Abstract
Nucleic acid testing for infectious diseases at the point of care is beginning to enter clinical practice in developed and developing countries; especially for applications requiring fast turnaround times, and in settings where a centralized laboratory approach faces limitations. Current systems for clinical diagnostic applications are mainly PCR-based, can only be used in hospitals, and are still relatively complex and expensive. Integrating sample preparation with nucleic acid amplification and detection in a cost-effective, robust, and user-friendly format remains challenging. This review describes recent technical advances that might be able to address these limitations, with a focus on isothermal nucleic acid amplification methods. It briefly discusses selected applications related to the diagnosis and management of tuberculosis, HIV, and perinatal and nosocomial infections.
Introduction
Nucleic acid testing (NAT) for infectious diseases is almost exclusively performed in centralized laboratories using high-end instrumentation and skilled personnel. However, point of care (POC) testing, which is defined as near-patient testing in a hospital, doctor’s office, clinic, or home, offers advantages when a rapid answer is required [1], or when suitable facilities or logistics chains are unavailable [2,3]. Compared to equivalent laboratory methods, POC testing is usually more expensive per test, although a true cost comparison needs to consider central laboratory overhead costs, better patient management and disease containment [4]. Under the United States Clinical Laboratory Improvement Amendments (CLIA), clinical diagnostic tests are categorized as high complexity, moderate complexity, or waived [5]. Most POC settings only allow the use of waived tests that are ‘so simple and accurate as to render the likelihood of erroneous results by the user negligible’. Currently, there are no waived POC NAT systems. High and moderate complexity tests are distinguished based on required operator training and experience, complexity of operational steps, system troubleshooting, and equipment maintenance. High complexity tests are limited to central laboratories, whereas moderate complexity tests can be performed near patients in a hospital. The Cepheid GeneXpert, a PCR-based system that performs fully automated ‘sample-in/answer-out’ testing is currently the only moderate complexity NAT platform approved in the USA.
POC NAT in developed countries must address patient needs that are insufficiently met by existing centralized laboratory methods, which includes rapid screening for perinatal and nosocomial infections. POC NAT in the developing world focuses on diagnosis and management of endemic infectious diseases, such as tuberculosis (TB) and HIV infection [2,3,6]. Developing countries have limited financial resources and cannot implement well-established, yet complex commercially available NAT systems or laboratory-developed tests through a network of centralized laboratories [7,8]. POC NAT can provide access to much-needed diagnostic methods in low-resource, high disease-burden areas, but appropriate devices should be affordable, robust and easy to use by minimally trained personnel, with stable, ready-to-use reagents, simple, maintenance-free instrumentation, and clear, actionable results, in addition to being suitably sensitive and specific. The World Health Organization (WHO) recently endorsed the Cepheid GeneXpert to facilitate diagnosis of multi-drug-resistant TB in high-burden, low-resource settings (http://www.who.int/mediacentre/news/releases/2010/tb_test_20101208/en/index.html). However, this system is still relatively expensive and complex. It most likely will penetrate into district hospitals and microscopy centers [2,9], but will not be appropriate for POC settings such as rural clinics with sub-optimal infrastructure [2]. POC NAT is still in the emerging stages, but technological advances are beginning to address these challenges.
Current NAT technologies
NAT involves three main steps: sample preparation, amplification, and detection. Examples of fully or partially integrated platforms that are commercially available or close to market are listed in Table 1. Integration of all three steps into a POC-compatible format has been demonstrated in the GeneXpert [10], the IQuum LIAT analyzer [11], and other real-time PCR-based benchtop systems (Table 1). Size, cost and complexity can be reduced by using isothermal amplification with real-time detection, as implemented in Lumora’s Bioluminescent Assay in Real-Time (BART) reader and in the TwistDX Twista. NAT with minimal instrumentation is facilitated by simple endpoint detection schemes, such as nucleic acid lateral flow (NALF), exemplified in the BioHelix BESt Cassette [12]. These examples also illustrate the spectrum of advances in systems integration: the GeneXpert and LIAT analyzer enable complete sample-in/answer-out testing; the BART and Twista perform amplification and detection; and the BESt Cassette performs detection only.
Table 1.
Example POC NAT platforms that are commercially available or close to market
| Platform | Manufacturer | Sample prep included? | Amplification | Detection | Time to result (min)a | Website |
|---|---|---|---|---|---|---|
| GeneXpert | Cepheid | Y | PCR | RTF | <120 | www.cepheid.com |
| Liat Analyzer | IQuum | Y | PCR | RTF | <60 | www.iquum.com |
| MDx | Biocartis | Y | PCR | RTF | Unknown | www.biocartis.com |
| FL/ML | Enigma | Y | PCR | RTF | <45 | www.enigmadiagnostics.com |
| FilmArray | Idaho technologies | Y | PCR | RTF | 60 | www.idahotech.com |
| Razor | Idaho technologies | N | PCR | RTF | <60 | www.idahotech.com |
| R.A.P.I.D. | Idaho technologies | N | PCR | RTF | <30 | www.idahotech.com |
| LA-200 | Eiken | N | Isothermal (LAMP) | RTT | < 60 | www.eiken.co.jp |
| Twista | TwistDX | N | Isothermal (RPA) | RTF | < 20 | www.twistdx.co.uk |
| BART | Lumora | N | Isothermal (LAMP) | RTB | < 60 | lumora.co.uk/ |
| Genie II | Optigene | N | Isothermal (LAMP) | RTF | < 20 | www.optigene.co.uk |
| SAMBA | Diagnostics for the Real World | N | Isothermal (similar to NASBA) | NALF | > 60 | Not available |
| BESt Cassetteb | BioHelix/ Ustar Biotech | N | Not included, but typically isothermal | NALF | N/A | www.biohelix.com; www.bioustar.com |
Time to result depends upon the particular assay. Longer times may be required for assays with a reverse transcriptase step.
Device sold by BioHelix in the USA; manufactured and sold by Ustar Biotech in China.Abbreviations: RTB real-time bioluminescence; RTF real-time fluorescence; RTT real-time turbidimetry.
Sample preparation
Sample preparation is a bottleneck in NAT, especially for POC applications [13], because it involves lengthy processes that are often manually performed. In clinical laboratories, sample preparation is typically automated on large instruments. For POC NAT, sample preparation starting from the clinical specimen needs to be integrated and coupled with amplification and detection in an inexpensive, automated, miniaturized, closed system format. Microfluidic devices have been developed to automate individual steps or the entire sample preparation process [14,15], but to date, none of the systems have become commercially viable products, possibly because of the challenges related to system complexity, manufacturability, and reproducible device performance. Most infectious disease applications further require extraction and concentration of target nucleic acids from sample input volumes >500 μl to reach a suitably low limit of detection (LOD). This requirement often cannot be accommodated in micro-fluidic devices. The GeneXpert therefore employs a macro-fluidic approach to sample preparation [10] (Figure 1a).
