Abstract
Experimental and computational evidence suggests that HLAs preferentially bind conserved regions of viral proteins, a concept we term “targeting efficiency,” and that this preference may provide improved clearance of infection in several viral systems. To test this hypothesis, T-cell responses to A/H1N1 (2009) were measured from peripheral blood mononuclear cells obtained from a household cohort study performed during the 2009–2010 influenza season. We found that HLA targeting efficiency scores significantly correlated with IFN-γ enzyme-linked immunosorbent spot responses (P = 0.042, multiple regression). A further population-based analysis found that the carriage frequencies of the alleles with the lowest targeting efficiencies, A*24, were associated with pH1N1 mortality (r = 0.37, P = 0.031) and are common in certain indigenous populations in which increased pH1N1 morbidity has been reported. HLA efficiency scores and HLA use are associated with CD8 T-cell magnitude in humans after influenza infection. The computational tools used in this study may be useful predictors of potential morbidity and identify immunologic differences of new variant influenza strains more accurately than evolutionary sequence comparisons. Population-based studies of the relative frequency of these alleles in severe vs. mild influenza cases might advance clinical practices for severe H1N1 infections among genetically susceptible populations.
Keywords: MHC, HLA association, T-cell epitope, Native American
The pandemic A/H1N1 influenza virus (pH1N1) spread globally during the influenza season of 2009–2010. An estimated 2,500–6,000 pH1N1-related deaths occurred between April and October 17, 2009 in the United States (www.cdc.gov/h1n1flu/estimates/April_March_13.htm). The pH1N1 virus was a triple-reassortant virus that had not been reported previously in animals or humans (1), and several epidemiologic characteristics differed substantially between this and other circulating H1N1 viruses. pH1N1-associated hospitalization rates and mortality among the elderly population were lower than those for seasonal influenza and higher for children and young adults (1); they also were higher for Hispanic, African American, Pacific Islander and American Indian/Alaska Native populations than for those of European/Caucasian descent (2–10). Early analyses found that vaccination with seasonal influenza vaccines was unlikely to provide protection from pH1N1 because of very low levels of cross-reactive antibody responses (11). Structural analysis showed that the pH1N1 HA protein antigenic regions are more similar to the H1N1 1918 pandemic strain than to other contemporary H1N1 strains (12, 13), and cross-reactive CD8+ T-cell responses exist between these viruses (14).
Although protection against influenza virus infection appears to be mediated by neutralizing antibodies directed at the HA surface protein, considerable evidence indicates the importance of CD4+ and CD8+ T-cell epitopes for controlling and clearing influenza infections (15–18). Influenza infection in humans induces proliferation of a large subset of HA-specific CD4+ T helper cells (19), resulting in the development of heterotypic antibody responses (20). Thus, both CD4+ and CD8+ T cells appear to play a role in the clearance of influenza (15, 21–23). Cross-reactive T-cell responses, specifically CD8+ T-cell responses to conserved influenza regions, appear to be an important factor in defining the severity of disease, especially disease caused by emerging new variants and subtypes (15, 17, 18, 24–28). A recent analysis reported that 69% of the previously characterized T-cell epitopes for H1N1 were conserved in pH1N1, and that memory T-cell responses to pH1N1 were present in the adult population with a magnitude similar to those for seasonal influenza H1N1 (26).
HLA alleles display differences in antigen-binding preferences and, in combination with the high diversity of viral sequences, directly influence the potential set of targets that become T-cell epitopes. Recently, we used both experimental and computational predictions of HLA binding to show that HLA alleles preferentially bind to conserved regions of viral proteins, a concept we term “targeting efficiency” (29). The HLA targeting efficiency score is defined as the Spearman correlation coefficient of the binding score of a given HLA allele and the conservation score for a given protein (or proteome) (SI Materials and Methods). A positive score indicates preferential targeting of conserved regions, and a negative score indicates preferential binding of variable regions. Efficiency score computation uses HLA binding predictors over a sequence of interest to infer binding targets for a given protein and correlates these with evolutionary conservation of the protein, computed using homologous sequence data (Fig. 1). In a previous analysis of 52 common human viruses, we found that HLA alleles preferentially bind to conserved regions of viral proteomes (29). We further showed that the efficiency scores of HLA alleles for the HIV Gag protein predicted disease progression, in that alleles associated with nonprogression had higher efficiency scores than alleles associated with rapid progression.
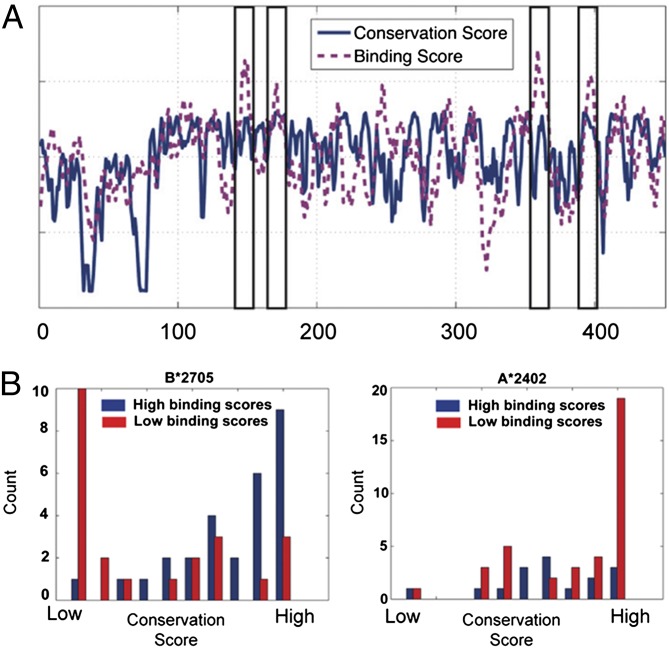
Fig. 1.
