Abstract
Break-induced replication (BIR) refers to recombination-dependent DNA synthesis initiated from one end of a DNA double-strand break and can extend for more than 100 kb. BIR initiates by Rad51-catalyzed strand invasion, but the mechanism for DNA synthesis is not known. Here, we used BrdU incorporation to track DNA synthesis during BIR and found that the newly synthesized strands segregate with the broken chromosome, indicative of a conservative mode of DNA synthesis. Furthermore, we show the frequency of BIR is reduced and product formation is progressively delayed when the donor is placed at an increasing distance from the telomere, consistent with replication by a migrating D-loop from the site of initiation to the telomere.
Keywords: translocation, Pol32, cell cycle
Homologous recombination (HR) is an important mechanism to repair DNA double-strand breaks (DSBs) that occur spontaneously during cell growth or following exposure to DNA damaging agents (1). HR relies on the presence of a homologous duplex to template repair of the broken chromosome and is generally considered to be an error-free mechanism. However, HR can lead to a local loss of heterozygosity (LOH) if the recombining sequences are not identical, and to extensive LOH if repair is associated with a crossover between chromosome homologs. Furthermore, if a repeated sequence at an ectopic site is used as the sequence donor and recombination is associated with crossing over, translocations can occur (2, 3). When both ends of the DSB share homology with the donor duplex sequence, HR proceeds by a two-ended mechanism, such as DSB repair or synthesis-dependent strand-annealing (SDSA) (4–6). However, if coordination of the two ends is not maintained or only one end of the break is available, such as at a critically short telomere, repair can occur by break-induced replication (BIR) (7). In this case, following strand invasion replication occurs to the end of the chromosome to generate a stable repaired product (8, 9). This process can cause very long gene conversion tracts and significant LOH, and nonreciprocal translocation if invasion occurs at a dispersed repeated sequence (10).
The repair of DSBs by HR requires the 5′–3′ nucleolytic degradation of the DNA ends to form invasive 3′ single-stranded DNA (ssDNA) tails (1). The 3′ ssDNA tail created by end resection is bound by Rad51 to form a nucleoprotein filament that searches for homology and promotes pairing between the ssDNA bound by Rad51 and complementary sequence in the donor duplex forming a D-loop intermediate. The invading 3′ end is then used to prime DNA synthesis templated by the donor sequence. If the invading 3′-tail is displaced by helicases and anneals with the other end of break, repair by SDSA results in noncrossover products. If the second end of the break is captured by the D-loop, a double Holliday junction can be generated after DNA repair synthesis and ligation. Double Holliday junctions can either be dissolved by the Sgs1 helicase and Top3 topoisomerase to form noncrossover products or resolved by endonucleases to generate crossovers or noncrossovers (2, 11, 12). To complete two-ended repair, two short tracts of leading strand DNA synthesis are required, whereas BIR requires extensive leading and lagging strand DNA synthesis (13, 14).
The mechanisms that limit the extent of DNA synthesis during gene conversion repair, but promote extensive DNA synthesis in BIR are poorly understood. Previous studies have shown that BIR can involve multiple rounds of strand invasion and dissociation, suggesting repair might occur by progressive extension of the invading 3′ end until the chromosome terminus is reached, with lagging strand synthesis initiating on the nascent displaced strand (Fig. 1) (10, 15, 16). Nonetheless, the requirement for the replicative minichromosome maintenance helicase and lagging strand synthesis components to detect early BIR intermediates suggests BIR might involve assembly of a replication fork for semiconservative synthesis, as in normal S-phase replication (13, 17). Here, we developed a chromosomal system to monitor BIR by genetic and physical methods. We show BIR is an efficient process if the donor sequence is located close to the telomere, but the frequency is reduced and product formation is progressively delayed when the donor is placed at an increasing distance from the telomere. By using BrdU incorporation to track DNA synthesis during BIR, we found that the newly synthesized strands segregate with the broken chromosome. These observations are most consistent with a conservative mode of DNA synthesis via a migrating D-loop.
