Abstract
Two histone marks, H3K27me3 and H3K9me3, are well known for their repressive roles in the genic and nongenic regions of metazoan genomes. Several protein complexes are known to be responsible for generating these marks, including polycomb repression complex 2 and several H3K9 methylases. Recent studies have shown that the targeting of these histone-modifying complexes within mammalian genomes may be mediated through several DNA-binding proteins, including AEBP2, JARID2, and YY1. In this review, we discuss the potential targeting mechanisms in light of the recent results that have been derived from genome-wide chromatin immunoprecipitation sequencing data and the in vivo functions of these two histone marks in light of the results derived from mouse and human genetic studies.
Keywords: AEBP2, H3K9me3, H3K27me3, JARID2, PRC2, YY1
Introduction
Two histone marks, H3K27me3 (trimethylation on Lys 27 of histone 3) and H3K9me3 (trimethylation on Lys 9 of histone 3), are known for their roles related to repression in the genic and nongenic regions of metazoan genomes. Several complexes are known to be responsible for generating these marks, including polycomb group complexes and H3K9 methylases. However, the mechanisms by which these complexes are targeted are not well understood, although a number of hypotheses have been proposed to explain these targeting mechanisms. One mechanism would be through DNA-binding proteins, such as JARID2 and AEBP2 for H3K27me3 and other DNA-binding proteins for H3K9me3, whereas the other mechanism recently discovered and proposed would be through noncoding RNA (ncRNA1), which has been recently discovered and proposed. In this review, we summarize and discuss the first mechanism, a DNA-binding protein–mediated targeting mechanism, with the recent results that have been derived from genome-wide chromatin immunoprecipitation sequencing (ChIP-Seq1) data, and we also discuss the in vivo functions of these histone modification marks with the data derived from mouse and human genetic studies.
Genome-Wide Distribution of Histone Modification Marks in Mammals
In the past several years, the genome-wide distribution of the histone marks H3K27me3 and H3K9me3 has been analyzed using ChIP techniques (Boyer et al. 2006; Lee et al. 2006; Mikkelsen et al. 2007). From these studies, the following conclusions about the genome-wide distributions of the two histone modification marks can be derived.
First, although these two marks are recognized as repressive signals, these marks are found in different chromosomal regions. H3K27me3 is detected preferentially in gene-rich regions, which are traditionally defined as R-banding regions by Giemsa staining (Pauler et al. 2009). In particular, H3K27me3 is closely associated with a set of development regulators, estimated to be about 500 genes, in embryonic stem cells (Boyer et al. 2006; Lee et al. 2006). The list of these genes includes Hox, Pax, and Sox gene family members, which are expressed during animal development, but not in pluripotent embryonic stem cells. In a given gene, the promoter region is usually marked with this histone modification, which shows CpG-rich sequence structures (Ku et al. 2008). In contrast, H3K9me3 is detected preferentially in gene-poor regions, which are traditionally defined as G-banding regions by Giemsa staining (Pauler et al. 2009). These regions include satellite repeats in telomeres and pericentromeres, which show tandem repeat sequence structures. H3K9me3 is also detected in several families of retrotransposons that have been amplified through RNA-mediated mechanisms in vertebrate genomes. These retrotransposons with H3K9me3 include long interspersed DNA elements and long terminal repeats (LTRs1) (Mikkelsen et al. 2007). H3K9me3 is also found in one particular gene family, Kruppel-type zinc finger genes, the coding regions of which show tandem repeat sequence structures (Blahnik et al. 2011). Thus, H3K9me3 marks are overall closely associated with tandem repeat sequences, whereas H3K27me3 marks are associated with CpG-rich sequences.
Second, these histone marks are much more prevalent during early embryonic stages, but they become less prevalent once embryonic stem cells are differentiated into somatic cells. In most genomic regions, these early histone modification signals are changed into more permanent repression signals, such as DNA methylation (Meissner et al. 2008). This transition from histone to DNA modifications also appears to be different between the two histone modification marks. In the case of H3K27me3, the majority of the genomic regions with this mark is usually protected from DNA methylation. On the other hand, the regions with H3K9me3 are methylated in somatic cells. In summary, H3K27me3 is regarded as a temporary repression signal that is designed for controlling a set of development regulators. In contrast, H3K9me3 is considered to be a permanent repression signal that is designed for the heterochromatin formation of chromosomal regions with tandem repeat structures.
