ABSTRACT
The importance of livestock as a source of bacterial pathogens with the potential for epidemic spread in human populations is unclear. In recent years, there has been a global increase in community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections of healthy humans, but an understanding of the different evolutionary origins of CA-MRSA clones and the basis for their recent expansion is lacking. Here, using a high-resolution phylogenetic approach, we report the discovery of two emergent clones of human epidemic CA-MRSA which resulted from independent livestock-to-human host jumps by the major bovine S. aureus complex, CC97. Of note, one of the new clones was isolated from human infections on four continents, demonstrating its global dissemination since the host jump occurred over 40 years ago. The emergence of both human S. aureus clones coincided with the independent acquisition of mobile genetic elements encoding antimicrobial resistance and human-specific mediators of immune evasion, consistent with an important role for these genetic events in the capacity to survive and transmit among human populations. In conclusion, we provide evidence that livestock represent a reservoir for the emergence of new human-pathogenic S. aureus clones with the capacity for pandemic spread. These findings have major public health implications highlighting the importance of surveillance for early identification of emergent clones and improved transmission control measures at the human-livestock interface.
IMPORTANCE
Animals are the major source of new pathogens affecting humans. However, the potential for pathogenic bacteria that originally were found in animals to switch hosts and become widely established in human populations is not clear. Here, we report the discovery of emergent clones of methicillin-resistant Staphylococcus aureus (MRSA) that originated in livestock and switched to humans, followed by host-adaptive evolution and epidemic spread in global human populations. Our findings demonstrate that livestock can act as a reservoir for the emergence of new human bacterial clones with potential for pandemic spread, highlighting the potential role of surveillance and biosecurity measures in the agricultural setting for preventing the emergence of new human pathogens.
Introduction
Emerging infections represent a major challenge to human and veterinary medicine. The great majority of new pathogens of humans originate in animal populations, but most are associated with episodic zoonotic infections that do not have the capacity to transmit to other individuals (1). Staphylococcus aureus is a major pathogen responsible for considerable human morbidity and mortality on a global scale (2). S. aureus is also a leading cause of infections of livestock such as cows and is a major economic burden on the global dairy industry (3). The results of population genetics studies have shown that most strains of S. aureus are host specific, indicating a low frequency of cross-species transmission (3). However, recent studies employing multilocus sequence typing (MLST) and whole-genome sequencing (WGS) have identified several S. aureus sequence types (ST) that are associated with multiple host species, implying either zoonotic transmission or a recent common ancestor. For example, the poultry-associated sequence type 5 (ST5) and livestock-associated, methicillin-resistant S. aureus (LA-MRSA) ST398 clones are descended from bacteria that recently made human-to-animal host jumps (4, 5). Importantly, LA-MRSA ST398 strains have acquired antibiotic resistance due to selective pressures in the pig farming industry and have the capacity to cause severe zoonotic infections of humans in contact with pigs on pig farms (5). However, carriage of LA-MRSA ST398 by pig farmers is transient, and LA-MRSA ST398 does not appear to be readily transmissible between humans, probably due to the loss of expression of proteins associated with the cell wall that are required for human host colonization and transmission (6). Loss of function of superfluous genes is a common feature of bacteria undergoing niche adaptation (7), which is likely to attenuate the capacity to infect the ancestral host species (6). Recently, Harrison et al. demonstrated that ST130 MRSA isolates with the mecC gene have spread from cows to humans resulting in clinical infections (8). However, the potential for animal-specialized strains of S. aureus to successfully cross the human species barrier and become epidemic in human populations is unclear. Previously, we reported that the clonal complex 59 (CC59) S. aureus clone that is endemic in Taiwan and has spread to other parts of the world may have originated in livestock about 500 years ago, and this has recently been corroborated by Shepheard et al. (9). However, the limited resolution of MLST has precluded rigorous examination of the occurrence of more-recent livestock-to-human host jump events leading to the emergence of new epidemic S. aureus clones (10).