Computing HLA targeting efficiency scores for the NA protein of pH1N1. (A) The HLA targeting efficiency score is defined as the Spearman correlation coefficient between the conservation score and binding score of a given HLA allele on a protein of interest. Scores for the pH1N1 proteome are computed by concatenating the binding and conservation scores of all 10 individual proteins (SI Materials and Methods). The conservation scores for pH1N1 NA are plotted in blue against the amino acid position. HLA-B*27:05 binding scores are plotted in magenta for the NA protein against the amino acid position. Areas marked by black rectangles show the positions with the highest binding scores. All these positions also have high conservation scores. (B) Comparison of the conservation scores of positions that have high binding scores with those that have low binding scores. The conservation scores for two sets of positions on the NA protein of pH1N1 (blue, positions with high binding scores; red, positions with low binding scores) are plotted as histograms. The distribution of conservation scores for the two binding score distributions of B*27:05 (Left) differs markedly. The two-tailed t test for the difference between the two binding score distributions is highly significant (P < 1.7e−5). The distributions of A*24:02 (Right) are not statistically significantly different (P = 0.084), and positions with high binding scores are less conserved than those with low binding scores.
To expand on our previous studies, we assessed the relationships between HLA targeting efficiency and pH1N1 outcome for a cohort of individuals infected with pH1N1 who were enrolled in a clinical trial examining T-cell responses in young children and their families. We found a correlation between T-cell responses and the efficiency scores of these patients. We then conducted a population-based analysis and found that the carriage frequencies of the alleles with the lowest targeting efficiencies, HLA A*24, were associated with pH1N1 mortality. Interestingly, these alleles are common in certain indigenous populations in which increased pH1N1 morbidity has been reported (2–10). These data suggest that HLA alleles may be useful markers for assessing the risk of potential pandemics.
Results
HLA Targeting Efficiency Correlates with CD8+ T-Cell Responses to pH1N1.
CD8+ T-cell responses to the pH1N1 variant differ markedly from prior circulating H1N1 strains of influenza A (30). We tested T-cell responses from a household cohort study performed during the 2009–2010 influenza season at St. Jude Children’s Research Hospital and Le Bonheur Children’s Hospital in Memphis, TN. The basic design of the study involved the enrollment of index cases that tested positive for pandemic influenza A infection by PCR, along with cohabiting family members, most of whom were sampled every 3–7 d for a total of 28–35 d. Ninety-eight participants were enrolled during the 2009–2010 influenza season. Of these, 35 tested positive for pH1N1 by RT-PCR. Peripheral blood mononuclear cell (PBMC) samples at late time points were available from 13 individuals (12 individuals, days 27–35; 1 individual, day 9) (Table 1).
Table 1.
pH1N1 specific T-cell responses by ELISpot for pH1N1 clinical cohort
| HLA efficiency |
Received vaccine? |
||||||||
| Participant | Age at enrollment, y, mo | Sex | Score ranking | Score | ELISpot, SFUs/million* | 2006–7 | 2007–8 | 2008–9 | 2009 pH1N1 |
| 1 | 27, 4 | F | 1 | 0.029 | 56.67 | No | No | No | No |
| 2 | 7, 10 | M | 2 | 0.0301 | 216.67 | Yes | Yes | Yes | No |
| 3 | 14, 6 | M | 3 | 0.0316 | 120.00 | Yes | Yes | Yes | Yes |
| 4 | 40, 6 | F | 6 | 0.0367 | 41.67 | Yes | Yes | Yes | No |
| 5 | 19, 0 | F | 7 | 0.0371 | 60.00 | No | No | No | No |
| 6 | 1, 6 | M | 8 | 0.0377 | 101.67 | No | No | Yes | No |
| 7 | 9, 1 | M | 14 | 0.0486 | 201.67 | No | No | No | No |
| 8 | 23, 6 | F | 15 | 0.0488 | 311.67 | No | No | No | No |
| 9 | 44, 11 | F | 17 | 0.0517 | 313.33 | Yes | Yes | Yes | No |
| 10 | 7, 10 | F | 18 | 0.0521 | 98.33 | No | No | No | No |
| 11 | 55, 11 | M | 20 | 0.0533 | 81.67 | No | No | No | No |
| 12 | 10, 10 | F | 23 | 0.0589 | 305.00 | No | No | No | No |
| 13 | 34, 1 | F | 28 | 0.0719 | 251.67 | No | No | No | No |
ELISpot response to A/CA/04/09 pH1N1.
We computed the HLA targeting efficiency scores of 95 common HLA-A and HLA-B alleles over the entire pH1N1 virus proteome (A/California/04/2009). Fig. 1 depicts the computed HLA targeting efficiency scores using the representative protein neuraminidase (NA) (SI Materials and Methods). We then ranked all infected study participants based on their average HLA-A and HLA-B allele targeting efficiency scores over the entire pH1N1 proteome (Table 1). This approach assumed that each allele contributes linearly to the T-cell response to a given antigen.
