Abstract
The carcinogenic heterocyclic amine (HA) 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is formed during the cooking of various meats. To enable structure/activity studies aimed at understanding how DNA damaged by a member of the HA class of compounds can ultimately lead to cancer, we have determined the first solution structure of an 11-mer duplex containing the C8-dG adduct formed by reaction with N-acetoxy-PhIP. A slow conformational exchange is observed in which the PhIP ligand either intercalates into the DNA helix by denaturing and displacing the modified base pair (main form) or is located outside the helix in a minimally perturbed B-DNA duplex (minor form). In the main base-displaced intercalation structure, the minor groove is widened, and the major groove is compressed at the lesion site because of the location of the bulky PhIP-N-methyl and phenyl ring in the minor groove; this distortion causes significant bending of the helix. The PhIP phenyl ring interacts with the phosphodiester-sugar ring backbone of the complementary strand and its fast rotation with respect to the intercalated imidazopyridine ring causes substantial distortions at this site, such as unwinding and bulging-out of the strand. The glycosidic torsion angle of the [PhIP]dG residue is syn, and the displaced guanine base is directed toward the 3′ end of the modified strand. This study contributes, to our knowledge, the first structural information on the biologically relevant HA class to a growing body of knowledge about how conformational similarities and differences for a variety of types of lesions can influence protein interactions and ultimately biological outcome.
Consumption of foods containing heterocyclic amines (HA) has been implicated in the etiology of human cancers, including cancer of the colon, lung, and breast (1–4). The mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most mass abundant of the HAs, which are formed in meat and fish by the condensation of amino acids with creat(in)ine during cooking (5, 6). PhIP has been shown to induce tumors in several organs in rodents (7–9) and form DNA adducts, which are considered initiating events in chemical carcinogenesis (10, 11). Treatment of cultured mammalian cells with PhIP results in predominantly single-base substitutions (12, 13). A −1 frame-shift hotspot also has been observed in a 5′-GGGA-3′ sequence in the Apc gene of PhIP-induced rat colon tumors and in the lacI gene of rat mammary glands (14–16).
The major DNA adduct formed in vivo, and the only one unequivocally identified to date, is derived from the binding of metabolically activated PhIP to the C8 position of guanine [C8-dG-PhIP {N2-(2′-deoxyguanosin-8-yl)-PhIP}; refs. 17 and 18]. The reactive form of PhIP, a nitrenium ion, arises as a consequence of N-hydroxylation, which is catalyzed primarily by cytochrome CYP1A2 in humans (19, 20), followed by N:O-sulfation or N:O-acetylation (21). Analysis of phage vectors containing a site-specific C8-dG-PhIP adduct replicated in mammalian cells has revealed preferential incorporation of the correct dC base opposite the adduct and mainly G → T transversions, with lesser amounts of G → A transitions and G → C transversions (22). The mutational spectra were similar whether the 5′-flanking base to the adduct was dC, dT, dA, or dG, but higher mutational frequencies (28–30%) were observed when dC or dG was 5′ to C8-dG-PhIP, whereas a mere 13% was observed when dA was 5′ to the lesion (22). Single-base deletions were detected also, but only when dG or dT flanked the adduct. The ability of this single site-specific C8-dG-PhIP adduct to induce a variety of mutations may be a result of the existence of multiple conformers, as was first described for the C8-dG-2-acetylaminofluorene (AAF) duplex adduct by O'Handley et al. (23). Moreover, the equilibrium between different structures may be governed by DNA sequence context, as was shown for the C8-dG-2-aminofluorene (AF) DNA adducts (24).
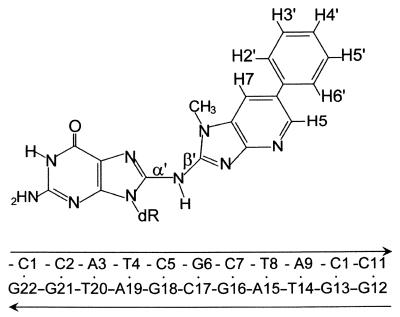
To date, no structural studies of HA-DNA adducts have been reported, because of difficulties in synthesizing the large quantities of highly purified samples needed. Recently, we optimized the synthesis of the C8-dG-PhIP adduct in an 11-mer DNA sequence (ref. 25; Fig. 1), and we report here the structural studies for a member of the HA class of compounds. The presence of exchange cross peaks in the NMR data for the [PhIP]dG⋅dC 11 oligomer indicates that there is a slow equilibrium between two distinct conformational states. A combined NMR–molecular mechanics computational study of the major conformer shows that the PhIP ligand intercalates between flanking dG⋅dC base pairs by denaturation and displacement of the [PhIP]dG6⋅dC17 pair. Chemical-shift perturbation studies of the minor conformer indicate that the PhIP ligand is located outside the helix and solvent-exposed in a minimally perturbed B-DNA duplex. The conformational distortions induced into DNA by the C8-dG-PhIP adduct is compared with those induced by the aromatic amines adducts (24). The unique chemical structure of the HA-PhIP molecule (the presence of ring nitrogens, N-methyl group and phenyl ring) governs the nature and extent of the distortions in the DNA helix as well as the population balance of the minor and major conformers.
Figure 1.
Schemes of the C8-dG-PhIP adduct and DNA sequence. PhIP-dG linkage is defined by torsion angles α′ = [PhIP]dG6(N9)-[PhIP]dG6(C8)-PhIP(N)-PhIP(C2) and β′ = [PhIP]dG6(C8)-PhIP(N)-PhIP(C2)-PhIP(C1).
Materials and Methods
Preparation of the Duplex Adduct.