S. aureus strains belonging to MLST CC97 are a leading cause of bovine mastitis in Europe, Asia, and North and South America (11, 12). Less commonly, CC97 has also been reported to cause infections of small ruminants, pigs, and humans (11, 13, 14). Importantly, there have been increasing reports of human infections caused by CC97 isolates in Europe, North and South America, Africa, and Asia (13, 15–18). However, the origin of human CC97 strains and their relatedness to livestock-associated CC97 strains are unknown. Here, we investigate the evolutionary history of the CC97 S. aureus lineage and identify clones that are epidemic in human populations that evolved through host jumps from cows followed by human host adaptation. Our findings highlight cows as a potential reservoir for the emergence of new clones with the capacity for pandemic spread in humans.
RESULTS AND DISCUSSION
CC97 is an emerging cause of human infections.
The earliest report in the literature of a CC97 isolate is the archetypal bovine reference strain of S. aureus, Newbould 305 (NCIMB 702892), isolated from a case of mastitis in Canada in 1958 (19), whereas the earliest report of human CC97 isolates relates to strains isolated 38 years later in 1996 (20). However, human CC97 S. aureus bacteria have since been identified in at least 35 countries (see Table S2 in the supplemental material). In order to investigate the possibility that infections due to CC97 strains are increasing in prevalence in human populations, we determined the number of CC97-associated strains isolated from MRSA and bacteremia infections in Denmark between 2007 and 2011. Uniquely, in Denmark, submission of all MRSA to the Danish National MRSA Reference Laboratory, Statens Serum Institut, has been mandatory since November 2006, and all MRSA and bacteremia S. aureus isolates submitted since 2007 have been genotyped by staphylococcal protein A (spa) typing. In Denmark, cases of MRSA caused by CC97-related spa types increased from a total of 2 in 2007 to 22 in 2011, equivalent to an 11-fold increase in 5 years. This represents a significant increase in prevalence from 0.3% to 1.7% of total annual MRSA in Denmark since 2007 (P < 0.01). Although surveillance systems for other countries are currently inadequate to identify trends in S. aureus genotype associated with human infections, a recent study of community-associated S. aureus isolated from humans in 16 countries in Europe reported that 3% of community-associated MRSA (CA-MRSA) and 8% of community-associated methicillin-sensitive S. aureus (CA-MSSA) were ST97 isolates (21). Taken together, these data suggest that CC97 is an emerging cause of human infections.
Human ST97 S. aureus clones originated from bovine-to-human host jumps.
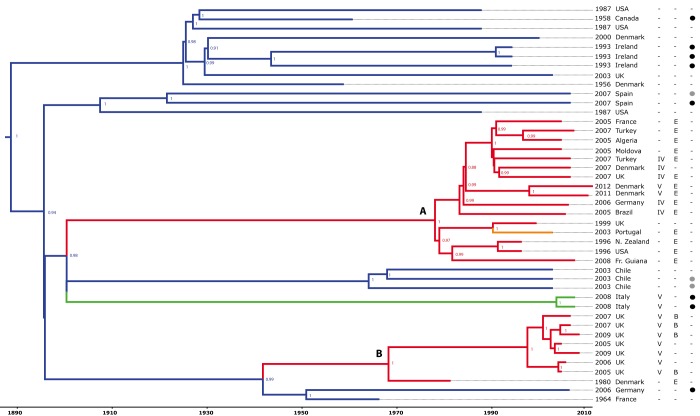
In order to investigate the evolutionary history of the CC97 clone, we obtained 220 CC97 S. aureus isolates of human, bovine, porcine, and caprine origin, isolated in 18 countries on four continents between 1956 and 2012. For whole-genome sequencing, we selected 43 CC97 S. aureus isolates (including 16 MRSA) which broadly represented the breadth of host, geographic, and temporal variation identified among strains reported in the literature, in addition to a single isolate of the closely related ST28 as a human-specific outgroup. The core genome of the 43 CC97 isolates consisted of 2,079,972 bp which included 5,425 high-quality single nucleotide polymorphisms (SNP). Phylogenetic reconstruction of the CC97 lineage was carried out by using the BEAST program (named BEAST for Bayesian evolutionary analysis by sampling trees) (22) (Fig. 1). The consistency index for parsimonious sites predicted using PAUP*4.0b10 (23) indicated a very low predicted frequency of homoplasic events (consistency index [CI] = 0.95), implying an unconflicted phylogenetic signal. Furthermore, a maximum likelihood tree constructed from the same core genome alignment resulted in a largely similar phylogeny with strong bootstrap support at most nodes (see Fig. S1 in the supplemental material). Overall, the high-resolution phylogenetic tree resolves S. aureus CC97 into distinct host-associated clades (Fig. 1). Of note, there is considerable genetic diversity among CC97 isolates