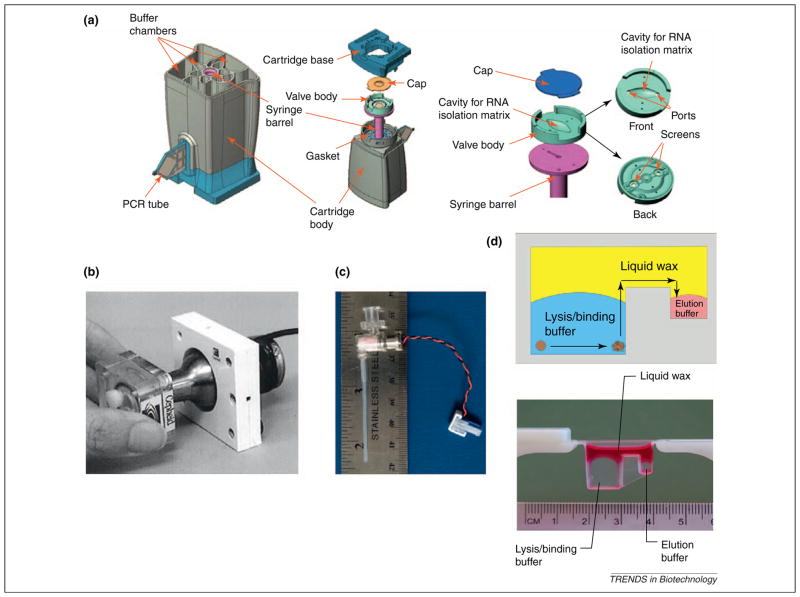
Figure 1.
Selected sample preparation technologies for POC NAT. (a) Detailed schematic of Cepheid’s GeneXpert cartridge reproduced with permission from [10], most of which is devoted to sample preparation and reaction setup. A plunger from the instrument engages with the syringe barrel to draw sample and lysis/binding, wash, and elution buffers through the rotating valve body at the bottom of the cartridge into the cavity that holds beads for NA SPE. Eluted, purified nucleic acids are combined with lyophilized mastermix reagents and transferred into the PCR chamber at the side of the cartridge for amplification with real-time fluorescence-based detection. (b) Miniaturized ultrasonic horn incorporated into the GeneXpert system reproduced with permission from [16], which engages with the bottom of the cartridge to facilitate pathogen lysis. (c) Miniaturized bead blender developed by Claremont BioSolutions [17] to mechanically disrupt lysis-resistant pathogens. (d) “No wash” sample preparation, based on moving nucleic acids bound to magnetic beads from the lysis/binding buffer on the left through a layer of liquid wax into the elution buffer chamber on the right, reproduced with permission from [19].
Lysis of most pathogens can be performed using chemical or enzymatic means. Certain microorganisms, such as Mycobacterium tuberculosis, however, are difficult to lyse. The GeneXpert utilizes a miniaturized sonicator for mechanical cell disruption (Figure 1b) [16], whereas Claremont BioSolutions has developed a miniaturized, battery-operated, bead beating system for mechanical pathogen lysis (Figure 1c) [17]. Following lysis, nucleic acids are typically purified through solid phase extraction (SPE), which involves capturing nucleic acids onto a solid support, followed by wash steps and elution for downstream amplification [15]. Nucleic acid purification is necessary because subsequent target amplification using polymerases can be inhibited by compounds found in clinical samples. Most automated nucleic acid sample preparation systems perform SPE on silica-coated magnetic beads, with lysis/binding buffers that contain chaotropic salts and ethanol, which also can inhibit polymerases. SPE using chitosan-coated beads [18] or other resins, such as the ChargeSwitch method (Invitrogen), do not require chaotropic salts and organic solvents [13]. SPE without wash steps has been demonstrated by moving DNA bound to magnetic beads through a layer of liquid wax (Figure 1d) [19]. Recently, PCR assays that are less-prone to inhibition have been described [20,21], which might require less rigorous sample preparation. Many isothermal technologies appear to be less affected by inhibitory compounds; especially those found in whole blood [22].
Amplification
NAT for infectious disease diagnosis often requires reaching an LOD of ≤100 copies/ml of the pathogen, which, at present, can only be accomplished through polymerase-based amplification. PCR requires thermocycling to mediate DNA melting, followed by primer annealing and extension. Fully integrated macrofluidic [10,11,23] and microfluidic [24,25] devices and many permutations of microfluidic lab-on-a-chip systems [26,27] have been developed for miniaturized PCR.
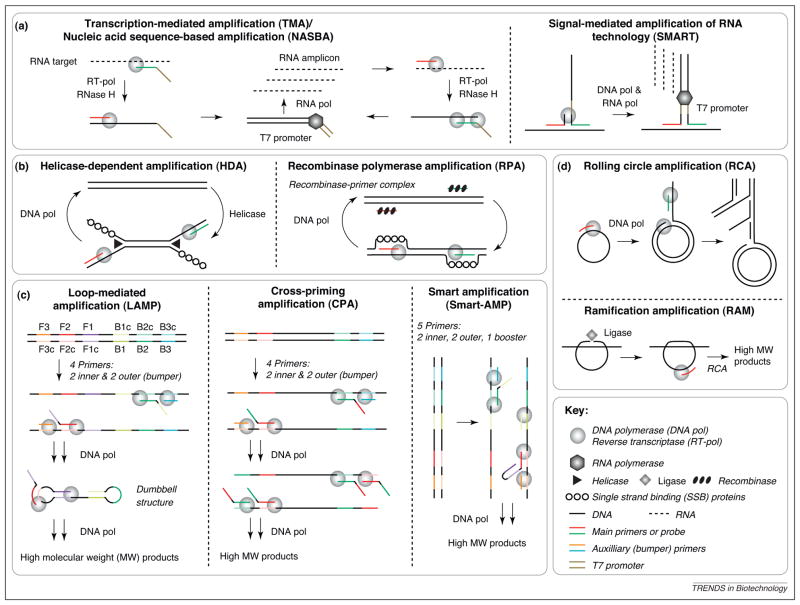
Conversely, isothermal nucleic acid amplification technologies use a single reaction temperature, which translates into less complex and less expensive instrumentation, more suited to applications in low-resource settings. Isothermal amplification can be performed in water baths, using simple resistive heaters, or via exothermic chemical heating [28]. Many isothermal amplification methods have been reported, which can be grouped based on the reaction principle (Figures 2 and 3; Table 2). Current methods include those based on RNA transcription (Figure 2a) [29–31]; DNA replication with enzymatic duplex melting and primer annealing (Figure 2b) [32–35]; strand displacement using polymerases only, with multiple linear primer sets (Figure 2c) [36–39]; and strand displacement from a circular or circularized target (Figure 2d) [40,41]. Many isothermal amplification methods use polymerase extension in conjunction with a single-strand cutting event (Figure 3). [42–50]. Transcription-mediated amplification (TMA; Gen-Probe) [29], nucleic acid sequence-based amplification (NASBA; BioMerieux) [30], and strand displacement amplification (SDA; Becton Dickinson) [42,43] are well-established and implemented in commercial laboratory-based NAT. Some methods, such as loop-mediated amplification (LAMP; Eiken) [36,37], are becoming mainstream, and many others, such as recombinase polymerase amplification (RPA, TwistDx) [34,35], are emerging.