A full account of the pioneering methodology used to synthesize and isolate large-scale quantities of the C8-dG-PhIP 11-mer adduct and the complete spectroscopic characterization of this adduct are reported in detail elsewhere (25). Briefly, the oligodeoxyribonucleotides d(CCATCGCTACC) and d(GGTAGCGATGG) were synthesized on an Applied Biosystems (model 394) DNA/RNA synthesizer and purified by reverse-phase HPLC. Equimolar concentrations of N-acetoxy-PhIP and d(CCATCGCTACC) were reacted for 1 h at 37°C in 40% methanol/60% 10 mM sodium citrate (pH 7.0) buffer containing 1 mM EDTA and 0.1 M NaCl. The C8-dG-PhIP single-stranded 11-mer adduct was purified by repeated HPLC on a semipreparative C18 column, using a 20 mM sodium phosphate, pH 7.0/methanol gradient. The modified 11-mer strand was annealed to its complementary strand at 70°C, and the stoichiometry followed by monitoring resolved single-proton resonances in each strand.
NMR Experiments.
NMR experiments were performed on a Varian INOVA 600 MHz spectrometer. A combination of through space nuclear Overhauser effect spectroscopy (NOESY; 50- and 300-ms mixing time), through rotational Overhauser effect spectroscopy (ROESY; 100-ms spin lock time), through bond correlated spectroscopy (COSY; phase-sensitive), and total correlated spectroscopy (TOCSY; 40- and 80-ms spin lock time) experiments were recorded in the States-time proportional phase incrementation (TPPI) mode on ≈9.1 mg of the [PhIP]dG⋅dC 11-mer duplex (and 9.0 mg of the control 11-mer duplex), dissolved in 0.6 ml of D2O buffer (10 mM Na2PO4/0.1 M NaCl/0.1 mM EDTA, pH 7.0) at 25 and 20°C. One-dimensional and NOESY (150-ms mixing time) spectra of the adduct duplex in 90% H2O/10% D2O (vol/vol) buffer were collected at 1°C by using a jump-return pulse sequence for solvent suppression. A set of 40-, 80-, 120-,160-, and 200-ms mixing time NOESY experiments were collected at 25°C to allow determination of distance restraints from volume buildup curves. Sweep widths of 6,000 and 12,000 Hz were used for the adduct duplex dissolved in D2O buffer and H2O buffer, respectively, with 1,024 complex data points and 300 t1 increments, each having 64 transients and a recycle delay of 2 s. Data sets were processed by using the software package vnmr (Varian), transferred to a Silicon Graphics (Mountain View, CA) workstation, and converted directly into felix (Accelrys, San Diego) matrixes for analysis.
Molecular Mechanics Computations.
Restrained conformational searches were carried out with duplex, a molecular mechanics program for nucleic acids that performs potential energy minimization in the reduced variable domain of torsional angle space (26). Full details of this approach are given in an earlier work (27). Parameters added to the force field for the C8-dG-PhIP adduct are given in ref. 12. The volume-buildup curves, and either the fixed 2.45-Å dC(H6) to dC(H5) distance or the averaged fixed 3.06-Å dT(H6) to dT(CH3) distance (for proton to methyl group distances), were used in the two spin approximation calculations. Many cross peaks exhibited broadening caused by conformational exchange, thus wide bounds were assigned (±20%). The DNA-PhIP distance bounds, in overlapped cases, also were estimated by the number of contour levels as weak (4.0–6.0 Å), medium (3.0–5.0 Å), and strong (2.0–4.0 Å). Because the conformational exchange is temperature-dependent, the exchangeable proton NOESY data at 1°C was not used in the structure calculations, with the exception of the DNA-PhIP NOEs, which were given very wide bounds (3.0–6.0 Å). Sixteen energy minimization trials were carried out by using NOE-derived distance restraints with initial linkage torsion angles α′ and β′ (Fig. 1) at 0°, 90°, 180°, and 270° in all combinations. The glycosidic torsion angle of the [PhIP]dG6 residue was started in the syn domain (χ = 60°), as indicated by the NMR data. All other glycosidic torsion angles were started in the anti domain (χ = 240°). The duplex hydrogen bond penalty function was used for Watson–Crick base-pairing at all residues except for [PhIP]dG6⋅dC17, in line with the NMR data. Initially, only three PhIP resonances (H5, H7, and N-CH3) provided unambiguous PhIP-DNA NOE restraints, because the PhIP phenyl protons are degenerate as a result of symmetry and rotation about the phenyl-imidazopyridine (IP) bond. The first set of structures afforded feedback to the NMR analysis and the reassessment of the ambiguous NOE restraints involving the PhIP phenyl protons, which were added in the next set of trials. A total of 265 NOE distant restraints were used (239 DNA–DNA and 26 DNA-PhIP). Helical parameters were calculated by using a nucleic acid structure analysis program developed by Babcock et al. (28), and ball-and-stick figures of a representative structure were generated by using MOLSCRIPT VI.1 (29).
Results
NMR Assignment and Chemical-Shift Studies.