of livestock origin, which is indicated by numerous deep branches in the phylogenetic tree, compared to human isolates which are restricted to two distinct clades of closely related isolates (Fig. 1). The majority of livestock-associated CC97 isolates lie basal to the human clades, and a continuous-time Markov model with host association as a discrete trait (10, 36) provided strong support that the most recent common ancestor of each human clade was of bovine origin (see Table S3 in the supplemental material). These data indicate that CC97 isolates circulating among human populations are the result of livestock-to-human host jumps which have occurred on at least 2 independent occasions. Human CC97 clade A consists of isolates of both MSSA and MRSA from 12 different countries on four different continents, indicating its global dissemination (Fig. 1). In contrast, human CC97 clade B is represented by a single MSSA isolate from a Danish patient in 1980 and several MRSA bacteria isolated within the last 10 years in the Midlands region of the United Kingdom, indicating a more limited geographic distribution among the strains sampled (13). In order to determine the time frame of the livestock-to-human host jump events, we determined mutation rates for the CC97 lineage, allowing for variation in rates associated with different clades. Initially, an uncorrelated lognormal relaxed molecular clock model was used to determine a mutation rate for the livestock strains only of 1.53 × 10−6 nucleotide substitutions per site per year (95% highest posterior densities [HPDs] of 1.35 × 10−6 to 1.72 × 10−6). This rate was then fixed for livestock strains, and a local rate clock model was applied to each of the human clades, resulting in estimates of 9.58 × 10−7 (7.80 × 10−7 to 1.15 × 10−6) for human clade A and 1.29 × 10−6 (1.05 × 10−6 to 1.54 × 10−6) for human clade B. These data indicate that closely related S. aureus clones can have different mutation rates which may reflect variations in selective pressures encountered in different ecological niches. The Bayesian analysis resulted in estimates of host jump events which occurred between 1894 and 1977 for human CC97 clade A and between 1938 and 1966 for human CC97 clade B (Fig. 1; Table S3). In addition, we estimated the date of the most-recent human ancestor (MRCA) of CC97 and the human outgroup ST28 to be AD 784 (BC 325 to AD 1460), which lies within the credibility intervals of the previous estimate of the most recent minimum date for the host jump event using MLST data (Table S3) (10).
FIG 1 .
Identification of human epidemic S. aureus clones descended from bacteria that made livestock-to-human host jumps. The Bayesian phylogenetic reconstruction of the CC97 lineage is shown. The tree is based on core genome alignment with branches color coded according to the host species association (blue, bovine; green, porcine; orange, caprine; red, human) and date and country of origin of each isolate indicated. The presence or absence (−) of β-toxin phage IEC variants B and E and SCCmec type IV or V is indicated by the appropriate letter, and the presence of S. aureus pathogenicity island (SaPI)-encoded vwb or phage-encoded lukM/lukF is denoted by black and gray circles, respectively. The branch lengths are scaled according to the time scale bar (years) and the posterior probability values are indicated at each node. Clades A and B are shown.
The results of previous studies employing MLST have suggested that livestock-associated strains of S. aureus originated from human ancestral strains through human-to-animal host jumps leading to host-adapted clones specialized for livestock with occasional host jumps back into humans (10). In particular, the CC59 S. aureus clone that is endemic in human populations in Taiwan may have originated in livestock about 500 years ago (10). Here, we provide an example of recently emerged clones of human S. aureus that evolved through independent host jumps from livestock. These data demonstrate that livestock represent a potential reservoir of pathogenic bacteria that may cross the species barrier and spread among global human populations.
Methicillin resistance was acquired by human CC97 clones subsequent to the host jumps from cows.