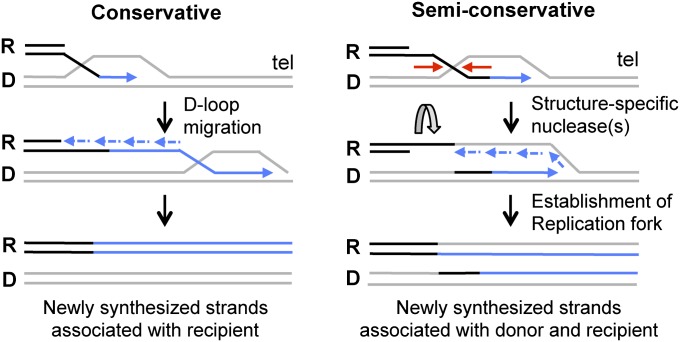
Fig. 1.
Model for conservative or semiconservative DNA synthesis during BIR. If the Rad51-catalyzed D-loop migrates to the chromosome end (tel) and lagging-strand synthesis initiates on the displaced nascent strand, both newly synthesized strands (shown in blue) will segregate with the recipient (R) chromosome. Cleavage of the strand invasion intermediate by a structure-selective nuclease and establishment of a replication fork is predicted to result in semiconservative synthesis detected by segregation of the newly synthesized strands to donor (D) and recipient chromosomes.
Results
Efficiency and Kinetics of BIR Depend on the Distance from the Site of Strand Invasion to the Telomere.
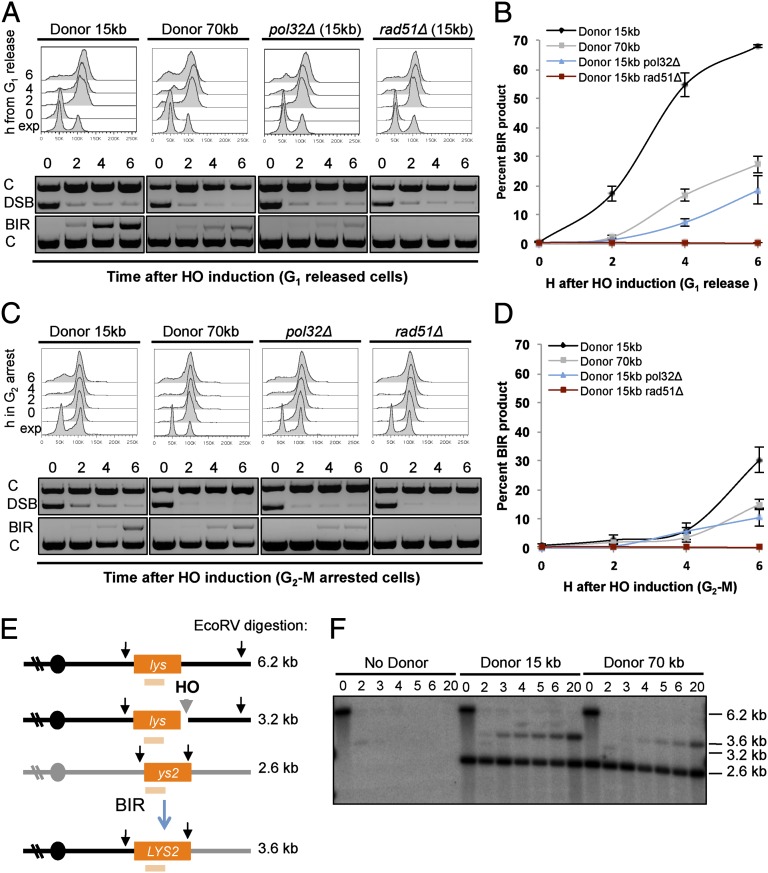
BIR events can best be detected by creating a DSB where just one of the two ends can undergo homology-dependent strand invasion (9, 13, 18). To model this reaction we designed a chromosomal substrate by inserting a recipient cassette with a 3′ truncated lys2 gene (lys), a 36-bp homothallic switching (HO) endonuclease cut site (HO cs), and KanMX to confer resistance to geneticin (G418), 34 kb from the left telomere of chromosome (Ch) V. There are no essential genes distal to the site of the insertion (13, 19). A donor cassette comprised of TRP1 and a 5′ truncation of lys2 (ys2) was inserted 15 kb from the telomere of Ch XI (Fig. 2A). The truncated lys2 genes share 2.1-kb homology. These donor and recipient cassettes were incorporated into a haploid strain capable of expressing the HO endonuclease from a galactose-regulated promoter, with the MATa-inc allele to prevent cleavage at the endogenous HO cs, and that constitutively expresses the Herpes Simplex Virus thymidine kinase gene to enable incorporation of the thymidine analog BrdU into DNA (20). An additional control strain was constructed with only the recipient cassette (no donor). After DSB formation, the lys sequence should invade the donor cassette and copy to the end of Ch XI, forming a Lys+ nonreciprocal translocation product. The other side of the Ch V DSB lacks homology to sequences in the yeast genome and is degraded, resulting in loss of the KanMX gene.
