Two Bruchpilot isoforms create a stereotypic arrangement of the cytomatrix that defines the size of the readily releasable pool of synaptic vesicles.
Abstract
Synaptic vesicles (SVs) fuse at a specialized membrane domain called the active zone (AZ), covered by a conserved cytomatrix. How exactly cytomatrix components intersect with SV release remains insufficiently understood. We showed previously that loss of the Drosophila melanogaster ELKS family protein Bruchpilot (BRP) eliminates the cytomatrix (T bar) and declusters Ca2+ channels. In this paper, we explored additional functions of the cytomatrix, starting with the biochemical identification of two BRP isoforms. Both isoforms alternated in a circular array and were important for proper T-bar formation. Basal transmission was decreased in isoform-specific mutants, which we attributed to a reduction in the size of the readily releasable pool (RRP) of SVs. We also found a corresponding reduction in the number of SVs docked close to the remaining cytomatrix. We propose that the macromolecular architecture created by the alternating pattern of the BRP isoforms determines the number of Ca2+ channel-coupled SV release slots available per AZ and thereby sets the size of the RRP.
Introduction
At the presynaptic side of synapses, neurotransmitter release is initiated by the depolarization-induced opening of voltage-gated presynaptic Ca2+ channels that are concentrated at defined release sites within the presynaptic active zone (AZ; Rettig and Neher, 2002; Gundelfinger et al., 2003; Murthy and De Camilli, 2003; Jin and Garner, 2008; Sigrist and Schmitz, 2011). Synchronous release can be explained by short-lived, so-called nanodomains of elevated [Ca2+], which build up and decay rapidly around these Ca2+ channels (Meinrenken et al., 2002; Bucurenciu et al., 2008; Neher and Sakaba, 2008). Whether discrete synaptic vesicle (SV) binding sites allowing for nanodomain-coupled release exist and what their molecular organization might look like remain unknown. Of note, an electron-dense cytomatrix referred to as presynaptic density or cytomatrix of the AZ (hereafter short cytomatrix; Gundelfinger et al., 2003), has been found to be associated with the intracellular face of the AZ membrane and is meant to coordinate SV fusion at the AZ (Südhof, 2012). Electron-dense cytomatrices directly superimpose on the intracellular parts of voltage-gated Ca2+ channels, as indicated by electron tomography of frog (Harlow et al., 2001) and Drosophila melanogaster (Fouquet et al., 2009) neuromuscular junctions (NMJs).
Clearly, genetic analysis of AZ components is critical for establishing precise causal relations between the molecular, structural, and functional features of AZs. Over the last years, an evolutionarily conserved complex of protein components highly enriched at AZs was identified: Munc13s (Wojcik and Brose, 2007), Rab3-interacting molecules (RIMs; Mittelstaedt et al., 2010), RIM-binding proteins (Liu et al., 2011), and α-Liprins (Spangler and Hoogenraad, 2007), as well as ELKS family proteins (Hida and Ohtsuka, 2010). Using superresolution light microscopy, we previously showed that ELKS family protein Bruchpilot (BRP) forms the electron-dense AZ cytomatrix (T bar) in Drosophila (Kittel et al., 2006; Fouquet et al., 2009). In brp-null mutants, T bars were lost, and AZ Ca2+ channels declustered (Kittel et al., 2006). In response, basal-evoked fusion of SVs was severely impaired. Importantly, the residual release was asynchronous, and short-term plasticity changed to atypical paired-pulse facilitation.
Here, we demonstrated a direct function of the BRP-based cytomatrix in controlling the number of readily releasable SVs. We characterized the two major isoforms of BRP with 190- and 170-kD apparent sizes by biochemical analysis and generated isoform-specific alleles and tagged genomic constructs. Electron and superresolution fluorescence microscopy showed that BRP-190 and BRP-170 contribute to T-bar cytomatrix assembly to a similar extent and alternate in a circular array. Mutants in both alleles still formed cytomatrices but of reduced size. These smaller cytomatrices still allowed normal Ca2+ channel clustering; however, baseline transmission was reduced nonetheless. The vesicular release probability determined by variance–mean analysis was also unchanged, indicating a different cause for the transmission defect in these alleles than in brp nulls. Instead, variance–mean analysis and back extrapolation of quantal contents both revealed a reduction in the size of the readily releasable pool (RRP) of SVs here. Correspondingly, the number of SVs docked close to the remaining cytomatrix found by EM was reduced. Thus, we suggest a novel role for BRP isoforms in creating a stereotypic arrangement of the cytomatrix that defines the number of Ca2+ nanodomain–coupled release slots available per AZ, a function which can be separated from Ca2+ channel clustering.
Results
The brp locus expresses two isoforms of 190 and 170 kD
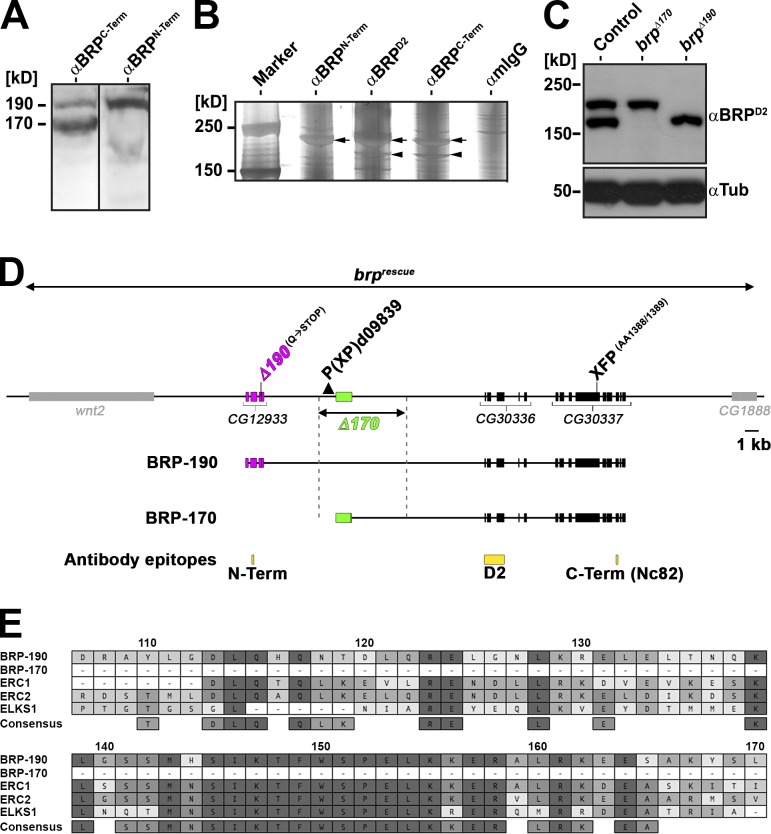
As previously reported (Wagh et al., 2006), Western blots of Drosophila adult head extracts probed with the monoclonal antibody (AB) NC82 (from here on termed BRPC-Term; epitope position shown in Fig. 1 D) show two bands of apparent 190 (BRP-190)- and 170 (BRP-170)-kD sizes (Fig. 1 A). Notably, another previously generated AB (Fouquet et al., 2009), anti-BRPN-Term, raised against a peptide encoded in the previously annotated exon cluster CG12933 (Fig. 1 D), recognized only BRP-190 (Fig. 1 A), indicating that BRP-170 might lack the CG12933 encoded protein sequence. To further address this, we used anti-BRPC-Term, anti-BRPN-Term, and a third AB, anti-BRPD2 (Fouquet et al., 2009), raised against a sequence encoded by CG30336 (Fig. 1 D), for immunoprecipitations (IPs). Silver staining of gels loaded with these IPs showed that anti-BRPN-Term AB only precipitated BRP-190 (Fig. 1 B, arrows), whereas anti-BRPD2 and anti-BRPC-Term precipitated both isoforms (Fig. 1 B, arrows and arrowheads, respectively). Mass spectrometry analysis of anti-BRPC-Term IPs found that the 190-kD band matched peptides encoded by CG12933, CG30336, and CG30337, identical to the protein to be expected from the reported cDNA (Wagh et al., 2006). In contrast, the 170-kD band contained the sequences of CG30336 and CG30337 but not of CG12933. Using cDNA library screening, we identified a partial clone defining a transcription start site for the BRP-170 encoding mRNA in between CG12933 and CG30336, indicating that the 170-kD isoform is transcribed from a second, independent promoter. This is compatible with the results from the IP, as the anti-BRPD2 and anti-BRPC-Term epitopes are shared by both isoforms, whereas the anti-BRPN-Term epitope is specific to BRP-190 (Fig. 1 D). The BRP-190–specific sequence encoded by CG12933 contains stretches highly homologous to other ELKS family proteins (Fig. 1 E).
Figure 1.
The brp locus encodes two major isoforms. (A) Western blots of Drosophila adult head extracts probed with anti-BRPC-Term AB (short AB) and anti-BRPN-Term AB. Black line indicates that intervening lanes have been spliced out. (B) Silver gel of IPs conducted on Drosophila adult head extracts with the indicated ABs. Arrows represent the two BRP isoforms (top arrows: BRP-190; bottom arrows: BRP-170). (C) Western blot of Drosophila adult head extracts of the indicated genotypes probed with anti-BRPD2 and anti-Tubulin (Tub). (D) Schematic representation of brp genomic locus. BRP-190 and BRP-170 have distinct N-terminal sequences (magenta, BRP-190; green, BRP-170) but share the same C-terminal exons (black). AB epitopes are indicated in yellow. brpΔ190 allele: ethyl methyl sulfonate–induced point mutation from CAG (Glutamine [Q]) to TAG, leading to a premature STOP at aa 261 (encoded by the BRP-190–exclusive exon 3). brpΔ170 was generated by imprecise excision of P element P(XP)d09839 deleting the region indicated by the dashed lines. Genomic rescue construct (brprescue) encompassing the whole locus including two neighboring genes. (E) Protein sequence alignment (MAFFT with BLOSUM62 scoring matrix) between Drosophila BRP-190 and BRP-170, mouse ERC1 (NCBI Nucleotide Database accession no. NP_835186) and ERC2 (NCBI Nucleotide Database accession no. NP_808482), and Caenorhabditis elegans ELKS1 (NCBI Nucleotide Database accession no. NP_500329). Numbering is based on BRP-190. Similarity in amino acid identity is indicated by a color code (dark gray, 100%; gray, 80–100%; light gray, 60–80%; white, <60% similarity). Term, terminal.
Generation of isoform-specific brp alleles
To investigate the functions of these two BRP isoforms, we produced isoform-specific mutants. Candidate chromosomes displaying reduced fitness and eclosion rate over previously established null alleles of brp (brp69 and brpΔ6.1) were screened by chemical mutagenesis (Fouquet et al., 2009). Thus, we isolated an allele (brpΔ190; Fig. 1 D) leading to a premature STOP codon at aa 261 of BRP-190 (Q to STOP). Western blots of brpΔ190 fly heads confirmed that this mutation eliminates BRP-190 but leaves BRP-170 intact (Fig. 1 C). Next, we mobilized the P element transposon line d09839, located 0.6 kb upstream of the first exon of the transcript encoding BRP-170 (Fig. 1 D). Imprecise excision led to an allele with a ∼5.9-kb deletion including the first exon of BRP-170 (brpΔ170; Fig. 1 D). Western blots of brpΔ170 fly heads confirmed that this mutant specifically removes BRP-170 without any major effect on the expression level of BRP-190 (Fig. 1 C). For further analysis, brpΔ170 and brpΔ190 were outcrossed into the wild-type background (Canton-S) for six generations. Different from brp-null alleles (Kittel et al., 2006; Fouquet et al., 2009), both isoform-specific alleles survived to adulthood when placed over deficiency.