Antibiotic susceptibility testing of CC97 isolates revealed that 7 of 17 bovine isolates were sensitive to all antimicrobial agents tested, with a further 6 resistant to only a single agent, demonstrating the low prevalence of antimicrobial resistance among isolates of the leading bovine clone of S. aureus (see Fig. S2 in the supplemental material). In contrast, 20 of 23 human isolates were resistant to at least one antimicrobial agent, with some strains resistant to β-lactam antimicrobials, lincosamides, erythromycin, and trimethoprim. Importantly, methicillin resistance is a characteristic of the human CC97 clades, with two distinct staphylococcal cassette chromosome mec element (SCCmec) types, types IV and V, which are associated with human CC97 clades A and B (Fig. 1). Of note, none of the isolates examined contained the novel mecC allele previously found among several bovine MRSA clones responsible for episodes of human zoonotic infection (8). In contrast, all of the bovine S. aureus isolates examined in the study are methicillin sensitive, suggesting that resistance was acquired after the host jump from cows to humans, presumably as a result of selective pressures imposed by prescription of antibiotics for treating human infections. Consistent with this, the earliest human isolate identified in clade B (isolated in Denmark in 1980) was sensitive to all antibiotics tested, whereas all other isolates of clade B (isolated since 2005) were resistant to multiple antimicrobial classes (see Fig. S2 in the supplemental material). Recently, it was demonstrated that methicillin and tetracycline resistance was likely acquired by LA-MRSA ST398 strains by antibiotic selective pressures encountered within the pig farming industry (5). Of note, the two ST97 isolates from pigs in the current study were resistant to both tetracycline and methicillin, in addition to ciprofloxacin (Fig. S2). Overall, the antimicrobial susceptibility profiles of the human and pig CC97 isolates demonstrated resistance to a much greater number of antimicrobials than the bovine CC97 S. aureus (Fig. S2). These data imply that the dairy industry does not strongly promote the emergence of antibiotic-resistant S. aureus, in spite of the widespread use of antibiotics for treating bovine mastitis. We speculate that this reflects the ability of bovine strains to invade and survive within bovine mammary epithelial cells, a niche which may have a markedly reduced antibiotic selective pressure (24).
The content of MGE among CC97 isolates correlates with host species.
Previous studies have highlighted the importance of mobile genetic elements (MGE) in adaptation of S. aureus to distinct host species (3). In the current study, of the 20 livestock isolates, 3 and 8 isolates contained MGE encoding the LukM/F leukotoxin and the von Willebrand binding protein (vWbp), respectively, both of which have been demonstrated to have ruminant host-specific activity (3) (Fig. 1). In contrast, none of the human isolates contained the livestock-associated MGE. The family of β-toxin-converting phages (φSa3) associated with human S. aureus typically contain an immune evasion cluster (IEC) of genes encoding secreted proteins such as staphylokinase (sak), staphylococcal complement inhibitor (scn), and chemotaxis inhibitory protein of S. aureus (chp) which contribute to immune evasion in a human host-specific manner (25). Consistent with previous reports, in the current study, we found that 19 of 23 human isolates and none of 19 bovine or pig isolates contained a φSa3 with an IEC, demonstrating a strong correlation with a human host association (25). Of the 16 human CC97 clade A isolates, 14 had an IEC containing genes sak and scn (IEC type E), and of the 8 human CC97 clade B isolates, 4 had an IEC containing sak, chp, and scn (IEC type B) and one isolate had an IEC type E (Fig. 1) (25). Of note, the single goat strain that cosegregates with human clade A contains a φSa3 phage which is highly similar to that of the human strains in clade A, implying that it is a human contaminant or the result of a very recent human-to-goat transmission event. In addition, the arginine catabolic mobile element (ACME) is a characteristic of some CA-MRSA clones, including the highly successful USA300 clone which may contribute to enhanced survival during community-associated infections (26). Of note, all seven United Kingdom isolates of the human CC97 clade B contained ACME linked to SCCmec type V (13). Taken together, the distribution of MGE among CC97 isolates reveals horizontal gene acquisition events which correlate with adaptation to different host-associated ecological niches.
Concluding comments.
