Abstract
Methamphetamine (METH) use can provoke psychotic reactions requiring immediate treatment, namely METH-induced psychosis. Although the distinction between METH-induced and primary psychosis is important for understanding their clinical courses, we do not have clear diagnostic procedure by their symptoms. Not only are there similarities between the clinical features of METH-induced psychosis and schizophrenia (SCZ), but there is also epidemiological evidence of a shared genetic risk between ‘METH-related' disorders and SCZ, which makes the differentiation of these two conditions difficult. In this study, we conducted a genome-wide association study (GWAS) targeting METH-dependent patients. The METH sample group, used in the METH-dependence GWAS, included 236 METH-dependent patients and 864 healthy controls. We also included a ‘within-case' comparison between 194 METH-induced psychosis patients and 42 METH-dependent patients without psychosis in a METH-induced psychosis GWAS. To investigate the shared genetic components between METH dependence, METH-induced psychosis, and SCZ, data from our previous SCZ GWAS (total N=1108) were re-analyzed. In the SNP-based analysis, none of the SNPs showed genome-wide significance in either data set. By performing a polygenic component analysis, however, we found that a large number of ‘risk' alleles for METH-induced psychosis are over-represented in individuals with SCZ (Pbest=0.0090). Conversely, we did not detect enrichment either between METH dependence and METH-induced psychosis or between METH dependence and SCZ. The results support previous epidemiological and neurobiological evidence for a relationship between METH-induced psychosis and SCZ. These also suggest that the overlap between genes scored as positive in these data sets can have higher probability as susceptibility genes for psychosis.
Keywords: substance-induced psychosis, genome-wide association study, schizophrenia, methamphetamine, substance use disorder, polygenic component analysis
INTRODUCTION
Illicit drug use, a major concern worldwide, can place a large burden both on individuals and on society. Methamphetamine (METH) use, in particular, is a growing problem; recent evidence from the United Kingdom suggests that METH is one of the most harmful drugs, with its overall harm (harm to users plus harm to others) ranking fourth out of 20 drugs (Nutt et al, 2010). It is of note that harm from METH is mainly associated with harm to the users (self-harm), and one reason for this finding is that METH can provoke psychotic reactions (METH-induced psychosis) requiring immediate medical treatment (Nutt et al, 2010).
The distinction between METH-induced psychosis and primary psychosis is critical for understanding the clinical courses of these disorders and planning appropriate treatment; however, we do not fully understand why some METH abusers develop schizophrenia (SCZ)-like psychosis (Grelotti et al, 2010) and others do not. Moreover, these two conditions are clinically similar partly because some patients do not remit psychotic symptoms for weeks or months after METH exposure, which suggests a specific phenotype induced by METH (ie, ‘prolonged type' of METH-induced psychosis) (Ujike and Sato, 2004). Clinical investigators in Japan have long suggested that exposure to METH may cause persistent SCZ-like psychosis, whereas this possibility is discounted in the Western literature: the ‘prolonged type' of METH-induced psychosis is recognized as a pre-existing psychotic state, such as SCZ (Callaghan et al, 2012).
Not only are there similarities in the clinical features between these conditions, but there are also several epidemiological studies that suggest a shared genetic risk in ‘METH-related' disorders (ie, METH use disorder, METH dependence, and/or METH-induced psychosis) and SCZ, which makes the distinction between the two conditions complex. A family study revealed an increased risk of SCZ in the relatives of METH-induced psychosis patients compared with non-psychotic METH abusers or the general population (Chen et al, 2005). More recently, although the authors left open the question regarding the clear classification of SCZ and the ‘prolonged type' of METH-induced psychosis, an epidemiological survey suggested that METH abuse patients were at the highest risk of developing SCZ compared with the users of other psychoactive drugs (eg, cocaine, alcohol, opioids, and cannabis; Callaghan et al, 2012).
In this investigation, we conducted a genome-wide association study (GWAS) of METH dependence to explore the relationship between METH dependence, METH-induced psychosis, and SCZ, by including a re-analysis of the following: (a) the METH data set by dividing it according to the presence of psychosis in each participant and (b) a previously reported Japanese SCZ GWAS (Ikeda et al, 2011).