BRP-190 and -170 coexpress within the AZs of neuromuscular terminals
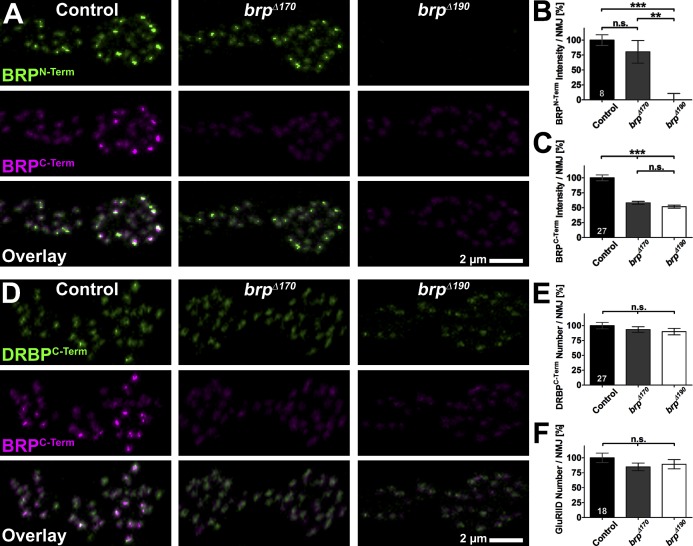
The glutamatergic NMJ synapses of Drosophila larvae are well suited for the integrated analysis of AZ function, and several brp alleles have been extensively studied here (Kittel et al., 2006; Fouquet et al., 2009; Hallermann et al., 2010). At control NMJs (muscle 6/7) costained with anti-BRPN-Term and anti-BRPC-Term, individual AZs were colabeled by both ABs (Fig. 2 A, left), as expected (Fouquet et al., 2009; Miśkiewicz et al., 2011). NMJs in brpΔ190 larvae lacked BRPN-Term AB staining completely (Fig. 2 A, right), consistent with Western results (Fig. 1 C). In contrast, brpΔ170 NMJs had rather normal BRPN-Term labeling intensity (Fig. 2, A [middle] and B [quantification]). Of note, BRPC-Term staining was reduced by ∼50% compared with controls in both mutants, indicating that both isoforms might contribute to the AZ cytomatrix in similar amounts (Fig. 2 C). Lack of either BRP isoform did not result in a change of NMJ size as quantified by HRP staining, which labels all neuronal membranes (control: 100.0 ± 5.7; brpΔ170: 88.8 ± 3.5, P = 0.1560 vs. control; brpΔ190: 100.6 ± 5.1, P = 0.9036 vs. control and P = 0.1368 vs. brpΔ170; n = 27 NMJs in nine larvae/genotype; unpublished data). For a BRP-independent estimate of AZ numbers, we stained for Drosophila RIM-binding protein (DRBP; Fig. 2 D), which closely associates with BRP at AZs (Liu et al., 2011), and glutamate receptors (GluRIID; Qin et al., 2005), which are postsynaptically matched to BRP-positive presynapses (not depicted; for an example, see Fig. S2 A). The number of both DRBPC-Term (Fig. 2, D and E, quantification) and GluRIID (Fig. 2 F [quantification] and not depicted) dots per NMJ were normal, indicating that there is no change in the AZ number in either of the BRP isoform mutants. Anti-DRBPC-Term staining intensity was slightly reduced in both isoform mutants (control: 100.0 ± 2.6; brpΔ170: 85.4 ± 2.7, P = 0.0004 vs. control; brpΔ190: 81.5 ± 3.0, P < 0.0001 vs. control and P = 0.2993 vs. brpΔ170; n = 27 NMJs in nine larvae/genotype), consistent with the partial dependency of DRBP on BRP presence we observed previously (compare with Fig. S7 in Liu et al., 2011). GluRIID label intensity was not changed compared with controls (control: 100.0 ± 7.2; brpΔ170: 100.0 ± 6.2, P = 0.7880 vs. control; brpΔ190: 99.7 ± 4.7, P = 0.3843 vs. control and P = 0.7397 vs. brpΔ170; n = 18 NMJs in six larvae/genotype), differing from brp nulls, which showed increased GluRIID intensity (Kittel et al., 2006).
Figure 2.
Confocal analysis of brp isoform mutants NMJs. (A and D) Two boutons of NMJs on muscles 6/7 from third instar larvae with the indicated genotypes labeled with the indicated ABs. (B) Quantification of BRPN-Term AB staining intensity measured over the whole NMJ (HRP mask). Control: 100.0 ± 9.1; brpΔ170: 80.4 ± 19.0, P = 0.8785 versus control; brpΔ190: 0.0 ± 10.7, P = 0.0002 versus control, P = 0.0047 versus brpΔ170; n = 8 NMJs in four larvae/genotype. (C) Quantification of BRPC-Term AB staining intensity measured over the whole NMJ (HRP mask). Control: 100.0 ± 5.0; brpΔ170: 58.2 ± 2.5, P < 0.0001 versus control; brpΔ190: 51.8 ± 2.4, P < 0.0001 versus control, P = 0.0667 versus brpΔ170; n = 27 NMJs in nine larvae/genotype. (E) Quantification of the number of DRBPC-Term dots measured over the whole NMJ (HRP mask). Control: 100.0 ± 5.5; brpΔ170: 93.5 ± 4.7, P = 0.4781 versus control; brpΔ190: 89.9 ± 5.4, P = 0.1800 versus control and P = 0.4889 versus brpΔ170; n = 27 NMJs in nine larvae/genotype. (F) Quantification of the number of GluRIID dots measured over the whole NMJ (HRP mask). Control: 100.0 ± 8.0; brpΔ170: 84.9 ± 6.4, P = 0.1455 versus control; brpΔ190: 89.3 ± 7.7, P = 0.3266 versus control, P = 0.6925 versus brpΔ170; n = 18 NMJs in six larvae/genotype. All quantifications were normalized to control. Error bars represent SEMs. **, P ≤ 0.01; ***, P ≤ 0.001; n.s., P > 0.05, Mann–Whitney U test. n is as indicated in the bars.
To explore the contribution of BRP-190 and BRP-170 to the overall BRP cytomatrix, we wanted to directly and individually visualize both BRP isoforms on the level of individual AZs. At its very N terminus, BRP-170 contains only 9 aa not included within BRP-190. Despite our efforts, we failed to produce a working AB against this short stretch. Thus, we generated a genomic rescue construct (brprescue) based on P(acman) technology (Venken et al., 2006), encompassing the whole locus, including two neighboring genes (Fig. 1 D). This construct rescued brp-null alleles to adult vitality and fertility. Because we could previously show that the N terminus of BRP binds to Ca2+ channels (Fouquet et al., 2009) and the C terminus is important for SV binding (Hallermann et al., 2010), we chose a low conserved region in exon 13 to insert an EGFP or mCherry tag (Fig. 1 D, XFP). We then introduced the STOP codon mutation giving rise to brpΔ190 into the brprescue,GFP construct (hereafter called BRP-170GFP). Similarly, a deletion mimicking brpΔ170 was introduced into the brprescue,Cherry transgene (hereafter called BRP-190Cherry). Both transgenes, when expressed in either wild-type or brp-null mutant background, appeared as bands of expected size (unpublished data) in Western blots. Expression of these constructs also rescued the lethality of brp-null alleles (unpublished data).
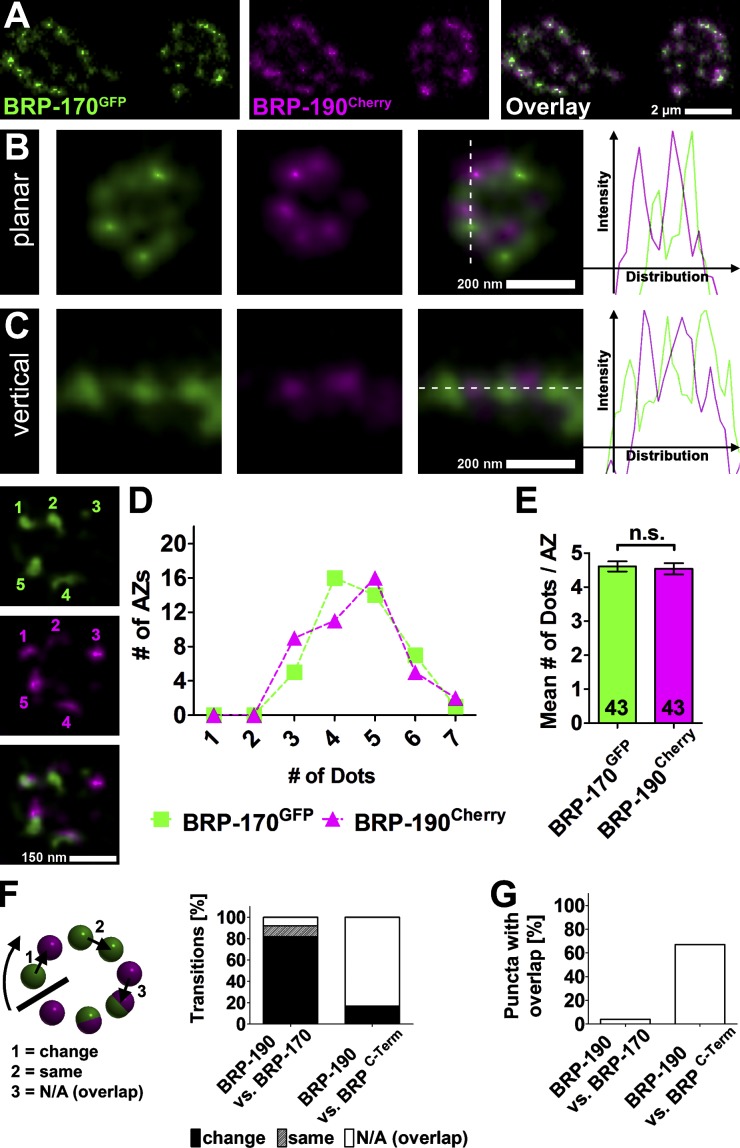
We previously developed protocols to in vivo live image individual NMJ terminals in intact larvae (Rasse et al., 2005; Schmid et al., 2008). Live imaging of larvae coexpressing BRP-170GFP and BRP-190Cherry (in wild-type background) revealed that all NMJ AZs contained both isoforms (Fig. 3 A). Western blots of larval body wall preparations (containing NMJs) indicated an equal abundance of both isoforms (Fig. S1, A and B, quantification). We sought to address the way these forms integrate into the AZ cytomatrix and what role they might play for structure and function there.
Figure 3.
Superresolution analysis of BRP isoform distribution at NMJ AZs. (A) Live imaging in intact third instar larvae expressing BRP-170GFP and BRP-190Cherry. (B and C) Images of individual AZs labeled with BRP-170GFP and BRP-190Cherry acquired with a two-color STED microscope (∼50 nm xy resolution compared with ≥200-nm resolution of a confocal microscope). Dotted lines define the position of the intensity distribution profiles (right). (D) The number of BRP-170GFP and BRP-190Cherry spots per AZs (x axis) is plotted against the number of AZs with that number of dots (y axis); counting example is shown on the left. (E) Quantification of mean number of BRP-170GFP and BRP-190Cherry dots per AZ. BRP-170GFP: 4.6 ± 0.1; BRP-190Cherry: 4.5 ± 0.2, P = 0.7663; Mann–Whitney U test; n (AZs) is indicated in the bars and is identical to D. Error bars represent SEMs. (F) Quantification of transitions determined via puncta to puncta line profiles of planar AZs. Transitions categorized into three categories as illustrated on the left. BRP-190 versus BRP-170: change, 82%; same, 10%; N/A (overlap), 8%; n (AZs) = 21; BRP-190 versus BRPC-Term: change, 17%; same, 0%; N/A (overlap), 83%; n (AZs) = 19. (G) Quantification of the fraction of overlapping punctae determined by line profiles of planar AZs. BRP-190 versus BRP-170: 4%; BRP-190 versus BRPC-Term: 67%; n is the same as in F.