Modifying Complexes
The H3K27me3 mark is established by a protein complex called polycomb repression complex 2 (PRC21). In the past decade, there have been numerous attempts to biochemically purify PRC2 (Cao and Zhang 2004; Li et al. 2010; Pasini et al. 2010; Peng et al. 2009; Shen et al. 2009). These studies have shown that the following proteins are considered core components for PRC2 based on their co-occurrence in the independent purification attempts: EZH2, EED, SUZ12, and RbAp46/48. EZH2 is the enzyme modifying H3K27, and EED is the adaptor protein connecting all the other components. The exact molecular functions of SUZ12 and RbAp46/48 are still unclear, but these two proteins are known to enhance the enzymatic activity of EZH2. These core components of PRC2 are conserved among all the higher eukaryotes, including insects, vertebrates, and even plants (Schuettengruber et al. 2007). This conservation is consistent with the fact that H3K27me3 is one of the earliest histone modification marks that have appeared during eukaryotes’ evolution. Besides the core components, recent studies have also identified two DNA-binding proteins, AEBP2 and JARID2, as PRC2-interacting components (Li et al. 2010; Pasini et al. 2010; Peng et al. 2009; Shen et al. 2009). Similarly, the two DNA-binding proteins are also conserved among all metazoans, ranging from insects to mammals (Kim et al. 2009; Liu and Montell 2001). According to the expression profiles for the core components of PRC2, the majority of proteins are highly expressed during early embryonic stages, which is also consistent with the fact that H3K27me3 is the most visible within the embryonic stem cells.
The H3K9me3 mark is known to be generated by several enzymes, including SETDB1 (or ESET), SUV39H1, SUV39H2, EHMT1 (GLP), and EHMT2 (G9A). This is in stark contrast to the dominant role played by a single protein, EZH2, for the modification of H3K27me3. This difference may be because H3K9me3 is more global and permanent than H3K27me3. Among these H3K9 methylases, SUV39H1 and SUV39H2 are responsible for establishing H3K9me3 in constitutive heterochromatic, pericentromeric, and telomeric regions (Peters et al. 2001). In contrast, EHMT1 and EHMT2 are responsible for setting up H3K9me3 in euchromatic regions (Tachibana et al. 2002). On the other hand, SETDB1 is highly expressed in ES cells and identified as a major regulator that is required for maintaining the pluripotency and self-renewal properties of ES cells (Yeap et al. 2009; Yuan et al. 2009).
The H3K9 methylases have also been found to interact with various proteins that may mediate recruitment to target genes. First, the interacting partners for SETDB1 include OCT4 in ES cells, suggesting that OCT4 might target SETDB1 to numerous genomic loci in ES cells (Yeap et al. 2009; Yuan et al. 2009). SETDB1 also interacts with KAP-1, which is also known as TRIM28 (tripartite motif–containing 28) (Schultz et al. 2002). KAP-1/TRIM28 is a transcriptional corepressor that interacts with a large number of Kruppel-type zinc finger proteins found in vertebrate genomes, estimated to be more than 500 genes per genome (Hamilton et al. 2003). Thus, the repressive function exerted by the vast majority of Kruppel-type zinc finger proteins is likely mediated through the interaction with KAP-1/TRIM28 and SETDB1, establishing H3K9me3 at the various target loci of these Kruppel-type zinc finger proteins. Second, although the main target loci of SUV39H1 and SUV39H2 are constitutive heterochromatic regions, these two methylases are also known to interact with several proteins, such as tumor suppressor RB and proto-oncoprotein EVI1, to repress individual gene loci (Ait-Si-Ali et al. 2004; Cattaneo et al. 2008). Third, EHMT2 interacts with several DNA-binding proteins, including BLIMP-1, a regulator of primordial germ cell and B cell development, and GFI1, a repressor of the cell cycle regulator p21Cip/EAF (Duan et al. 2005; Gyory et al. 2004). The major expression stages of these H3K9 methylases are also early embryonic stages similar to the core components of PRC2 for H3K27me3. Similar to PRC2, the homologues of these H3K9 methylases are also found in species ranging from insects to mammals, confirming the evolutionary conservation of these methylases, as seen for the H3K27me3 mark.
As described earlier, the histone marks H3K27me3 and H3K9me3 are very closely associated with DNA methylation, although a causal relationship between these two modifications has not been well understood. Consistent with this close association with DNA methylation, several histone methylases are known to interact with DNA methylation machineries. First, EZH2 is known to interact with DNMT3A, DNMT3B, and DNMT1 (Viré et al. 2006). This interaction appears to be critical for the repression of several PcG target genes. Second, EHMT2 is also known to interact with DNMT3A and DNMT3B through a protein subdomain called ankyrin, independent from the SET domain that is responsible for histone methylation (Epsztejn-Litman et al. 2008). Several studies involving biochemical purifications indeed confirmed that isolated multiprotein complexes usually contain both histone and DNA methylases (Cedar and Bergman 2009). This suggests that these complexes may have dual functions for nucleosomes—methylation on both histones and DNA (Cedar and Bergman 2009).