Figure 2.
Overview of isothermal nucleic acid amplification reactions. (a) Methods based on RNA transcription. In TMA (Gen-Probe) [29] and NASBA (BioMerieux) [30], an RNA target is converted to ds cDNA with a promoter region through reverse transcription, followed by RNase H degradation of the original strand and DNA polymerization initiated by a second primer. RNA polymerase (pol) amplification creates products that feed back into the original reaction. TMA and NASBA involve the same reaction scheme, but NASBA requires three enzymes (RT-DNA pol, RNase H, and RNA pol); TMA requires only two enzymes, because the RT-DNA pol has intrinsic RNase H activity. SMART (Cytocell) [31] utilizes a three-way junction with target and extension probes to initiate linear RNA polymerization based amplification (no exponential feedback). (b) Methods based on DNA replication with enzymatic duplex melting/primer annealing. In HDA (Biohelix) [32,33], a helicase enzymatically “melts” dsDNA. In RPA (TwistDx) [34,35], a recombinase–primer complex scans dsDNA for the target site and facilitates primer binding. In both HDA and RPA, single strand binding proteins stabilize the separated strands; the rest of the reaction sequence is analogous to PCR. (c) Methods based on strand displacement using polymerases only from a linear target, through use of sacrificial outer bumper primers. LAMP (Eiken) [36,37] involves six recognition sites on the target DNA; the 5′ overhangs of the inner primers recognize sequences in the amplicon, which leads to the generation of a dumbbell structure. CPA (Ustar) [38] involves four recognition sites on the target DNA, and the inner primers lead to cross-priming after the first round. SMART-AMP (Riken Institute) [39] includes five recognition sites on the target DNA as well as a fifth booster primer; single nucleotide polymorphism discrimination is facilitated through use of the MutS protein. (d) Methods based on strand-displacing polymerization from inherently circular targets in RCA (Molecular Staging) [40], or from padlock probes that are circularized through action of a ligase in RAM (Thorne Diagnostics) [41]. In both cases, branched amplification can be initiated through a second primer.
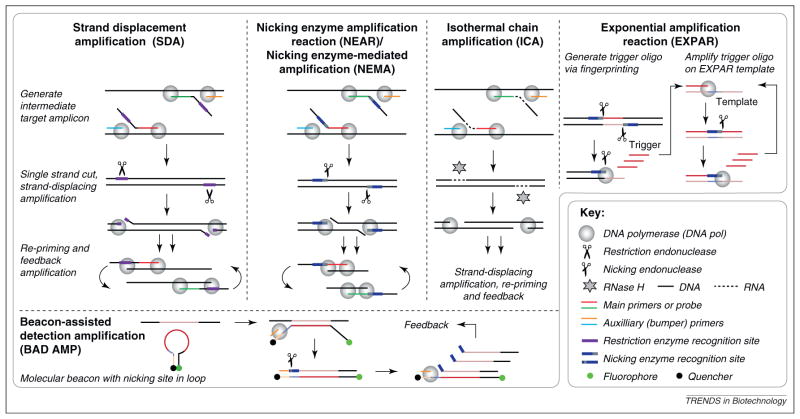
Figure 3.
Isothermal nucleic acid amplification methods that are based on polymerase extension plus a single-strand cutting event. In SDA (Becton Dickinson) [42,43], NEMA (Ustar) [45] and ICA (RapleGene) [46], an intermediate target is generated using strand-displacing amplification via sacrificial outer bumper primers. NEAR (Ionian Technologies) [44] does not use bumper primers and involves shorter amplicons than the other methods. In SDA, phosphothioates are incorporated into the amplicon during polymerization so that a restriction endonuclease only cuts one strand. NEAR and NEMA both use nicking endonucleases that are inherently single strand cutting. In ICA, a single strand cut is facilitated through RNase H and DNA–RNA–DNA chimeric primers. After the single-strand cutting event, amplicons are generated through strand-displacing amplification, for short amplicons further facilitated by thermal denaturation. These amplicons re-prime, and lead to exponential feedback amplification. EXPAR [47] amplifies short trigger oligonucleotides, which can be generated via the so-called “fingerprinting” reaction [48] from adjacent nicking enzyme recognition sites in genomic DNA. This is followed by rapid exponential amplification, mediated by a template sequence that contains two copies of the trigger complement that are separated by the nicking enzyme recognition site complement. BAD-AMP [49] uses a molecular beacon for signal generation and as a template for single-strand nicking and re-priming.
Table 2.
Current isothermal nucleic acid amplification methods
| Assay | Reaction temperature (°C)a | Reaction duration (min)a | Multiplexb | Rapid detection formatsc | Target | Amplification product | Refs. |
|---|---|---|---|---|---|---|---|
| Methods based on RNA transcription | |||||||
| NASBA | 41d | 105 | Y | RTF, NALF | RNA (DNA) | RNA, DNA | [30] |
| TMA | 60d | 140 | Y | RTF | RNA (DNA) | RNA, DNA | [29] |
| SMART | 41d | 180 | N/A | RTF | RNA, DNA | RNA | [31] |
| Methods based on DNA replication with enzymatic duplex melting/primer annealing | |||||||
| HDA | 65 | 75–90 | Y | RTF, NALF | DNAe | DNA | [32,33] |
| RPA | 30–42 | 20 | Y | RTF, NALF | DNAe | DNA | [34,35] |
| Methods based on DNA-polymerase-mediated strand displacement from linear or circular targets | |||||||
| LAMP | 60–65d | 60–90 | N/A | RTF, NALF, RTT, TE | DNAe | DNA | [36,37] |
| CPA | 65 | 60 | N/A | RTF, NALF | DNA | DNA | [38] |
| SMART-AMP | 60 | 45 | N/A | RTF | DNAe | DNA | [39] |
| RCA | 65 | 60 | N/A | RTF | DNAe | DNA | [40] |
| RAM | 63d | 120–180 | N/A | RTF | DNAe | DNA | [41] |
| Methods based on polymerase extension/strand displacement, plus a single strand cutting event | |||||||
| SDA | 37 | 120 | Y | RTF, NALF | DNAe | DNA | [42,43] |
| NEAR | 55 | 10 | Y | RTF, NALF | DNAe | DNA | [44] |
| NEMA | 65 | 30 | N/A | NALF | DNA | DNA | [45] |
| ICA | 60 | 60 | N/A | RTF | DNA | DNA | [46] |
| EXPAR | 55 | 10–20 | Y | RTF, NALF | DNA | DNA | [47,48] |
| BAD AMP | 40 | 40 | N/A | RTF | DNA | DNA | [49] |
| PG-RCA | 60 | 60–120 | N/A | RTF | DNA | DNA | [50] |
Typical incubation temperature and reaction time, variability might exist.