A “circular array” of alternating BRP-190 and -170 clusters forms the AZ cytomatrix
Because of the small size of AZ cytomatrices (200–300 nm; diffraction-limited resolution of conventional microscopy > 200 nm), the distribution of both isoforms within each individual AZ could not be further resolved with conventional confocal microscopy. Previously, we had been using stimulated emission depletion (STED) microscopy (Hell, 2007) to show that BRP takes up an elongated conformation with its N terminus closer to the AZ membrane than its C terminus (Fouquet et al., 2009). The C terminus appeared donut shaped when imaged at an ∼100-nm resolution (Kittel et al., 2006). Using higher STED resolution (<70 nm), this further dissolved into a circular array of single “clusters,” each cluster probably representing a bundle of BRP filaments (Fouquet et al., 2009; Liu et al., 2011). Here, we used a two-color STED microscope providing 50-nm resolution in the focal plane (Bückers et al., 2011) to image individual AZs labeled with BRP-170GFP and BRP-190Cherry in a wild-type background. Both isoforms were found in an alternating pattern of discrete, nonoverlapping clusters in planar (Fig. 3 B) and vertical (Fig. 3 C) views, also apparent in intensity distribution profiles. We counted the number of BRP-170GFP and BRP-190Cherry dots at planar-imaged AZs (also with reversed secondary ABs as a control; Fig. S1, C and D) to get a more quantitative view of the AZ composition (Fig. 3 D, counting example shown on the left). This revealed that most AZs contain four to five dots of each BRP isoform (Fig. 3 E). For quantification of the alternating pattern, we counted the transitions from one puncta to the next going clockwise around planar-imaged AZs with line profiles shifted in such a way as to achieve clear separation of peaks. Here, transitions with color changes (BRP-170GFP to BRP-190Cherry and BRP-190Cherry to BRP-170GFP) were counted as change, transitions without color change (BRP-170GFP to BRP-170GFP and BRP-190Cherry to BRP-190Cherry) were counted as same, and transitions to or from punctae with overlapping signals (inseparable BRP-170GFP and BRP-190Cherry peaks) were counted as not applicable (N/A; overlap; Fig. 3 F, left). In BRP-190Cherry versus BRP-170GFP, we observed a rate of 82% change transitions and only 10% same transitions (Fig. 3 F, right). 8% of the transitions were N/A (Fig. 3 F, transition 3). As a control for more overlap, we used BRP-190Cherry versus BRPC-Term (example AZs shown in Fig. S1 F). Here, the same quantification led to only 17% of transitions changing color but 83% N/A. Similarly, BRP-190Cherry versus BRP-170GFP had 4% of all punctae overlapping, whereas BRP-190Cherry versus BRPC-Term had 67% overlap (Fig. 3 G). Also, when put in brp-null background, BRP-170GFP and BRP-190Cherry showed an alternating pattern, excluding a major influence of unlabeled endogenous BRP on the pattern (Fig. S1 E). Thus, BRP-170GFP and BRP-190Cherry seem to integrate into the AZ cytomatrix in a stoichiometric manner forming a circular array of alternating clusters.
Smaller AZ cytomatrices after eliminating either BRP-190 or -170
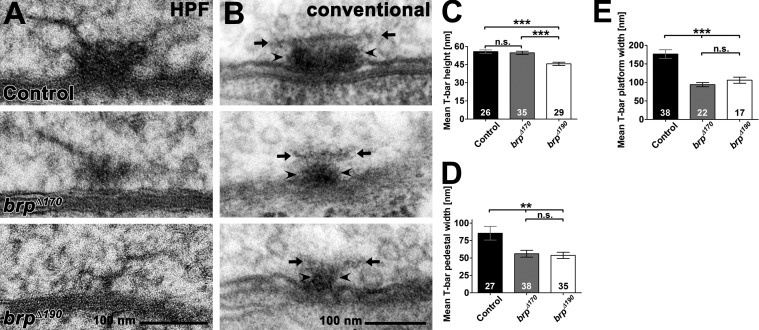
We previously showed using EM that the T bar found at Drosophila AZs is absent in brp-null mutants (Kittel et al., 2006; Fouquet et al., 2009). To preserve physiological ultrastructure, we first applied high-pressure freezing (HPF)/freeze substitution (FS) EM (HPF EM; Fouquet et al., 2009; Jiao et al., 2010; Südhof, 2012). In both brp isoform mutants, T bars appeared clearly smaller when compared with controls (Fig. 4 A; see also tomography Videos 1, 2, and 3). HPF EM displays T bars as filamentous structures, whereas in conventional embedded samples, including an initial aldehyde fixation step, T bars appear in a more compact manner and therefore easier to quantify (Südhof, 2012). In random sections of conventional embedded NMJ synapses (Fig. 4 B), T-bar height was slightly reduced only in brpΔ190 mutants (Fig. 4, B and C, quantification), whereas the widths of pedestal (Fig. 4, B [arrowheads] and D [quantification]) and platform (Fig. 4, B [arrows] and E [quantification]) were reduced in both mutants. Thus, both isoforms are needed to form proper-sized T bars.
Figure 4.
Ultrastructural analysis of brp isoform mutants. (A) Representative T bars taken from random ultrathin sections from controls, brpΔ170, and brpΔ190 mutant third instar larvae after HPF embedding. (B) Examples of electron micrographs of T bars in controls, brpΔ170, and brpΔ190 mutants third instar larvae after conventional embedding. T-bar pedestal (arrowheads) and platform (arrows) are indicated. (C) The T-bar height (pedestal plus platform) was reduced in brpΔ190 mutants compared with controls, whereas no difference could be detected in brpΔ170 mutants (control: 55.8 ± 1.4 nm; brpΔ170: 54.7 ± 1.4, P = 0.98 vs. control; brpΔ190: 45.7 ± 1.4 nm, P < 0.001 vs. control and P < 0.001 vs. brpΔ170; n is as indicated in the bars). (D) T-bar pedestals were significantly thinner in both isoform mutants compared with controls (control: 85.5 ± 9.8 nm; brpΔ170: 56.0 ± 4.9, P < 0.01 vs. control; brpΔ190: 53.7 ± 4.4 nm, P < 0.01 vs. control and P = 0.93 vs. brpΔ170; n is as indicated in the bars). (E) T-bar platforms were significantly thinner in both isoform mutants compared with controls (control: 176.5 ± 11.8 nm; brpD170: 94.0 ± 5.8 nm, P < 0.0001 vs. control; brpD190: 105.8 ± 8.2 nm, P = 0.0004 vs. control and P = 0.3573 vs. brpD170; n is as indicated in the bars). All panels show mean values and errors bars representing SEMs. **, P ≤ 0.01; ***, P ≤ 0.001; n.s., P > 0.05, Mann–Whitney U test.
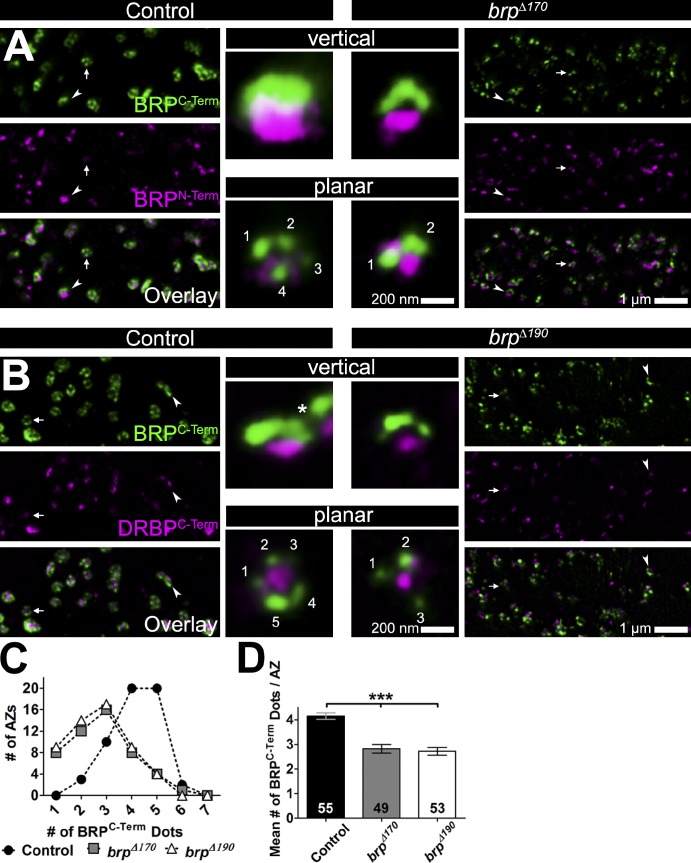
AZs of brpΔ170 mutants were costained with anti-BRPN-Term and anti-BRPC-Term and subjected to STED. As expected, N- and C-terminal labels were clearly separated in vertical views at the control AZs (Fig. 5 A, top left inset). Although N- and C-terminal epitopes remained separated, AZs in brpΔ170 mutants appeared smaller than those of the controls in both vertical and planar views (Fig. 5 A, insets and example AZs marked with arrowheads [vertical] and arrows [planar] in overviews). Because brpΔ190 mutants have no BRPN-Term staining left (Fig. 2 A), we instead used an AB against the C terminus of DRBP, which localizes very similarly to the N terminus of BRP (Fig. 5 B). AZs in brpΔ190 mutants, similar to brpΔ170 mutants, appeared smaller compared with the controls in both vertical and planar orientation (Fig. 5 B, insets and example AZs marked with arrowheads [vertical] and arrows [planar] in overviews).
Figure 5.
STED analysis of brp isoform mutants. (A and B) Overviews of two-color STED images of control and brpΔ170 (A) or brpΔ190 (B) mutant NMJs stained with the indicated antibodies. (insets) Magnifications of individual vertical (arrowheads)- and planar (arrows)-oriented AZs. The asterisk in the vertical inset in B marks beginning of a second AZ close to the centered one. (C) Number of BRPC-Term dots per AZs (x axis) plotted against the number of AZs with that number of dots (y axis) in control, brpΔ170, and brpΔ190 mutants. (D) Quantification of the mean number of BRPC-Term dots per AZ. Control: 4.1 ± 0.1; brpΔ170: 2.8 ± 0.2, P < 0.0001 versus control; brpΔ190: 2.7 ± 0.2, P < 0.0001 versus control and P = 0.7506 versus brpΔ170; Mann–Whitney U test; n (AZs) is indicated in the bars and is identical to C. Error bars represent SEMs.
In planar orientation, both brp isoform mutants showed a reduced number of BRPC-Term dots per AZ (Fig. 5, A and B, planar insets). Although the majority of wild-type AZs had four and five dots per AZ, in both isoform mutants, this was shifted to two to three dots per AZ (Fig. 5 C). Regularly, AZs with just one cluster of BRPC-Term were found in mutants but not controls. Accordingly, mean numbers of dots per AZ were reduced from approximately four to less than three in both isoform mutants (Fig. 5 D). Interestingly, the mean number of dots per wild-type AZ obtained for BRP-190 and BRP-170 each (mean number: 4.5/AZ; Fig. 3, D and E) is similar to the number of dots obtained by BRPC-Term AB staining (mean number: 4.1/AZ), which recognizes both isoforms. Together with the alternating pattern observed for BRP-170GFP and BRP-190Cherry, this suggests that each BRP dot recognized by the BRPC-Term AB is built up of one cluster of BRP-170GFP and BRP-190Cherry.
Overall, STED and EM analyses agree that both BRP isoforms localize to AZs independently of each other but cannot fill in for each other in the formation of a proper AZ molecular architecture. After losing one of the isoforms, the number of BRP clusters per AZ dropped, and the typical pentagonal gestalt no longer formed. Next, we were interested in which functional consequences these ultrastructural changes might have.