DNA-Binding Proteins and Targeting Mechanisms
In Drosophila, small genomic fragments, that are 60 to 100 base pairs in length are known to bind and recruit PRC2, and these fragments contain a cluster of DNA-binding sites for several transcription factors—Pho (pleiohomeotic), Trl/GAF (Trithorax-like, also known as GAGA factor), Psq (Pipsqueak), Grh (Grainhead), and Zeste (Müller and Kassis 2006). Thus, it is believed that these DNA-binding proteins may work together to recruit PRC2. Similar mechanisms have been proposed for the targeting of vertebrate PRC2, and two DNA-binding proteins have been identified as the targeting components for PRC2—AEBP2, and JARID2.
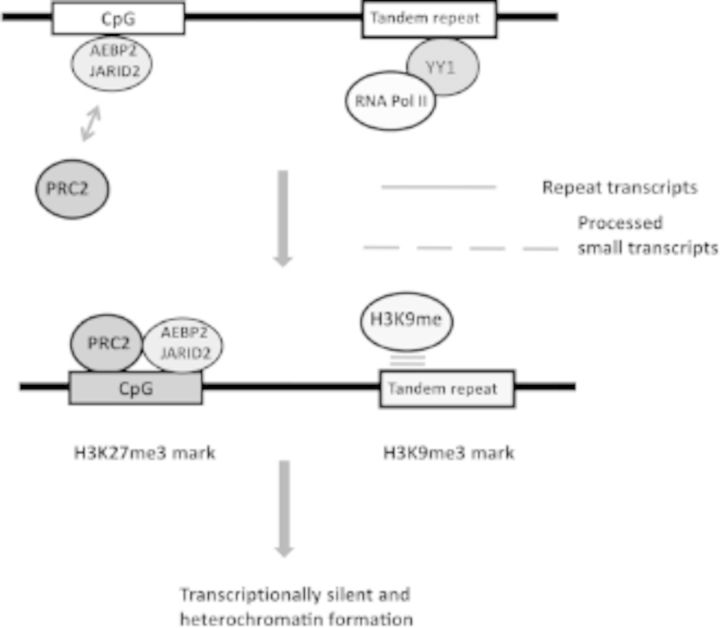
According to the results from ChIP experiments, both proteins occupy a set of genomic loci that are bound by the components of PRC2, further confirming that these two proteins are components of PRC2 (Kim et al. 2009; Li et al. 2010; Pasini et al. 2010; Peng et al. 2009; Shen et al. 2009). Because these two proteins have DNA-binding capability, these two proteins are predicted to function as targeting proteins for PRC2 (Figure 1). Both proteins are known to bind to the genomic regions that contain CpG-rich and GAGA motif–rich sequences (Kim et al. 2009; Peng et al. 2009). The binding of the two proteins to CpG-rich sequences is consistent with the fact that many H3K27me3-marked regions are in the CpG-rich promoter regions of development regulators. The binding to GAGA motifs is also noteworthy because similar motifs have been frequently identified as part of polycomb responsive elements in Drosophila.
Figure 1.
Potential targeting mechanisms for polycomb repression complex 2 (PRC2) and H3K9 methylases. The CpG-rich promoters of development regulators are first recognized by AEBP2 and/or JARID, and subsequently PRC2 is recruited to these target loci, resulting in trimethylation on H3K27. In contrast, the tandem repeats or retrotransposons are first recognized by YY1 and transcribed by Pol II. The repeat-driven transcripts are further processed and later used for the targeting of H3K9 methylases, resulting in trimethylation on H3K9 in the tandem repeats and retrotransposons. Once a given genomic region is marked by H3K27me3 or H3K9me3, that region becomes transcriptionally silent by a series of other follow-up events, including recruitment of PRC1 (H3K27me3) and HP1 (H3K9me3).