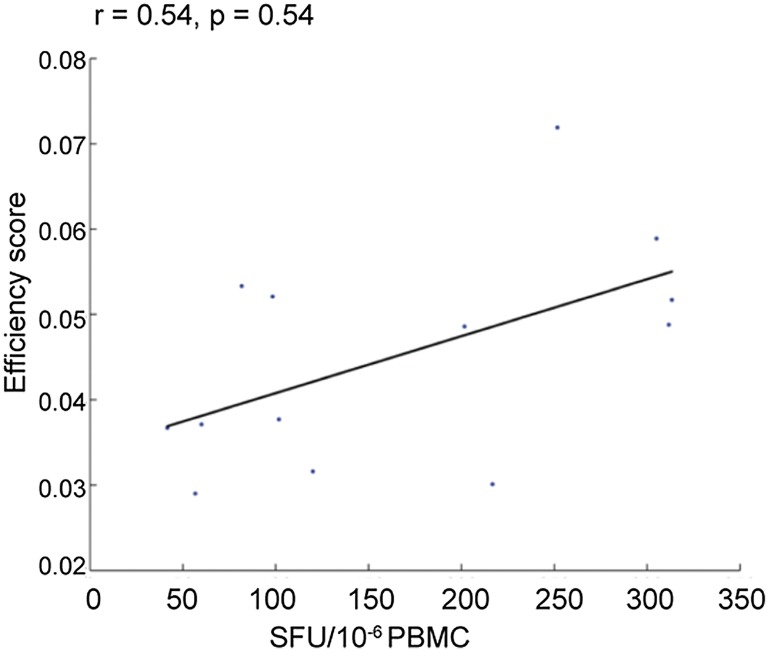
To elucidate overall T-cell responses, we measured IFN-γ secretion patterns from unfractionated PBMCs (from the 13 participants) exposed to β-propiolactone–inactivated A/California/04/2009 (pH1N1), A/Brisbane/59/2007 (2007–2008 seasonal H1N1), or A/Brisbane/10/2007 (seasonal H3N2) virus by enzyme-linked immunosorbent spot (ELISpot). We found that participants’ HLA targeting efficiency scores correlated with IFN-γ spot-forming units (SFUs) to pH1N1 (r = 0.54, P = 0.054) (Fig. 2); neither enrollment age nor sex significantly correlated with SFUs. Previous studies showed that vaccination increases the magnitude of influenza-specific T-cell responses (31, 32). We therefore computed the correlation between the efficiency scores and ELISpot responses after adjusting for receipt of the 2008–2009 influenza vaccine. We found that the frequency of IFN-γ–producing cells was significantly related to efficiency score (r = 0.60, P = 0.042; multivariate regression). We found no evidence for an interaction between the efficiency scores and vaccination status (P = 0.34, multivariate regression).
Fig. 2.
pH1N1 T-cell response correlates with HLA targeting efficiency. PBMCs were collected from 13 study participants who had confirmed pH1N1 infection, stimulated with A/California/04/09 (H1N1), and analyzed by ELISpot (SFUs per million PBMCs; x-axis). The y-axis depicts HLA targeting efficiency for each subject based on their HLA-A and HLA-B alleles. After controlling for vaccination status in multivariate regression, the P value for the correlation between targeting efficiency and the ELISpot response is reduced to 0.042.
HLA A*24 Allele Frequencies Correlate with pH1N1 Mortality Rates.
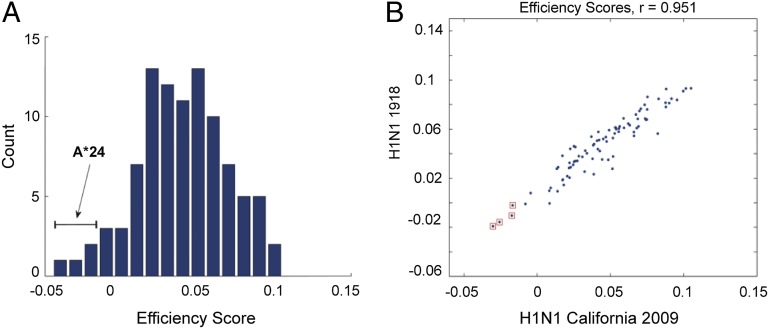
To assess the broader implications of targeting efficiency at the population level, we evaluated whether HLA binding affinities for conserved viral regions could influence pH1N1 disease severity. Because it is not feasible to characterize HLA-A and -B haplotypes in individuals across large populations, we instead ranked HLA alleles individually based on targeting efficiency. These scores were computed for the 95 most prevalent HLA class I alleles against a wide variety of influenza subtypes, including pH1N1 (2009), previously circulating H1N1 and currently circulating H3N2 subtypes, H5N1, and the 1918 H1N1 subtype (Table S1). Although most alleles had positive targeting efficiency scores, indicating a preference to bind to conserved regions of pH1N1, those for A*24 alleles (A*24:01, A*24:02, A*24:07, and A*23:01) were negative, forming the tail of the efficiency distribution (Fig. 3A). This tendency is not unique to pH1N1; it also was observed for the A/Brevig Mission/1/1918 (H1N1) strain (Fig. 3B), as well as for other circulating strains (Fig. S1).
Fig. 3.
A*24 alleles have the lowest HLA targeting efficiency scores for pH1N1. (A) Histogram of HLA targeting efficiency scores for 95 alleles analyzed in this study on pH1N1. HLA-A*24 alleles all lie in the tail of this distribution, and all have negative targeting efficiency scores, indicating an overall slight preference for binding to variable rather than conserved regions on the pH1N1 proteome. (B) HLA targeting efficiency scores for A/Brevig Mission/1918 (H1N1) (y-axis) plotted against scores for the pH1N1 2009 strain (x-axis). A*24 alleles are marked by red □. A*24 alleles have the lowest targeting efficiency scores for both viruses, as well as for circulating H1N1 strains, suggesting an overall tendency of these alleles to target the less-conserved regions of H1N1 viruses.