The recent global increase in CA-MRSA infections reflects the expansion of an array of S. aureus clones with distinct evolutionary histories. Here, we demonstrate that livestock is one potential reservoir of pathogenic bacteria with the capacity to cross the species barrier, undergo host-adaptive evolution, and become established in global human populations. Furthermore, the data suggest that a limited number of genetic events may be sufficient to transform an S. aureus strain which has coevolved with bovine hosts over several thousand years into a successful human epidemic lineage.
The importance of hygiene in prevention of hospital transmission of nosocomial pathogens such as MRSA is widely appreciated, and the recent reduction in hospital MRSA infections is likely due in part to improved hygiene measures for controlling transmission (27). Improved biosecurity and hygiene control measures which prevent the spread of bacterial flora between livestock and human hosts may limit opportunities for successful livestock-to-human transmission. Furthermore, regular surveillance of the microbiota in livestock and humans may facilitate the early identification of emergent clones with the capacity to transmit and cause disease among human populations.
MATERIALS AND METHODS
Bacterial isolates.
A total of 220 S. aureus isolates of CC97 from bovine, human, porcine, and caprine hosts isolated between 1956 and 2012 in 18 different countries on four continents were obtained. For whole-genome sequencing, a total of 43 CC97 strains were selected to represent the breadth of host, clinical, spatial, and temporal variation (see Table S1 in the supplemental material). Strains were grown for 16 h on tryptic soy agar (TSA) at 37°C or in tryptic soy broth (TSB) at 37°C with shaking at 200 rpm. Genomic DNA was isolated using the PurElute bacterial genomic kit (Edge BioSystems, MD), with an amended protocol as previously described (4). MLST was conducted to confirm the sequence type (ST) prior to whole-genome sequencing using methods described previously (28).
Genome sequencing, mapping assembly, and SNP calling.
Paired-end Illumina sequencing of bacterial strains was carried out on an Illumina genome analyzer IIx or Miseq, following standard Illumina protocols. Nucleotide distribution and quality scores of raw reads were assessed and filtered for quality using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) (genome assembly metrics are provided in Table S4 in the supplemental material). High-quality reads were aligned against the reference genomes of S. aureus strains MW2 (accession number NC_003923) or Mu50 (accession number NC_002758.2), variant sites were detected, and consensus sequences were called as described previously (29), with extended base alignment quality (BAQ) computation employed in SAMtools v.0.1.16 (30). The core genome was defined as all nucleotide sites shared by all isolates, and putative recombinant regions were removed after breakpoint detection with the suite of programs included in the Recombination Detection Program v4.13 (RDP) (31). For comparisons of gene content, de novo assemblies of short reads was carried out using Velvet v1.0.15 (32) and the VelvetOptimiser.pl script within VelvetOptimiser v-2.1.7 (http://bioinformatics.net.au/software.velvetoptimiser.shtml).
Phylogenetic analysis.
Core genome and in-frame protein-coding sequences were extracted from the genome consensus sequences to construct alignments using custom scripts. The maximum likelihood phylogeny was reconstructed using RAxML-7.2.6 (33) implementing a generalized time reversible (GTR) model with gamma correction for rate heterogeneity, and 1,000 bootstrap replicates (see Fig. S1 in the supplemental material). Bayesian phylogenetic analysis was conducted using BEAST v1.7.1 (34) implementing the Hasegawa-Kishino-Yano model of sequence evolution with gamma correction for rate heterogeneity. The rates were calculated by the dated tip method using a three-rate local clock model, with the livestock rate constrained to 1.53 × 10−6 nucleotide substitutions per site per year as determined using an uncorrelated relaxed molecular clock model with a constant coalescent prior (35). For the date of the MRCA with the human outgroup ST28, an in-frame coding sequence (CDS) alignment of CC97 and ST28 sequences was employed which allowed the estimation of mutation rate over all sites and third codon positions only. In each case, the mutation rates were comparable and resulted in similar estimates of the host jump date (Table S3). Ancestral host states were predicted using a model of discrete trait evolution (36), adapting the method by substituting geographic location with host state as described previously (10). The consistency index for parsimony informative sites was determined using PAUP v4.0b10 (23).