Although another DNA-binding protein called Trl/GAF(GAGA factor) is known to bind to the GAGA motif in flies, it will be interesting to test whether the insect homologues of AEBP2 and JARID2 also bind to the known polycomb responsive elements (Kim et al. 2009; Liu and Montell 2001). Several independent groups recently tested the potential targeting function of JARID2 in mouse ES cells. Interestingly, the results are somewhat inconsistent and controversial. RNA interference (RNAi1)–based reduction of JARID2 resulted in either no change or somewhat increased levels of H3K27me3 at the genomic loci that are targeted by PRC2 (Li et al. 2010; Peng et al. 2009; Shen et al. 2009). Although more detailed studies remain to be done, this might be an indication that JARID2 fine-tunes the levels of H3K27me3 for a given locus depending upon the development contexts of cells and tissues. In mutant mice in which the transcription of Aebp2 was disrupted by a knock-in allele, the heterozygotes provided a similar result as seen in the RNAi-based experiments of JARID2, causing increased levels of H3K27me3 (Kim et al. 2011). This unexpected outcome is also controversial at the moment. Nevertheless, there is no doubt that the most obvious role of these two proteins for PRC2 should be DNA binding, although the exact functional context of this DNA binding remains to be investigated.
Several DNA-binding proteins are known to be associated with H3K9 methylases, including OCT4, RB, EVI1, and GFI1, but the targeting function of these DNA-binding proteins is likely limited to a small number of genes because the vast majority of genomic loci marked with H3K9me3 are tandem repeats. Also, the sequences of tandem repeats are quite different from the binding motifs for these proteins. If some DNA-binding proteins are involved in the targeting of H3K9me3 to the tandem repeats, those proteins should have binding capability with tandem repeats and also some connection to H3K9me3 modification.
The protein YY1 fits these criteria based on the following reasons. First, YY1 is known to bind to g-satellite repeats, which are found as part of the pericentromeric repeats of the mouse genome (Shestakova et al. 2004). Also, many families of retrotransposons that are marked with H3K9me3, such as long interspersed DNA elements and LTRs, have the DNA-binding sites for YY1 (Khan et al. 2006; Satyamoorthy et al., 1993). In fact, because many repeats tend to have the DNA-binding sites for YY1, YY1 has been recognized as a surveillance gene that represses transcriptional noise from the vertebrate genomes (Shi et al. 1997). Second, according to the results of YY1 ChIP-Seq data (Mendenhall et al. 2010), many binding sites for YY1 overlap with the peaks of H3K4me3, but not with those of H3K27me3, which is somewhat contradictory to the long-standing prediction that YY1 may be a targeting protein for PRC2 (Atchison et al. 2003; Wilkinson et al. 2006). It is, however, important to note that two duplicated copies of YY1—REX1 and YY2 (Kim et al. 2007)—are known to interact with EED, a component of PRC2. Thus, it is still possible that REX1 or YY2 might recruit PRC2 in ES cells (Garcia-Tuñon et al. 2011). Nevertheless, some YY1 binding sites overlap very well with the peaks of H3K9me3. Furthermore, these genomic loci are g-satellites or retrotransposons, further supporting the possibility that YY1 might be involved in the targeting of H3K9me3 to repetitive regions. Third, although YY1 has long been predicted to be a targeting protein for PRC2, recent studies revealed that YY1 is a component of a newly discovered polycomb complex, termed PhoRC (Pho repressive complex) (Klymenko et al. 2006). The main components of this complex are Pho (YY1 homologue in flies) and another PcG protein called dSFMBT (Scm-related gene containing four MBT domains). SFMBT is a well-known repressor that recognizes H3K9me1 or K3K9me2 (Klymenko et al. 2006). This again supports the potential connection between YY1 and H3K9me3. Fourth, conditional knockdown and knockout of YY1 during spermatogenesis result in a complete depletion of H3K9me3 in developing sperm in the mouse (Wu et al. 2009). These in vivo data suggest that YY1 may be involved in establishing H3K9me3 in germ cells. In summary, the observations described above strongly support the idea that YY1 may be involved in the establishment of H3K9me3.