Capability to multiplex, defined as the ability to amplify simultaneously at least two different targets (or one target and an internal control): yes (Y) or data not available (N/A).
Most commonly reported formats only. Other formats might exist, but are not included here.
Initial incubation at a higher temperature is sometimes recommended.
RNA can be the target if a reverse transcriptase step to generate cDNA is first included.
Abbreviations: RTF, real-time fluorescence; RTT, real-time turbidity; TE, turbidity-related endpoint, including fluorescence and colorimetric enhancement.
Comparing the reaction schemes reveals a set of underlying principles to accomplish three basic goals: isothermal generation of ssDNA; exponential amplification; and intermediate target generation. To make DNA polymerase-based amplification isothermal, target ssDNA must be obtained at the same temperature that is required for annealing and extension. Different isothermal amplification methods require reaction temperatures between 30 and 65 °C (Table 2), which is mainly determined by the stability and activity of the polymerase and other enzymes in the reaction. Isothermal generation of ssDNA can be achieved through enzymatic duplex melting and primer annealing (Figure 2b), or by using a strand-displacing polymerase to ‘peel off’ ssDNA through a bumper primer, through extension around a circular target, or by creating a priming site through a single-strand cutting event (Figures 2c,d and 3). Very short amplicons thermally melt off the target, in addition to strand displacement. Isothermal duplex melting can be highly efficient, as reflected by the rapid amplification achievable in RPA and other methods (Table 2).
To obtain sensitive target amplification, amplicons must feed back into the reaction. In NASBA and TMA (Figure 2a), the RNA amplicon is converted into dsDNA with a promoter region. In most DNA polymerase-based methods, the amplicon either re-primes or self-primes. In isothermal methods, exponential amplification occurs asynchronously, as opposed to PCR, in which annealing, extension and duplex melting are synchronized through thermal cycling. Asynchronous reaction progression in some methods facilitates effective isothermal amplification in ≤20 min (Table 2), which is difficult to accomplish via PCR. Other isothermal reactions require ≥1 h to achieve sufficient amplification, which is similar to standard PCR, but not ideal for POC applications. Slower, more controlled reaction progression, however, often translates into less non-specific background amplification, and is better suited for target quantification.
If polymerase-mediated exponential amplification does not directly start from the genomic target, then steps have to be incorporated to generate a suitable intermediate target. In NASBA and TMA, target viral RNA is converted into dsDNA with a promoter region. Detection of RNA through DNA-polymerase-based amplification requires a reverse transcriptase step. Other methods that require the generation of intermediate targets include LAMP, which starts from a dumbbell structure (Figure 2c), and the ramification amplification (RAM) reaction, which requires generation of a circularized probe (Figure 2d) [41]. All methods based on single strand cutting events employ often sophisticated methods to generate the intermediate targets.
Another consideration when choosing an amplification method is the ability to multiplex, or at least co-amplify the target and an internal control (Table 2). Amplification reactions used for clinical diagnostics should incorporate an internal control, co-amplified with the target in the same reaction, to eliminate false negatives due to failed amplification or failed sample preparation.
Detection
Nucleic acid amplification can be analyzed after the reaction (endpoint detection) or while the reaction is progressing (real-time detection). Endpoint detection requires less complex instrumentation and provides simpler outputs for interpretation. Real-time methods integrate amplification with detection, and are superior for quantitative analyte detection with a large dynamic range. In all cases, detection methods are preferable that can differentiate target-specific amplicons from non-specific amplification products and that minimize the risk of carry-over contamination.
Real-time fluorescence detection for PCR or isothermal amplification reactions can be mediated by intercalating dyes, by oligonucleotide probes that are cleaved during the reaction, or by using probes or primers that change conformation upon target amplification. Most current POC NAT platforms are based on PCR amplification with real-time fluorescence detection (Table 1), which requires relatively expensive and complex instrumentation. Simpler and cheaper instruments enable real-time monitoring of isothermal amplification reactions. The Loopamp Realtime Turbidimeter (LA-200; Eiken) monitors LAMP reactions: pyrophosphate ions, a byproduct of DNA synthesis, precipitate in the presence of metal cations, which leads to an increase in turbidity. BART uses a bioluminescent readout, which couples pyrophosphate generation with luciferase-mediated light emission. Twista is a portable real-time fluorimeter that can detect twin fluorescent probes for monitoring RPA reactions.
Lateral flow (LF) devices perform endpoint detection sandwich assays using passive fluidics, and are well-established for POC diagnostics [2]. Sensitivity in NALF is provided through upstream nucleic acid amplification. Simple NALF devices without a reader can detect <10 copies of target nucleic acid per amplification reaction, which is equivalent to less than attomolar analyte concentrations. NALF has been implemented in antibody-dependent (Figure 4a) and antibody-independent (Figure 4b) configurations. Antibody-dependent NALF is similar to LF immunoassays, but antigenic labels have to be incorporated into the amplicon via the primers or labeled dNTPs, and a second labeled probe is often required. The BESt Cassette (Table 1), which detects labeled amplicons via antibody-dependent NALF, is designed to minimize the risk of carryover contamination [12]. Antibody-independent NALF has a simplified reaction scheme, but requires nucleotide immobilization on the LF strip and colored microparticles. In a device called simple amplification-based assay (SAMBA) (Table 1), isothermal nucleic acid amplification and antibody-independent NALF are integrated into a disposable cartridge in conjunction with a benchtop instrument [51]. NALF is amenable to low-level multiplexing, and has been coupled with PCR [52,53], NASBA [54], helicase dependent amplification (HDA) [12], RPA [35], LAMP [55], cross-priming amplification (CPA) [38], and other isothermal amplification methods.
Figure 4.
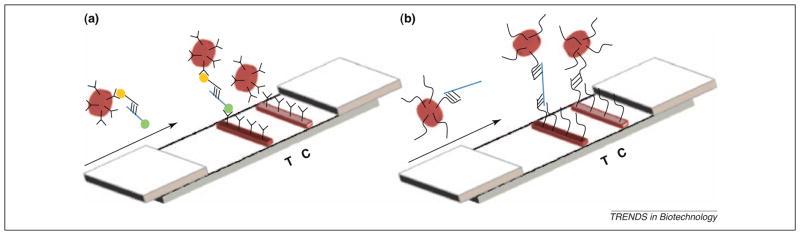
(a) Antibody-dependent NALF, which uses antibody-conjugated colored particles and LF strips, and also requires an antigen-functionalized capture oligonucleotide and a target amplicon (blue) that contains an antigen. (b) Antibody-independent NALF involves direct hybridization of unlabeled target amplicon (blue) with oligonucleotide-functionalized colored particles and with oligonucleotides deposited on the LF strip.