Reduction of baseline transmission in the absence of BRP-190 or -170
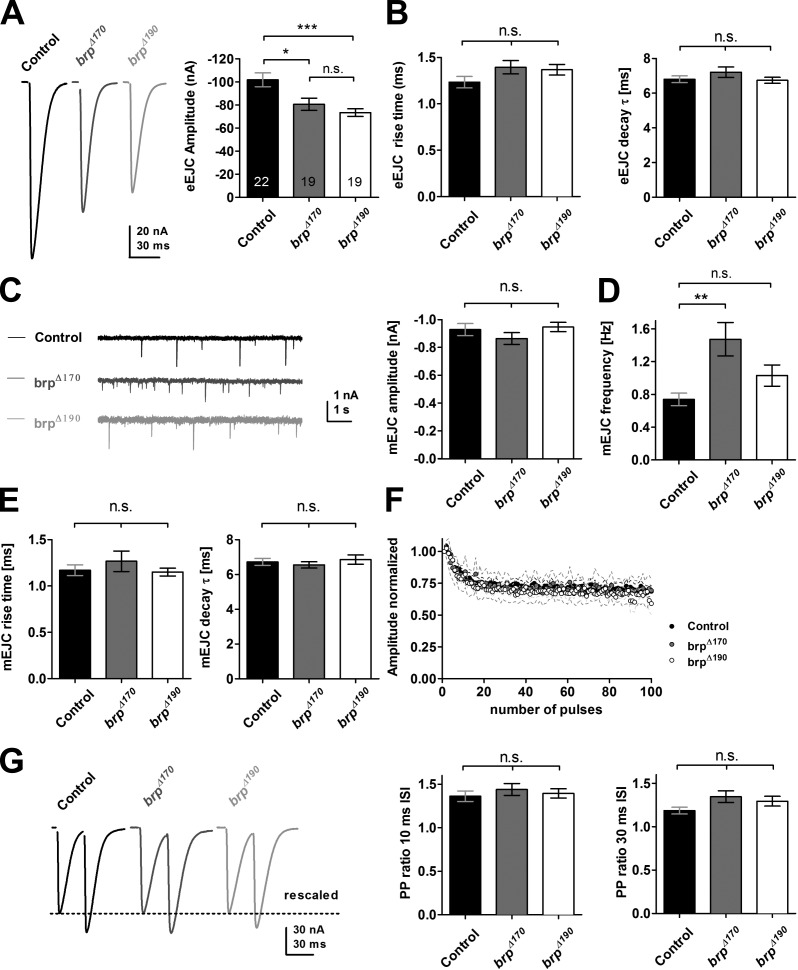
We were curious whether synaptic transmission might be affected by the reduced cytomatrix size. We performed two-electrode voltage clamp (TEVC) recordings on larval body wall muscles of third instar larvae to analyze potential physiological effects after losing either of the BRP isoforms. The mean evoked excitatory junctional current (eEJC) amplitude was decreased in both mutants by 20–30% (control: −101.7 ± 6.0, n = 22; brpΔ170: −80.5 ± 5.3, P < 0.01, n = 19; brpΔ190: −73.3 ± 3.3, n = 19, P < 0.001; one-way analysis of variance [ANOVA] Tukey’s posttest; Fig. 6 A) with a more pronounced effect in the brpΔ190 mutant. At the same time, kinetics of evoked synaptic responses were not altered (Fig. 6 B).
Figure 6.
Electrophysiological characterization of brp isoform mutant NMJs. (A) Example traces and quantification of evoked excitatory junctional currents (eEJC) recorded in 1.5 mM Ca2+ at 0.2 Hz. n is as indicated in the bars. (B) Rise time and the decay time constants (τ) of eEJCs were unchanged (rise time: control, 1.23 ± 0.06, n = 21; brpΔ170, 1.40 ± 0.07, P > 0.05, n = 19; brpΔ190, 1.37 ± 0.06, P > 0.05, n = 19; τ: control, 6.74 ± 0.17; brpΔ170, 6.80 ± 0.19, P > 0.05; brpΔ190, 7.21 ± 0.30, P > 0.05; one-way ANOVA Tukey’s posttest). (C, left) Example traces of mEJCs. (right) mEJC amplitude was unchanged in both brp isoform mutants (control: −0.93 ± 0.04, n = 19; brpΔ170: −0.86 ± 0.04, P > 0.05, n = 16; brpΔ190: −0.95 ± 0.03, P > 0.05, n = 16; one-way ANOVA Tukey’s posttest). (D) mEJC frequency was increased in brpΔ170mutants (control: 0.74 ± 0.08, n = 19; brpΔ170: 1.47 ± 0.20, P < 0.01, n = 16; brpΔ190: 1.03 ± 0.13, P > 0.05, n = 16; one-way ANOVA Tukey’s posttest). (E) mEJC kinetics were not changed in both brp isoform mutants (mEJC rise time: control, 1.17 ± 0.06, n = 19; brpΔ170, 1.27 ± 0.11, P > 0.05, n = 15; brpΔ190, 1.15 ± 0.04, n = 16, P > 0.05; mEJC τ: control, 6.73 ± 0.20, P > 0.05; brpΔ170, 6.55 ± 0.18, P > 0.05; brpΔ190, 6.86 ± 0.27, P > 0.05; one-way ANOVA Tukey’s posttest). (F) Normalized amplitudes at a 10-Hz stimulation for 10 s. Control and both mutants showed no difference in depression kinetics (control, n = 14; brpΔ170, n = 6; brpΔ190, n = 8). (G) Paired-pulse ratio at two different ISIs of 10 and 30 ms was not different between control and mutants (ISI 10 ms: control, 1.36 ± 0.06, n = 19; brpΔ170, 1.44 ± 0.07, P > 0.05, n = 15; brpΔ190, 1.40 ± 0.05, P > 0.05, n = 16; ISI 30 ms: control, 1.19 ± 0.04, n = 15; brpΔ170, 1.35 ± 0.07, P > 0.05, n = 13; brpΔ190, 1.29 ± 0.06, P > 0.05, n = 18; one-way ANOVA Tukey’s posttest). Example traces for 30-ms ISI rescaled to the amplitude of the first pulse (dotted line) are shown. All panels show mean values and errors bars representing SEMs (unless otherwise noted). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., P > 0.05.
The size of spontaneous vesicle fusion events (miniature excitatory junctional currents [mEJCs]) was unaltered (Fig. 6 C); however, mEJCs were more frequent in brpΔ170 (Fig. 6 D). mEJC kinetics were identical to controls for both mutants (Fig. 6 E).
Notably, although basal synaptic transmission was reduced in brpΔ170 and brpΔ190 (Fig. 6 A), short-term plasticity analyzed with standard protocols was not changed, unlike other brp alleles analyzed up to now (Kittel et al., 2006; Hallermann et al., 2010). Depression kinetics at 10-Hz stimulation for 10 s were identical to controls, as was obvious from normalized response amplitudes (Fig. 6 F). Additionally, no difference in paired-pulse ratios at a 10- or 30-ms interstimulus interval (ISI) was detected (Fig. 6 G).
The release deficit in brp-null mutants can likely be attributed mainly to Ca2+ channel declustering (Kittel et al., 2006). Given that we did not detect any changes in short-term plasticity in either of the isoform mutants with standard protocols, we wondered whether Ca2+ channel mislocalization could be observed. The α1 subunit Cacophony (Cac) is the only representative of the mammalian Cav2.1/2.2 family in Drosophila. In cac-null mutants, evoked SV release at the NMJ is almost absent, leading to embryonic lethality (Kawasaki et al., 2002; Hou et al., 2008). We generated a genomic Cac construct (Venken et al., 2008) labeled with GFP at the C terminus, similar to the upstream activator sequence-CacGFP previously used (Kawasaki et al., 2004). In brpnull background, CacGFP signals appeared declustered as expected (unpublished data). In neither of the brp isoform mutants (Fig. S2 A), however, were any Ca2+ channel clustering defects observed. Instead, CacGFP intensities measured over the whole NMJ, number of dots, and mean dot size were all similar to controls (Fig. S2, B–D). Thus, the release deficit observed in brp isoform mutants cannot be attributed to declustering of Ca2+ channels. As mentioned earlier in this paper, tight local organization of Ca2+ channels and the release apparatus is crucial for the spatiotemporal coupling of Ca2+ influx to SV release and thus determinative of the release probability of single SVs. Ca2+ channel declustering in both brp- and drbp-null mutants goes along with delayed synaptic transmission that is consequential of imperfect synchronization of AP-triggered SV release. Lack of either of the two main BRP isoforms does not decluster Ca2+ channels and, therefore, does not influence the tight spatiotemporal control of release. This is reflected by the unchanged kinetics of evoked release (Fig. 6 B) as well as the normal short-term plasticity (Fig. 6, F and G) found in both mutants.
BRP isoforms define the number of readily releasable SVs
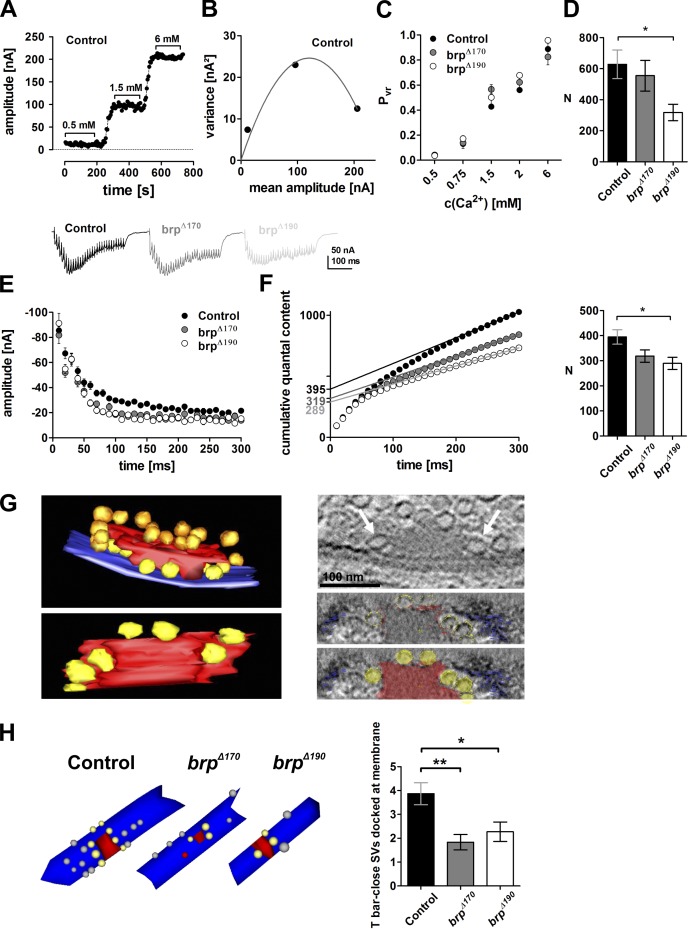
To further address the question how the reduced size of the BRP scaffold affects synaptic transmission, we made use of variance–mean analysis of synaptic responses (Clements and Silver, 2000; Sakaba et al., 2002; Silver, 2003). Thus, we determined the mean amplitude and variance of eEJCs in TEVC mode at Ca2+ concentrations between 0.5 and 6 mM and calculated vesicular release probabilities at the used Ca2+ concentrations applying parabolic fits (see Materials and methods; Fig. 7, A [an example recording of a control cell] and B [the according fit]; and Fig. S3, A and B, examples for all genotypes). The vesicular release probability was indeed unchanged in both isoform mutants compared with controls over the whole range of concentrations tested (Fig. 7 C).
Figure 7.
BRP isoform mutants have normal vesicular release probability but harbor fewer release-ready vesicles. (A) Control example of eEJC amplitudes recorded at the indicated Ca2+ concentrations. (B) Parabolic fit of variance–mean analysis of example cell in A. (C) Vesicular release probabilities of brpΔ170 and brpΔ190 mutants at various Ca2+ concentrations do not differ from controls (P > 0.05 for all Ca2+ concentrations; one-way ANOVA Tukey’s posttest). (D) Number of release-ready vesicles determined by variance–mean analysis. (E) Example traces (top) and quantification (bottom) of high-frequency trains for control, brpΔ170, and brpΔ190 recorded in 1.5 mM extracellular Ca2+. eEJCs decreased in both mutants and control to a steady-state level with a faster decay in the mutants. (F) Release-ready vesicle pool sizes determined by back extrapolation of cumulative quantal contents (extrapolated straight lines are indicated in the graph). (G, left) Rendered tomogram in side and bottom view of a wild-type NMJ T bar. SVs tethered distally at the T bar (red) indicated in gold, and SVs docked at the membrane (blue) proximal to the T bar indicated in yellow. (right) Virtual section showing two SVs next to the T-bar matrix (arrows). (right bottom) Bottom view of the T bar with membrane/T bar close SVs using the slicer option of 3dmod. SVs are arranged around the T-bar pedestal. (H, left) Examples of serial section EM reconstructions of AZs from controls, brpΔ170, and brpΔ190 mutants. Red, electron-dense structures; gray, docked vesicles; yellow, docked, T bar proximal SVs. (right) Quantifications of SV numbers obtained from AZ reconstructions after serial sectioning EM. T bar–proximal SVs docked at the plasma membrane were significantly reduced in both isoform mutants (control, 3.9 ± 0.5; brpΔ170, 1.8 ± 0.3, P < 0.01 vs. control; brpΔ190, 2.3 ± 0.4, P < 0.05 vs. control, Mann–Whitney U test). All panels show mean values and errors bars representing SEMs (unless otherwise noted). *, P ≤ 0.05; **, P ≤ 0.01.