In terms of detailed mechanisms, YY1's role in H3K9me3 is predicted to be quite different from the mechanism proposed for PRC2, by which DNA-binding proteins, such as AEBP2 and JARID2, bind and recruit the histone-modifying complex. This model is impossible in the case of H3K9me3 because there are a greater number of tandem repeats that need to be marked by H3K9me3 than available protein molecules of YY1. One feasible scenario is illustrated in Figure 1. YY1 might bind to a small number of the tandem repeats, but not all of them, and trigger the modification process of H3K9me3, which in turn spreads over all tandem repeats or retrotransposons. One candidate molecule for transforming the initial marking to genome-wide marking could be small RNAs. It is well known that small RNAs transcribed from pericentromeric repeats are used for recruiting epigenetic machinery in plants, including H3K9 methylases and DNA methylases (Martienssen et al. 2008). Although similar mechanisms have not been identified so far for the vertebrate genomes, recent studies revealed that small RNAs derived from LTRs, such as Piwi-interacting RNA, are responsible for the transcriptional repression of LTRs and other retrotransposons through DNA methylation (Saito et al. 2006; Watanabe et al. 2006). It is still unclear how these small RNAs are generated. However, given the ubiquitous presence of YY1 binding sites in many retrotransposons, it is feasible to predict that YY1 might trigger this initial transcription from the tandem repeats and LTRs, which are then processed into Piwi-interacting RNAs. Finally, these Piwi-interacting RNAs might be used to recruit H3K9 methylases to the tandem repeats or retrotransposons. Consistent with this, some of the pericentromeric repeats in the mouse are known to be transcribed in dividing cells (Lu and Gilbert 2007). In summary, several lines of evidence are consistent with the hypothesis that YY1 is involved in establishing H3K9me3, and thus it will be of great interest to test the mechanism of YY1 as a triggering factor for the establishment of H3K9me3.
Besides the proposed targeting mechanisms for PRC2 and H3K9 methylases, several other mechanisms are also likely, based on the following reasons. First, many loci with H3K9me3 show tandem repeat sequence structures, but they do not show any sequence similarity among themselves. This suggests that the repeat structure itself, not the sequence, might be important for the recognition by H3K9 methylases. Second, recent studies have identified ncRNAs as possible targeting molecules for PRC2. These exemplary studies have been derived from the Hox and Xist loci (Rinn et al. 2007; Zhao et al. 2008). The ncRNAs from these loci are required for the targeting of PRC2 and subsequent H3K27me3 establishment. In fact, EZH2 has been shown to be the protein that interacts with these ncRNAs (Zhao et al. 2008). Similar observations have also been derived for one H3K9 methylase, EHMT2. According to the results from two imprinted loci, Igf2r and Kcnq1, the ncRNAs from these imprinted loci are also required for the targeting of EHMT2 and subsequent establishment of the allele-specific H3K9me3 (Nagano et al. 2008; Redrup et al. 2009). In summary, these observations suggest that there are likely multiple potential targeting mechanisms for PRC2 and H3K9 methylases and they potentially act through both DNA-binding proteins and ncRNAs.
Functional Consequences Associated with Defects in Histone-Modifying Complexes
Genome-wide profiling of the histone marks H3K27me3 and H3K9me3 clearly identify the two different genomic regions that are marked by these modifications, providing immediate hints for the potential functions of these marks at the genomic level. On the other hand, the biological processes in which these marks are important can be inferred from the cellular- and organism-level phenotypes produced by mutations of the histone-modifying complexes responsible for these marks. According to the results derived from PRC2, complete or near-complete depletion of the core components of PRC2 in ES cells usually causes unscheduled differentiation of ES cells, suggesting that PRC2 plays a role in maintaining the pluripotency and self-renewal properties of ES cells (Margueron and Reinberg 2011; Schuettengruber and Cavalli 2009). Similar experiments using ES cells were conducted to test the function of H3K9 methylases. Depletion of SETDB1 causes the induction of several key genes for the trophectoderm lineage in ES cells, resulting in differentiation into trophectoderm cells (Yeap et al. 2009; Yuan et al. 2009). These results confirm that, besides constitutive heterochromatic regions, many developmental regulators located in euchromatic regions are also subject to the H3K9-mediated repression (Bilodeau et al. 2009; Lohmann et al. 2010). Deletion of SETDB1 in ES cells also causes de-repression of a large number of endogenous retroviruses, supporting the fact that many retrotransposons are repressed by H3K9me3 (Matsui et al. 2010). This is also the case for KAP-1, an interacting partner of SETDB1: deletion of KAP-1 in ES cells de-represses a large number of endogenous retroviruses in the mouse genome (Rowe et al. 2010).