Changes in turbidity owing to pyrophosphate precipitation have been utilized for visual endpoint detection of LAMP reactions [37]. By including calcein dye, turbidity is enhanced via fluorescence. Precipitates of different colors can be obtained through the addition of cationic polymers in conjunction with fluorescently labeled primers and probes [56]. Colorimetric detection can be achieved through the addition of hydroxyl napthol [57]. These approaches minimize the risk for amplicon carryover, because the reaction tube remains sealed after incubation. Many other optical and electronic biosensor technologies enable endpoint nucleic acid detection [58]. For example, on-chip isothermal amplification has been coupled with microarray-based readout for multiplexed pathogen detection [59], and electrochemical biosensors can be miniaturized and integrated into microfluidic devices [60].
A unique combination of sample preparation, amplification and detection technologies might facilitate fully integrated, low-cost POC NAT. Isothermal amplification with endpoint detection can be implemented in a non- or minimally instrumented format. Recently, several single-use, sample preparation devices have been described that interface with various isothermal amplification methods, including LAMP, RPA and HDA [34,61,62]. Ultimately, moving NAT to the POC is driven by the clinical applications, some of which are presented in the next section.
Selected applications
Tuberculosis
TB is a disease of high public health priority owing to the increase in multi-drug-resistant strains and the high incidence of complications associated with HIV/TB co-infection (http://www.who.int/tb/publications/global_report/2009/en/index.html). Rapid diagnosis and drug susceptibility testing can limit the spread of TB and facilitate effective treatment. Commercial systems for TB NAT using large analyzers with manual sample preparation steps, followed by amplification based on PCR (Roche), TMA (GenProbe) and SDA (Beckton Dickinson), are available [63]. Sample preparation is particularly challenging for TB NAT because pre-processing is required to liquefy sputum and concentrate the pathogen; mycobacteria are lysis-resistant; and sputum contains polymerase inhibitors. Fully integrated, automated, POC-compatible TB NAT starting from raw sputum has been demonstrated in <2 h using the GeneXpert M. tuberculosis complex/rifampicin resistance (MTB/RIF) test [64,65]. In this method, chemically liquefied and decontaminated sputum samples are introduced into the GeneXpert cartridge, where intact mycobacteria are captured, concentrated, and purified. Following sonication lysis to liberate mycobacterial genomic material, the system performs a real-time, hemi-nested PCR assay with five molecular beacons that target the rifampicin resistance core region of the rpoB gene, plus one molecular beacon that detects a process/internal control. A multi-site evaluation has demonstrated that the MTB/RIF assay exhibits excellent sensitivity and specificity not only for TB diagnosis, but also for detection of rifampicin resistance as a marker for multi-drug-resistant tuberculosis (MDR-TB) [66]. The system has been endorsed by the WHO; however, the cost and complexity of the GeneXpert is a concern for broad implementation in low-resource settings. Inexpensive TB NAT in low-resource settings has been demonstrated using LAMP with visual endpoint detection [67], but the method is performed manually with separate sample preparation steps and additional instrument requirements. A need exists for simple, inexpensive, yet fully integrated POC NAT systems with performance characteristics sufficient to diagnose MDR-TB.
HIV viral load monitoring
Improved access to antiretroviral therapy in low-resource settings has decreased AIDS-related morbidity and mortality, but the emergence of drug-resistant HIV strains, coupled with the increasing rates of treatment failure, might undermine long-term therapeutic success [68–70]. Monitoring HIV viral load is routinely performed in developed countries to identify virological treatment failure, which prompts enhanced adherence counseling and possibly a change to second-line therapy. The WHO recognizes that increased access to viral-load monitoring in low-resource settings can help prevent emergence of drug-resistant HIV strains and patient health decline; however, the WHO does not recommend routine monitoring owing to the expense, complexity, and required infrastructure to implement current technologies [71].
NAT remains the preferred method for HIV viral-load monitoring in developed and developing countries [72], and commercial systems are available for high-volume centralized laboratory testing based on real-time quantitative PCR (e.g. Roche and Abbott), NASBA (BioMerieux), or a branched sandwich assay called bDNA (Bayer/Siemens) [68]. Several African countries are utilizing viral-load monitoring from dried blood spots sent to centralized laboratories, which poses logistical challenges and cannot reach all patients [7,68]. Technologies are in development that can move HIV viral-load monitoring out of the central laboratory. IQuum recently has demonstrated POC-compatible sample-in/answer-out HIV viral-load monitoring in 1.5 h using the LIAT analyzer [11], with analytical performance comparable to, and quantitative results concordant with, US Food and Drug Administration (FDA)-approved central laboratory systems.
Infant HIV diagnosis
Early diagnosis and timely intervention with antiretroviral drugs are essential to extend the life expectancy of HIV-1-infected infants [73]. Infant HIV diagnosis using standard serological assays is compromised owing to the presence of maternal HIV antibodies [74]. The WHO and the United Nations Children’s Fund (www.unicef.org) therefore recommend virological testing for infant HIV diagnosis, which can be accomplished through HIV NAT that targets viral RNA or proviral DNA using commercially available assays or laboratory-developed tests [75,76]. Central laboratories enable large-volume NAT [77], but transporting specimens and reporting results can result in turnaround times of 1–3 months [78]. Several POC-compatible platforms are under development. The SAMBA system (Table 1) currently performs HIV diagnosis through an isothermal nucleic acid amplification method similar to NASBA (Figure 2a), coupled with LF detection in an integrated cartridge combined with a benchtop instrument [51]. Inclusion of sample preparation is planned for a future version of this device. BioHelix has developed an HDA-based assay for HIV diagnosis with NALF detection using the BESt Cassette [78,79]; however, sample preparation, isothermal amplification, and NALF detection are performed as separate steps.
Group B streptococcus
Maternal carriage of group B streptococcus (GBS) in the vagino-cervical tract can cause severe infections in the neonate. Maternal carriage might be asymptomatic, therefore, rapid diagnosis of GBS during labor (intrapartum) is needed to minimize transmission risk at birth [1]. Screening for GBS colonization at 35–37 weeks via culture is insufficient because not all cases of GBS are detected [80]. The GeneXpert GBS enables testing directly from a vaginal swab during labor in 75 min, with sensitivity and specificity similar to GBS culture methods [80,81].