MATERIALS AND METHODS
Samples
A total of 236 patients with METH dependence (185 males, 51 females), most of whom were analyzed in a previous GWAS using the pooling method (Uhl et al, 2008), and 864 healthy controls (410 males, 454 females) were included in this study (we will refer to this case-control analysis as the ‘METH-dependence GWAS'). These controls had been used as the comparison subjects in other GWASs for narcolepsy (Miyagawa et al, 2008), panic disorder (Otowa et al, 2009), and SCZ (Hashimoto et al, unpublished data).
The METH-dependence GWAS data set was re-analyzed within case samples based on the presence (or absence) of psychotic symptoms (we will refer to this analysis as the ‘METH-induced psychosis GWAS'). This METH-induced psychosis sample group consisted of 194 METH-dependent patients with psychosis (METH-induced psychosis: 155 males and 39 females) and 42 METH-dependent patients without psychosis (METH non-psychosis: 30 males and 12 females).
All subjects were unrelated Japanese subjects and were recruited from the same relatively small geographical area of Japan. Consensus diagnoses were made by at least two experienced psychiatrists according to ICD-10 criteria on the basis of unstructured interviews with patients and their families, as well as a review of medical records. Patients were excluded if they had a history of SCZ, bipolar disorder, or known intellectual disability. The Japan SCZ sample consisted of 560 SCZ cases and 548 controls, and results from the GWAS of this sample were published previously (‘SCZ GWAS') (Ikeda et al, 2011). Healthy controls reported no personal history of mental disorders, but they were not screened using standard diagnostic procedures.
After providing a complete description of the study to the subjects, written informed consent was obtained. This study was approved by the ethics committees of each university participating in this project.
Genotyping and Quality Control (QC)
Genotyping for the METH-dependence GWAS was performed using the Affymetrix Genome-Wide Human SNP Array 5.0 or 6.0 (Affymetrix, Santa Clara, CA) according to the manufacturer's protocol. Of the 236 subjects with METH dependence, 169 of them (all METH-induced psychosis) were genotyped using the 5.0 chip, whereas 67 subjects (25 METH-induced psychosis and 42 METH non-psychosis) were genotyped using the 6.0 chip. The healthy controls were genotyped using the Affymetrix 6.0 chip. Genotypes were called from the CEL files using the BRLMM-P algorithm for the 5.0 chip and Birdseed v2 for the 6.0 chip implemented in the Genotyping Console software (Affymetrix). To correct for hidden confounding factors introduced by different genotyping platforms, only SNPs that are in common were selected (total of 436 213 SNPs). We then applied the following QC criteria to exclude samples: (1) arrays with a low QC (<86% for 5.0 chip or <0.4 for 6.0 chip) according to the BRLMM-P or Birdseed v2 algorithm (n=0) and (2) samples for which <95% of genotypes were called (n=0). Next, we excluded SNPs that (1) had low call rates (<0.95), (2) were duplicated, (3) localized to sex chromosomes, (4) deviated from Hardy–Weinberg equilibrium in controls (P<0.0001), or (5) had low minor allele frequencies (<0.05). Finally, 244 224 QC-ed SNPs were used in the subsequent analyses.
To test for the presence of genetic structure in the data, we performed a principal component analysis (PCA) using EIGENSTRAT 3.0 (Price et al, 2006). Ten Eigenvectors were calculated. Genotype information from the JPT, CHB, CEU, and YRI in HapMap phase III was compared with our data set to check for population stratification (Supplementary Figure S1). All Japanese samples from our case-control sample were in a separate cluster from the non-Japanese HapMap samples; however, two samples lay outside the main Japanese cluster, and those samples were excluded. Therefore, the final analysis consisted of 234 METH-dependent (193 METH-induced psychosis and 41 non-psychosis) subjects and 864 healthy comparison subjects.
The genotyping platform for the Japanese SCZ Study was Affymetrix 5.0, and SNPs were evaluated using the same procedures used in our previous study (Ikeda et al, 2011).
Statistical Analysis
To assess the association between individual SNPs, we used genomic control (GC)-adjusted P-values derived from allele-wise analysis based on median chi-square statistics (Purcell et al, 2007).