In the past decade, the core components of PRC2 and several H3K9 methylases have been mutated in the mouse, and subsequently the in vivo roles of these genes have been analyzed. Results from breeding experiments have shown that embryos homozygous for the mutant alleles of each PRC2 component are usually lethal around the implantation stage, mainly because of a failure to establish the three germ layers. This suggests that PRC2 is required for lineage specification (Faust et al. 1995; O’Carroll et al. 2001; Pasini et al. 2004). The homozygotes for some H3K9 methylase mutants are also embryonic lethal but at different stages of embryonic development. The homozygotes for SetDB1 die between 3.5 and 5.5 days postcoitum, whereas the homozygotes for Ehmt2 survive up to 8.5 days postcoitum (Dodge et al. 2004; Tachibana et al. 2002). In the case of Suv39h1 and Suv39h2, the homozygotes for single-gene knockouts are viable, but the double homozygotes for both genes are lethal around 13.5 days postcoitum (Peters et al. 2001). DNA methylation analyses with these surviving embryos revealed that the pericentromeric repeats are hypomethylated, further supporting the close association between these H3K9 methylases and DNA methylation machineries. The homozygotes for Ehmt2, Suv39h1, and Suv39h2 show embryonic lethality at relatively later stages than those for PRC2, potentially caused by functional redundancy between different H3K9 methylases. On the other hand, the heterozygotes for the mutants described above are usually viable within the Mendelian ratios, but some fraction of these heterozygotes also show visible phenotypes, such as reduced growth rates and defects in neurulation (Miró et al. 2009). However, the actual causes for these phenotypes are currently unknown.
In placental mammals, H3K27me3 and H3K9me3 histone modification marks are also part of the main repression mechanisms for genomic imprinting and X chromosome inactivation. First, in genomic imprinting, small genomic regions function as imprinting control regions, which control the allele-specific expression and DNA methylation of the surrounding imprinted genes (Bartolomei 2009). Interestingly, the known imprinting control regions tend to show tandem repeat structures (Kim 2008) and, furthermore, are marked by H3K9me3 in ES cells (Mikkelsen et al. 2007). This is very unusual because many CpG-rich promoter regions are usually marked by H3K4me3 and/or H3K27me3 in ES cells (Mikkelsen et al. 2007). Nevertheless, it is interesting to point out the presence of a similar pattern, tandem repeats with H3K9me3, between the imprinting control regions of imprinted regions and the tandem repeats of heterochromatic regions. It is unclear which proteins are responsible for establishing this histone modification on the known imprinting control regions. However, two H3K9 methylases are likely involved based on the following observations. RNAi-based knockdown of SETDB1 results in the removal of H3K9me3 and subsequent induction of several imprinted genes in ES cells, suggesting potential roles for SETDB1 in genomic imprinting (Yuan et al. 2009). Also, EHMT2 has been shown to be involved in setting up H3K9me3 on the Igf2r- and Kcnq1-imprinted domains, which is again required for maintaining the imprinting of these two domains (Nagano et al. 2008; Redrup et al. 2009). Second, a large fraction of imprinted genes are imprinted only in the placenta, yet these genes are usually marked with allele-specific H3K27me3, not with DNA methylation (Lewis et al. 2004; Umlauf et al. 2004). This observation was first noticed in the mutant mice targeting Eed (Mager et al. 2003). Many imprinted genes in the Eed mutant are deregulated in terms of their allele-specific expression, consistent with the fact that EED is the core component for PRC2 responsible for H3K27me3. Besides this involvement, PRC2 is also required for X chromosomal inactivation in female mammals. During the early stages of X chromosomal inactivation, PRC2 has been shown to be recruited to the inactivating X chromosome for the establishment of H3K27me3 (Plath et al. 2004).
In humans, there are several diseases or disease states that are closely associated with defects in the components of PRC2 and H3K9 methylases. First, human EZH2 has been identified as an overexpressed gene in metastatic prostate cancers, suggesting its involvement in the progression of prostate cancers (Varambally et al. 2002). EZH2 is also considered to be a tumor suppressor based on the observation that loss-of-function type mutations are frequently associated with myeloid disorders (Ernst et al. 2010). Second, human SUZ12 is frequently identified as part of chimeric transcripts resulting from the recurrent translocation between human chromosomes 7 and 17 in endometrial stromal tumors (Koontz et al. 2001). Third, recent results indicate that improper establishment of H3K9me3 on tandem repeats might be responsible for human diseases. Facioscapulohumeral dystrophy is usually caused by the contraction of a repeat, termed D4Z4 located in human chromosome 4q. In a minor group of facioscapulohumeral dystrophy, a loss of SUV39H1-mediated H3K9me3 is likely related to the misexpression of the nearby genes that are involved in muscle development (Zeng et al. 2009). There are many human diseases that are associated with tandem repeat expansion, such as the GAA repeat expansion in Friedreich's ataxia, and H3K9me3 involvement in the etiology of these diseases is currently unknown. However, the expansion of tandem repeats and subsequent spreading of H3K9me3 to nearby genes could be one feasible path for these repeat-associated diseases (Al-Mahdawl et al. 2008).