Methicillin-resistant Staphylococcus aureus surveillance and diagnosis
The incidence of hospital-acquired Staphylococcus aureus infections is increasing, particularly in the USA. S. aureus is a common hospital-acquired infection and is the leading cause of ventilator-associated pneumonia and surgical site infections [82]. Methicillin-resistant S. aureus (MRSA) accounts for 64% of all hospital-acquired S.aureusinfections [83]; is resistant to vancomycin as well as most β-lactam antibiotics; and can lead to severe complications once the infection spreads within the body, particularly in immuno-compromised individuals. NAT has been recommended recently by the Clinical and Laboratory Standards Institute (www.CLSI.org) for active MRSA surveillance of patients admitted to hospitals and other healthcare facilities [84]. Roche, Becton Dickinson and Cepheid market FDA-approved PCR-based MRSA NATs. The Roche and Becton Dickinson tests are conducted in central laboratories, but the GeneXpert MRSA test enables POC MRSA surveillance in hospital wards and outpatient clinics [85]. GeneXpert MRSA tests performed in hospital wards have been shown to provide results >10 h faster than GeneXpert MRSA tests performed in a central laboratory [86]. Isothermal MRSA NAT using HDA [12] and RPA [35] have also been reported. RPA integrated into a ‘lab on a foil’ system has enabled detection of the MRSA mecA gene at low copy numbers in under 20 min [34]. This system consists of a microfluidic centrifugal disk that contains all reagents on-board and a benchtop analyzer for disk rotation, incubation, and real-time fluorescence detection. The system can perform up to 30 amplification/detection reactions in parallel, but does not currently include sample preparation.
Conclusions
The progress in developing POC-compatible NAT technologies has increased dramatically in the past 5 years; driven by applications that require rapid turnaround in developed countries, and by expanding markets related to endemic diseases in the developing world. Cepheid has demonstrated that POC NAT is possible: the GeneXpert, originally developed to detect biothreat agents, has successfully crossed over to clinical diagnostic applications. After receiving FDA clearance for the GBS assay as the first moderate complexity NAT, Cepheid has rapidly expanded the GeneXpert test menu to a range of nosocomial infections, including MRSA, and has demonstrated, through the MTB/RIF test, that fully integrated sample-in/answer-out NAT can meet crucial needs that are related to infectious disease diagnosis in the developing world. Other integrated platforms are in the late stages of development (Table 1).
Technical obstacles remain that are related to achieving appropriate integration of multiple crucial components in a robust, user-friendly format, while minimizing complexity and cost. Combination of sample preparation with amplification and detection in one integrated system is the greatest challenge. New sample preparation technologies are needed with simplified workflow and high nucleic acid recovery, which are capable of processing a large sample input volume and purifying nucleic acids from a variety of sample matrices, without introducing additional inhibitory compounds. Isothermal amplification methods often require less-stringent sample preparation, and can facilitate rapid, sensitive, and specific target amplification via single-temperature incubation, which reduces system complexity and cost compared to PCR-based methods. Established and emerging isothermal amplification methods vary in terms of complexity (e.g. use of multiple enzymes and/or complex primer design), reaction speed, attainable sensitivity and specificity, and ability to multiplex. To date, no method is clearly superior, and different methods will probably find their way to market. Real-time detection offers superior performance, especially for analyte quantification. However, system cost and complexity can be reduced significantly through simple visual endpoint detection methods, such as NALF. Through a combination of such new technologies, POC NAT might be able to advance towards CLIA waived status and beyond testing near patients in hospital. Systems developed for non-clinical applications related to biothreat detection, food-safety, agricultural and veterinary diagnostics require the same inherent characteristics, but have an easier path to market, and serve as a stepping stone towards clinical diagnostic POC NAT.
Sustained implementation of POC NAT requires that new technologies truly address the needs of the targeted settings, which is especially true in developing countries [87]. Once a suitable method has been developed, stakeholders within the countries need to be convinced through rigorous evaluation, demonstration, and assessment studies that the new diagnostic method leads to better patient outcomes and has a positive impact on society overall, while being cost-effective and implementable in the existing healthcare system.
Acknowledgments
The authors acknowledge NIH NIAID funding through awards R01AI090831 and R01AI076247, which supports ongoing collaborative efforts to develop integrated POC NAT systems based on isothermal amplification.
Footnotes
Disclosure statement
Keck Graduate Institute, the employer of A.N., owns intellectual property related to EXPAR/Fingerprinting. In 2009, A.N. spent a 6-month sabbatical at Roche Molecular Diagnostics. Claremont BioSolutions, the employer of T.M.F., is developing sample preparation methods described herein, and has rights to or has filed U.S. Patent Application Nos. 12/052,950, 12/732,070, and 12/823,081. D.B. is currently developing and validating RPA assays with TwistDX. PATH and Lumora are working together on a product development project.
References
- 1.Clerc O, Greub G. Routine use of point-of-care tests: usefulness and application in clinical microbiology. Clin Microbiol Infect. 2010;16:1054–1061. doi: 10.1111/j.1469-0691.2010.03281.x. [DOI] [PubMed] [Google Scholar]
- 2.Yager P, et al. Point-of-care diagnostics for global health. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 3.Peeling RW, Mabey D. Point-of-care tests for diagnosing infections in the developing world. Clin Microbiol Infect. 2010;16:1062–1069. doi: 10.1111/j.1469-0691.2010.03279.x. [DOI] [PubMed] [Google Scholar]
- 4.Gutierres SL, Welty TE. Point-of-care testing: an introduction. Ann Pharmacother. 2004;38:119–125. doi: 10.1345/aph.1D212. [DOI] [PubMed] [Google Scholar]
- 5.Ehrmeyer SS, Laessig RH. Point-of-care testing, medical error, and patient safety: a 2007 assessment. Clin Chem Lab Med. 2007;45:766–773. doi: 10.1515/CCLM.2007.164. [DOI] [PubMed] [Google Scholar]
- 6.Weigl BH, et al. Simplicity of use: a critical feature for widespread adoption of diagnostic technologies in low-resource settings. Expert Rev Med Devices. 2009;6:461–464. doi: 10.1586/erd.09.31. [DOI] [PubMed] [Google Scholar]
- 7.Puren A, et al. Laboratory operations, specimen processing, and handling for viral load testing and surveillance. J Infect Dis. 2010;201:S27–S36. doi: 10.1086/650390. [DOI] [PubMed] [Google Scholar]
- 8.Stevens WS, Marshall TM. Challenges in implementing HIV load testing in South Africa. J Infect Dis. 2010;201:S78–S84. doi: 10.1086/650383. [DOI] [PubMed] [Google Scholar]
- 9.Small PM, Pai M. Tuberculosis diagnosis – time for a game change. N Engl J Med. 2010;363:1070–1071. doi: 10.1056/NEJMe1008496. [DOI] [PubMed] [Google Scholar]
- 10.Raja S, et al. Technology for automated, rapid, and quantitative PCR or reverse transcription-PCR clinical testing. Clin Chem. 2005;51:882–890. doi: 10.1373/clinchem.2004.046474. [DOI] [PubMed] [Google Scholar]