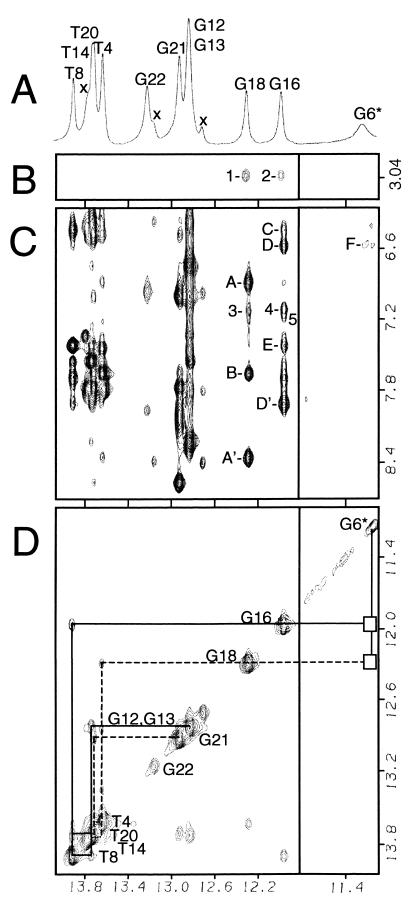
The proton NMR spectrum of the [PhIP]dG⋅dC 11-mer adduct duplex in H2O buffer at 1°C exhibits three up-field-shifted imino resonances in addition to the partially resolved resonances between 12.8–13.9 ppm (Fig. 2A). Several small unassigned resonances also are observed that correspond to a minor conformer of the adduct duplex (Xs, Fig. 2A). The narrow imino resonances of dG16 (11.97 ppm) and dG18 (12.29 ppm) in the major conformer are shifted up-field by −0.77 and −0.38 ppm, respectively, relative to the corresponding residues in the control 11-mer duplex (Fig. 6, which is published as supplemental data on the PNAS web site, www.pnas.org). Both dG16 and dG18 imino protons show NOE cross peaks to their partner C7 (peaks A and A′, Fig. 2C) and C5 (peaks D and D′, Fig. 2C) exposed and hydrogen-bonded amino protons, establishing the formation of stable dC5⋅dG18 and dC7⋅dG16 base pairs on either side of the [PhIP]dG lesion site. The broad up-field-shifted [PhIP]dG6 imino proton at 11.19 ppm (Fig. 2A) is not hydrogen-bonded to its partner dC17 and exhibits one broad very weak NOE to the 3′ side C7-exposed amino proton (peak F, Fig. 2C). Connectivities between the imino protons of adjacent base pairs can be followed from the dC2⋅dG21 base pair located at one end of the helix, to the dC5⋅dG18 pair located 5′ to the [PhIP]dG6 residue, and from the 3′ neighbor dC7⋅dG16 pair to the dC10⋅G13 pair at the opposite end of the helix (Fig. 2D). The NOE connectivities between the imino protons of dG16, [PhIP]dG6, and dG18 are not observed (boxes, Fig. 2D), although these residues are adjacent along the duplex sequence. The exchangeable PhIP(NH) resonance could not be assigned.
Figure 2.
Imino proton spectrum (11.1–14.1 ppm) (A) and expanded NOESY (150-ms mixing time) contour plots (B–D) of the [PhIP]dG⋅dC 11-mer duplex adduct in H2O buffer at 1°C. The right panels are plotted at 10 times lower level relative to the left panels. The assignments for the imino resonances in the major conformer are given in A and D. Resonances belonging to the minor conformer are designated by Xs in A. (B) NOE connectivities between the 1 [dG18(NH1)] and 2 [dG16(NH1)] imino protons and PhIP(N-CH3). (C) PhIP-DNA cross peaks are 3, dG18(NH1)-PhIP(H5); 4, dG16(NH1)-PhIP(H5); and 5, dG16(NH1)-PhIP(H7). DNA–DNA cross peaks are lettered as follows: A and A′, dG18(NH1)-dC5(NH2-4e,b), where e and b designate exposed and hydrogen-bonded, respectively; B, dG18(NH1)-dA19(H2); C, dG16(NH1)-dA15(NH2-6); D and D′, dG16(NH1)-dC7(NH2-4e,b); E, dG16(NH1)-dA15(H2); and F, [PhIP]dG6(NH1)-dC7(NH2-4e). (D) Imino–imino connectivities are traced by the solid line from dG13 to dG16 and by the dashed line from dG18 to dG21. The boxes indicate the loss of connectivities between the [PhIP]dG6 imino and the dG16 and dG18 iminos.
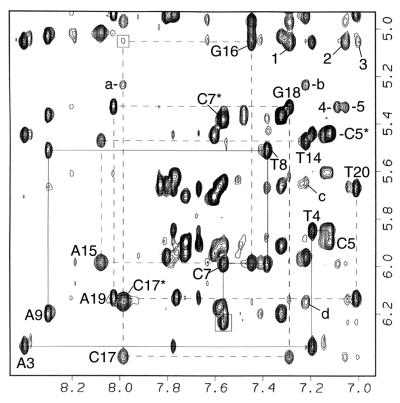
Nonexchangeable proton data for the adduct duplex dissolved in D2O was collected at two temperatures (25 and 20°C) to enable assignment of key H3′ proton resonances that overlapped with the H2O signal at 4.76 ppm (25°C) or 4.81 ppm (20°C). Exchange cross peaks are observed in the NMR data between two distinct [PhIP]dG⋅dC 11-mer conformers (see peaks a–d, Fig. 3). The exchange cross peaks were identified from a 100-ms spin lock time rotational Overhauser effect spectroscopy experiment at 25°C (Fig. 7, which is published as supplemental data on the PNAS web site). Assignments of the PhIP proton chemical shifts in the major and minor conformations are compared with those in the C8-dG-PhIP nucleoside adduct in DMSO (ref. 17; Table 1). The assignments of the base H8/H6 and H1′ protons for the 6 residues in the central 3-bp segment of the adduct duplex also are given for the major and minor conformers and are compared with those in the unmodified 11-mer control duplex (Table 1). The amount of minor conformer present is temperature-dependent and ranges from ≈10% at 25°C, to 15% at 20°C, and to 32% at 1°C, as determined by the ratios of the volume integrals of well resolved cytosine H5/H6 cross peaks belonging to either conformer. Although NOE connectivities are too weak or overlapped for the minor conformer, which prevented a detailed determination of its structure, the PhIP and DNA chemical shifts (Table 1) indicate that the PhIP ligand is most likely located externally in a minimally disturbed B-DNA duplex. In contrast, the chemical-shift patterns observed for the major conformer (Fig. 2, Table 1, and described in detail below) are indicative of a structure in which the PhIP ligand intercalates among intact flanking base pairs by denaturing and displacing the modified base-pair residues into a DNA groove (for examples, see refs. 24 and 30).