Apart from the vesicular release probability, the amount of release-ready vesicles determines how many quantal units get exocytosed per arriving action potential. By the performed variance–mean analyses, we could also determine the size (n) of the RRP of SVs. In fact, RRP sizes were reduced by half in brpΔ190 and, to a lesser extent, in brpΔ170 mutants (control: 627.6 ± 92.0, n = 8; brpΔ170: 554.4 ± 99.0, n = 8, P > 0.05; brpΔ190: 317.7 ± 53.3, n = 8, P < 0.05; one-way ANOVA Tukey’s posttest; Fig. 7 D). Mean quantal size was unchanged (Fig. S3 C).
We used a high-frequency stimulation protocol of 100 Hz for 300 ms to assess the RRP size with an additional independent assay (Fig. 7 E). Here, RRP sizes were determined by back extrapolation of cumulative quantal contents of responses. Again, both mutants showed fewer release-ready SVs, again with a significant difference of pool size in brpΔ190 compared with the control (control: 395 ± 29, n = 21; brpΔ170: 319 ± 25, n = 20 , P > 0.05; brpΔ190: 289 ± 24, n = 14 , P < 0.05; one-way ANOVA Tukey’s posttest; Fig. 7 F).
T-bar pedestal close SVs specifically reduced in brp isoform mutants
It appears that the reduction in the size of the AZ cytomatrix we observed scales with the reduction in the number of RRP SVs, as determined by electrophysiological analysis. As mentioned previously in this paper, physiological data show that Ca2+ sensors on the SVs get into a very close physical proximity to the Ca2+ channels. Our previous analysis had shown that Ca2+ channels are clustered directly underneath the BRP N termini, at the T-bar pedestal (Fouquet et al., 2009). Fig. 7 G shows a tomography (Fig. 7 G, left) of a particularly large AZ cytomatrix (Fig. 7 G, red) where SVs tethered to the distal parts of the cytomatrix can be clearly detected (Fig. 7 G, gold). Most importantly, we regularly observed SVs clustered directly at the base of the T bar (Fig. 7 G, right, yellow and arrows in virtual section). Thus, the BRP cytomatrix might define SV binding and release sites (“slots”) in tight proximity to the Ca2+ channels, allowing the coupling of SVs to Ca2+ nanodomains. In EM serial section reconstructions, both mutants showed a significant decrease in those SVs found in close proximity to both the plasma membrane and the T-bar pedestal (Fig. 7 H).
In brpnude mutants, lacking the last 17 aa, T bars are bare of SVs in EM (Hallermann et al., 2010). When we counted the distally tethered SVs and SVs lateral to T bars using three 50-nm concentric shells in random ultrathin sections (similar to Hallermann et al., 2010), no reduction could be observed in the brp isoform mutants (Fig. S4). Collectively, these findings suggest a reduced amount of RRP SVs in the AZs with reduced size of the BRP cytomatrix, probably reflecting a decreased number of slots in which SVs could bind to achieve nanodomain coupling to Ca2+ channels.
Discussion
An elaborate protein cytomatrix covering the AZ membrane is meant to facilitate and control the SV release process. Quantitative analysis of neurotransmitter release has provided evidence that the number of SV release sites per AZ might be fixed (Clements and Silver, 2000). Although these sites are thought to be located in close proximity to presynaptic Ca2+ channels, ultrastructural and molecular information is largely missing here (Haucke et al., 2011). Potentially, specific interactions between SVs and certain cytomatrix components might be involved. Here, we provide evidence that the BRP-based cytomatrix plays a role in defining the number of readily releasable SVs, possibly by offering morphological and molecular-determined “release slots.”
BRP alleles isolate discrete functionalities of the presynaptic cytomatrix
Previously, we characterized the role of BRP based on null alleles, which result in a complete absence of AZ cytomatrix (T bar), partially declustered Ca2+ channels, and likely as a direct consequence, reduced vesicular release probability (Kittel et al., 2006). In contrast, in the analysis of BRP isoform–specific mutants, we here neither observed any Ca2+ channel clustering deficits nor changes in vesicular release probability.
We have previously found a binding site between the intracellular C terminus of the Cac Ca2+ channel and an N-terminal stretch of BRP, which is unique to BRP-190 (Fouquet et al., 2009). That solely losing BRP-190 is not sufficient to affect Ca2+ channel clustering could possibly be explained by the presence of redundant binding sites within BRP-170. Ca2+ channel clustering might well be a collective feature of the cytomatrix, and Ca2+ channels likely use multiple simultaneous interactions with several cytomatrix proteins to anchor within the AZ membrane.
In fact, RIM-binding protein family proteins at rodent and Drosophila AZs bind Ca2+ channels, and loss of the only RIM-binding protein in Drosophila results in partial loss of Ca2+ channels from AZs (Liu et al., 2011). RIM-binding protein levels at AZ were slightly but significantly reduced in the BRP isoform mutants. Clearly, it remains a possibility that RIM-binding protein is a major scaffold determinant of the release slots and that e.g., subtle mislocalizations of RIM-binding protein might in part contribute to the BRP isoform mutant phenotype. The brp-null phenotype we now interpret as a “catastrophic event” in which a complete loss of this large scaffold protein leads to a severe decrease of cytomatrix avidity (potentially mediated via a loss of RIM-binding protein) below a critical level, resulting in a “collapse” of the normal cytomatrix architecture. Thus, functionalities associated with discrete regions of BRP and RIM-binding protein can apparently be masked when the BRP-based AZ scaffold is completely eliminated.
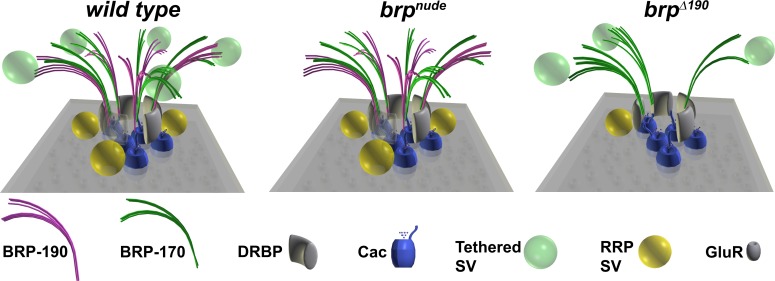
As mentioned previously in this paper, the distal cytomatrix in brpnude is bare of SVs (model in Fig. 8, brpnude) in EM, and SV replenishment is defective, resulting in short-term depression (and not facilitation as in brp nulls). However, no change of short-term plasticity could be detected in the brp isoform alleles with the same analyses, consistent with neither a change in Ca2+ channel clustering (Fig. S2) nor in SV clustering at the distal cytomatrix (Fig. S4). Nevertheless, a basal release deficit was observed, which can be explained by a reduction in the size of the readily releasable vesicle pool (Fig. 7), assigning an additional function to the BRP cytomatrix.
Figure 8.
Model of AZ organization in different brp mutants. Wild-type AZs contain BRP filaments made of both BRP isoforms in an alternating pattern (Fig. 3), which create release slots for RRP SVs. In brpnude mutants, lacking the very C terminus, SVs tethered to the BRP cytomatrix are absent (Hallermann et al., 2010). In brpΔ190 mutants, the number of BRP filaments is reduced (Fig. 5) because of the lack of the BRP-170 isoform. This leads to a ∼50% decrease in the number of SVs docked close to the T bar, which is also reflected by a decrease in the number of RRP vesicles (Fig. 7).
Cytomatrix architecture and local environments for SV release slots
Release-ready SVs are meant to be molecularly and positionally primed for release. Important factors are the equipment with or the attachment to the proteins of the core release machinery (Rizo and Südhof, 2012) and the localization of the SV in proximity to the Ca2+ source. At the Drosophila NMJ, SV release is insensitive to slow Ca2+ buffers such as EGTA (Kittel et al., 2006); therefore, SVs are thought to be spatially tightly coupled to Ca2+ channels (nanodomain coupling; Eggermann et al., 2012). As we find Ca2+ channels localized directly underneath the T-bar pedestal composed of the N-terminal region of BRP (Fouquet et al., 2009), release-ready SVs might well correspond to the SVs that we find docked at the pedestal of the T bar and thus in very close proximity to the Ca2+ channels. This in turn is in agreement with BRP itself being important for defining the number of release-ready SVs determined by electrophysiology and EM (Fig. 7).
Light microscopic inspection of an AB directed against the C terminus of BRP, common to both isoforms (Fig. 1), with 50-nm STED resolution, typically revealed approximately five dots arranged as a circle or regular pentagon (Fig. 5). We labeled both isoforms individually and found that (a) both isoforms seem to localize with their C termini similarly toward the distal edge of the cytomatrix and (b) both isoforms typically form an identical number of dots per AZ (Fig. 3) similar to the number of dots observed with the BRPC-Term AB recognizing both isoforms. Thus, the BRP isoforms seem to be arranged in neighboring but not overlapping clusters, forming a circular array (Fig. 8, wild-type model). Consistent with both BRP isoforms not overlapping in space, there was neither efficient co-IP between them (Fig. 1 B, N-term AB lane) nor did elimination of one isoform substantially interfere with the AZ localization of the respective other isoform. Thus, BRP-190 and -170 seem to form discrete oligomers. The alternating pattern of BRP-190 and -170 appears to set a typical cytomatrix size, as both isoform mutants had a reduced T-bar width in EM (Fig. 4) and a reduced mean number of BRP dots per AZ (Fig. 5 and Fig. 8, brpΔ190 model). As this corresponded with a similar reduction in the number of SVs in the RRP (Fig. 7), this AZ architecture could set a typical number of Ca2+ nanodomain-coupled RRP slots possibly located between BRP clusters. However, beyond providing a discrete morphological architecture, the two BRP isoforms described here might harbor additional functionalities. The brpΔ190 phenotype was more pronounced than the brpΔ170, leaving the possibility that the highly conserved N terminus of BRP-190 (Fig. 1 E) promotes release by further mechanisms going beyond the points analyzed in this study. Future analysis will also have to address whether localization and regulation of additional cytomatrix and release components, such as RIM-binding protein (Liu et al., 2011), Unc-13 family proteins (Aravamudan et al., 1999), or RIM (Müller et al., 2012), contribute to the formation of release slots as well.
Scaling between cytomatrix size and evoked release
Ultimately, functional differences between individual synaptic sites must be defined by variances in their molecular organization. Functional features of a synapse can be extracted electrophysiologically. Thereby, the number of Ca2+ channels was recently identified as a major determinant of the release probability of single vesicles, Pvr, in rat calyces (Sheng et al., 2012). Furthermore, AZ size seems to scale with the overall likelihood of release from a given AZ (Holderith et al., 2012). Our results suggest that the BRP-based cytomatrix should be a general determinant of the release likelihood per AZ by establishing Pvr, through Ca2+ channel clustering, as shown previously (Kittel et al., 2006), and, as shown here, by determining the size of the RRP. Our genetic results show that the cytomatrix can, in principle, control the RRP size independent of Ca2+ channel clustering. A coupled increase in the size of the T-bar cytomatrix together with increasing SV release was previously observed at NMJs compensating for loss of the glutamate receptor subunit glurIIA (Reiff et al., 2002). Moreover, an increase in the number of release-ready SVs together with an increase in the amount of BRP was recently described as part of a homeostatic presynaptic response after pharmacological block of postsynaptic GluRIIA (Weyhersmüller et al., 2011). In line with this scenario, it was recently shown that lack of acetylation of BRP in elp3 mutants led to an increase in the complexity of the AZ cytomatrix along with an increase in RRP size (Miśkiewicz et al., 2011). Furthermore, in vivo imaging of synaptic transmission with single synapse resolution revealed that the likelihood of release correlates with the amount of BRP present at an individual AZ (Peled and Isacoff, 2011). This cytomatrix size–SV release scaling might be a general principle, as a correlation between the amount of SV exocytosis, measured by an optical assay, and the amount of the AZ protein Bassoon at individual synapses of cultured rat hippocampal neurons has also been observed (Matz et al., 2010). Our results suggest that not only the mere size, but also the distinct architecture of the cytomatrix influence release at individual synapses through determining RRP size.