We performed a polygenic component analysis following the method described by the International Schizophrenia Consortium (ISC) (Purcell et al, 2009). This analysis is based on the idea that common SNPs collectively contribute to a substantial proportion of the heritability of complex diseases. To test this hypothesis, in essence, the authors defined sets of putative ‘risk' alleles as being those that surpassed more liberal thresholds for association (eg, P<0.5) in a discovery case-control sample set (‘discovery' sample). These sets (ie, putative ‘risk' alleles) were used to construct polygenic scores, which represent the number of ‘risk' alleles carried by individuals in a second test data set (‘target' sample). The analysis showed that the cases had on average significantly higher polygenic scores than controls. After discounting the influence of potential sources of bias, the authors in ISC concluded that the findings were best explained by the existence of polygenic component to the disorder comprised of a large number of common ‘risk' alleles with small and cumulative effect.
Although several approaches are now available to examine the polygenic effects on the complex diseases/phenotypes (eg, GCTA: Yang et al, 2011), the polygenic component analysis which we used in this study is suitable to evaluate the genetic overlap within the same phenotype (eg, in the ISC, they extracted ‘risk' from their ‘discovery' SCZ GWAS and applied these to the independent ‘target' SCZ GWASs to examine whether cases in the target samples had higher polygenic score) or between different diseases (extracted risk from their ‘discovery' SCZ GWAS and applied to the ‘target' bipolar disorder GWASs). Therefore, we applied this method to evaluate the genetic risk overlap with relaxed thresholds in METH dependence, METH-induced psychosis, and SCZ. The same criteria used by the ISC (Purcell et al, 2009) for LD pruning were applied to METH samples (r2 threshold of 0.25 and window size of 200 SNPs) and 64 815 LD-independent SNPs based on linkage equilibrium were analyzed. The polygenic score was calculated using PLINK ver.1.07 (Purcell et al, 2007). Nagelkerke's pseudo R2 was calculated by logistic regression with covariation for ‘non-missing SNPs'. A one-tailed test was applied under a directional hypothesis that expects higher scores to be associated with an increased risk of disease.