- 11.Tanriverdi S, et al. A rapid and automated sample-to-result HIV load test for near-patient application. J Infect Dis. 2010;201:S52–S58. doi: 10.1086/650387. [DOI] [PubMed] [Google Scholar]
- 12.Goldmeyer J, et al. Identification of Staphylococcus aureus and determination of methicillin resistance directly from positive blood cultures by isothermal amplification and a disposable detection device. J Clin Microbiol. 2008;46:1534–1536. doi: 10.1128/JCM.02234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dineva MA, et al. Sample preparation: a challenge in the development of point-of-care nucleic acidbased assays for resource-limited settings. Analyst. 2007;132:1193–1199. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, et al. Microfluidic sample preparation: cell lysis and nucleic acid purification. Integr Biol. 2009;1:574–586. doi: 10.1039/b905844c. [DOI] [PubMed] [Google Scholar]
- 15.Price CW, et al. Nucleic acid extraction techniques and application to the microchip. Lab Chip. 2009;9:2484–2494. doi: 10.1039/b907652m. [DOI] [PubMed] [Google Scholar]
- 16.Belgrader P, et al. A minisonicator to rapidly disrupt bacterial spores for DNA analysis. Anal Chem. 1999;71:4232–4236. doi: 10.1021/ac990347o. [DOI] [PubMed] [Google Scholar]
- 17.Doebler RW, et al. Continuous-flow, rapid lysis devices for biodefense nucleic acid diagnostic systems. J Assoc Lab Automation. 2009;14:119–125. [Google Scholar]
- 18.Cao WD, et al. Chitosan as a polymer for pH-induced DNA capture in a totally aqueous system. Anal Chem. 2006;78:7222–7228. doi: 10.1021/ac060391l. [DOI] [PubMed] [Google Scholar]
- 19.Sur K, et al. Immiscible phase nucleic acid purification eliminates PCR inhibitors with a single pass of paramagnetic particles through a hydrophobic liquid. J Mol Diagn. 2010;12:620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kermekchiev MB, et al. Mutants of Taq DNA polymerase resistant to PCR inhibitors allow DNA amplification from whole blood and crude soil samples. Nucleic Acids Res. 2009;37:e40. doi: 10.1093/nar/gkn1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, et al. Direct DNA amplification from crude clinical samples using a PCR enhancer cocktail and novel mutants of Taq. J Mol Diagn. 2010;12:152–161. doi: 10.2353/jmoldx.2010.090070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis KA, et al. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP) J Virol Methods. 2008;151:264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Zhou P, et al. Weak solvent based chip lamination and characterization of on-chip valve and pump. Biomed Microdevices. 2010;12:821–832. doi: 10.1007/s10544-010-9436-z. [DOI] [PubMed] [Google Scholar]
- 24.Easley CJ, et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci USA. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns MA, et al. An integrated nanoliter DNA analysis device. Science. 1998;282:484–487. doi: 10.1126/science.282.5388.484. [DOI] [PubMed] [Google Scholar]
- 26.Zhang YH, Ozdemir P. Microfluidic DNA amplification – a review. Anal Chim Acta. 2009;638:115–125. doi: 10.1016/j.aca.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 27.Zhang CS, et al. PCR microfluidic devices for DNA amplification. Biotechnol Adv. 2006;24:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 28.LaBarre P, et al. 32nd Annual International Conference of the IEEE. 2010. EMBS; 2010. Non-instrumented nucleic acid amplification (NINA): instrument-free molecular malaria diagnostics for low-resource settings; pp. 1097–1099. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann WP, et al. Comparison of transcription mediated amplification (TMA) and reverse transcription polymerase chain reaction (RT-PCR) for detection of hepatitis C virus RNA in liver tissue. J Clin Virol. 2005;32:289–293. doi: 10.1016/j.jcv.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Gracias KS, McKillip JL. Nucleic acid sequence-based amplification (NASBA) in molecular bacteriology: a procedural guide. J Rapid Methods Autom Microbiol. 2007;15:295–309. [Google Scholar]
- 31.Wharam SD, et al. Detection of virus mRNA within infected host cells using an isothermal nucleic acid amplification assay: marine cyanophage gene expression within Synechococcus sp. Virol J. 2007;4:52. doi: 10.1186/1743-422X-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong YJ, et al. Isothermal DNA amplification in vitro: the helicase-dependent amplification system. Cell Mol Life Sci. 2009;66:3325–3336. doi: 10.1007/s00018-009-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent M, et al. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutz S, et al. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA) Lab Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 35.Piepenburg O, et al. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curtis KA, et al. Sequence-specific detection method for reverse transcription, loop-mediated isothermal amplification of HIV-1. J Med Virol. 2009;81:966–972. doi: 10.1002/jmv.21490. [DOI] [PubMed] [Google Scholar]
- 37.Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang RD, et al. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol. 2009;47:845–847. doi: 10.1128/JCM.01528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitani Y, et al. Rapid and cost-effective SNP detection method: application of SmartAmp2 to pharmacogenomics research. Pharmacogenomics. 2009;10:1187–1197. doi: 10.2217/pgs.09.39. [DOI] [PubMed] [Google Scholar]
- 40.Johne R, et al. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009;17:205–211. doi: 10.1016/j.tim.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Yao B, et al. Quantitative analysis of zeptomole microRNAs based on isothermal ramification amplification. RNA. 2009;15:1787–1794. doi: 10.1261/rna.1555209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hellyer TJ, Nadeau JG. Strand displacement amplification: a versatile tool for molecular diagnostics. Expert Rev Mol Diagn. 2004;4:251–261. doi: 10.1586/14737159.4.2.251. [DOI] [PubMed] [Google Scholar]
- 43.Mchugh TD, et al. Prospective evaluation of BDProbeTec strand displacement amplification (SDA) system for diagnosis of tuberculosis in non-respiratory and respiratory samples. J Med Microbiol. 2004;53:1215–1219. doi: 10.1099/jmm.0.45780-0. [DOI] [PubMed] [Google Scholar]
- 44.Maples BK, et al. Ionian Technologies Inc. Nicking and extension amplification reaction for the exponential amplification of nucleic acids. 2009081670-A1 US. 2009
- 45.You Q, et al. Hagzhou Yousida Biotechnology Co. Ltd. Method for amplifying target nucleic acid sequence by nickase, and kit for amplifying target nucleic acid sequence and its use. 2006:CN1850981-A, CN100489112-C. [Google Scholar]
- 46.Jung C, et al. Isothermal target and signaling probe amplification method, based on a combination of an isothermal chain amplification technique and a fluorescence resonance energy transfer cycling probe technology. Anal Chem. 2010;82:5937–5943. doi: 10.1021/ac100606m. [DOI] [PubMed] [Google Scholar]
- 47.Van NJ, et al. Isothermal reactions for the amplification of oligonucleotides. Proc Natl Acad Sci USA. 2003;100:4504–4509. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan E, et al. Isothermal DNA amplification with gold nanosphere-based visual colorimetric readout for herpes simplex 2 virus detection. Clin Chem. 2007;53:2017–2020. doi: 10.1373/clinchem.2007.091116. [DOI] [PubMed] [Google Scholar]
- 49.Connolly AR, Trau M. Isothermal detection of DNA by beacon-assisted detection amplification. Angewandte Chemie-International Edition. 2010;49:2720–2723. doi: 10.1002/anie.200906992. [DOI] [PubMed] [Google Scholar]