Accessory genome analysis.
De novo contigs and reference assembly sequences were interrogated for specific MGE using BLASTn (37), and MGE sequences were aligned using Mauve v2.3.1 (38). Immune evasion cluster (IEC) type was defined as described previously (25).
Antimicrobial sensitivity testing.
All isolates in the study were tested using the Vitek 2 system (AST-P620 card) (bioMérieux United Kingdom Limited, Basingstoke, United Kingdom) with a panel of antimicrobial agents, including cefoxitin, benzylpenicillin, oxacillin, gentamicin, ciprofloxacin, inducible clindamycin resistance, erythromycin, clindamycin, linezolid, daptomycin, teicoplanin, vancomycin, tetracycline, tigecycline, nitrofurantoin, fusidic acid, mupirocin, chloramphenicol, rifampicin, and trimethoprim. Results for antibiotic susceptibility were interpreted using standards defined by the Clinical and Laboratory Standards Institute (CLSI) (39).
Statistical analysis.
For analysis of the increase in CC97-associated infections in Denmark, the Cochran-Armitage trend test was employed (GraphPad Prism 5.0).
Nucleotide sequence accession numbers.
The Illumina sequences generated and used in this study are deposited and available in the Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under the study accession number PRJEB1411, located at http://www.ebi.ac.uk/ena/data/view/PRJEB1411. The S. aureus isolates are under sample accession numbers ERS212248 to ERS212287 and ERS249844 to ERS249847.
SUPPLEMENTAL MATERIAL
Maximum likelihood tree of CC97 S. aureus lineage based on core genome alignment rooted with an outgroup strain of ST28 (18_1997). The branches are color coded according to the host species from which the strain was isolated (blue, bovine; green, porcine; orange, caprine; red, human). The scale bar represents the number of nucleotide substitutions per site. Bootstrap values (based on 1,000 replicates) are shown for each node. Download
Antimicrobial sensitivity profiles of CC97 S. aureus. The antibiogram for each strain is shown as a horizontal panel against the corresponding taxa as depicted in the Bayesian phylogeny. Sensitivity to each antimicrobial agent is indicated as follows: S, susceptible; R, resistant; I, intermediate resistance; NS, nonsusceptible (likely indicating a heterogenous population containing a resistant subpopulation). Antimicrobial abbreviations are as follows: OXSF, cefoxitin; P, benzylpenicillin; OX, oxacillin; GM, gentamicin; CIP, ciprofloxacin; ICR, inducible clindamycin resistance; E, erythromycin; CM, clindamycin; LNZ, linezolid; DAP, daptomycin; TEC, teicoplanin; VA, vancomycin; TE, tetracycline; TGC, tigecycline; FT, furantoin; MUP, mupirocin; C, chloramphenicol; RA, rifampin; TMP, trimethoprim. Download
Bacterial strains used in this study.
Countries in which human CC97 S. aureus has been identified.
Estimated dates of host jump events associated with CC97 S. aureus.
Assembly metrics for genome sequences included in the study (BWA reference based).
ACKNOWLEDGMENTS
This work was funded by a project grant (BB/I013873/1) and institute strategic grant funding from the Biotechnology and Biological Sciences Research Council (United Kingdom) to J.R.F., and by a Riddell-Swan, Ker memorial Ph.D. studentship to L.E.S.
We are grateful to R. Cartwright, A. Battisti, E. Smith, L. Fox, J. Kruze, R. de Campo, J. Lindsay, H. de Lencastre, R. Miller, S. Monecke, F. Layer, B. Strommenger, W. Witte, M. Sudagidan, K. R. N. dos Santos, R. Ruimy, H. Westh, A. Kearns, M. Ellington, and E. Feil for providing isolates, ARK Genomics, Roslin Institute for sequencing services, and K. Templeton and G. McAllister for performing antimicrobial sensitivity testing.
L. E. Spoor and J. R. Fitzgerald designed research. L. E. Spoor, L. A. Weinert, P. R. McAdam, A. R. Larsen, and J. R. Fitzgerald performed research and analyzed data. R. E. Skov, H. Hasman, F. M. Aarestrup, and A. M. Kearns contributed new reagents or analytic tools. L. E. Spoor and J. R. Fitzgerald wrote the paper.