We also performed gene-based testing of the associations, because this method can provide more statistical power than single SNP-based analysis (Liu et al, 2010). To conduct the analysis, the software package VEGAS (Liu et al, 2010) was used by applying individual SNP-based P-values of the GC adjustment. VEGAS can perform analyses by taking into account not only gene size but also linkage disequilibrium patterns based on the HapMap East Asia (JPT and CHB) panel. SNPs were assigned to one or more defined boundaries of autosomal genes by extending the genomic sequence corresponding to each gene by 50 kb in the 5′ and 3′ directions. Gene-based P-values for all genes were calculated in this analysis, and 17 049 genes were assigned for both disorders (genomic positions were based on hg18). To include genes with nominal associations, we set the type I error rate for the gene-based test to 0.05. The enrichment was assessed by hyper-geometric analysis.
RESULTS
SNP-Based Association Results of ‘METH-Dependence' and ‘METH-Induced Psychosis' GWASs
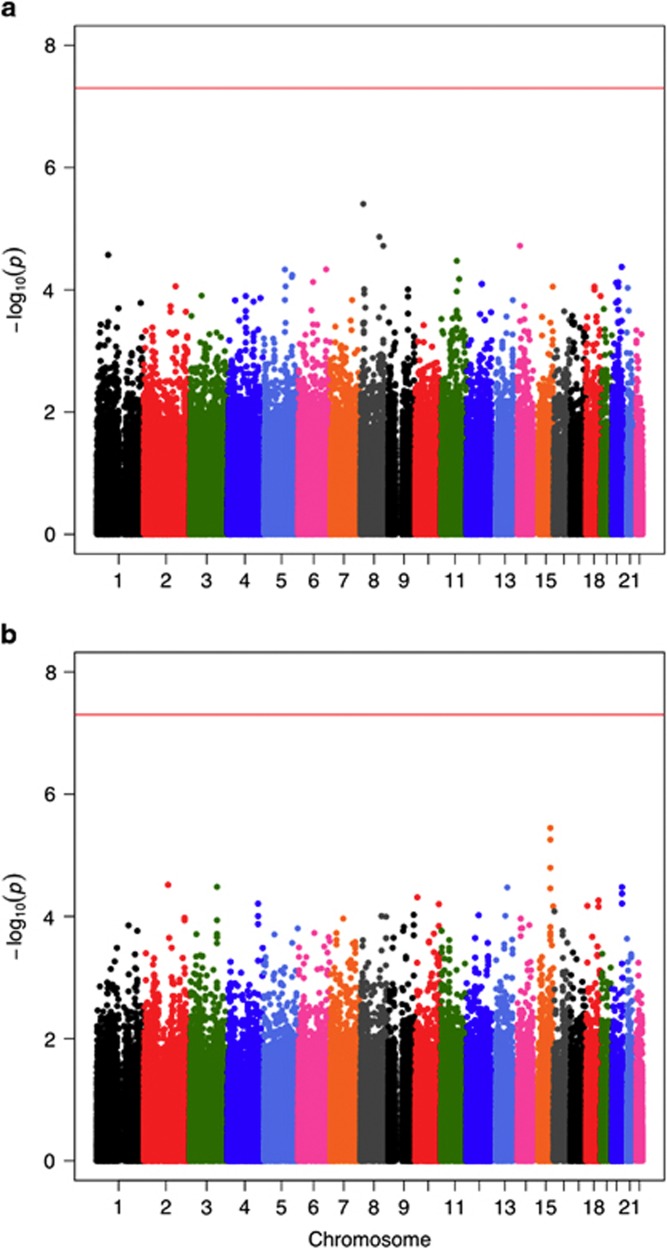
The Manhattan plots for the ‘METH-dependence' and ‘METH-induced psychosis' GWASs are shown in Figure 1, and the Q-Q plots are shown in Supplementary Figures S2 and S3. The GC inflation factors (λ) were 1.024 and 1.016 for the ‘METH-dependence' and ‘METH-induced psychosis' GWASs, respectively. We did not detect any SNPs with genome-wide significance (5 × 10−8), which is widely used as a benchmark of association, in either data set. The strongest association with METH dependence was observed for the rs4427170 SNP in the sarcoglycan zeta gene (SGCZ) (P=3.9 × 10−6, two-tailed test; Table 1), and the strongest association with METH-induced psychosis was observed for rs12591257, an intronic SNP in ATP/GTP binding protein-like1 (AGBL1) (P=3.6 × 10−6, two-tailed test; Table 1). The 100 SNPs most associated with these disorders are shown in Supplementary Tables S1 and S2 (Figure 1 and Table 1).
Figure 1.
The Manhattan plots for (a) METH dependence and (b) METH-induced psychosis GWASs. METH, methamphetamine.
Table1. Individual SNP-Based Analyses of METH-Dependence and METH-Induced Psychosis GWASs.
| Phenotype | SNP | Rank | CHRa | BPb | A1c | Frequency of A1 in case | Frequency of A1 in control | A2d | ORe | PGCf | Geneg |
|---|---|---|---|---|---|---|---|---|---|---|---|
| METH-dependence GWAS | rs4427170 | 1 | 8 | 14853781 | T | 0.515 | 0.395 | A | 1.63 | 3.9 × 10−6 | SGCZ |
| rs7826857 | 2 | 8 | 99436341 | A | 0.081 | 0.162 | G | 0.45 | 1.4 × 10−5 | KCNS2 | |
| rs12894058 | 3 | 14 | 33532016 | A | 0.222 | 0.140 | G | 1.75 | 1.9 × 10−5 | NPAS3 | |
| rs2326193 | 4 | 8 | 120029709 | A | 0.338 | 0.239 | C | 1.63 | 1.9 × 10−5 | ||
| rs4915748 | 5 | 1 | 61954169 | T | 0.150 | 0.242 | C | 0.55 | 2.7 × 10−5 | ||
| rs617231 | 6 | 11 | 84837898 | C | 0.131 | 0.070 | T | 1.99 | 3.4 × 10−5 | DLG2 | |
| rs6022102 | 7 | 20 | 51425077 | C | 0.209 | 0.133 | T | 1.73 | 4.2 × 10−5 | ||
| rs6940190 | 8 | 6 | 148644731 | A | 0.280 | 0.383 | T | 0.63 | 4.6 × 10−5 | SASH1 | |
| rs2416305 | 9 | 5 | 112483604 | G | 0.197 | 0.293 | A | 0.59 | 4.7 × 10−5 | MCC | |