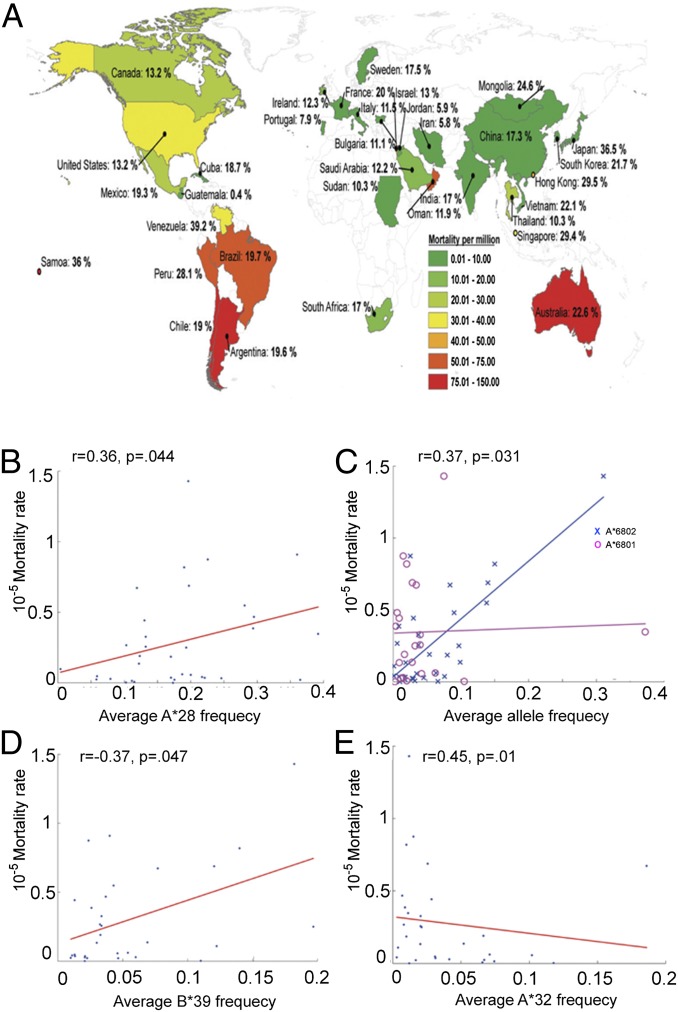
If the relatively poor ability of A*24 alleles to target conserved regions of pH1N1 results in less efficient viral control and clearance upon infection, we reasoned that populations enriched in A*24 frequency may have a higher risk for developing severe pH1N1 disease. We therefore compared the average A*24 carriage frequencies to the pH1N1 mortality rates computed for 34 countries (33, 34) (Fig. 4A). We found that the average A*24 carriage frequency was positively correlated with mortality rate (Fig. 4B, r = 0.37, P = 0.031 and Fig. S2A). Our analysis is supported by several reports following the pH1N1 outbreaks in the spring of 2009 in North America and in Australia and New Zealand (2–11).
Fig. 4.
Comparisons between HLA allele frequencies and mortality from pH1N1 for various populations. (A) World map overlaying pH1N1 mortality ratios per million and HLA-A*24 frequencies for 34 countries. Analyses were performed on all countries for which both types of information, allele frequency and mortality from pH1N1, are publicly available. (B) pH1N1 mortality rates (y-axis) plotted against average HLA-A*24 frequencies (x-axis) for 34 countries. Mortality rates correlate with A*24 frequencies (r = 0.37, P = 0.031). (C) pH1N1 mortality rates (y-axis) are plotted against average HLA-A*68:01 (blue ×) and HLA-A*68:02 frequencies (magenta ○) (x-axis) for 32 and 22 countries, respectively. Mortality rates are correlated with A*68:01 frequencies (r = 0.45, P = 0.01), but not with A*6802 frequencies (P = 0.77). (D) pH1N1 mortality rates (y-axis) are plotted against average HLA-B*39 frequencies (x-axis) for 31 countries. Mortality rates correlate with B*39 frequencies (r = 0.36, P = 0.044). (E) pH1N1 mortality rates (y-axis) are plotted against average HLA-A*32 frequencies (x-axis) for 30 countries. Mortality rates negatively correlate with A*32 frequencies (r = −0.37, P = 0.047).
To determine whether we could find an association with mortality for any other HLA allele, we computed the Spearman correlation coefficient for all two-digit HLA families within the set of 95 HLA alleles previously analyzed; this was done without correction of P values for multiple comparisons to maximize our ability to detect other associations. Only 3 of the 95 additional two-digit families—A*68, B*39, and A*32—had a statistically significant correlation with pH1N1 mortality data, indicating that the association between HLA type and influenza mortality is relatively uncommon.
Further analyses showed that A*68 carriage frequencies positively correlated with pH1N1 mortality (r = 0.35, P = 0.044). Our HLA dataset contains two A*68 alleles that are known to belong to different supertypes: A*68:01 to the A3 supertype and A*68:02 to the A2 supertype (35). Interestingly, A*68:01 frequency was strongly associated (r = 0.45, P = 0.0096, Fig. 4C and Fig. S2B) and A*68:02 was not significantly associated (P = 0.77) with pH1N1 mortality (Fig. 4C). A*68:01 also is highly prevalent in some Native American groups (33) (Table S2) and has a low targeting efficiency score, supporting the hypothesis that low efficiency scores correlate with pH1N1 mortality.
B*39 carriage frequencies also positively correlated with pH1N1 mortality rates (r = 0.36, P = 0.044, Fig. 4D and Fig. S2D). B*39 alleles are highly prevalent in Taiwan and Japan (B*39:01) and numerous Native American populations (B*39:01–B*39:10) and are found in unison with A*24:02 (Taiwan) and A*24:02–B*39:01 (Native American) (Table S2) (33). Although the two B*39 alleles in our dataset (B*39:01 and B*39:06) possess high targeting efficiency scores (ranked 69 and 80 of 95, respectively), they are in linkage disequilibrium with A*24:02, and as such we cannot exclude the possibility that the HLA-B*39 correlation with mortality is driven by HLA-A*24:02.