Fig. 2.
BIR efficiency is determined by the length of DNA to be synthesized. (A) Schematic showing the locations of the recipient and donor cassettes on Ch V and XI, respectively, and the PCR primer pairs used to detect DSB formation by HO endonuclease (D1, D2), BIR products (P1, P2), and control locus (C1, C2). (B) BIR efficiency determined by CFU Lys+ YPGal/CFU YPGlu for each of the indicated strains from three independent trials; error bars show SD. (C) Schematic of the donor chromosomes used to measure BIR efficiency showing the distance from the donor homology to the telomere. The TRP1-ys2 cassette and primers used to detect BIR are the same for all of the donor chromosomes.
The frequency of BIR was determined by the plating efficiency of cells on galactose-containing medium (YPGal, HO on) relative to medium with glucose (YPD, HO off). The plating efficiency of cells with the 15-kb donor cassette was high (∼90%), and >99% of the colonies formed on YPGal plates were Lys+ and sensitive to G418, as predicted from formation of the BIR product (Fig. 2 A and B). In contrast, the plating efficiency of the no donor strain was <1% and Lys+ colonies were not recovered. No Lys+ products were formed from a rad51Δ derivative of the 15 kb donor strain, and the frequency of BIR was reduced by 18-fold in the pol32Δ mutant, consistent with previous studies (Fig. 2B) (8, 13, 21, 22).
We constructed a strain with the donor cassette inserted 70 kb from the Ch XI telomere with the expectation this would provide a more sensitive system for BrdU incorporation because of the additional amount of DNA to be synthesized (Fig. 2C). However, the BIR frequency of the 70-kb donor strain (∼70%) was significantly lower (P < 0.03) than the 15-kb donor (Fig. 2B). The difference in the BIR frequency between the 15- and 70-kb donor strains could be because of the amount of DNA synthesis required to reach the telomere, or because of different sequence contexts of the donor cassette. To determine whether the extent of synthesis is correlated with BIR frequency, the donor cassette was inserted on different chromosomes at varying distances from the telomere (Fig. 2C). Insertion of the donor 20 kb from the Ch VI telomere yielded a similar BIR frequency to the 15-kb Ch XI donor, the 60-kb Ch I donor behaved similarly to the 70-kb Ch XI donor, and the lowest BIR frequency was obtained for the Ch I 128-kb donor (Fig. 2B). These results support the notion that BIR frequency is length-dependent, as suggested previously (13).
BIR Is Optimal in Cells Released from a G1 Arrest.