| rs17111695 | 10 | 5 | 150432446 | C | 0.186 | 0.115 | T | 1.77 | 5.7 × 10−5 | TNIP1 | |
| METH-induced psychosis GWASh | rs12591257 | 1 | 15 | 87064089 | C | 0.041 | 0.183 | A | 0.19 | 3.6 × 10−6 | AGBL1 |
| rs2346713 | 2 | 15 | 87078337 | C | 0.067 | 0.232 | A | 0.24 | 5.6 × 10−6 | AGBL1 | |
| rs16977267 | 3 | 15 | 86993464 | T | 0.073 | 0.232 | G | 0.26 | 1.6 × 10−5 | AGBL1 | |
| rs13414154 | 4 | 2 | 129012073 | A | 0.145 | 0.342 | G | 0.33 | 3.1 × 10−5 | HS6ST1 | |
| rs6767236 | 5 | 3 | 143758399 | G | 0.189 | 0.402 | T | 0.35 | 3.3 × 10−5 | ||
| rs6091985 | 6 | 20 | 53445939 | C | 0.044 | 0.171 | G | 0.22 | 3.3 × 10−5 | ||
| rs7333069 | 7 | 13 | 82458858 | A | 0.394 | 0.646 | C | 0.36 | 3.4 × 10−5 | ||
| rs8026683 | 8 | 15 | 86825255 | A | 0.098 | 0.268 | C | 0.30 | 3.5 × 10−5 | AGBL1 | |
| rs6064117 | 9 | 20 | 53458592 | G | 0.047 | 0.175 | A | 0.23 | 4.2 × 10−5 | ||
| rs2768428 | 10 | 10 | 12523129 | T | 0.052 | 0.183 | C | 0.24 | 4.9 × 10−5 | CAMK1D |
CHR: chromosome.
BP: base position based on hg19.
A1: minor allele name based on whole sample.
A2: major allele name.
OR: odds ratio (for A1: A2 is reference).
PGC: P-value-adjusted genomic control (two-sided).
Gene: gene (±20 kb).
This comparison was based on the presence (or absence) of psychotic symptoms in the METH-dependence samples.
Polygenic Component Analysis and Gene-Based Analysis
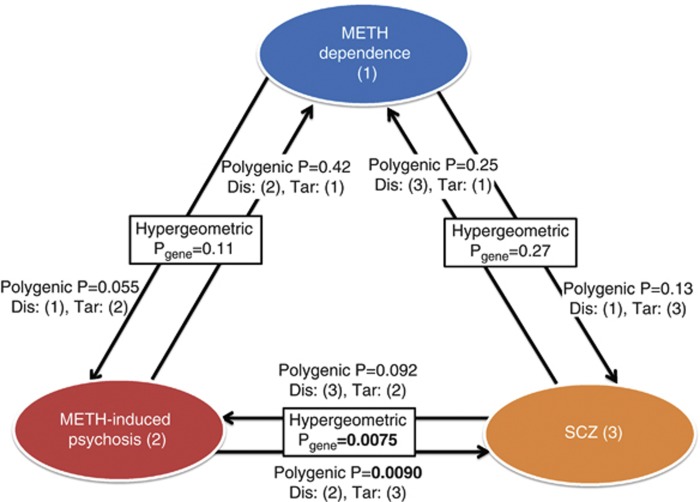
In the polygenic component analysis, we detected statistically significant enrichment of alleles scored in the ‘discovery' METH-induced psychosis GWAS sample in the ‘target' SCZ GWAS sample at P-thresholds (PTs) of <0.3, 0.4, and 0.5 (Pbest=0.0090, Supplementary Figure S4; Figure 2). However, the variances in SCZ liability explained by the ‘risk' SNPs of METH-induced psychosis were smaller (R2∼0.7%, Supplementary Figure S4) than those found in previous studies when only SCZ samples were used in the polygenic component analysis (R2∼3%) (Purcell et al, 2009; Ikeda et al, 2011). The reciprocal analysis (discovery/target SCZ/METH-induced psychosis pair) revealed a non-significant P-value of statistical enrichment of the ‘risk' SCZ alleles in the ‘target' METH-induced psychosis samples (Pbest=0.092, Supplementary Figure S5). Although statistical evidence was not obtained from this analysis (likely due to the small ‘target' sample), it is of note that the variances explained were higher (R2∼1.2%, Supplementary Figure S5) than that detected in the discovery/target METH-induced psychosis/SCZ pair, suggesting that larger METH-induced psychosis sample sizes will be essential for obtaining conclusive results. By contrast, no statistical overlap was observed among other combinations of discovery/target pairs (Figure 2, Supplementary Figures S6–S9).