One allele, HLA-A*32, was negatively correlated with pH1N1-associated mortality (r = −0.37, P = 0.047, Fig. 4E and Fig. S2C), suggesting that carriers of this allele may develop less severe pH1N1 disease. The efficiency score of A*32:01 is the sixth highest score among the 95 alleles examined.
HLA A*24 Alleles Are Highly Prevalent in Indigenous Populations, Which Had Higher Hospitalization and Mortality Rates During the pH1N1 Pandemic.
Although indigenous populations represent less than 5% of the general population, during the 2009 pandemic they accounted for 17.6% of all pH1N1 hospitalized cases in Canada and 17.5% in Arizona (2). A follow-up analysis by the Centers for Disease Control and Prevention in 12 states in which more than 50% of the American Indian/Alaska Natives reside found an H1N1 mortality rate four times higher than for persons in all other racial/ethnic populations combined (4). Another study reported higher rates of increased disease severity and intensive care unit admission in First Nation residents. Mortality rates were three to eight times higher for indigenous populations in Australia and New Zealand (3). Importantly, these aboriginal populations have an unusually high prevalence of A*24 alleles (33) (Table 2). Interestingly, indigenous populations also had a high case-fatality rate compared with the general population during the H1N1 1918–1919 pandemic (8.5% vs. 2.5%, respectively) (2).
Table 2.
Comparison of HLA frequency and increased risk factors for pH1N1 as reported by La Ruche et al. (2) for indigenous populations of the Americas and Australia
| Population | A*24 frequency (%) (28) | Indigenous/nonindigenous risk factors (odds ratio) (2) |
| Americas | ||
| USA Alaska Yupik Native | 0.603 | 3.4 |
| USA Arizona Pima | 0.360 | 4.3* |
| USA New Mexico Canoncito Navajo | 0.318 | 4.3* |
| USA North American Native | 0.305 | 4.3* |
| USA South Dakota Lakota Sioux | 0.235 | 4.3* |
| USA Hawaii Okinawa | 0.343 | N/A |
| Australia | ||
| Australian Aborigine Cape York Peninsula | 0.223 | 5.1 |
| Australian Aborigine Groote Eylandt | 0.293 | 5.1 |
| Australian Aborigine Kimberly Australian | 0.097 | 5.1 |
| Aborigine Yuendumu | 0.329 | N/A |
N/A, not applicable.
American Indians.
Using HLA Targeting Efficiency Scores to Measure the Similarity of pH1N1 to Other Influenza A Strains.
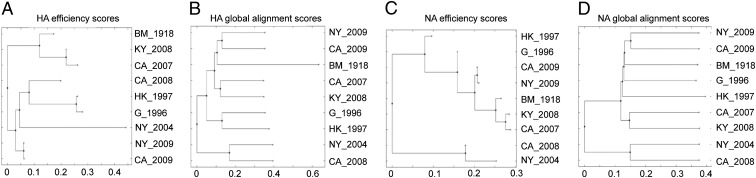
The HLA targeting efficiency score of a given allele can be computed for an entire viral proteome, measuring the overall ability of that HLA to target the conserved regions of a given viral strain, or for a specific protein of interest. We computed efficiency scores of the 95 alleles for the HA and NA proteins of the influenza strains analyzed in this study (Fig. S3 A and B and Table S1). The efficiency profile of a set of alleles for a given protein may be used as a representation of that strain in an HLA-specific immunological space. It measures similarity as viewed through the prism of HLA binding preferences. In generating phylogenetic trees of the HA and NA proteins based on this distance measure (Fig. 5 A and C), we found for HA that the pH1N1 strains are related more closely to the human H5N1 and to the H3N2 2008 strains than to recently circulating H1N1 strains. As previously shown, the H1N1 1918 strain is closer to pH1N1 than to circulating H1N1 strains. This immunological assessment differs markedly from classical genetic alignment (Fig. 5 B and D), in which pH1N1 is more homologous to circulating H1N1 than to all other strains (36). We also found that the NA protein of pH1N1 is more similar to an H5N1 avian isolate than it is to previously circulating seasonal H1N1 strains.
Fig. 5.
A comparison of efficiency-based phylogenetic trees to sequence-based phylogenetic trees. A phylogenetic tree based on targeting efficiencies of the HA (A) and NA (C) proteins of influenza strains analyzed in this study, clustered using the neighbor-joining method, based on sequence distances computed using the HLA targeting efficiency profiles of the 95 class I alleles used in this study. See Table S1 for strain names. pH1N1 strains are closer to the H3N2 2008 strain and the human H5N1 strain (A) and the H5N1 avian strain (C) than to the circulating H1N1 2007 and 2008 strains. The H3N2 2004 strain from New York shows a distinct separation from the H3N2 California 2008 strain. In 2004, the circulating strain in New York was the Fujian strain, which caused an unusually severe influenza season in 2003–2004 (A). A phylogenetic tree based on genetic sequences of HA (B) and NA (D) proteins of the influenza strains analyzed in this study clustered using the neighbor-joining method based on alignment distances computed using the Needleman–Wunsch global alignment algorithm. As previously shown, the H1N1 1918 strain is closer to the pH1N1 strains than it is to circulating H1N1 strains (B). The pH1N1 strains are more similar to the H1N1 1918 strain and to the H5N1 strain than to circulating H1N1 strains from 2008 and 2007 (D).
Discussion
In this study, we used the HLA targeting efficiency score to analyze the 2009 pandemic influenza A strain (pH1N1) and to compare it with other influenza strains, including circulating H3 and H1 strains, several historical strains of interest, and an avian H5 strain. We analyzed data from a household cohort study of pH1N1-infected children and their family members and found that ranking individuals by their HLA targeting efficiency scores, based on their HLA alleles, was correlated with the magnitude of CD8+ T-cell responses, as measured using an ELISpot assay. We then ranked a set of 95 common HLA-A and HLA-B alleles by their efficiency scores to the pH1N1 virus and identified a set of alleles that had poor efficiency scores, all from the HLA A*24 family. We found that the prevalence of HLA A*24 alleles correlated with mortality rates from pH1N1.