To identify optimal conditions for BIR, we arrested cells in G1 with α-factor and released them into S-phase at the time of HO induction or measured BIR in G2-M arrested cells. BIR was monitored physically using PCR primers unique to Ch V and Ch XI (P1 and P2) to detect the strand invasion product, which was normalized to the product obtained from control (C) primers that amplify sequences 66-kb centromere proximal to the DSB (Fig. 2A). The efficiency of HO cutting was determined using primers flanking the HO cs (D1 and D2). Cells released into S-phase exhibited BIR products 2–4 h after HO induction, whereas cells arrested in G2-M showed a delay and reduced yield of BIR products (Fig. 3 A and C). Consistent with the genetic assay, BIR products were not detected in the rad51Δ mutant, and the yield of BIR products was considerably higher for the 15-kb donor compared with the 70-kb donor (Fig. 3 B and D). The frequency of BIR determined by PCR was lower at the 6-h time point than detected by the genetic assay for the 15- and 70-kb donor strains, but showed the same trend. However, the yield of PCR product from the pol32Δ mutant was higher than predicted from the genetic assay and could correspond to aborted products. The reduced efficiency of BIR observed for G2-M arrested cells could be a result of lower levels of replication proteins and nucleotides at this phase of the cell cycle. For G1-released cells, there could be higher levels of these factors present when BIR initiates, even though cells are arrested in G2 by the time BIR products are detected. Alternatively, if HO cuts the two sister chromatids asynchronously and the cut sister engages in unproductive repair with the uncut chromatid, this might result in delayed repair by BIR (23).
Fig. 3.
BIR product formation is delayed for the 70-kb donor strain compared with the 15-kb donor strain and occurs with highest efficiency in G1 released cells. (A) FACS profiles and PCR to detect DSB (D primers) and BIR product (P primers) from cells of the indicated strains that were arrested in G1 and released at the time of HO induction. (B) Kinetics of BIR product formation in G1-released cells determined by the ratio of BIR to control (C) PCR products from three trials; error bars show the mean ± SD. (C) FACS profiles and PCR to detect DSB and BIR product from cells that were arrested in G2-M at the time of HO induction. (D) Kinetics of BIR product formation in G2-M arrested cells; error bars show the mean from three trials ± SD. (E) Schematic showing sizes of digestion products for the recipient chromosome before and after HO cutting, the donor chromosome and BIR product. The pale orange bar indicates the position of the hybridization probe and the vertical arrows show the location of EcoRV sites. (F) Southern blot of EcoRV-digested genomic DNA of the indicated strains before and after HO induction.
In principle, amplification of genomic DNA with the P primers should detect extension of the invading strand by only 278 nucleotides; thus, we expected the initiation of BIR to occur with the same kinetics for the 15- and 70-kb donor strains because the recipient and donor cassettes are the same. However, the initial detection of the BIR PCR product for the 70-kb donor strain was delayed by 1–2 h compared with the 15-kb donor (Fig. 3A). To verify the PCR results, genomic DNA from the 15- and 70-kb donor strains that had been released from G1 arrest at the time of HO induction was digested with EcoRV to detect BIR products by Southern blot hybridization (Fig. 3 E and F). To detect BIR products by restriction digestion, both strands must be synthesized. The kinetics of product formation mirrored the results with the PCR assay, suggesting that the BIR intermediate can only be amplified by PCR once it has become double-stranded.