Figure 3.
Expanded NOESY (300-ms mixing time) contour plot of the [PhIP]dG⋅dC 11-mer adduct duplex in D2O buffer at 25°C showing the NOE connectivities between the base protons and their own, and 5′-flanking sugar H1′ protons for dA3 to dA9 (solid lines) and dT14 to dT20 (dashed lines). The connectivity is disrupted in the modified strand at the dC5-[PhIP]dG6 step. The dC7(H6)-[PhIP]dG6(H1′) cross peak is boxed. The connectivity is very weak at the dG16-dC17 step (boxed). dC5*, dC7*, and dC17* designate the H6 to H5 cross peaks. PhIP-DNA cross peaks are 1, PhIP(H3′, H5′)-dG16(H1′); 2, PhIP(H2′, H6′)-dG16(H1′); 3, PhIP(H5)-dG16(H1′); 4, PhIP(H7)- dG18(H1′); and 5, PhIP(H2′, H6′)-dG18(H1′). The dC17 exchange cross peaks and minor conformer cross peaks are a, C17(H6) major to C17(H5) minor; b, C17(H6)-C17(H5) minor; c, C17(H6)-C17(H1′) minor; and d, C17(H6) minor to C17(H5) major.
Table 1.
Chemical shifts in the central 3-bp segment of the major and minor conformations of the adduct duplex, C8-dG-PhIP nucleoside adduct (17), and the control 11-mer
| Major conformer adduct duplex, ppm | Minor conformer adduct duplex, ppm | C8-dG-PhIP/control 11-mer, ppm | ||||
|---|---|---|---|---|---|---|
| PhIP(N-CH3) | 3.04 | 3.74 | 3.65 | |||
| PhIP(H7) | 7.09 | 7.99 | 8.01 | |||
| PhIP(H5) | 7.00 | 8.40 | 8.41 | |||
| PhIP(H2′,H6′) | 7.06 | 7.80 | 7.76 | |||
| PhIP(H3′,H5′) | 7.29 | 7.61 | 7.49 | |||
| PhIP(H4′) | 7.29 | 7.50 | 7.38 | |||
| H8/H6 | H1′ | H8/H6 | H1′ | H8/H6 | H1′ | |
| dC5 | 7.12 | 5.90 | 7.28 | 5.99 | 7.48 | 5.65 |
| [PhIP]dG6 | — | 6.23 | — | 5.80 | 7.86 | 5.85 |
| dC7 | 7.57 | 5.99 | 7.49 | 6.07 | 7.32 | 5.83 |
| dG16 | 7.46 | 5.06 | 7.63 | 5.72 | 7.63 | 5.67 |
| dC17 | 7.99 | 6.38 | 7.22 | 5.63 | 7.18 | 5.58 |
| dG18 | 7.29 | 5.33 | 7.84 | 5.61 | 7.83 | 5.61 |
An expanded plot of the 300-ms mixing time NOESY spectrum of the adduct duplex in D2O buffer at 25°C correlating the base H8/H6 proton to their own, and 5′-flanking sugar H1′ protons for the central 7-mer segment of the major conformer is shown in Fig. 3. The sequential NOE connectivity is missing at the dC5-[PhIP]dG6 step in the modified strand because of the absence of the [PhIP]dG6(H8) proton. Several DNA protons in the central 3-bp region of the adduct duplex exhibit unusual chemical shifts relative to those observed in the control 11-mer duplex (Fig. 6). The dG18(H5′,H5′′) protons exhibit dramatically up-field-shifted resonances, as do most of the dG16 and dG18 protons (Figs. 3 and 6). In contrast, all of the dC17 protons are shifted down-field. Other unusual chemical shifts relative to the control 11-mer duplex are observed also. In particular, the [PhIP]dG6(H2′) proton is shifted dramatically down-field by +0.98 ppm, whereas the [PhIP]dG6(H2′′) proton is shifted up-field by −0.47 ppm (Fig. 6). This large splitting and inversion of the H2′/H2′′ chemical shifts are useful markers for identifying syn glycosidic torsion angles when the NOE connectivity between a base proton and its own H1′ proton is missing (31), such as in the case of the C8-dG-PhIP adduct. The down-field shift of the [PhIP]dG6(H1′) sugar proton (+0.38 ppm; Fig. 6), loss of hydrogen-bonding with the partner dC17 residue (Fig. 2C), and missing connectivities between the imino proton and flanking dG16 and dG18 imino protons (Fig. 2D) provide experimental evidence that the guanine base of the [PhIP]dG6 residue is not stacking within the helix.
Structure Determination of the Major Conformer.