Our analyses provide a potential immunological explanation for the observed differences in mortality from recent pH1N1 outbreaks and a framework for several testable predictions regarding the evolution and immunogenetics of influenza A. In seasonal influenza infections, B cells can provide sterilizing immunity, with T cells playing a less accentuated role. However, during the initial spread of a pandemic strain, which differs significantly from circulating strains and to which large segments of the population have no cross-reactive antibody responses, the ability of memory T cells to expand and reduce or control viral titer until novel antibodies are generated becomes an important factor in controlling infection (37). Indeed, we found that A*24 alleles have low targeting efficiency scores for a wide range of H1N1 viruses, including the 1918 Brevig Mission strain as well as seasonal circulating H1N1 strains (Fig. 3B and Fig. S1), but this may have clinical relevance only in pandemic settings in which there is a lack of cross-reactive antibody responses.
We hypothesized that in such settings, individuals who make T-cell responses to the variable regions of influenza viruses are likely to have less cross-reactive memory T-cell responses to the pandemic strain, which may allow for increased viral loads and more severe clinical symptoms. We found that ranking pH1N1 patients from the 2009–2010 pandemic by their ability to target conserved regions of pH1N1 correlated with the magnitude of their T-cell responses to this virus. Considering the small sample size (n = 13), the correlation coefficient obtained was relatively high (r = 0.54), suggesting that the patients’ overall HLA targeting efficiency may be a critical factor for determining the magnitude of ensuing T-cell responses. However, additional follow-up studies are required to further assess the power of the HLA targeting efficiency scores for predicting T-cell responses following influenza infection.
Alleles important for the control of HIV infection, such as B*57:01, bind to several conserved epitopes in Gag; viral escape by mutation in these conserved regions likely would incur a high fitness cost (38, 39). Previously, we showed that the efficiency scores of HLA alleles for the HIV Gag were associated with disease progression, in that alleles associated with nonprogression had higher efficiency scores than alleles associated with rapid progression (29). Recent work on human T-lymphotropic virus type 1 (HTLV-1) found that the strength of HLA binding to a specific HTLV-1 protein was associated with a reduced risk of developing chronic inflammatory syndromes and reduced proviral load, and that this was not restricted to a small set of alleles previously associated with disease status in HTLV-1 infection (40). In other diseases, such as HIV and dengue, HLA alleles have been associated with increased susceptibility or disease progression (38, 41). Several other studies have shown that viruses mutate in regions that contain HLA binders (42–44), and in some specific cases, such escape from T-cell responses has been shown to affect viral fitness (45, 46). Although the mechanisms of HLA-associated protection or susceptibility to disease have not been fully elucidated yet, these studies demonstrate that HLA binding properties may have a multifarious effect on disease progression and viral control, as well as other properties of the T-cell receptor (TCR) repertoire.
In our previous work, we found that HLA alleles preferentially target the conserved regions of double-stranded DNA viruses (HLA-A) and RNA viruses (HLA-B) (29). We found that for pH1N1, HLA-B alleles had efficiency scores significantly higher than those of HLA-A alleles (P < 0.005, Fig. S4), indicating an improved ability to target conserved regions of pH1N1. This finding is in accordance with a previous study that found that CD8+ T-cell responses to influenza infection that were restricted by HLA-A alleles, in comparison with HLA-B, tended to be monofunctional (47).
Our data suggest that HLA A*24 and HLA A*68 alleles are risk factors associated with severe pH1N1 infection and other common circulating influenza strains from recent years and coincide with an ecological analysis that found pH1N1 mortality correlated with the frequency of these HLA alleles. This also provides a potential explanation for the increased pH1N1 disease severity and morbidity in indigenous populations worldwide (2–10), populations that are all enriched for A*24 alleles despite their widespread geographic divergence.
We note that other factors associated with increased pH1N1 mortality most likely exist, including demographic, metabolic, and immunological elements. Associations between metabolic diseases such as diabetes and the A*24 allele have been reported, indicating that certain HLA alleles may influence pH1N1 mortality directly and indirectly (48–51). Furthermore, indigenous populations have a high prevalence of comorbidities such as lung and heart disease, which are directly related to influenza severity. These are mediated predominantly by environmental factors and may strongly confound the ecologic association reported here. One additional limitation of these analyses is that the mortality rates are based on the number of laboratory-confirmed deaths reported by ministries of health. A recent report estimated that pH1N1 mortality rates might be 15 times higher than the confirmed deaths (52). In countries with different HLA A*24 prevalence, variations between testing and reporting may result in biased mortality estimates.
Using the HLA targeting efficiency scores, we suggested a unique representation of viral strains defined by an HLA-specific embedding into an immunological space based on potential T-cell epitopes. We then defined a sequence similarity measure in this space. Our analysis is in agreement with a previous finding showing that the functional activity of the H5N1 NA protein is lower than that of several circulating H1N1 strains (53). Interestingly, the immunological distance between the two H3N2 strains (H3N2 2004 New York and H3N2 California 2008) is distinctly separate, unlike in the evolutionary alignment tree (Fig. 5 A and B). This is in agreement with the unusually severe influenza that a novel H3N2 strain caused in 2003 (54). Our computational approach may be applied to any influenza strain, and the limited number of strains chosen for this study is a mere demonstration of the ability to identify components of the host immune response important to containing viral infections, as reflected by changes in the HLA binding patterns to these influenza strains.