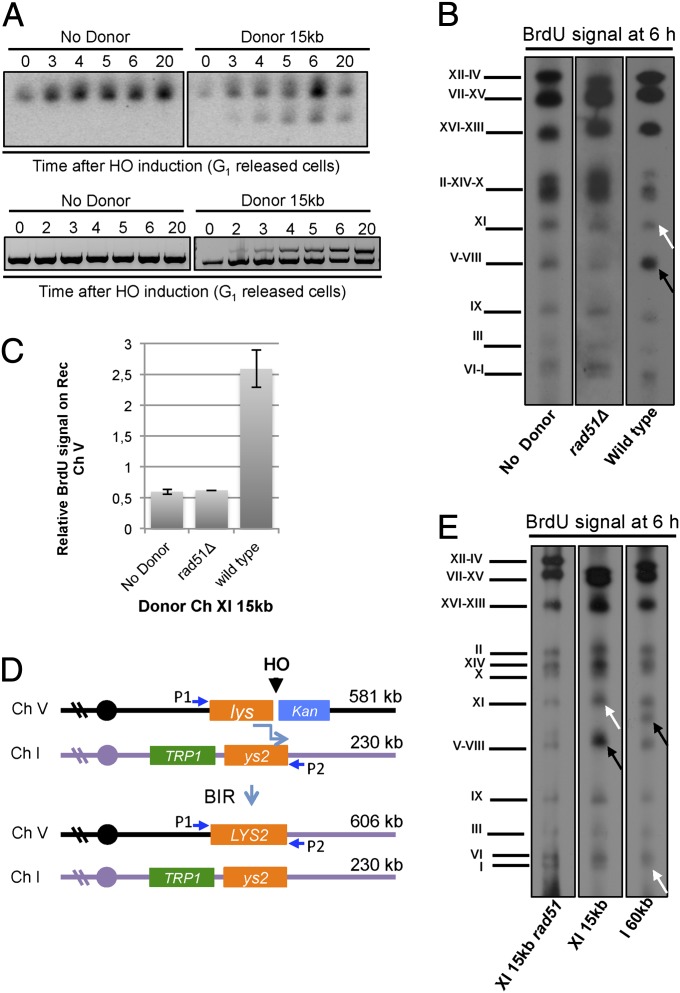
To confirm formation of a full-length nonreciprocal translocation product by BIR, intact chromosomes from cells with the 15-kb donor were separated by pulsed-field gel electrophoresis (PFGE) for Southern blot analysis using a probe to a sequence 9.8 kb from the telomere of Ch XI. Only the donor Ch XI (667 kb) and BIR-repaired Ch V (561 kb) are able to hybridize with the probe (Figs. 2A and 4A). The repaired product was detected at 3–4 h for the G1-released cells and at 6 h for the G2-M arrested cells (Fig. 4A and Fig. S1). Repaired products were not detected from the donorless strain.
Fig. 4.
Newly synthesized DNA strands are associated with the recipient chromosome. (A) Detection of the BIR translocation product in G1-released cells by PFGE and Southern blot hybridization using a probe specific to Ch XI. Lower panel shows the BIR product detected by PCR for the same samples. (B) Western blot of the pulsed field gel of the indicated strains 6 h after HO induction shows greater BrdU incorporation into the recipient chromosome compared with the donor chromosome. (C) Quantification of the relative BrdU signal (Ch V–VIII/Ch XI) from three independent trials; error bars show SD. (D) Schematic showing the locations of the recipient and donor cassettes on Ch V and I, respectively, and the PCR primer pair used to detect BIR products (P1, P2). BIR generates a novel-sized chromosome of 606 kb that can be resolved from the donor and recipient chromosomes by PFGE. (E) BrdU was added to the culture 2 h after HO induction and used to track incorporation into recipient or donor chromosomes by PFGE. The black arrows indicate the positions of the recipient chromosomes and white arrows mark the positions of the donor chromosomes.
Preferential Incorporation of BrdU into the Recipient Chromosome.
Because BIR was optimal for cells released from G1, these conditions were used to monitor BrdU incorporation for the Ch XI 15-kb donor strain. HO was induced at the same time cells were released from G1 and BrdU was added to the culture 2 h after HO induction at the time bulk DNA synthesis had completed and BIR synthesis had initiated (Fig. S2). The DSB maintains a G2-M arrest for 4–6 h after HO induction and cells progress to G1 after repair (Fig. 3A). Because early-repairing cells entering the next cell cycle would incorporate BrdU during normal DNA synthesis, α-factor was added to the culture 2 h after BrdU to arrest cells at the subsequent G1 phase. Semiconservative DNA synthesis would be expected to result in BrdU incorporation in both donor and recipient chromosomes, whereas by a conservative mode of DNA synthesis BrdU would be associated with only the recipient chromosome. Following PFGE to resolve intact chromosomes, we found that BrdU was incorporated preferentially into the recipient chromosome (Ch V and Ch VIII comigrate by PFGE) (Fig. 4B). To control for background incorporation into Ch V and XI because of some late-replicating cells in the population, the rad51Δ and no donor strains were used. The ratio of BrdU incorporation for Ch V-VIII/Ch XI was ∼0.5 for the control strains and 2.5 for the experimental strain, the fivefold increase in BrdU incorporation into the recipient chromosome is most consistent with a conservative mode of DNA synthesis (Fig. 4C). When the donor cassette was located 60 kb from the telomere of Ch I, we also detected preferential incorporation of BrdU into the recipient chromosome, indicating that conservative synthesis is a general feature of BIR and not restricted to subtelomeric regions (Fig. 4 D and E). The detection of BIR products by PCR was delayed for the 60-kb donor strain compared with the 15-kb donor, and final yield lower, consistent with data for the 70-kb donor strain (Fig. S3).