A set of 26 PhIP-DNA (Table 2) and 239 DNA–DNA NOE cross peaks have been identified for the major conformer. Caution was used to ensure that the exchange cross peaks and those belonging to the minor conformer were not included as NOE distance restraints used for the major conformer. The PhIP N-CH3 and H7 protons, which are located along one edge of the IP ring (Fig. 1), show NOEs predominately to [PhIP]dG6, dC7, and dG18 protons, whereas PhIP(H5), which is located on the opposite edge of the IP ring, shows NOEs predominately to dG16 protons. The PhIP phenyl H2′,H6′ and H3′,H5′ proton pairs are degenerate because of symmetry, and the appearance of a single resonance corresponding to each pair indicates that the ring undergoes a rapid rotation with respect to the IP ring. The PhIP aromatic ring clearly shows NOEs predominantly to dG16 and dG18 protons (Table 2). Cross-peak patterns involving base to sugar (NOESY) and sugar to sugar protons (NOESY and correlated spectroscopy) were qualitatively evaluated to differentiate between syn and anti glycosidic torsion angles (32) and between the C3′-endo and C2′-endo families of DNA sugar pucker conformations (33).
Table 2.
DNA-PhIP NOE distance restraints compared with distances in the six structures of the [PhIP]dG⋅dC 11-mer
| DNA-PhIP NOE | Bounds, Å | Distance, Å |
|---|---|---|
| Exchangeable DNA protons | ||
| dC5(NH4-b)-PhIP(H5) | 3.0–6.0 | 5.51 ± 0.37 |
| dC7(NH4-b)-PhIP(H5) | 3.0–6.0 | 5.35 ± 0.56 |
| dG16(NH1)-PhIP(N-CH3) | 3.0–6.0 | 5.45* ± 0.11 (2†) |
| dG16(NH1)-PhIP(H5) | 3.0–6.0 | 4.00 ± 0.21 (4†) |
| dG16(NH1)-PhIP(H7) | 3.0–6.0 | 4.30 ± 0.21 (5†) |
| dG18(NH1)-PhIP(N-CH3) | 3.0–6.0 | 5.39* ± 0.16 (1†) |
| dG18(NH1)-PhIP(H5) | 3.0–6.0 | 5.45 ± 0.11 (3†) |
| Nonexchangeable DNA protons | ||
| [PhIP]dG6(H1′)-PhIP(N-CH3) | 3.0–5.0 | 3.86* ± 0.12 |
| [PhIP]dG6(H2")-PhIP(N-CH3) | 4.0–6.0 | 4.94* ± 0.37 |
| dC7(H1′)-PhIP(N-CH3) | 3.0–5.0 | 3.94* ± 0.32 |
| dC7(H2′)-PhIP(N-CH3) | 4.0–6.0 | 5.39* ± 0.14 |
| dC7(H2")-PhIP(N-CH3) | 4.0–6.0 | 5.82* ± 0.21 |
| dC7(H3′)-PhIP(N-CH3) | 4.0–6.0 | 6.12* ± 0.23 |
| dC7(H5)-PhIP(N-CH3) | 3.0–5.0 | 5.02* ± 0.45 |
| dC7(H6)-PhIP(N-CH3) | 3.0–5.0 | 4.22* ± 0.29 |
| dG18(H1′)-PhIP(N-CH3) | 4.0–6.0 | 5.53 ± 0.36 |
| dG16(H1′)-PhIP(H5) | 4.0–6.0 | 4.69 ± 0.70 (3‡) |
| dG16(H1′)-PhIP(H3′,H5′) | 3.0–6.0 | 5.74* ± 0.22 (1‡) |
| dG16(H1′)-PhIP(H2′,H6′) | 3.0–6.0 | 5.44* ± 0.42 (2‡) |
| dC17(H4′)-PhIP(H2′,H6′) | 3.0–6.0 | 4.15* ± 0.44 |
| dG18(H1′)-PhIP(N-CH3) | 3.0–6.0 | 5.53* ± 0.36 |
| dG18(H1′)-PhIP(H7) | 3.0–6.0 | 4.78 ± 0.75 (4‡) |
| dG18(H1′)-PhIP(H2′,H6′) | 3.0–6.0 | 6.11* ± 0.85 (5‡) |
| dG18(H4′)-PhIP(H2′,H6′) | 3.0–6.0 | 6.01* ± 0.52 |
| dG18(H5′)-PhIP(H2′,H6′) | 3.0–6.0 | 6.45* ± 0.21 |
| dG18(H5")-PhIP(H2′,H6′) | 3.0–6.0 | 4.97* ± 0.19 |
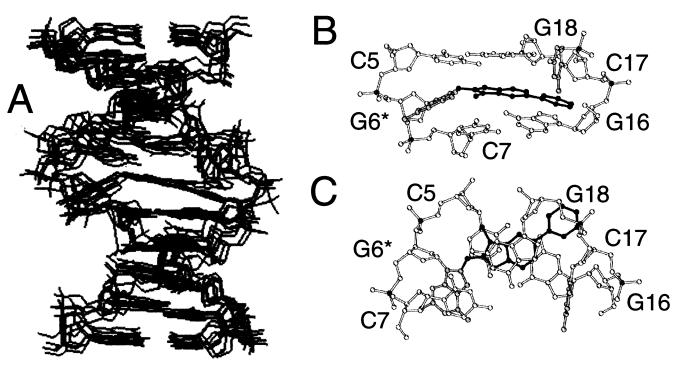
The program duplex was used to calculate the structure of the major conformer of the adduct duplex. Six of the structures obtained (Fig. 4A) exhibited low energy (−509.6 kcal/mol ± 12.2) and good fit to the NMR data with a pairwise rms deviation of 1.20 ± 0.56 Å for all atoms. Views looking into the major groove (Fig. 4B) and down the helix axis (Fig. 4C) of the central 3-bp segment of a representative structure show that the heterocyclic IP ring stacks with the dG18 purine ring and the edge of the dG16 ring, in agreement with the up-field chemical shifts observed for the PhIP H5 and H7 protons (Table 1) and dG16 and dG18 imino protons (Fig. 2). The PhIP phenyl ring is inclined out of plane relative to the IP ring. (The C7-C6-C1′-C2′ torsion angle is 21.8° ± 4.0.) The phenyl ring and N-CH3 and H7 edge of the IP ring are positioned toward the minor groove edge of the DNA, with the methyl group directed toward the modified strand and the phenyl group directed toward the complementary strand dC17-dG18 step phosphorus, dG18(H5′,H5′′), and dG18 and dG16 sugar ring protons (Figs. 6 and 8, which are published as supplemental data on the PNAS web site). The phenyl ring's location inbetween the dG16 and dC17 residues explains the very weak NOE connectivities between the dC17 base and dG16 sugar protons (Fig. 3). The PhIP-dG6 linkage site is defined by torsion angles α′ = 221.3° ± 3.0 and β′ = 132.5° ± 8.0 (Fig. 1), which displace the modified guanine into the major groove, with the glycosidic torsion angle rotated to a syn orientation (χ = 68.5° ± 4.2), and the base directed toward the 3′ end of the modified strand (peak F, Fig. 2C).