Footnotes
Citation Spoor LE, McAdam PR, Weinert LA, Rambaut A, Hasman H, Aarestrup FM, Kearns AM, Larsen AR, Skov RL, Fitzgerald JR. 2013. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. mBio 4(4):e00356-13. doi:10.1128/mBio.00356-13.
REFERENCES
- 1. Woolhouse M, Gaunt E. 2007. Ecological origins of novel human pathogens. Crit. Rev. Microbiol. 33:231–242 [DOI] [PubMed] [Google Scholar]
- 2. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 3. Fitzgerald JR. 2012. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 20:192–198 [DOI] [PubMed] [Google Scholar]
- 4. Lowder BV, Guinane CM, Ben Zakour NL, Weinert LA, Conway-Morris A, Cartwright RA, Simpson AJ, Rambaut A, Nübel U, Fitzgerald JR. 2009. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 106:19545–19550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3(1):e00520-12. 10.1128/mBio.00305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uhlemann A-C, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, Lipkin WI, Lindsay JA, DeLeo FR, Lowy FD. 2012. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 3(2):e00027-12. 10.1128/mBio.00027-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lerat E, Ochman H. 2005. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 33:3125–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrison EM, Paterson GK, Holden MTG, Larsen J, Stegger M, Larsen AR, Petersen A, Skov RL, Christensen JM, Bak Zeuthen A, Heltberg O, Harris SR, Zadoks RN, Parkhill J, Peacock SJ, Holmes MA. 2013. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol. Med. 5:505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shepheard MA, Fleming VM, Connor TR, Corander J, Feil EJ, Fraser C, Hanage WP. 2013. Historical zoonoses and other changes in host tropism of Staphylococcus aureus, identified by phylogenetic analysis of a population dataset. PLoS One 8:e62369. 10.1371/journal.pone.0062369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weinert LA, Welch JJ, Suchard MA, Lemey P, Rambaut A, Fitzgerald JR. 2012. Molecular dating of human-to-bovid host jumps by Staphylococcus aureus reveals an association with the spread of domestication. Biol. Lett. 8:829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smyth DS, Feil EJ, Meaney WJ, Hartigan PJ, Tollersrud T, Fitzgerald JR, Enright MC, Smyth CJ. 2009. Molecular genetic typing reveals further insights into the diversity of animal-associated Staphylococcus aureus. J. Med. Microbiol. 58:1343–1353 [DOI] [PubMed] [Google Scholar]
- 12. Smith EM, Green LE, Medley GF, Bird HE, Fox LK, Schukken YH, Kruze JV, Bradley AJ, Zadoks RN, Dowson CG. 2005. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol. 43:4737–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ellington MJ, Yearwood L, Ganner M, East C, Kearns AM. 2008. Distribution of the ACME-arcA gene among methicillin-resistant Staphylococcus aureus from England and Wales. J. Antimicrob. Chemother. 61:73–77 [DOI] [PubMed] [Google Scholar]
- 14. Battisti A, Franco A, Merialdi G, Hasman H, Iurescia M, Lorenzetti R, Feltrin F, Zini M, Aarestrup FM. 2010. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet. Microbiol. 142:361–366 [DOI] [PubMed] [Google Scholar]
- 15. Ruimy R, Armand-Lefevre L, Barbier F, Ruppé E, Cocojaru R, Mesli Y, Maiga A, Benkalfat M, Benchouk S, Hassaine H, Dufourcq JB, Nareth C, Sarthou JL, Andremont A, Feil EJ. 2009. Comparisons between geographically diverse samples of carried Staphylococcus aureus. J. Bacteriol. 191:5577–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Udo EE, Aly NY, Sarkhoo E, Al-Sawan R, Al-Asar AS. 2011. Detection and characterization of an ST97-SCCmec-V community-associated meticillin-resistant Staphylococcus aureus clone in a neonatal intensive care unit and special care baby unit. J. Med. Microbiol. 60:600–604 [DOI] [PubMed] [Google Scholar]