Discussion
We show BIR can be an efficient mechanism of DSB repair when it initiates in S-phase cells and the extent of synthesis is less than 20 kb. The delayed repair noted in a previous study might be because of the location of the donor sequence ∼100 kb from the telomere (24) or use of G2-M arrested cells (13). DNA is synthesized by a conservative mechanism during BIR, consistent with formation of a migrating D-loop and lagging strand synthesis initiating on the nascent strand, similar to a classic mechanism envisioned for bacteriophage T4 recombination-initiated DNA synthesis (25). The need for the D-loop to migrate to the chromosome terminus could explain the decreased BIR efficiency when the donor cassette is placed at increasing distance from the telomere. Chromatin remodeling is likely to be required for the D-loop to traverse long distances and initiation of lagging strand synthesis could be limiting to complete BIR synthesis. We cannot rule out the possibility that sequences within 30 kb from the telomere have greater mobility increasing the frequency of pairing; however, this does not explain the decreased BIR frequency for the 128-kb donor compared with the 70-kb donor.
Gene conversion repair also occurs by a conservative mode of synthesis, suggesting BIR bears closer resemblance to other recombination events than to normal DNA synthesis (26, 27). DNA synthesis during BIR and gene conversion is more mutagenic than normal S-phase synthesis, and the mutational spectrum is similar for both processes, consistent with a common mechanism. Any errors generated during BIR leading-strand synthesis would become fixed by second-strand synthesis with no opportunity for mismatch correction because of the absence of a heteroduplex intermediate. BIR is a rare outcome for repair of a two-ended DSB (11, 28, 29), presumably because of the possibility of increased mutagenesis over tens of kilobases, in addition to the potential for extensive LOH or nonreciprocal translocation. Mechanisms to ensure repair of two-ended DSBs by gene conversion could be defective in aging or cancer cells, contributing to genome instability (30).
For gene conversion repair, the nascent strands are primed from the two 3′ ends. However, during BIR there is only one end to prime the nascent leading strand and the second strand must be generated by a different mechanism. It is currently unclear how lagging-strand synthesis initiates on the nascent ssDNA extruded from the tail end of the D-loop during BIR. If priming were a stochastic rate-limiting step it could explain the length-dependent delay in the detection of BIR products. Repriming of DNA polymerase can occur at random sites downstream of a lesion on the leading-strand template in an Escherichia coli reconstituted system (31), suggesting a similar mechanism might operate during BIR with only a few priming events on the nascent displaced strand. If the nascent strand extruding from the D-loop remains ssDNA for some time, it might be susceptible to degradation leading to a reduction in repair efficiency, particularly for longer templates. Depletion of replication protein A (RPA) from cells results in decreased stability of ssDNA because of formation of secondary structure and cleavage by endonucleases (32). The three genes encoding RPA are periodically transcribed with a peak at the G1/S boundary (33); thus, the greater abundance of RPA in S phase could contribute to the increased efficiency of BIR when it initiates in S compared with G2-arrested cells.
Previous studies showed BIR could occur by several cycles of displacement of the extended invading strand from the D-loop and reinvasion (10, 16). The observation of conservative DNA synthesis during BIR is in agreement with these studies. The displaced extended end could be subject to degradation, resulting in reduced BIR frequency, particularly in assays that use donor and recipient cassettes with limited homology (13, 24). The assay described by the Deem et al. uses a Ch III disome with extensive homology at the centromere proximal side of the HO-induced DSB and BIR occurs with greater efficiency than limited-homology assays, even though the donor is >100 kb from the telomere (21). A URA3 marker inserted 3-kb upstream of the HO-induced DSB on the recipient chromosome was frequently lost during BIR in agreement with the proposal that degradation of the 3′ extended invading strand may limit BIR (9). The frequency of BIR could be increased in cells that overexpress Rad51, consistent with the need to protect the displaced ssDNA from nucleases and to promote reinvasion (34).