Figure 4.
(A) Superposition of five structures of the major conformer of the [PhIP]dG⋅dC 11-mer adduct duplex. Views of the central 3-bp segment of the best structure looking (B) into the major groove and normal to the helix axis and (C) down the helix axis with the modified [PhIP]G6⋅C17 in gray and the PhIP in black.
Discussion
The covalent binding of bulky carcinogens such as PhIP to DNA can alter biological processing of the DNA by cellular proteins governing replication, transcription, and repair, and thereby cause mutations and ultimately cancer, especially if the lesion is located in an oncogene or tumor-suppressor gene (10, 11). Consequently, considerable efforts are being made to understand how adduct conformation affects cellular responses to DNA damage. Our studies show that the [PhIP]dG⋅dC 11-mer adduct duplex undergoes a conformational exchange between a major base-displaced intercalative structure and a minor external groove-binding structure. Similar distortions have been observed for other DNA adducts, in which the guanine is modified by activated polycyclic aromatic hydrocarbons (30) and aromatic amines (24, 34). Here we report that the subtle differences in the structural details and in the population ratios of each conformer of the C8-dG-PhIP-DNA adduct reveal the importance of the chemical structure of the PhIP molecule in governing its DNA adduct conformation.
A comparison of the structures of the major conformers of the C8-dG-PhIP and C8-dG-AF (31) 11mer duplexes (Fig. 5) shows that although both DNA adducts adopt overall similar structural motifs, several important differences are observed. In contrast to the planarity of the AF ligand, the out of plane geometry of the phenyl ring with respect to the IP ring in the PhIP-DNA adduct contributes to a dramatically larger unwinding and twisting of the DNA helix (Figs. 5 and 9, which is published as supplemental data on the PNAS web site). However, unlike the higher propensity for formation of the base-displaced intercalative conformer for the PhIP-DNA adduct, the structure of the C8-dG-4-aminobiphenyl (ABP) DNA adduct, in which the ligand also has an out of plane geometry, adopts predominantly a major groove-binding structure (35). These differences in the population ratio of conformers is most likely because of a greater ability of the heterocyclic PhIP IP ring, compared with that of a ABP phenyl ring, to intercalate into DNA. Differences in the shapes and relative sizes of the phenyl and IP rings may be factors in promoting stacking interactions with the flanking DNA guanine purine rings. For example, the larger aromatic surface area of the pyrene ring resulted in obtaining 100% of the base-displaced intercalative structure for the C8-dG-1-aminopyrene (AP)-DNA adduct (36). However, because the sizes of the phenyl and IP rings do not differ that significantly, the presence of nitrogen atoms in the IP ring and thus the greater similarity to DNA purine bases, may promote stacking.
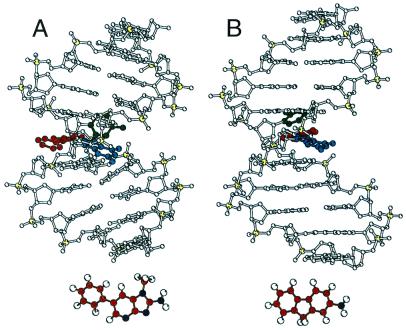
Figure 5.
Comparison of the [PhIP]dG⋅dC and [AF]dG⋅dC (31) 11-mer adduct duplexes with [PhIP]dG6 in blue, dC17 in green, and PhIP and AF ligands in red. The PhIP and AF structures are shown below.
Another difference between the base-displaced intercalative structures of the PhIP and aromatic amine DNA adducts is that the bulky N-methyl and phenyl ring of PhIP are pushed completely into the minor groove with only the planar IP ring forming stacking interactions with the flanking dG⋅dC base pairs (Fig. 5). The subsequent widening of the minor groove and compression of the major groove induce a significant bend in the helix (Figs. 4 and 5). More importantly, protein recognition and binding that would normally occur in the major groove of the DNA may be hindered dramatically. In contrast, the planar aromatic amine rings, such as the fluorene ring of AF, do not result in a compression of the major groove (Fig. 5; ref. 24). A further major difference between the PhIP and the aromatic amine DNA adduct structures is the rapid rotation of the PhIP phenyl ring in the minor groove, which results in the phosphodiester backbone of the complementary strand bulging out significantly (Fig. 4A).