- 17. Diep BA, Perdreau-Remington F, Sensabaugh GF. 2003. Clonal characterization of Staphylococcus aureus by multilocus restriction fragment typing, a rapid screening approach for molecular epidemiology. J. Clin. Microbiol. 41:4559–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schuenck RP, Nouér SA, de Oliveira Winter C, Cavalcante FS, Scotti TD, Ferreira ALP, Giambiagi-de Marval M, Netto dos Santos KR. 2009. Polyclonal presence of non-multiresistant methicillin-resistant Staphylococcus aureus isolates carrying SCCmec IV in health care-associated infections in a hospital in Rio de Janeiro, Brazil. Diagn. Microbiol. Infect. Dis. 64:434–441 [DOI] [PubMed] [Google Scholar]
- 19. Prasad LB, Newbould FH. 1968. Inoculation of the bovine teat duct with Staph. aureus: the relationship of teat duct length, milk yield and milking rate to development of intramammary infection. Can. Vet. J. 9:107–115 [PMC free article] [PubMed] [Google Scholar]
- 20. Zinn CS, Westh H, Rosdahl VT, Sarisa Study Group 2004. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb. Drug Resist. 10:160–168 [DOI] [PubMed] [Google Scholar]
- 21.Rolo J, Miragaia M, Turlej-Rogacka A, Empel J, Bouchami O, Faria NA, Tavares A, Hryniewicz W, Fluit AC, De Lencastre H, the CONCORD . Working Group; 2012. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLoS One 7:e34768. doi:10.1371/journal.pone.0034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 24. Garzoni C, Kelley WL. 2009. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 17:59–65 [DOI] [PubMed] [Google Scholar]
- 25. van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Jag JA. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wyllie DH, Walker AS, Miller R, Moore C, Williamson SR, Schlackow I, Finney JM, O’Connor L, Peto TE, Crook DW. 2011. Decline of meticillin-resistant Staphylococcus aureus in Oxfordshire hospitals is strain-specific and preceded infection-control intensification. BMJ Open 1:e000160. 10.1136/bmjopen-2011-000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McAdam PR, Templeton KE, Edwards GF, Holden MT, Feil EJ, Aanensen DM, Bargawi HJ, Spratt BG, Bentley SD, Parkhill J, Enright MC, Holmes A, Girvan EK, Godfrey PA, Feldgarden M, Kearns AM, Rambaut A, Robinson DA, Fitzgerald JR. 2012. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 109:9107–9112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup; 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 34. Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. 10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5:e1000520. 10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 38. Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clinical and Laboratory Standards Institute 2007. Performance standards for antimicrobial susceptibility testing. CLSI Document M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood tree of CC97 S. aureus lineage based on core genome alignment rooted with an outgroup strain of ST28 (18_1997). The branches are color coded according to the host species from which the strain was isolated (blue, bovine; green, porcine; orange, caprine; red, human). The scale bar represents the number of nucleotide substitutions per site. Bootstrap values (based on 1,000 replicates) are shown for each node. Download
Antimicrobial sensitivity profiles of CC97 S. aureus. The antibiogram for each strain is shown as a horizontal panel against the corresponding taxa as depicted in the Bayesian phylogeny. Sensitivity to each antimicrobial agent is indicated as follows: S, susceptible; R, resistant; I, intermediate resistance; NS, nonsusceptible (likely indicating a heterogenous population containing a resistant subpopulation). Antimicrobial abbreviations are as follows: OXSF, cefoxitin; P, benzylpenicillin; OX, oxacillin; GM, gentamicin; CIP, ciprofloxacin; ICR, inducible clindamycin resistance; E, erythromycin; CM, clindamycin; LNZ, linezolid; DAP, daptomycin; TEC, teicoplanin; VA, vancomycin; TE, tetracycline; TGC, tigecycline; FT, furantoin; MUP, mupirocin; C, chloramphenicol; RA, rifampin; TMP, trimethoprim. Download
Bacterial strains used in this study.
Countries in which human CC97 S. aureus has been identified.
Estimated dates of host jump events associated with CC97 S. aureus.
Assembly metrics for genome sequences included in the study (BWA reference based).