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308: 428–433 [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Nickoloff BJ, Chaturvedi V, Bacon P, Qin JZ, Denning MF, Diaz MO (2000) Id-1 delays senescence but does not immortalize keratinocytes. J Biol Chem 275: 27501–27504 [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E (2001) Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409: 1067–1070 [DOI] [PubMed] [Google Scholar]
- Park C, Lee I, Kang WK (2006) E2F-1 is a critical modulator of cellular senescence in human cancer. Int J Mol Med 17: 715–720 [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5: 603–614 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Lange M, Simboeck E, Di Croce L (2010) Setting and resetting of epigenetic marks in malignant transformation and development. Bioessays 32: 669–679 [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G (2011) Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol 12: 799–814 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA (2001) Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413: 86–91 [DOI] [PubMed] [Google Scholar]
- Simboeck E, Ribeiro JD, Teichmann S, Di Croce L (2011) Epigenetics and senescence: learning from the INK4-ARF locus. Biochem Pharmacol 82: 1361–1370 [DOI] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A (2006) Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13: 852–854 [DOI] [PubMed] [Google Scholar]
- Tenney K, Shilatifard A (2005) A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J Cell Biochem 95: 429–436 [DOI] [PubMed] [Google Scholar]
- Tournay O, Benezra R (1996) Transcription of the dominant-negative helix-loop-helix protein Id1 is regulated by a protein complex containing the immediate-early response gene Egr-1. Mol Cell Biol 16: 2418–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT (2013) Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol 10: 2067–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brandenstein M, Schlosser M, Richter C, Depping R, Fries JW (2012) ETS-dependent p16(INK4a) and p21(waf1/cip1) gene expression upon endothelin-1 stimulation in malignant versus and non-malignant proximal tubule cells. Life Sci 91: 562–571 [DOI] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, Ge K, Krumlauf R, Shilatifard A (2009) Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Pan K, Chen Y, Huang C, Zhang X (2012) The acetylation of transcription factor HBP1 by p300/CBP enhances p16INK4A expression. Nucleic Acids Res 40: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J, Duboule D (1999) Hox genes in digit development and evolution. Cell Tissue Res 296: 19–25 [DOI] [PubMed] [Google Scholar]
- Zebedee Z, Hara E (2001) Id proteins in cell cycle control and cellular senescence. Oncogene 20: 8317–8325 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu WG (2009) The changing face of HDAC inhibitor depsipeptide. Curr Cancer Drug Targets 9: 91–100 [DOI] [PubMed] [Google Scholar]
- Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, Jurkin J, Rembold M, Meunier D, Egger G, Lagger S, Chiocca S, Propst F, Weitzer G, Seiser C (2010) The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol 30: 1171–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.