DNA sequence context plays an important role in the amount of minor and major conformer obtained, as has been observed for the AF-DNA adducts (37), which in turn may affect protein-binding reactions as reported for the 2-acetylaminofluorene-DNA adducts (38). Studies of the mutations induced by the C8-dG-PhIP adduct replicated in simian kidney cells (22) reveal that the identity of the 5′-flanking residue to the lesion does affect the mutational frequency. Twice the amount of mutations (28–30%) were detected when a dC or dG was 5′ to the PhIP adduct compared with when dA was 5′, suggesting the lower thermostability of a flanking dA⋅dT base pair may afford more flexibility in the polymerase reaction to allow the proper dC base to be more readily inserted. A small number of single-base deletions also was observed when dG or dT flanked the adduct (22), suggesting that the relative ability to form bulged-out intermediates that may cause polymerase blockage is determined by DNA base-sequence context (39). The conformational equilibrium observed for the PhIP-DNA adduct is also highly temperature-dependent, with the minor external binding-conformer population increasing with lower temperature. Interestingly, the opposite trend has been reported for the two cis-benzo[a]pyrene-DNA adducts, in which the population of the major base-displaced intercalative structure increases with lower temperature (30). The factors that influence this conformational exchange are the relative stabilities of DNA base pairing vs. those of ligand DNA-stacking interactions. However, in the case of the N6-dA-benzo[c]phenanthrenyl-DNA adducts, both types of interactions could be accommodated simultaneously without any conformational exchange (40), suggesting that the allowed topology of the ligand at the base-linkage site is also an important determinant (30).
The alterations in the DNA structure induced by the covalent binding of PhIP would directly influence how cellular proteins interact with this type of damaged DNA substrate. Thus, knowledge of this structure is crucial for future structure/activity studies that compare PhIP with other DNA-damaged substrates. Moreover, the determination of the structure of the C8-dG-PhIP-DNA adduct is a very important first step in elucidating the mechanisms by which DNA damaged by PhIP can lead to cancer.
Supplementary Material
Acknowledgments
We thank Lihua Wang (New York University) for the helical parameter analyses. This work was carried out under the auspices of the U.S. Department of Energy by the Lawrence Livermore National Laboratory Grant W-7405-ENG-48, by the Oak Ridge National Laboratory Grant DE-AC0500-OR22725, and with support from National Institutes of Health Grants CA55861 and RR13461 (to K.W.T.), and by National Institutes of Health Grant CA75449 and Department of Energy Grant DE-FG0290ER60931 (to S.B.). Computations were carried out at the Department of Energy National Research Computer Center (Berkeley, CA) and the National Science Foundation Partnership for Advanced Computational Infrastructure (San Diego).
Abbreviations
- HA
heterocyclic amine
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- C8-dG-PhIP
N2-(2′-deoxyguanosin-8-yl)-PhIP
- IP
imidazopyridine
- NOE
nuclear Overhauser effect
- AF
2-aminofluorene
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1HZ0).
References
- 1.Wakabayashi K, Nagao M, Esumi H, Sugimura T. Cancer Res. 1992;52:2092S–2098S. [PubMed] [Google Scholar]
- 2.Gooderham N J, Murray S, Lynch A M, Edwards R J, Yadollahifarsani M, Bratt C, Rich K J, Zhao K, Murray B P, Bhadresa S, et al. Br J Clin Pharmacol. 1996;42:91–98. doi: 10.1046/j.1365-2125.1996.37513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinha R, Gustafson D R, Kulldorff M, Wen W Q, Cerhan J R, Zheng W. J Natl Cancer Inst. 2000;92:1352–1354. doi: 10.1093/jnci/92.16.1352. [DOI] [PubMed] [Google Scholar]
- 4.Sinha R, Kulldorff M, Swanson C A, Curtin J, Brownson R C, Alavanja M C. Cancer Res. 2000;60:3753–3756. [PubMed] [Google Scholar]
- 5.Layton D W, Bogen K T, Knize M G, Hatch F T, Johnson V M, Felton J S. Carcinogenesis. 1995;16:39–52. doi: 10.1093/carcin/16.1.39. [DOI] [PubMed] [Google Scholar]
- 6.Felton J S, Knize M G, Shen N H, Lewis P R, Andresen B D, Happe J, Hatch F T. Carcinogenesis. 1986;7:1081–1086. doi: 10.1093/carcin/7.7.1081. [DOI] [PubMed] [Google Scholar]
- 7.Ito N, Hasegawa R, Sano M, Tamano S, Esumi H, Takayama S, Sugimura T. Carcinogenesis. 1991;12:1503–1506. doi: 10.1093/carcin/12.8.1503. [DOI] [PubMed] [Google Scholar]