Our analysis suggests that appropriately designed population-based studies of the relative frequency of select alleles in severe cases or deaths associated with influenza A compared with mild cases or asymptomatic infections might lead to more directed clinical treatment practices for severe H1N1 influenza infections among genetically susceptible populations and guide public health interventions. Additional studies using larger cohorts and testing other pathogens are warranted, but these data suggest that HLA targeting efficiency might be a useful clinical correlate in emerging pandemics and an effective tool for determining the pathogenic potential of emerging variant viruses.
Materials and Methods
Information on the following may be found in SI Materials and Methods: HLA predictor accuracy, T-cell enrichment, IFN-γ ELISpot assay, statistical analyses, conservation scores, binding scores, viruses, HLA allele determination, sequence similarity measures, pH1N1 mortality rates, HLA frequency data, HLA targeting efficiency, and participants.
Supplementary Material
Acknowledgments
We thank Jonathan Sugimoto for help in generating the world map figure, Chen Yanover for fruitful discussions and comments, and Dr. Rachel Tompa and Dr. Mindy Miner for editing the manuscript. We also thank Dr. Victoria Turner, the St. Jude Children’s Research Hospital HLA Laboratory, Dr. Daniel Geraghty, and Dr. Quyen Vu for determining participant HLA alleles. We acknowledge the clinical work of Lisa Harrison, Peter Snell, Lauren Lazar, and Magan Stephens at the University of Tennessee Health Science Center, Le Bonheur Children’s Hospital. This work was funded, in part, by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, under contract number HHSN266200700005C and American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
The authors declare no conflict of interest.
†This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221555110/-/DCSupplemental.
References
- 1.Jhung MA, et al. Epidemiology of 2009 pandemic influenza A (H1N1) in the United States. Clin Infect Dis. 2011;52(Suppl 1):S13–S26. doi: 10.1093/cid/ciq008. [DOI] [PubMed] [Google Scholar]
- 2.La Ruche G, et al. epidemic intelligence team at InVS The 2009 pandemic H1N1 influenza and indigenous populations of the Americas and the Pacific. Euro Surveill. 2009;14(42):19366. doi: 10.2807/ese.14.42.19366-en. [DOI] [PubMed] [Google Scholar]
- 3.Webb SA, et al. ANZIC Influenza Investigators Critical care services and 2009 H1N1 influenza in Australia and New Zealand. N Engl J Med. 2009;361(20):1925–1934. doi: 10.1056/NEJMoa0908481. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) Deaths related to 2009 pandemic influenza A (H1N1) among American Indian/Alaska Natives—2 states, 2009. MMWR Morb Mortal Wkly Rep. 2009;58(48):1341–1344. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) pandemic influenza A (H1N1) virus infections—Chicago, Illinois, April-July 2009. MMWR Morb Mortal Wkly Rep. 2009;58(33):913–918. [PubMed] [Google Scholar]
- 6.Brooks EG, et al. 2009 H1N1 fatalities: The New Mexico experience. J Forensic Sci. 2012;57(6):1512–1518. doi: 10.1111/j.1556-4029.2012.02163.x. [DOI] [PubMed] [Google Scholar]
- 7.Lindstrom S, et al. Human infections with novel reassortant influenza A(H3N2)v viruses, United States, 2011. Emerg Infect Dis. 2012;18(5):834–837. doi: 10.3201/eid1805.111922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaffer A, et al. The impact of influenza A(H1N1)pdm09 compared with seasonal influenza on intensive care admissions in New South Wales, Australia, 2007 to 2010: A time series analysis. BMC Public Health. 2012;12:869. doi: 10.1186/1471-2458-12-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trauer JM, et al. Australia, New Zealand and Singapore Pandemic Serosurveillance Study Group Seroepidemiologic effects of influenza A(H1N1)pdm09 in Australia, New Zealand, and Singapore. Emerg Infect Dis. 2013;19(1):92–101. doi: 10.3201/eid1901.111643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarychanski R, et al. Correlates of severe disease in patients with 2009 pandemic influenza (H1N1) virus infection. CMAJ. 2010;182(3):257–264. doi: 10.1503/cmaj.091884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention (CDC) Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58(19):521–524. [PubMed] [Google Scholar]
- 12.Xu R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328(5976):357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, et al. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell. 2010;1(5):459–467. doi: 10.1007/s13238-010-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gras S, et al. Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc Natl Acad Sci USA. 2010;107(28):12599–12604. doi: 10.1073/pnas.1007270107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMichael AJ, Gotch FM, Noble GR, Beare PA. Cytotoxic T-cell immunity to influenza. N Engl J Med. 1983;309(1):13–17. doi: 10.1056/NEJM198307073090103. [DOI] [PubMed] [Google Scholar]
- 16.Wahl A, et al. T-cell tolerance for variability in an HLA class I-presented influenza A virus epitope. J Virol. 2009;83(18):9206–9214. doi: 10.1128/JVI.00932-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunham EJ, et al. Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J Virol. 2009;83(11):5485–5494. doi: 10.1128/JVI.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Madan R, Karp CL, Braciale TJ. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med. 2009;15(3):277–284. doi: 10.1038/nm.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104(12):R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshall D, Sealy R, Sangster M, Coleclough C. TH cells primed during influenza virus infection provide help for qualitatively distinct antibody responses to subsequent immunization. J Immunol. 1999;163(9):4673–4682. [PubMed] [Google Scholar]
- 21.Yap KL, Ada GL, McKenzie IF. Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature. 1978;273(5659):238–239. doi: 10.1038/273238a0. [DOI] [PubMed] [Google Scholar]
- 22.Yap KL, Braciale TJ, Ada GL. Role of T-cell function in recovery from murine influenza infection. Cell Immunol. 1979;43(2):341–351. doi: 10.1016/0008-8749(79)90178-3. [DOI] [PubMed] [Google Scholar]