Figure 2.
Relationship between METH-dependence (1), METH-induced psychosis (2), and schizophrenia (3). METH, methamphetamine; SCZ, schizophrenia; Dis, Discovery sample; Tar, Target sample. ‘Polygenic P' indicates the best P-value calculated by logistic regression analysis in the polygenic component analysis. ‘Hyper-geometric P' indicates the P-value calculated by hyper-geometric analysis to assess the enrichment of the risk genes that showed significance in both the two conditions (based on gene-based analysis).
We also performed gene-based testing of the associations, because this method can provide more statistical power than individual SNP-based analysis (Liu et al, 2010). Seventy-four of the 17 049 genes showed significant associations at Pgene<0.05, both in METH-induced psychosis and SCZ, suggesting significant enrichment by hyper-geometric analysis (P=0.0075: Supplementary Table S3). Supplementary Table S4 lists the genes that overlapped. Notably, NOTCH4, a promising candidate gene for SCZ (Stefansson et al, 2009; Ikeda et al, 2012) was found to be associated with both disorders (Figure 2).
DISCUSSION
In the current study, we detected shared genetic risk between METH-induced psychosis and SCZ but failed to detect an overlap between METH dependence and SCZ. These results highlight that the comparison between the ‘presence' and ‘absence' of psychosis within METH-dependent subjects, in our case, is preferable and important (even for polygenic component analysis) because the effect size of genetic markers associated with drug response (ie, psychosis as a response to METH exposure) is considered to be larger than that expected in the susceptibility to common and complex diseases (ie, SCZ and METH dependence) (Cirulli and Goldstein, 2010). Although the division of the METH-dependent subjects according to psychotic status will result in lowering the statistical power due to the reduced sample size, this type of research, especially in the psychiatry field where substantial disease heterogeneity is assumed, may benefit from the larger effect size following by this pharmacogenetic/genomic concept (Bousman et al, 2009; Cirulli and Goldstein, 2010).
However, our SNP-based analysis, even for METH-induced psychosis, did not show any associations with genome-wide significance. The presence of type II errors in this study are inevitable given our sample size; it is of note, however, that AGBL1 (rs12591257, P=3.6 × 10−6) showed a trend for an association with METH-induced psychosis. ABGL1 was one of the candidate genes for SCZ based on the CATIE GWAS (Sullivan et al, 2008), as well as for the side effect by antidepressant treatment in the STAR*D study (Clark et al, 2012). In addition, several reports suggested an association between SCZ and the genes listed in our top hits, such as NPAS3 (Pieper et al, 2005; Lavedan et al, 2009; Pickard et al, 2009; Huang et al, 2010; Macintyre et al, 2010) and DLG2 (Kristiansen et al, 2006; MacLaren et al, 2011). Therefore, although we failed to find associations in this study, the non-genome-wide level of statistical significance should be interpreted with caution and be validated by independent replication study.