- 8.Takayama K, Yamashita K, Wakabayashi K, Sugimura T, Nagao M. Jpn J Cancer Res. 1989;80:1145–1148. doi: 10.1111/j.1349-7006.1989.tb01644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esumi H, Ohgaki H, Kohzen E, Takayama S, Sugimura T. Jpn J Cancer Res. 1989;80:1176–1178. doi: 10.1111/j.1349-7006.1989.tb01651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garner R C. Mutat Res. 1998;402:67–75. doi: 10.1016/s0027-5107(97)00283-2. [DOI] [PubMed] [Google Scholar]
- 11.Perera F P, Weinstein I B. Carcinogenesis. 2000;21:517–524. doi: 10.1093/carcin/21.3.517. [DOI] [PubMed] [Google Scholar]
- 12.Carothers A M, Yuan W, Hingerty B E, Broyde S, Grunberger D, Snyderwine E G. Chem Res Toxicol. 1994;7:209–218. doi: 10.1021/tx00038a015. [DOI] [PubMed] [Google Scholar]
- 13.Yadollahi-Farsani M, Gooderham N J, Davies D S, Boobis A R. Carcinogenesis. 1996;17:617–624. doi: 10.1093/carcin/17.4.617. [DOI] [PubMed] [Google Scholar]
- 14.Kakiuchi H, Watanabe M, Ushijima T, Toyota M, Imai K, Weisburger J H, Sugimura T, Nagao M. Proc Natl Acad Sci USA. 1995;92:910–914. doi: 10.1073/pnas.92.3.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okochi E, Watanabe N, Shimada Y, Takahashi S, Wakazono K, Shirai T, Sugimura T, Nagao M, Ushijima T. Carcinogenesis. 1999;20:1933–1938. doi: 10.1093/carcin/20.10.1933. [DOI] [PubMed] [Google Scholar]
- 16.Burnouf D Y, Fuchs R P P. Mutat Res. 2000;462:281–291. doi: 10.1016/s1383-5742(00)00010-7. [DOI] [PubMed] [Google Scholar]
- 17.Frandsen H, Grivas S, Andersson R, Dragsted L, Larsen J C. Carcinogenesis. 1992;13:629–635. doi: 10.1093/carcin/13.4.629. [DOI] [PubMed] [Google Scholar]
- 18.Lin D X, Kaderlik K R, Turesky R J, Miller D W, Lay J O, Kadlubar F F. Chem Res Toxicol. 1992;5:691–697. doi: 10.1021/tx00029a016. [DOI] [PubMed] [Google Scholar]
- 19.Boobis A R, Lynch A M, Murray S, Delatorre R, Solans A, Farre M, Segura J, Gooderham N J, Davies D S. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- 20.Kadlubar F, Kaderlik R K, Mulder G J, Lin D, Butler M A, Teitel C H, Minchin R F, Ilett K F, Friesen M D, Bartsch H, et al. Princess Takamatsu Symp. 1995;23:207–213. [PubMed] [Google Scholar]
- 21.Buonarati M H, Turteltaub K W, Shen N H, Felton J S. Mutat Res. 1990;245:185–190. doi: 10.1016/0165-7992(90)90048-o. [DOI] [PubMed] [Google Scholar]
- 22.Shibutani S, Fernandes A, Suzuki N, Zhou L, Johnson F, Grollman A P. J Biol Chem. 1999;274:27433–27438. doi: 10.1074/jbc.274.39.27433. [DOI] [PubMed] [Google Scholar]
- 23.O'Handley S F, Sanford D G, Xu R, Lester C C, Hingerty B E, Broyde S, Krugh T R. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 24.Patel D J, Mao B, Gu Z, Hingerty B E, Gorin A, Basu A K, Broyde S. Chem Res Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 25.Brown K, Guenther E A, Dingley K H, Cosman M, Harvey C A, Shields S J, Turteltaub K W. Nucleic Acids Res. 2001;29:1951–1959. doi: 10.1093/nar/29.9.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hingerty B E, Figueroa S, Hayden T L, Broyde S. Biopolymers. 1989;28:1195–1222. doi: 10.1002/bip.360280703. [DOI] [PubMed] [Google Scholar]
- 27.Cosman M, Hingerty B E, Geacintov N E, Broyde S, Patel D J. Biochemistry. 1995;34:15334–15350. doi: 10.1021/bi00046a043. [DOI] [PubMed] [Google Scholar]
- 28.Babcock M S, Pednault E P, Olson W K. J Mol Biol. 1994;237:125–156. doi: 10.1006/jmbi.1994.1213. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 30.Geacintov N E, Cosman M, Hingerty B E, Amin S, Broyde S, Patel D J. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 31.Mao B, Hingerty B E, Broyde S, Patel D J. Biochemistry. 1998;37:81–94. doi: 10.1021/bi972257o. [DOI] [PubMed] [Google Scholar]
- 32.Patel D J, Kozlowski S A, Nordheim A, Rich A. Proc Natl Acad Sci USA. 1982;79:1413–1417. doi: 10.1073/pnas.79.5.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Ven F J, Hilbers C W. Eur J Biochem. 1988;178:1–38. doi: 10.1111/j.1432-1033.1988.tb14425.x. [DOI] [PubMed] [Google Scholar]
- 34.Eckel L M, Krugh T R. Biochemistry. 1994;33:13611–13624. doi: 10.1021/bi00250a012. [DOI] [PubMed] [Google Scholar]
- 35.Cho B P, Beland F A, Marques M M. Biochemistry. 1992;31:9587–9602. doi: 10.1021/bi00155a011. [DOI] [PubMed] [Google Scholar]
- 36.Mao B, Vyas R R, Hingerty B E, Broyde S, Basu A K, Patel D J. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 37.Mao B, Hingerty B E, Broyde S, Patel D J. Biochemistry. 1998;37:95–106. doi: 10.1021/bi972258g. [DOI] [PubMed] [Google Scholar]
- 38.Seeberg E, Fuchs R P P. Proc Natl Acad Sci USA. 1990;87:191–194. doi: 10.1073/pnas.87.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baynton K, Fuchs R P P. Trends Biochem Sci. 2000;25:74–79. doi: 10.1016/s0968-0004(99)01524-8. [DOI] [PubMed] [Google Scholar]
- 40.Cosman M, Laryea A, Fiala R, Hingerty B E, Amin S, Geacintov N E, Broyde S, Patel D J. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.