Materials and methods
IPs and silver staining
Wild-type adult fly heads were mechanically homogenized in a buffer (B) containing 50 mM Tris/HCl, pH 8, 1% Na-deoxycholate, 150 mM NaCl, and protease inhibitor (Complete; Roche) followed by incubation at 36°C for 30 min and, after addition of 0.1% Triton X-100, by incubation for another 30 min at 4°C. Tissue debris were isolated from the supernatant by repeated centrifugation (Owald et al., 2010). Supernatant was subsequently precleared with random IgGs coupled to protein A–Sepharose beads (Bio-Rad Laboratories). For IPs, protein A–Sepharose beads were coupled to either mouse monoclonal Nc82 (BRPC-Term; Wagh et al., 2006), rabbit polyclonal BRPN-Term (Fouquet et al., 2009), rabbit polyclonal BRPD2 (Fouquet et al., 2009), or polyclonal mouse IgGs (Dianova). After incubation of beads and supernatant at slow rotation, the beads were washed in B, and proteins were eluted by boiling in SDS sample buffer. Samples were subjected to SDS-PAGE using NuPAGE 4–12% gradient gels and subsequently stained with silver (Blum et al., 1987).
Western blot analysis of BRP isoform mutants
For fly head protein extraction, 20 heads per genotype were sheared manually in 20 µl of 2% SDS aqueous solution using a micropistil fitting tightly into a 1.5-ml cup. 20 µl of 2× sample buffer (Laemmli, 1970) was added, and samples were heated at 95°C for 5 min. After centrifugation for 5 min at 16,000 g, to pellet fly head debris, 10 µl of each sample (equivalent to five heads) was subjected to denaturing SDS-PAGE using a 6% Tris-Cl gel. Proteins were transferred onto a nitrocellulose membrane and probed with affinity-purified primary AB rabbit anti-BRPD2 (1:5,000). For detection, a secondary anti–rabbit IgG HRP-conjugated AB (Dianova) and an ECL detection system (GE Healthcare) were used.
Western blot analysis of larval NMJs
Larval body wall protein extraction was performed as quadruplicate as follows: 15 wild-type larva each were dissected as described previously (Qin et al., 2005) with minor modifications. Internal organs including the larval central nervous system were removed, and body walls were sheared manually in 30 µl of 2% SDS aqueous solution using a micropistil fitting tightly into a 1.5-ml cup. 30 µl of 2× sample buffer (Laemmli, 1970) was added, and samples were heated at 95°C for 10 min. After centrifugation for 5 min at 16,000 g, to pellet debris, 4, 8, 12, and 16 µl of one replicate sample (equivalent to one, two, three, and four larval body walls) were subjected to denaturing SDS-PAGE using a 6% Tris-Cl gel. Proteins were transferred onto a nitrocellulose membrane and probed with affinity-purified primary AB rabbit anti–BRP-D2 (1:5,000). For detection, a secondary anti–rabbit IgG HRP-conjugated AB (Dianova) and an ECL detection system with Hyperfilm ECL (GE Healthcare) were used. As a loading control, α-Tubulin was probed (mouse monoclonal DM1A; Sigma-Aldrich).
For densitometry analysis of BRP-190 and -170 bands, films were scanned in transmission mode (V770; Epson) and measured using the gel analyzer tool of ImageJ (v1.46r; National Institutes of Health) for relative quantification. In brief, lane profile plots were generated, and peak areas were determined.
Fly stocks
All fly strains were reared under standard laboratory conditions. For larval genotypes, see Immunostainings.
Isolation of brpΔ190 mutant
The brpΔ190 allele was identified in a chemical mutagenesis screen with 25 mM ethyl methyl sulfonate by reduced fitness and eclosion rate when placed over previously established null alleles of brp (brp69 and brp6.1).
Generation of brpΔ170 mutant
To get access to a specific elimination of the 170-kD isoform, we mobilized the P element transposon line d09839, located 0.6 kb upstream of the first exon of the transcript encoding BRP-170. The brpΔ170 allele was generated through imprecise excision of the P element. P element transposon line d09839 was obtained from the Exelixis Collection at the Harvard Medical School.
Molecular cloning
Generation of P(acman)-BRP genomic constructs.
The P(acman) constructs for BRP have a size of 83 kb and were cloned according to Venken et al. (2006). In brief, the bacterial artificial chromosome (BAC) clone RP98-14J24 obtained from BACPAC Resources Center was used as a template for cloning of the BRP genomic P(acman) construct. 5′ homology arm (left arm) flanked by AscI–NotI was produced by PCR using primers 5′-GGCGCGCCCAAAGTAGCAACCTGAGTGTG-3′ and 5′-GCGGCCGCGGAAACAAACAGTTTAAGTAGCTG-3′. 3′ homology arm (right arm) flanked by NotI–PacI was produced by PCR using the primers 5′-GCGGCCGCCATTCATTGCAAACTCGCTCG-3′ and 5′-TTAATTAACCGAATTATCCGCGATAATG-3′.
The left arm and right arm PCR products were cut by PacI and AscI and ligated into attB-P(acman)-ApR. A recombination event between the BAC and the attB-P(acman)-ApR-LARA entailing the complete brp locus inclusive of the neighboring genes with a size of 70.31 kb (genomic region 5,367,010–5,437,720) resulted in a P(acman)-BRP construct of 83.4-kb size.
Generation of modified and tagged P(acman)-BRP constructs.
The insertion of the Δ190 point mutation and the Δ170 deletion of the alternative promoter and first exon were cloned according to the Counter Selection BAC Modification kit obtained from Gene Bridges GmbH company. The Δ190 point mutation was introduced at aa 261 of the brp locus corresponding to the brpΔ190 allele. To delete the BRP-170, the 170 kD–specific promoter and the first exon were removed resulting in a 1,111-bp deletion. The rpsL-neomycin (neo) template DNA was used to generate selectable cassettes. The primers consisted of 50-bp homology regions and a sequence for amplification of the rpsL-neo counter selection cassette. The selectable cassettes were generated in a proof reading PCR with Vent Polymerase (New England Biolabs, Inc.) and the following primer pairs: EGFP/mCherry, 5′-CAGAAGCTCCAAGAGTCGGAGCGCGCCCTGCAAGCCGCCGGTCCCCAACAGGGCCTGGTGATGATGGCGGGATCG-3′ and 5′-CCATTGCTCGATCTCCTCGCGACTCGCTCCTGCCGCCGCTGCAGCCTGTGCTCAGAAGAACTCGTCAAGAAGGCG-3′; Δ190, 5′-CAAACACTGATTGCCCGCGACGAGAGTATCAAGAAGCTGCTGGAAATGCTGGGCCTGGTGATGATGGCGGGATCG-3′ and 5′-GACAATGCTTTATGCTTAATCCATTCAACGACTCACCCATTCCCTTGGCCTTCAGAAGAACTCGTCAAGAAGGCG-3′; and Δ170, 5′-CTGAGTGGCCTCGTGGTCGCAAAAGCTCCGCAGGCGCGAACAGTTGTACAGGCCTGGTGATGATGGCGGGATCG-3′ and 5′-CTGACTGGATGGGGTGACTGTTTCCGGTGGAAGCAGTGGGCGGTACAATACTCAGAAGAACTCGTCAAGAAGGCG-3′. The XFP tags were inserted into the exon13 between aa 1,388 and 1,389.
EGFP or mCherry was flanked by an AscI restriction site and a linker region, based on the Tn5 mosaic end linkers described in Sheridan et al. (2002). For the generation of the EGFP and the mCherry nonselectable cassettes, the proof reading polymerase Vent was used. For the EGFP nonselectable cassette, the following primer pair was used: 5′-CAGAAGCTCCAAGAGTCGGAGCGCGCCCTGCAAGCCGCCGGTCCCCAACAGTGTCTCTTATACACATCTGGCGCGCC-3′ and 5′-CCATTGCTCGATCTCCTCGCGACTCGCTCCTGCCGCCGCTGCAGCCTGTGCTCTGTCTCTTATACACATCTGCCCGGGCGCGCC-3′. For the mCherry cassette, the following primer pair was used: 5′-CAGAAGCTCCAAGAGTCGGAGCGCGCCCTGCAAGCCGCCGGTCCCCAACAGGGAAATTGTCTCTTATACACATCTG-3′and 5′-CCATTGCTCGATCTCCTCGCGACTCGCTCCTGCCGCCGCTGCAGCCTGTGCTCTGTCTCTTATACACATCTGCCCG-3′.
For generating a nonselectable cassette for the Δ170 construct, two independent PCR fragments were generated with Vent proofreading polymerase, with the following primers: for fragment one, 5′-CAAATGAGTGCCAAAAGTTG-3′ and 5′-CTGTACAACTGTTCGCGCCTGCGGAGC-3′; and for fragment two, 5′-CGCGAACAGTTGTACAGTATTGTACCGC-3′ and 5′-GAAGGTTCTGTTCTGTTCTTG-3′. In independent PCRs using the Elongase Enzyme Mix (Invitrogen), these two fragments were fused and introduced into the P(acman)-BRP construct via recombineering, exchanging the selectable cassette for Δ170.
The same was performed for the nonselectable cassette of Δ190. For this, a single point mutation was introduced through a primer. To generate a nonselectable cassette for the Δ190 construct, two independent PCR fragments were generated, with the following primers: for fragment one, 5′-GACCTGCAGCGGGAGCTGGGGAAC-3′ and 5′-CCTTGGCCTACAGCATTTCCAGCAGCTTC-3′; and for fragment two, 5′-GGAAATGCTGTAGGCCAAGGGAATGGGTG-3′ and 5′-CTGATGTTGCATCAAATGTATCGAACTC-3′. In independent PCRs using the Elongase Enzyme Mix, these two fragments were fused introduced into the P(acman)-BRP construct via recombineering, exchanging the selectable cassette for Δ190.
Transgenic flies were produced by PhiC31 integration system. All constructs were inserted at the same genomic position 75A10 (24862 fly strain; Bloomington Drosophila Stock Center).
Generation of genomic CacGFP flies.
The attB-P(acman) BAC clone CH321-60D21 containing the genomic region of Cac was obtained from the BACPAC Resources Center. The C-terminal EGFP tag was incorporated according to Venken et al. (2008) using PL452-C-EGFP-KanR (Addgene plasmid 19178) and the following primers: 5′-AGAGTGGTCTGAACCCTCGACACTCCGATTCGGATGAGGAGGATTGGTGCGCAGCCCAATTCCGATCATATTC-3′ (forward) and 5′-ACCCAGATCTGATCCTTTCGATTACGCGATCGCAATCAAAGCCTTTGCTATTACTTGTACAGCTCGTCCATG-3′ (reverse). After sequencing, the construct was injected into an attP site containing fly strain (PBac {y[+]-attP-3B}VK00033; Bloomington Drosophila Stock Center line #9750) using the services of BestGene Inc.
Immunostainings
Dissections and immunostainings were performed as described before (Qin et al., 2005). All larvae were raised at 25°C on semidefined medium (Bloomington Drosophila Stock Center recipe) and the following genotypes were used: control, Canton-S; brpΔ170, brpΔ170/Df(2R)BSC29; brpΔ190, brpΔ190/Df(2R)BSC29; BRP-170GFP/190Cherry, brp genomic rescue BRP-170GFP/brp genomic rescue BRP-190Cherry (III); CacGFP control, +/+; genomic CacGFP; brpΔ170; CacGFP, brpΔ170/Df(2R)BSC29; genomic CacGFP; and brpΔ190; CacGFP, brpΔ190/Df(2R)BSC29; genomic CacGFP. Primary antibodies were used in the following concentrations: for mouse monoclonal AB Nc82, anti-BRPC-Term at 1:250 (provided by E. Buchner, Universitätsklinikum Würzburg, 97078 Würzburg, Germany); and for mouse monoclonal AB 3E6, anti-GFP at 1:500, rabbit anti-DGluRIID at 1:500 (Qin et al., 2005), rabbit antiDRBPC-Term at 1:500, and rabbit anti-dsRed at 1:500 (Takara Bio Inc.). Except for staining against CacGFP, in which larvae were fixed for 5 min with ice-cold ethanol, all fixations were performed for 10 min with 4% PFA in 0.1 mM PBS.
Secondary antibodies for standard immunostainings were used in the following concentrations: goat anti–HRP-Cy5 at 1:200, goat anti–rabbit-Cy3 at 1:500, and goat anti–mouse Alexa Fluor-488 at 1:500. Secondary antibodies for STED were used in the following concentrations: goat anti–mouse Atto594 at 1:250, goat anti–rabbit at Atto594 1:250, goat anti–mouse Atto647N at 1:100, and goat anti–rabbit Atto647N at 1:100.