Materials and Methods
Construction of Yeast Strains.
All strains are derivative of W303 (leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15) (Table S1). To generate the BIR assay system, the native LYS2 locus (Ch II 474,781–469,689) was first replaced with a NatMX4 cassette in a strain containing MATa-inc, GAL-HO integrated at the ade3 locus and URA3::GPD-TK7 (20). The recipient and donor cassettes that had been amplified by PCR from engineered lys2 recipient and donor plasmids were then integrated at the desired chromosome locations. The PCR products were designed with 40-bp flanking homology to direct homologous integration. The recipient plasmid contains an XmaI-BglII 36-bp synthetic HO cut site (HOcs) (AGTTTCAGCTTTCCGCAACAGTATAATTTTATAAAC) cloned upstream of the KanMX6 marker in a pfA6aKanMX6 plasmid (35). A fragment of the LYS2 gene lacking the 3′ end was then cloned upstream of HOcs-KanMX6. The donor plasmid contains a fragment of the LYS2 gene lacking 5′ sequences inserted downstream of the TRP1 marker in pRS404 (36). All targeted chromosomes have the recipient and donor lys2 fragments with the same polarity on the left chromosome arms.
Determination of BIR Frequency.
Cells were grown to exponential phase in 1% yeast extract, 2% peptone, 2% raffinose (YPR), and then plated on rich medium (YP) containing 2% glucose or 2% galactose (wt/vol). Colonies were counted after 3 d and were then replica plated onto synthetic complete (SC) medium lacking lysine or YPD containing geneticin. Cell viability after HO induction was determined by dividing the number of CFUs on YPG by that on YPD. The percentage of cells repairing by BIR was determined by dividing the number of Lys+ by the number of YPG CFU, and then normalized to the number of CFU on YPD. Approximately 1% of cells remained Lys− geneticinr following growth on YPG because of repair by nonhomologous end-joining. BIR frequencies were determined at least three times for each strain.
Cell Synchronization and Flow Cytometry.
For G1 release experiments, cells were presynchronized in G1 with α-factor (0.5 μg/mL) and then treated with pronase-E (0.1 mg/mL) for 5 min at room temperature, washed twice with YP, and then released in fresh YPR + 2% galactose medium. Cells were arrested in G2/M with 15 μg/mL nocodazole. DNA content was analyzed using a LSR Fortessa (Becton-Dickinson) cell analyzer and FlowJo software.
Detection of BIR Products by PCR and Southern Blot Hybridization.
For PCR analysis, 25 ng of genomic DNA was amplified for 25 cycles (determined to be within the linear range). PCR reactions were subject to electrophoresis through 1% agarose and quantified using Bio-Rad Quantity One software. The percentage of BIR product formed (primers P1 and P2) was determined by dividing the BIR product signal to that amplified from an independent locus 66-kb centromere proximal to the DSB on chromosome V (primers C1 and C2) obtained in the same reaction and then normalized to the ratio from a Lys+ colony (100% BIR) and plotted against time. At least three PCR reactions were performed for each strain. DSB formation was monitored using primers D1 and D2. The percentage of DSB formation was obtained by dividing the DSB signal to that amplified from the control primers obtained in the same reaction and then normalized to the ratio obtained before galactose addition. Genomic DNA was digested with EcoRV for Southern blot hybridization.
Analysis of BIR by PFGE.