- 50.Murakami T, et al. Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res. 2009;37:e19. doi: 10.1093/nar/gkn1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HH, et al. Simple amplification-based assay: a nucleic acid based point-of-care platform for HIV-1 testing. J Infect Dis. 2010;201:S65–S72. doi: 10.1086/650385. [DOI] [PubMed] [Google Scholar]
- 52.Dineva MA, et al. Simultaneous visual detection of multiple viral amplicons by dipstick assay. J Clin Microbiol. 2005;43:4015–4021. doi: 10.1128/JCM.43.8.4015-4021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mens PF, et al. Molecular diagnosis of malaria in the field: development of a novel 1-step nucleic acid lateral flow immunoassay for the detection of all 4 human Plasmodium spp. and its evaluation in Mbita, Kenya. Diagn Microbiol Infect Dis. 2008;61:421–427. doi: 10.1016/j.diagmicrobio.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Mugasa CM, et al. Nucleic acid sequence-based amplification with oligochromatography for detection of Trypanosoma brucei in clinical samples. J Clin Microbiol. 2009;47:630–635. doi: 10.1128/JCM.01430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puthawibool T, et al. Detection of shrimp infectious myonecrosis virus by reverse transcription loop-mediated isothermal amplification combined with a lateral flow dipstick. J Virol Methods. 2009;156:27–31. doi: 10.1016/j.jviromet.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 56.Mori Y, et al. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 2006;6:3. doi: 10.1186/1472-6750-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goto M, et al. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009;46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 58.Sassolas A, et al. DNA biosensors and microarrays. Chem Rev. 2008;108:109–139. doi: 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- 59.Andresen D, et al. Helicase dependent OnChip-amplification and its use in multiplex pathogen detection. Clin Chim Acta. 2009;403:244–248. doi: 10.1016/j.cca.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 60.Mir M, et al. Integrated electrochemical DNA biosensors for lab-on-a-chip devices. Electrophoresis. 2009;30:3386–3397. doi: 10.1002/elps.200900319. [DOI] [PubMed] [Google Scholar]
- 61.Fang X, et al. Loop-mediated isothermal amplification integrated on microfluidic chips for point-of-care quantitative detection of pathogens. Anal Chem. 2010;82:3002–3006. doi: 10.1021/ac1000652. [DOI] [PubMed] [Google Scholar]
- 62.Mahalanabis M, et al. An integrated disposable device for DNA extraction and helicase dependent amplification. Biomed Microdevices. 2010;12:353–359. doi: 10.1007/s10544-009-9391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ling DI, et al. Commercial nucleic-acid amplification tests for diagnosis of pulmonary tuberculosis in respiratory specimens: meta-analysis and meta-regression. PLoS ONE. 2008;3:e1536. doi: 10.1371/journal.pone.0001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helb D, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blakemore R, et al. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol. 2010;48:2495–2501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boehme CC, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boehme CC, et al. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol. 2007;45:1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stevens WS, et al. Quantifying HIV for monitoring antiretroviral therapy in resource-poor settings. J Infect Dis. 2010;201:S16–S26. doi: 10.1086/650392. [DOI] [PubMed] [Google Scholar]
- 69.Gupta RK, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect Dis. 2009;9:409–417. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]
- 70.Harries AD, et al. Diagnosis and management of antiretroviral-therapy failure in resource-limited settings in sub-Saharan Africa: challenges and perspectives. Lancet Infect Dis. 2010;10:60–65. doi: 10.1016/S1473-3099(09)70321-4. [DOI] [PubMed] [Google Scholar]
- 71.Bennett DE, et al. The World Health Organization’s global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13:1–13. [PubMed] [Google Scholar]
- 72.Rouet F, Rouzioux C. The measurement of HIV-1 viral load in resource-limited settings: How and where? Clin Lab. 2007;53:135–148. [PubMed] [Google Scholar]
- 73.Violari A, et al. Early antiretroviral therapy and mortality among HIV-infected infants. N Engl J Med. 2008;359:2233–2244. doi: 10.1056/NEJMoa0800971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chantry CJ, et al. Seroreversion in human immunodeficiency virus-exposed but uninfected infants. Pediatr Infect Dis J. 1995;14:382–387. doi: 10.1097/00006454-199505000-00009. [DOI] [PubMed] [Google Scholar]
- 75.Lambert JS, et al. Performance characteristics of HIV-1 culture and HIV-1 DNA and RNA amplification assays for early diagnosis of perinatal HIV-1 infection. J Acquir Immune Defic Syndr. 2003;34:512–519. doi: 10.1097/00126334-200312150-00011. [DOI] [PubMed] [Google Scholar]
- 76.Leelawiwat W, et al. Dried blood spots for the diagnosis and quantitation of HIV-1: stability studies and evaluation of sensitivity and specificity for the diagnosis of infant HIV-1 infection in Thailand. J Virol Methods. 2009;155:109–117. doi: 10.1016/j.jviromet.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 77.Stevens W, et al. Role of the laboratory in ensuring global access to ARV treatment for HIV-infected children: consensus statement on the performance of laboratory assays for early infant diagnosis. AIDS J. 2008;2:17–25. doi: 10.2174/1874613600802010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khamadi S, et al. Rapid identification of infants for antiretroviral therapy in a resource poor setting: the Kenya experience. J Trop Pediatr. 2008;54:370–374. doi: 10.1093/tropej/fmn036. [DOI] [PubMed] [Google Scholar]
- 79.Tang W, et al. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J Infect Dis. 2010;201:S46–S51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Tejada BM, et al. Intrapartum group B streptococcus detection by rapid polymerase chain reaction assay for the prevention of neonatal sepsis. Clin Microbiol Infect. 2010 doi: 10.1111/j.1469-0691.2010.03378.x. in press. [DOI] [PubMed] [Google Scholar]
- 81.El Helali N, et al. Diagnostic accuracy of a rapid real-time polymerase chain reaction assay for universal intrapartum group B streptococcus screening. Clin Infect Dis. 2009;49:417–423. doi: 10.1086/600303. [DOI] [PubMed] [Google Scholar]
- 82.Hidron AI, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29:996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 83.Gorwitz RJ, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 84.McDonald G. New CLSI report provides guidance on methicillin-resistant Staphylococcus aureus (MRSA) surveillance. Labmedicine. 2010;41:375–376. [Google Scholar]
- 85.Wolk DM, et al. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J Clin Microbiol. 2009;47:758–764. doi: 10.1128/JCM.01714-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brenwald NP, et al. Feasibility study of a real-time PCR test for meticillin-resistant Staphylococcus aureus in a point of care setting. J Hosp Infect. 2010;74:245–249. doi: 10.1016/j.jhin.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 87.Stop TB Partnership. Pathways to better diagnostics for tuberculosis: a blueprint for the development of TB diagnostics. 2009 http://www.stoptb.org/wg/new_diagnostics/assets/documents/BluePrintTB_annex_web.pdf [Websites should be cited in the text rather than the reference list]