Image acquisition
Image acquisition of confocal and live microscopy was obtained with a confocal microscope (TCS SP5; Leica). STED microscopy was performed with a custom-built STED microscope (see STED microscopy). Images of fixed and live samples were acquired at RT. Confocal imaging of NMJs and whole brains was performed using a z step of 0.5 µm. The following objectives were used: 63×, 1.4 NA oil immersion for NMJ confocal imaging and a 100×, 1.4 NA oil immersion STED objective for STED imaging (all obtained from Leica). All confocal images were acquired using the LCS AF software (Leica). All STED images were acquired using Imspector Software (Max Planck Innovation GmbH). For previous descriptions, see Fouquet et al. (2009), Owald et al. (2010), and Liu et al. (2011).
Live imaging, confocal imaging, and analysis
Live imaging, confocal imaging, and image processing were essentially performed as previously described (Fouquet et al., 2009; Liu et al., 2011). Images from fixed samples were taken from the third instar larval NMJ (segments A3 and A4) of either muscles 6 and 7 (Fig. 2) or muscle 4 (Fig. S2). Images for figures were processed with ImageJ software to enhance brightness using the brightness/contrast function and smoothened (1–2-pixel σ radius) using the Gaussian blur function. Quantifications of AZ number, NMJ size, and fluorescence intensity were performed as previously described (Owald et al., 2010). In short, a mask was created from the HRP signal using the thresholding function of ImageJ. Then, the number of AZs, NMJ size, and fluorescence intensity were determined by semiautomatic thresholding within the HRP mask. For details on image acquisition, see the section Image acquisition. Data were analyzed using the Mann–Whitney U test for linear independent data groups. Means are annotated ±SEM. Asterisks are used to denote significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., P > 0.05.
STED microscopy
Two-color STED images were recorded with a custom-built STED microscope, which combines two pairs of excitation and STED laser beams, all derived from a single supercontinuum laser source (as described in Bückers et al., 2011). STED images were processed using a linear deconvolution function integrated into Imspector Software. The point spread function for deconvolution was generated by using a 2D Lorentz function with its half-width and half-length fitted to the half-width and half-length of each individual image. The numbers of BRP isoform or BRPC-Term dots per AZ were counted manually on planar-imaged AZs of deconvolved images with the help of the Find Maxima function of ImageJ using default settings; obvious background punctae were manually removed. The quantification of transitions was performed with line profiles shifted in such a way as to achieve clear separation of peaks from one puncta to the next going clockwise around planar-imaged AZs. Punctae were defined with the Find Maxima function of ImageJ with default setting. Transitions with color changes (green to magenta and magenta to green) were counted as change, transitions without color change (green to green and magenta to magenta) were counted as same, and transitions to or from puncta with overlapping signal (inseparable green and magenta peaks) were counted as N/A (overlap).
Electrophysiology
TEVC recordings were performed at restricted temperature between 18 and 20°C on male, third instar, larval NMJs (muscle 6 and segments A2 and A3) essentially as previously reported (Kittel et al., 2006). The composition of the extracellular hemolymph-like saline (HL-3; Stewart et al., 1994) was (mM) NaCl 70, KCl 5, MgCl2 20, NaHCO3 10, trehalose 5, sucrose 115, Hepes 5, and CaCl2 as indicated, pH adjusted to 7.2. Recordings were made from cells with an initial membrane potential (Vm) between −50 and −70 mV and input resistances of ≥4 MΩ, using intracellular electrodes with resistances of 8–20 MΩ, filled with 3 M KCl. Both eEJCs, which reflect the compound excitatory junctional current of both motoneurons innervating muscle 6 (voltage clamp at −60 mV) and mEJCs (voltage clamp at −80 mV) were low pass filtered at 1 kHz. 0.2-Hz stimulation protocols included 20 traces/cell, and high-frequency stimulation followed after 30-s rest. Paired-pulse recordings consisted of 10 traces/interval/cell in which a 4-s rest was left between paired pulses. The rise time and decay time constants (τ) were obtained from the mean event of the corresponding recording. The rise time was measured from 10 to 90% of the maximum amplitude, and the decay was fit with a single exponential function from 60% of the peak. The recordings were analyzed with pClamp 10 (Molecular Devices) and MATLAB R2010b (MathWorks). Stimulation artifacts of eEJCs were removed for clarity. The data are reported as means ± SEM, n indicates the number of cells examined and included, and p-value denotes the significance according to one-way ANOVA with Tukey’s multiple comparison posttest. In the figures, the level of significance is marked with asterisks: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
Variance–mean analysis
The current amplitude at different extracellular Ca2+ concentrations of 0.5–6 mM (≥20 eEJCs each) was recorded. For a binomial model, the mean amplitude of synaptic responses is given by I = NPvrq (Sakaba et al., 2002; Silver, 2003), with q being the quantal size, N being the number of release-ready vesicles, and Pvr representing the vesicular probability of release at a certain extracellular Ca2+ concentration. The variance Var(I) = Nq2Pvr (1 − Pvr) was calculated by Var(I) = (1/(n – 1)) Σ (Ii − Imean)2. The variance–mean plots were fitted for each individual cell with Var(I) = I2/N+qI. Vesicular release probability (Pvr) and quantal size (q) were adjusted with the experimentally determined coefficient of variation of 0.3 (Pawlu et al., 2004) according to Clements and Silver (2000). The data are reported as means ± SEM, n indicates the number of cells examined and included, and p-value denotes the significance according to one-way ANOVA with Tukey’s multiple comparison posttest. In the figures, the level of significance is marked with asterisks: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
Cumulative postsynaptic current analysis
30 consecutive postsynaptic responses were recorded at a stimulation frequency of 100 Hz. eEJC amplitudes were measured from peak to baseline directly before the onset of the response. Quantal content of each response was calculated by division of the amplitude by the mean quantal size of the respective genotype. Release-ready vesicles were determined by back extrapolation of cumulative quantal contents. The data are reported as means ± SEM, n indicates the number of cells examined and included, and p-value denotes the significance according to one-way ANOVA with Tukey’s multiple comparison posttest. In the figures, the level of significance is marked with asterisks: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001.
EM
For conventional embedding, third instar larvae were dissected and fixed in 4% PFA and 0.5% glutaraldehyde in 0.1 M PBS, pH 7.2, for 10 min and then for 60 min in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2, washed three times for 5 min in sodium cacodylate buffer, and postfixed on ice for 1 h with 1% osmium tetroxide and 0.8% KFeCn in 0.1 M sodium cacodylate buffer. After washing in sodium cacodylate buffer and distilled water (three 5-min washing steps), the samples were stained en bloc with 1% uranyl acetate in distilled water on ice. Samples were dehydrated in ascending ethanol concentrations at RT and infiltrated in Epon resin. After infiltration, the muscles 6/7 of segment A2/3 of two to five larvae per genotype were cut out and placed parallel to each other in an embedding mold. Blocks were polymerized for 48 h at 70°C. After conventional embedding, 65–70-nm serial sections were obtained and collected on Formvar-coated copper slot grids. Sections were postfixed and poststained with uranyl acetate/lead citrate following standard protocols. Micrographs were taken with an electron microscope (JEM 1011; JEOL) equipped with a camera (Orius 1200A; Gatan) using the DigitalMicrograph software package (Gatan). Serial 3D reconstructions were conducted using the 3D reconstruct software (Fiala, 2005).
For HPF embedding, about three to five Drosophila late second/early third instar larvae were placed in aluminum specimen carrier of 200-µm depth (type A; Bal-Tec), filled with yeast paste, and covered with a lid. The samples were frozen immediately in an HPF machine (HPM100; Leica) and rapidly transferred to liquid nitrogen for storage. Cryosubstitution was performed in an AFS (Leica) in anhydrous acetone with 1% EMD Millipore water, 1% glutaraldehyde, and 1% osmiumtetroxide at −90°C for 10 h. The samples were slowly (5°C/h) warmed to −20°C and incubated for an additional 12 h before being warmed (10°C/h) to 20°C. The samples were washed with acetone and incubated with 0.1% uranylacetate in anhydrous acetone for 1 h at RT. After washing, the samples were incubated in 30% Epon/70% acetone for 4 h followed by 70% Epon/30% acetone overnight. They were incubated twice in 100% Epon for 2 h before being embedded. 60–65-nm sections were cut using an ultramicrotome (RMC Power Tome XL; Boeckeler). Sections were collected on Formvar-coated 100 mesh grids. Sections were poststained with 2% uranylacetate for 30 min and lead citrate for 3 min. Micrographs were acquired on a microscope (Tecnai Spirit; FEI).
For electron tomography on conventional embedded samples, 250-nm sections were applied to Formvar-coated 100 copper mesh grids and stained with 4% uranyl acetate and Reynold’s lead citrate; subsequently, 10-nm gold particles were applied to the grid. A single tilt series was acquired at an electron microscope (JEM 2100; JEOL) operated at 200 kV and equipped with a camera (Orius 1200A) at 200 kV from −58 to 58° with a 1° increment using SerialEM software. The tomogram was generated using the IMOD package eTomo, and the model was rendered using 3dmod (The Boulder Lab for 3D Electron Microscopy).
For electron tomography on HPF samples, 200-nm embedded sections of second instar larvae were applied to Formvar-coated copper 100 mesh grids and stained with 2% uranyl acetate and Reynold’s lead citrate. 10-nm gold beads were applied on both sides of the grid. Tilt series were acquired on a microscope (Tecnai Spirit) operated at 120 kV equipped with a charge-coupled device camera (2k Eagle; FEI). Tilt series (±56° with 1 or 2° tilt increments) were recorded at a nominal magnification of 30,000 at 3–5-µm defocus using Xplore3D (FEI). 3D reconstruction was performed using the Inspect 3D software package (FEI), and rendering was performed using the IMOD 3dmod software (Kremer et al., 1996).
Online supplemental material
Fig. S1 shows an additional Western blot of BRP isoforms with quantifications and more examples of the alternating pattern of BRP-170GFP versus BRP-190GFP in direct comparison to a BRP-190Cherry versus BRPC-Term (NC82) control staining. Fig. S2 shows the analysis of Ca2+ channel clustering, which is not defective in BRP isoform mutants. In Fig. S3, examples of the variance–mean analysis for all genotypes are presented. Fig. S4 displays the distribution of cytomatrix distal and lateral SV. Videos 1–3 show HPF tomographies that display the T bar phenotypes of the isoform mutants for control (Video 1), brpΔ170 (Video 2), and brpΔ190 (Video 3). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201301072/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201301072.dv.
Supplementary Material
Acknowledgments
We would like to thank Janine Lützkendorf and Christine Quentin for technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to S.J. Sigrist (Exc 257, SI849/4-1, TP A3 SFB 958, and TP B9/SFB665). M. Siebert was supported by a Ph.D. fellowship from the Max Delbrück Center for Molecular Medicine and a Boehringer Ingelheim Fonds Ph.D. fellowship. E. Knoche was supported by a Ph.D. fellowship from the graduate school (GRK 1123) funded by the Deutsche Forschungsgemeinschaft. C. Wichmann was supported by the German Research Foundation through the Collaborative Research Center 889 (TP A7). A. Sickmann was supported by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, and the Bundesministerium für Bildung und Forschung. T. Matkovic was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 740 TP Z1).