Samples for PFGE were obtained from 30-mL aliquots of cultures. Plugs containing lysed cells were prepared as described previously (37). Chromosomes were separated by electrophoresis through 1% agarose at 6 V in 0.5× Tris-borate-EDTA at 14 °C for 24 h (initial time = 45 s, final time = 95 s) using a CHEF-DR II Pulsed-Field Electrophoresis system (Bio-Rad). Gels were stained with SYBR gold (Molecular Probes), and the chromosomes were then transferred to nylon membranes. To assay for the completion of BIR, membranes were probed with a 1.3-kb PCR product generated by amplification of sequences 9.8–11.1 kb from the left telomere of Ch XI.
Western Blot to Detect BrdU Incorporation.
BrdU incorporation was detected by overnight incubation of the membrane with monoclonal antibodies sc-56255 (2B1; Sigma) diluted 1:100 in 1× PBS1; 0.1% X-tween 20; 5% dried milk. The BrdU intensity was calculated by comparing the ratio of the repaired recipient Ch V signal to the corresponding donor Ch XI signal, using Bio-Rad Quantity One software.
Supplementary Material
Acknowledgments
We thank A. Aguilera and P. Pasero for generous gifts of yeast strains, and W.K. Holloman and members of the L.S.S. laboratory for comments on the manuscript. This study was supported by a Grant GM094386 from the National Institutes of Health. R.A.D. is supported by an International Fellowship in Cancer Research cofunded by Associazione Italiana per la Ricerca sul Cancro and Marie Curie Actions.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309800110/-/DCSupplemental.
References
- 1.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66(4):630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115(4):401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robert T, Dervins D, Fabre F, Gangloff S. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 2006;25(12):2837–2846. doi: 10.1038/sj.emboj.7601158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson DO, Holloman WK. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93(11):5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 6.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14(3):1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llorente B, Smith CE, Symington LS. Break-induced replication: What is it and what is it for? Cell Cycle. 2008;7(7):859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 8.Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24(6):2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25(3):933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447(7140):102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 11.Ho CK, Mazón G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell. 2010;40(6):988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 13.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448(7155):820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, et al. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24(16):6891–6899. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pardo B, Aguilera A. Complex chromosomal rearrangements mediated by break-induced replication involve structure-selective endonucleases. PLoS Genet. 2012;8(9):e1002979. doi: 10.1371/journal.pgen.1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz JF, Gómez-González B, Aguilera A. Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol Cell Biol. 2009;29(20):5441–5454. doi: 10.1128/MCB.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lydeard JR, et al. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24(11):1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosco G, Haber JE. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics. 1998;150(3):1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23(1):81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 20.Lengronne A, Pasero P, Bensimon A, Schwob E. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 2001;29(7):1433–1442. doi: 10.1093/nar/29.7.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deem A, et al. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics. 2008;179(4):1845–1860. doi: 10.1534/genetics.108.087940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CE, Lam AF, Symington LS. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol. 2009;29(6):1432–1441. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazón G, Lam AF, Ho CK, Kupiec M, Symington LS. The Rad1-Rad10 nuclease promotes chromosome translocations between dispersed repeats. Nat Struct Mol Biol. 2012;19(9):964–971. doi: 10.1038/nsmb.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain S, et al. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23(3):291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Formosa T, Alberts BM. DNA synthesis dependent on genetic recombination: Characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47(5):793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 26.Arcangioli B. Fate of mat1 DNA strands during mating-type switching in fission yeast. EMBO Rep. 2000;1(2):145–150. doi: 10.1093/embo-reports/kvd023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ira G, Satory D, Haber JE. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol Cell Biol. 2006;26(24):9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93(14):7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickoloff JA, Sweetser DB, Clikeman JA, Khalsa GJ, Wheeler SL. Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics. 1999;153(2):665–679. doi: 10.1093/genetics/153.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurray MA, Gottschling DE. An age-induced switch to a hyper-recombinational state. Science. 2003;301(5641):1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 31.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439(7076):557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell. 2013;50(4):589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5(9):1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 34.Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS Genet. 2010;6(5):e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 36.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hyppa RW, Smith GR. Using Schizosaccharomyces pombe meiosis to analyze DNA recombination intermediates. Methods Mol Biol. 2009;557:235–252. doi: 10.1007/978-1-59745-527-5_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.