- 23.Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. Compromised influenza virus-specific CD8(+)-T-cell memory in CD4(+)-T-cell-deficient mice. J Virol. 2002;76(23):12388–12393. doi: 10.1128/JVI.76.23.12388-12393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan WS, Jr, Badger GF, Dingle JH. A study of illness in a group of Cleveland families. XVI. The epidemiology of influenza, 1948-1953. Am J Hyg. 1958;68(2):169–189. doi: 10.1093/oxfordjournals.aje.a119961. [DOI] [PubMed] [Google Scholar]
- 25.Lee LY-H, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008;118(10):3478–3490. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenbaum JA, et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc Natl Acad Sci USA. 2009;106(48):20365–20370. doi: 10.1073/pnas.0911580106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan PT, et al. Conservation and diversity of influenza A H1N1 HLA-restricted T cell epitope candidates for epitope-based vaccines. PLoS One. 2010;5(1):e8754. doi: 10.1371/journal.pone.0008754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan PT, Khan AM, August JT. Highly conserved influenza A sequences as T cell epitopes-based vaccine targets to address the viral variability. Hum Vaccin. 2011;7(4):402–409. doi: 10.4161/hv.7.4.13845. [DOI] [PubMed] [Google Scholar]
- 29.Hertz T, et al. Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J Virol. 2011;85(3):1310–1321. doi: 10.1128/JVI.01966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agrati C, et al. Association of profoundly impaired immune competence in H1N1v-infected patients with a severe or fatal clinical course. J Infect Dis. 2010;202(5):681–689. doi: 10.1086/655469. [DOI] [PubMed] [Google Scholar]
- 31.Lambe T, et al. T-cell responses in children to internal influenza antigens, 1 year after immunization with pandemic H1N1 influenza vaccine, and response to revaccination with seasonal trivalent-inactivated influenza vaccine. Pediatr Infect Dis J. 2012;31(6):e86–e91. doi: 10.1097/INF.0b013e318255e443. [DOI] [PubMed] [Google Scholar]
- 32.Richards KA, Chaves FA, Alam S, Sant AJ. Trivalent inactivated influenza vaccines induce broad immunological reactivity to both internal virion components and influenza surface proteins. Vaccine. 2012;31(1):219–225. doi: 10.1016/j.vaccine.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Middleton D, Menchaca L, Rood H, Komerofsky R. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens. 2003;61(5):403–407. doi: 10.1034/j.1399-0039.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- 34.European Centre for Disease Prevention and Control 2009. ECDC daily update: 2009 influenza A (H1N1) pandemic. Available at http://ecdc.europa.eu/en/healthtopics/Documents/100118_Influenza_AH1N1_Situation_Report_0900hrs.pdf. Accessed October 25, 2009.
- 35.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha N, Roy A, Das B, Das S, Basak S. Evolutionary complexities of swine flu H1N1 gene sequences of 2009. Biochem Biophys Res Comm. 2009;390(3):349–351. doi: 10.1016/j.bbrc.2009.09.060. [DOI] [PubMed] [Google Scholar]
- 37.Valkenburg SA, et al. Immunity to seasonal and pandemic influenza A viruses. Microbes Infect. 2011;13(5):489–501. doi: 10.1016/j.micinf.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 39.Kiepiela P, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13(1):46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 40.Macnamara A, et al. HLA class I binding of HBZ determines outcome in HTLV-1 infection. PLoS Pathog. 2010;6(9):e1001117. doi: 10.1371/journal.ppat.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaturvedi U, Nagar R, Shrivastava R. Dengue and dengue haemorrhagic fever: Implications of host genetics. FEMS Immunol Med Microbiol. 2006;47(2):155–166. doi: 10.1111/j.1574-695X.2006.00058.x. [DOI] [PubMed] [Google Scholar]
- 42.Moore CB, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296(5572):1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 43.Vider-Shalit T, Raffaeli S, Louzoun Y. Virus-epitope vaccine design: Informatic matching the HLA-I polymorphism to the virus genome. Mol Immunol. 2007;44(6):1253–1261. doi: 10.1016/j.molimm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharya T, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315(5818):1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 45.Miura T, et al. HLA-associated viral mutations are common in human immunodeficiency virus type 1 elite controllers. J Virol. 2009;83(7):3407–3412. doi: 10.1128/JVI.02459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miura T, et al. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J Virol. 2009;83(6):2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harari A, et al. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc Natl Acad Sci USA. 2007;104(41):16233–16238. doi: 10.1073/pnas.0707570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuji S, et al. HLA-A*24-B*07-DRB1*01 haplotype implicated with genetic disposition of peak bone mass in healthy young Japanese women. Hum Immunol. 1998;59(4):243–249. doi: 10.1016/s0198-8859(98)00009-3. [DOI] [PubMed] [Google Scholar]
- 49.Qu H-Q, Polychronakos C. The effect of the MHC locus on autoantibodies in type 1 diabetes. J Med Genet. 2009;46(7):469–471. doi: 10.1136/jmg.2009.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karan MA, Tascioglu NE, Ozturk AO, Palanduz S, Carin M. The role of HLA antigens in chronic hepatitis B virus infection. J Pak Med Assoc. 2002;52(6):253–256. [PubMed] [Google Scholar]
- 51.Nakajima S, Kobayashi S, Nohara M, Sato S. HLA antigen and susceptibility to leprosy. Int J Lepr Other Mycobact Dis. 1977;45(3):273–277. [PubMed] [Google Scholar]
- 52.Dawood FS, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis. 2012;12(9):687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 53.Rawangkhan A, et al. Comparison of neuraminidase activity of influenza A virus subtype H5N1 and H1N1 using reverse genetics virus. Southeast Asian J Trop Med Public Health. 2010;41(3):562–569. [PubMed] [Google Scholar]
- 54.Ghedin E, et al. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437(7062):1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.