Footnotes
Abbreviations used in this paper:
- AB
- antibody
- ANOVA
- analysis of variance
- AZ
- active zone
- BAC
- bacterial artificial chromosome
- BRP
- Bruchpilot
- Cac
- Cacophony
- DRBP
- Drosophila RIM-binding protein
- eEJC
- evoked excitatory junctional current
- FS
- freeze substitution
- HPF
- high-pressure freezing
- IP
- immunoprecipitation
- ISI
- interstimulus interval
- mEJC
- miniature excitatory junctional current
- N/A
- not applicable
- NMJ
- neuromuscular junction
- RIM
- Rab3-interacting molecule
- RRP
- readily releasable pool
- STED
- stimulated emission depletion
- SV
- synaptic vesicle
- TEVC
- two-electrode voltage clamp
References
- Aravamudan B., Fergestad T., Davis W.S., Rodesch C.K., Broadie K. 1999. Drosophila UNC-13 is essential for synaptic transmission. Nat. Neurosci. 2:965–971 10.1038/14764 [DOI] [PubMed] [Google Scholar]
- Blum H., Beier H., Gross H.J. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 8:93–99 10.1002/elps.1150080203 [DOI] [Google Scholar]
- Bückers J., Wildanger D., Vicidomini G., Kastrup L., Hell S.W. 2011. Simultaneous multi-lifetime multi-color STED imaging for colocalization analyses. Opt. Express. 19:3130–3143 10.1364/OE.19.003130 [DOI] [PubMed] [Google Scholar]
- Bucurenciu I., Kulik A., Schwaller B., Frotscher M., Jonas P. 2008. Nanodomain coupling between Ca2+ channels and Ca2+ sensors promotes fast and efficient transmitter release at a cortical GABAergic synapse. Neuron. 57:536–545 10.1016/j.neuron.2007.12.026 [DOI] [PubMed] [Google Scholar]
- Clements J.D., Silver R.A. 2000. Unveiling synaptic plasticity: a new graphical and analytical approach. Trends Neurosci. 23:105–113 10.1016/S0166-2236(99)01520-9 [DOI] [PubMed] [Google Scholar]
- Eggermann E., Bucurenciu I., Goswami S.P., Jonas P. 2012. Nanodomain coupling between Ca²+ channels and sensors of exocytosis at fast mammalian synapses. Nat. Rev. Neurosci. 13:7–21 10.1038/nrn3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala J.C. 2005. Reconstruct: a free editor for serial section microscopy. J. Microsc. 218:52–61 10.1111/j.1365-2818.2005.01466.x [DOI] [PubMed] [Google Scholar]
- Fouquet W., Owald D., Wichmann C., Mertel S., Depner H., Dyba M., Hallermann S., Kittel R.J., Eimer S., Sigrist S.J. 2009. Maturation of active zone assembly by Drosophila Bruchpilot. J. Cell Biol. 186:129–145 10.1083/jcb.200812150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E.D., Kessels M.M., Qualmann B. 2003. Temporal and spatial coordination of exocytosis and endocytosis. Nat. Rev. Mol. Cell Biol. 4:127–139 10.1038/nrm1016 [DOI] [PubMed] [Google Scholar]
- Hallermann S., Kittel R.J., Wichmann C., Weyhersmüller A., Fouquet W., Mertel S., Owald D., Eimer S., Depner H., Schwärzel M., et al. 2010. Naked dense bodies provoke depression. J. Neurosci. 30:14340–14345 10.1523/JNEUROSCI.2495-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow M.L., Ress D., Stoschek A., Marshall R.M., McMahan U.J. 2001. The architecture of active zone material at the frog’s neuromuscular junction. Nature. 409:479–484 10.1038/35054000 [DOI] [PubMed] [Google Scholar]
- Haucke V., Neher E., Sigrist S.J. 2011. Protein scaffolds in the coupling of synaptic exocytosis and endocytosis. Nat. Rev. Neurosci. 12:127–138 10.1038/nrn2948 [DOI] [PubMed] [Google Scholar]
- Hell S.W. 2007. Far-field optical nanoscopy. Science. 316:1153–1158 10.1126/science.1137395 [DOI] [PubMed] [Google Scholar]
- Hida Y., Ohtsuka T. 2010. CAST and ELKS proteins: structural and functional determinants of the presynaptic active zone. J. Biochem. 148:131–137 10.1093/jb/mvq065 [DOI] [PubMed] [Google Scholar]
- Holderith N., Lorincz A., Katona G., Rózsa B., Kulik A., Watanabe M., Nusser Z. 2012. Release probability of hippocampal glutamatergic terminals scales with the size of the active zone. Nat. Neurosci. 15:988–997 10.1038/nn.3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Tamura T., Kidokoro Y. 2008. Delayed synaptic transmission in Drosophila cacophonynull embryos. J. Neurophysiol. 100:2833–2842 10.1152/jn.90342.2008 [DOI] [PubMed] [Google Scholar]
- Jiao W., Masich S., Franzén O., Shupliakov O. 2010. Two pools of vesicles associated with the presynaptic cytosolic projection in Drosophila neuromuscular junctions. J. Struct. Biol. 172:389–394 10.1016/j.jsb.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Jin Y., Garner C.C. 2008. Molecular mechanisms of presynaptic differentiation. Annu. Rev. Cell Dev. Biol. 24:237–262 10.1146/annurev.cellbio.23.090506.123417 [DOI] [PubMed] [Google Scholar]
- Kawasaki F., Collins S.C., Ordway R.W. 2002. Synaptic calcium-channel function in Drosophila: analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony. J. Neurosci. 22:5856–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F., Zou B., Xu X., Ordway R.W. 2004. Active zone localization of presynaptic calcium channels encoded by the cacophony locus of Drosophila. J. Neurosci. 24:282–285 10.1523/JNEUROSCI.3553-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel R.J., Wichmann C., Rasse T.M., Fouquet W., Schmidt M., Schmid A., Wagh D.A., Pawlu C., Kellner R.R., Willig K.I., et al. 2006. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 312:1051–1054 10.1126/science.1126308 [DOI] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Liu K.S., Siebert M., Mertel S., Knoche E., Wegener S., Wichmann C., Matkovic T., Muhammad K., Depner H., Mettke C., et al. 2011. RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science. 334:1565–1569 10.1126/science.1212991 [DOI] [PubMed] [Google Scholar]
- Matz J., Gilyan A., Kolar A., McCarvill T., Krueger S.R. 2010. Rapid structural alterations of the active zone lead to sustained changes in neurotransmitter release. Proc. Natl. Acad. Sci. USA. 107:8836–8841 10.1073/pnas.0906087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinrenken C.J., Borst J.G., Sakmann B. 2002. Calcium secretion coupling at calyx of held governed by nonuniform channel-vesicle topography. J. Neurosci. 22:1648–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miśkiewicz K., Jose L.E., Bento-Abreu A., Fislage M., Taes I., Kasprowicz J., Swerts J., Sigrist S., Versées W., Robberecht W., Verstreken P. 2011. ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron. 72:776–788 10.1016/j.neuron.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Mittelstaedt T., Alvaréz-Baron E., Schoch S. 2010. RIM proteins and their role in synapse function. Biol. Chem. 391:599–606 10.1515/bc.2010.064 [DOI] [PubMed] [Google Scholar]
- Müller M., Liu K.S., Sigrist S.J., Davis G.W. 2012. RIM controls homeostatic plasticity through modulation of the readily-releasable vesicle pool. J. Neurosci. 32:16574–16585 10.1523/JNEUROSCI.0981-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy V.N., De Camilli P. 2003. Cell biology of the presynaptic terminal. Annu. Rev. Neurosci. 26:701–728 10.1146/annurev.neuro.26.041002.131445 [DOI] [PubMed] [Google Scholar]
- Neher E., Sakaba T. 2008. Multiple roles of calcium ions in the regulation of neurotransmitter release. Neuron. 59:861–872 10.1016/j.neuron.2008.08.019 [DOI] [PubMed] [Google Scholar]
- Owald D., Fouquet W., Schmidt M., Wichmann C., Mertel S., Depner H., Christiansen F., Zube C., Quentin C., Körner J., et al. 2010. A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. J. Cell Biol. 188:565–579 10.1083/jcb.200908055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlu C., DiAntonio A., Heckmann M. 2004. Postfusional control of quantal current shape. Neuron. 42:607–618 10.1016/S0896-6273(04)00269-7 [DOI] [PubMed] [Google Scholar]
- Peled E.S., Isacoff E.Y. 2011. Optical quantal analysis of synaptic transmission in wild-type and rab3-mutant Drosophila motor axons. Nat. Neurosci. 14:519–526 10.1038/nn.2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Schwarz T., Kittel R.J., Schmid A., Rasse T.M., Kappei D., Ponimaskin E., Heckmann M., Sigrist S.J. 2005. Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. J. Neurosci. 25:3209–3218 10.1523/JNEUROSCI.4194-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasse T.M., Fouquet W., Schmid A., Kittel R.J., Mertel S., Sigrist C.B., Schmidt M., Guzman A., Merino C., Qin G., et al. 2005. Glutamate receptor dynamics organizing synapse formation in vivo. Nat. Neurosci. 8:898–905 10.1038/nn1484 [DOI] [PubMed] [Google Scholar]
- Reiff D.F., Thiel P.R., Schuster C.M. 2002. Differential regulation of active zone density during long-term strengthening of Drosophila neuromuscular junctions. J. Neurosci. 22:9399–9409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettig J., Neher E. 2002. Emerging roles of presynaptic proteins in Ca++-triggered exocytosis. Science. 298:781–785 10.1126/science.1075375 [DOI] [PubMed] [Google Scholar]
- Rizo J., Südhof T.C. 2012. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—guilty as charged? Annu. Rev. Cell Dev. Biol. 28:279–308 10.1146/annurev-cellbio-101011-155818 [DOI] [PubMed] [Google Scholar]
- Sakaba T., Schneggenburger R., Neher E. 2002. Estimation of quantal parameters at the calyx of Held synapse. Neurosci. Res. 44:343–356 10.1016/S0168-0102(02)00174-8 [DOI] [PubMed] [Google Scholar]
- Schmid A., Hallermann S., Kittel R.J., Khorramshahi O., Frölich A.M., Quentin C., Rasse T.M., Mertel S., Heckmann M., Sigrist S.J. 2008. Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat. Neurosci. 11:659–666 10.1038/nn.2122 [DOI] [PubMed] [Google Scholar]
- Sheng J., He L., Zheng H., Xue L., Luo F., Shin W., Sun T., Kuner T., Yue D.T., Wu L.G. 2012. Calcium-channel number critically influences synaptic strength and plasticity at the active zone. Nat. Neurosci. 15:998–1006 10.1038/nn.3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan D.L., Berlot C.H., Robert A., Inglis F.M., Jakobsdottir K.B., Howe J.R., Hughes T.E. 2002. A new way to rapidly create functional, fluorescent fusion proteins: random insertion of GFP with an in vitro transposition reaction. BMC Neurosci. 3:7 10.1186/1471-2202-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S.J., Schmitz D. 2011. Structural and functional plasticity of the cytoplasmic active zone. Curr. Opin. Neurobiol. 21:144–150 10.1016/j.conb.2010.08.012 [DOI] [PubMed] [Google Scholar]
- Silver R.A. 2003. Estimation of nonuniform quantal parameters with multiple-probability fluctuation analysis: theory, application and limitations. J. Neurosci. Methods. 130:127–141 10.1016/j.jneumeth.2003.09.030 [DOI] [PubMed] [Google Scholar]
- Spangler S.A., Hoogenraad C.C. 2007. Liprin-alpha proteins: scaffold molecules for synapse maturation. Biochem. Soc. Trans. 35:1278–1282 10.1042/BST0351278 [DOI] [PubMed] [Google Scholar]
- Stewart B.A., Atwood H.L., Renger J.J., Wang J., Wu C.F. 1994. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 175:179–191 10.1007/BF00215114 [DOI] [PubMed] [Google Scholar]
- Südhof T.C. 2012. The presynaptic active zone. Neuron. 75:11–25 10.1016/j.neuron.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J., He Y., Hoskins R.A., Bellen H.J. 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 314:1747–1751 10.1126/science.1134426 [DOI] [PubMed] [Google Scholar]
- Venken K.J., Kasprowicz J., Kuenen S., Yan J., Hassan B.A., Verstreken P. 2008. Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 36:e114 10.1093/nar/gkn486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh D.A., Rasse T.M., Asan E., Hofbauer A., Schwenkert I., Dürrbeck H., Buchner S., Dabauvalle M.C., Schmidt M., Qin G., et al. 2006. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 49:833–844 10.1016/j.neuron.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Weyhersmüller A., Hallermann S., Wagner N., Eilers J. 2011. Rapid active zone remodeling during synaptic plasticity. J. Neurosci. 31:6041–6052 10.1523/JNEUROSCI.6698-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik S.M., Brose N. 2007. Regulation of membrane fusion in synaptic excitation-secretion coupling: speed and accuracy matter. Neuron. 55:11–24 10.1016/j.neuron.2007.06.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.