Conclusion
Owing to technical advances, such as next-generation sequencing technology, a large amount of genomic data about the epigenetic modification of human and other mammalian genomes has been obtained. These results indicate that the histone modification marks H3K27me3 and H3K9me3 play very important roles in mammalian genome regulation. Also, we have been able to identify protein complexes that are responsible for producing these histone modification marks, including PRC2 and several H3K9 methylases. Intriguingly, some of the components for these complexes have been identified as disease genes that are closely associated with cancers or genetic disorders, further supporting the significant roles played by these histone modification marks. So far, much progress has been made in understanding these repression signals, but the following aspects remain to be investigated. First, the targeting mechanisms for both PRC2 and H3K9 methylases are not fully understood. In that regard, it would be very interesting to focus on the two newly identified DNA-binding proteins for PRC2—AEBP2 and JARID2—and also on YY1 for possible roles in the targeting of H3K9me3. The most important questions for these proteins are: (1) Are these proteins responsible for the targeting of PRC2 and H3K9 methylases? If so, then (2) what are the exact functions of each of these proteins for the histone-modifying complexes? Second, although the two histone marks have been mainly recognized as transcription repressors, these marks might have some functions other than transcription. For instance, H3K9me3 is mainly detected in the genomic regions with tandem repeats, which are well known for duplication through crossover. The H3K9me3-mediated heterochromatin formation might be designed for blocking potential illegitimate crossover to preserve genomic stability. This possibility was previously noticed in the phenotype of the Suv39h1 and Suv39h2 mutant mice, which exhibit a high degree of genomic instability (Peters et al. 2001). Third, recent data from personal genome sequencing indicate that many human disorders tend to have mutations on the genes that are involved in epigenetic setting (Ernst et al. 2010). Although the homozygotes for any mutation in PRC2 and H3K9 methylases are not viable, as demonstrated in several mouse models, the heterozygotes for these mutations are likely viable and display altered phenotypes. In this regard, it is important to note that there are many adult stem cells that require the function of PRC2 for their pluripotency and self-renewal properties. In a heterozygous individual with a mutation, suboptimal levels of enzymatic activity for either H3K27me3 or H3K9me3 modification could easily cause disease states for this very vulnerable population of adult stem cells. In summary, although we need to wait for more data from the future experiments described above, it is very clear that these two histone modification marks are major repression signals in the development processes of humans and other mammals.
Acknowledgments
We thank Dr. Michelle Thiaville, Wesley Frey, and Muhammad Ekram for their careful reading and discussion of this manuscript. This work was supported by the National Institute of Health (R01-GM066225 and R15-ES019118).
Biography
Joomyeong Kim, PhD, is a George C. Kent Professor and Hana Kim, PhD, is a postdoctoral student in the Department of Biological Sciences at Louisiana State University, Baton Rouge.
Footnotes
Abbreviations that appear ≥3x throughout the article: ChIP-Seq, chromatin immunoprecipitation sequencing; LTR, long terminal repeat; ncRNA, noncoding RNA; PRC2, polycomb repression complex 2; RNAi, RNA interference.
References
- Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. 2004. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J 23:605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mahdawl S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. 2008. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet 17:735–746 [DOI] [PubMed] [Google Scholar]
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J 22:1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS. 2009. Genomic imprinting: Employing and avoiding epigenetic processes. Genes Dev 23:2124–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. 2009. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23:2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blahnik KR, Dou L, Echipare L, Iyengar S, O’Geen H, Sanchez E, Zhao Y, Marra MA, Hirst M, Costello JF, Korf I, Farnham PJ. 2011. Characterization of the contradictory chromatin signatures at the 3 #57401; exons of zinc finger genes. PLoS One 6:e17121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. 2004. Suz12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15:57–67 [DOI] [PubMed] [Google Scholar]
- Cattaneo F, Nucifora G. 2008. EVI1 recruits the histone methyltransferase SUV39H1 for transcription repression. J Cell Biochem 105:344–352 [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 10:295–304 [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. 2004. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24:2478–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol 25:10338–10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nature Struct Mol Biol 15:1176– 1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, Cross NC. 2010. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genet 42:722–726 [DOI] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T. 1995. The eed mutation disrupts anterior mesoderm production in mice. Development 121:273–285 [DOI] [PubMed] [Google Scholar]
- Garcia-Tuñon I, Guallar D, Alonso-Martin S, Benito AA, Benítez-Lázaro A, Pérez-Palacios R, Muniesa P, Climent M, Sánchez M, Vidal M, Schoorlemmer J. 2011. Association of Rex-1 to target genes supports its interaction with polycomb function. Stem Cell Res 7:1–16 [DOI] [PubMed] [Google Scholar]