To address the issue of the small sample size in the current study, we applied the powerful method of polygenic component analysis described by ISC, which showed that common SNPs collectively contribute to a substantial proportion of the heritability of common complex diseases. Our main finding supports previous epidemiological evidence linking between METH-induced psychosis and SCZ (Chen et al, 2005). Nevertheless, the variance-explained connecting these conditions was modest (R2∼0.7%), and it showed similar magnitude reported in the comparison of SCZ between Japanese and the UK populations (R2∼0.8%, Ikeda et al, 2011). This suggests that at least some of the liberal ‘risk' alleles of the METH-induced psychosis GWAS are likely to be SCZ risk alleles, but these disorders have specific risk alleles as well. Furthermore, the results obtained by gene-based analysis, which showed a significant enrichment of the risk genes between these two conditions, are also in agreement with the results of the polygenic component analysis (Figure 2). Therefore our results based on the polygenic component and the gene-based analyses are the first molecular genetic evidence for overlap between METH-induced psychosis and SCZ in humans: this, in turn, supports the proposed role of the METH exposure, where mice treated by METH have been used as a mouse model of SCZ (Machiyama, 1992). In the gene-based analysis, although it involves multiple comparison issues, it is also of note that we detected a significant association of NOTCH4 (Stefansson et al, 2009; Ikeda et al, 2012) both with the METH-induced psychosis and SCZ samples. Other overlap genes are possible candidate genes for ‘psychosis', thus these genes should be examined with different sets of samples, especially for SCZ, because our results indicate a higher previous likelihood that these genes are susceptibility genes for SCZ.
Several limitations should be noted to interpret our results. Firstly, the polygenic score aggregates evidence from multiple weakly associated genomic loci, which is important in the situation where most of the true-positive association signals fall below the genome-wide association threshold due to a lack of statistical power. In this context, there are several confounding problems associated with the interpretation of polygenic score analysis results. One critical factor is population stratification. To check for the possibility of this effect, we performed ‘discovery/target METH-induced psychosis/SCZ' analysis with the major four principle components included as covariates. In this analysis, however, we still found statistically significant enrichment at PT<0.3, 0.4, and 0.5, despite the decreased variance described above (Supplementary Figure S10). Another concern is the difference between cases and controls in terms of gender ratio, which is biased substantially towards males in the METH-dependent subjects (>3 : 1). We have not conducted the analysis of X-linked loci, as there is no gold standard so far established in analyzing the genotypes on chromosome X but included the ‘discovery/target METH-induced psychosis/SCZ' analysis using gender-adjusted P-values for discovery statistics (Supplementary Figure S11). Again no large change was observed in this explorative analysis.
In summary, a large number of ‘risk' SNPs selected from a METH-induced psychosis GWAS are enriched in individuals with SCZ. This result suggests that the overlap between genes scored as positive in both the data sets can have higher probability as susceptibility genes for psychosis. In the future, the shared genetic risk component between these disorders may provide insights into disease processes and the diagnosis and may open up new avenues for drug development in terms of pharmacological modeling of psychosis. Based on the current study, however, it is difficult to clarify the question of whether the ‘transient' type (remitting a psychiatric state immediately or a couple of weeks after METH exposure) or ‘prolonged' type of psychosis has more genetic similarity with SCZ. To obtain conclusive results, further studies with much larger samples are required.
FUNDING AND DISCLOSURE
The authors declare no conflict of interest.
Acknowledgments
This work was supported by research grants from Grants-in-Aid for Scientific Research of the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan; Grants-in-Aid for Scientific Research on Innovative Areas (Comprehensive Brain Science Network) from MEXT of Japan; Ministry of Health, Labor and Welfare of Japan; Academic Frontier Project for Private Universities, Comparative Cognitive Science Institutes; Core Research for Evolutional Science and Technology; Uehara Memorial Foundation; SENSHIN Medical Research Foundation; Takeda Science Foundation; the Novartis Foundation, Japan; Naito Foundation, Japan; and Strategic Research Program for Brain Sciences of the MEXT of Japan.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Bousman CA, Glatt SJ, Everall IP, Tsuang MT. Genetic association studies of methamphetamine use disorders: a systematic review and synthesis. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:1025–1049. doi: 10.1002/ajmg.b.30936. [DOI] [PubMed] [Google Scholar]
- Callaghan RC, Cunningham JK, Allebeck P, Arenovich T, Sajeev G, Remington G, et al. Methamphetamine use and schizophrenia: a population-based cohort study in California. Am J Psychiatry. 2012;169:389–396. doi: 10.1176/appi.ajp.2011.10070937. [DOI] [PubMed] [Google Scholar]
- Chen CK, Lin SK, Sham PC, Ball D, Loh el W, Murray RM. Morbid risk for psychiatric disorder among the relatives of methamphetamine users with and without psychosis. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:87–91. doi: 10.1002/ajmg.b.30187. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- Clark SL, Adkins DE, Aberg K, Hettema JM, McClay JL, Souza RP, et al. Pharmacogenomic study of side-effects for antidepressant treatment options in STAR*D. Psychol Med. 2012;42:1151–1162. doi: 10.1017/S003329171100239X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelotti DJ, Kanayama G, Pope HG., Jr. Remission of persistent methamphetamine-induced psychosis after electroconvulsive therapy: presentation of a case and review of the literature. Am J Psychiatry. 2010;167:17–23. doi: 10.1176/appi.ajp.2009.08111695. [DOI] [PubMed] [Google Scholar]
- Huang J, Perlis RH, Lee PH, Rush AJ, Fava M, Sachs GS, et al. Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Kinoshita Y, Okochi T, Kawashima K, Kushima I, et al. Genome-wide association study of schizophrenia in a Japanese population. Biol Psychiatry. 2011;69:472–478. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Aleksic B, Yamada K, Iwayama-Shigeno Y, Matsuo K, Numata S, et al. 2012Genetic evidence for association between NOTCH4 and schizophrenia supported by a GWAS follow-up study in a Japanese population Mol Psychiatry(in press). [DOI] [PubMed]
- Kristiansen LV, Beneyto M, Haroutunian V, Meador-Woodruff JH.2006Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia Mol Psychiatry 11737–747.705. [DOI] [PubMed] [Google Scholar]
- Lavedan C, Licamele L, Volpi S, Hamilton J, Heaton C, Mack K, et al. Association of the NPAS3 gene and five other loci with response to the antipsychotic iloperidone identified in a whole genome association study. Mol Psychiatry. 2009;14:804–819. doi: 10.1038/mp.2008.56. [DOI] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiyama Y. Chronic methamphetamine intoxication model of schizophrenia in animals. Schizophr Bull. 1992;18:107–113. doi: 10.1093/schbul/18.1.107. [DOI] [PubMed] [Google Scholar]
- Macintyre G, Alford T, Xiong L, Rouleau GA, Tibbo PG, Cox DW. Association of NPAS3 exonic variation with schizophrenia. Schizophr Res. 2010;120:143–149. doi: 10.1016/j.schres.2010.04.002. [DOI] [PubMed] [Google Scholar]
- MacLaren EJ, Charlesworth P, Coba MP, Grant SG. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol Cell Neurosci. 2011;47:93–99. doi: 10.1016/j.mcn.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa T, Kawashima M, Nishida N, Ohashi J, Kimura R, Fujimoto A, et al. Variant between CPT1B and CHKB associated with susceptibility to narcolepsy. Nat Genet. 2008;40:1324–1328. doi: 10.1038/ng.231. [DOI] [PubMed] [Google Scholar]
- Nutt DJ, King LA, Phillips LD. Drug harms in the UK: a multicriteria decision analysis. Lancet. 2010;376:1558–1565. doi: 10.1016/S0140-6736(10)61462-6. [DOI] [PubMed] [Google Scholar]
- Otowa T, Yoshida E, Sugaya N, Yasuda S, Nishimura Y, Inoue K, et al. Genome-wide association study of panic disorder in the Japanese population. J Hum Genet. 2009;54:122–126. doi: 10.1038/jhg.2008.17. [DOI] [PubMed] [Google Scholar]
- Pickard BS, Christoforou A, Thomson PA, Fawkes A, Evans KL, Morris SW, et al. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol Psychiatry. 2009;14:874–884. doi: 10.1038/mp.2008.24. [DOI] [PubMed] [Google Scholar]
- Pieper AA, Wu X, Han TW, Estill SJ, Dang Q, Wu LC, et al. The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc Natl Acad Sci USA. 2005;102:14052–14057. doi: 10.1073/pnas.0506713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Liu QR, Johnson C, Walther D, Komiyama T, et al. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008;65:345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- Ujike H, Sato M. Clinical features of sensitization to methamphetamine observed in patients with methamphetamine dependence and psychosis. Ann N Y Acad Sci. 2004;1025:279–287. doi: 10.1196/annals.1316.035. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.