- Gyory I, Wu J, Fejér G, Seto E, Wright KL. 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5:299–308 [DOI] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Kim J, Branscomb E, Stubbs L. 2003. Lineage-specific expansion of KRAB zinc-finger transcription factor genes: implications for the evolution of vertebrate regulatory networks. Cold Spring Harb Symp Quant Biol 68:131–140 [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. 2006. Molecular evolution and temporal amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16:78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Ekram MB, Roh TY, Kim J. 2011. Aebp2 as an epigenetic regulator for neural crest cells. PLoS One 6:e25174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. 2009. AEBP2 as a potential targeting protein for polycomb repression complex PRC2. Nucleic Acids Res 37:2940–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. 2008. Multiple YY1 and CTCF binding sites in imprinting control regions. Epigenetics 3:115–118 [DOI] [PubMed] [Google Scholar]
- Kim JD, Faulk C, Kim J. 2007. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res 35:3442–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Köcher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J. 2006. A polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20:1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz JI, Soreng AL, Nucci M, Kuo FC, Pauwels P, van Den Berghe H, Dal Cin P, Fletcher JA, Sklar J. 2001. Frequent fusion of the JAZF1 and JJAZ1 genes in endometrial stromal tumors. Proc Natl Acad Sci U S A 98:6348–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. 2006. Control of developmental regulators by polycomb in human embryonic stem cells. Cell 125:301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nature Genet 36:1291–1295 [DOI] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24:368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Montell DJ. 2001. Jing: a downstream target of slbo required for developmental control of border cell migration. Development 128:321–330 [DOI] [PubMed] [Google Scholar]
- Lohmann F, Loureiro J, Su H, Fang Q, Lei H, Lewis T, Yang Y, Labow M, Li E, Chen T, Kadam S. 2010. KMT1E mediated H3K9 methylation is required for the maintenance of embryonic stem cells by repressing trophectoderm differentiation. Stem Cells 28:201–212 [DOI] [PubMed] [Google Scholar]
- Lu J, Gilbert DM. 2007. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J Cell Biol 179:411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP, Magnuson T. 2003. Genome imprinting regulated by the mouse polycomb group protein Eed. Nat Genet 33:502–507 [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The polycomb complex PRC2 and its mark in life. Nature 469:343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Kloc A, Slotkin RK, Tanurdzić M. 2008. Epigenetic inheritance and reprogramming in plants and fission yeast. Cold Spring Harb Symp Quant Biol 73:265–271 [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464:927–931 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE. 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miró X, Zhou X, Boretius S, Michaelis T, Kubisch C, Alvarez-Bolado G, Gruss P. 2009. Haploinsufficiency of the murine polycomb gene Suz12 results in diverse malformations of the brain and neural tube. Dis Model Mech 2:412–418 [DOI] [PubMed] [Google Scholar]
- Müller J, Kassis JA. 2006. Polycomb response elements and targeting of polycomb group proteins in Drosophila. Curr Opin Genet Dev 16:476–484 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322:1717–1720 [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21:4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23:4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. 2010. JARID2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature 464:306–310 [DOI] [PubMed] [Google Scholar]
- Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. 2009. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139:1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. 2001. Loss of the suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131–135 [DOI] [PubMed] [Google Scholar]
- Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. 2009. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463:237–240 [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20:2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyamoorthy K, Park K, Atchison ML, Howe CC. 1993. The intracisternal A-particle upstream element interacts with transcription factor YY1 to activate transcription: Pleiotropic effects of YY1 on distinct DNA promoter elements. Mol Cell Biol 13:6621–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. 2009. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136:3531–3542 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. 2007. Genome regulation by polycomb and trithorax proteins. Cell 128:735–745 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. 3rd. 2002. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16:919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. 2009. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139:1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Mansuroglu Z, Mokrani H, Ghinea N, Bonnefoy E. 2004. Transcription factor YY1 associates with pericentromeric gamma-satellite DNA in cycling but not in quiescent (G0) cells. Nucleic Acids Res 32:4390–4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. 1997. Everything you have ever wanted to know about Yin Yang 1...... Biochim Biophys Acta 1332:F49–66 [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 16:1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of polycomb group complexes. Nat Genet 36:1296–1300 [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624–629 [DOI] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. 2006. The polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871–874 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20:1732–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson FH, Park K, Atchison ML. 2006. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A 103:19296–19301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Hu YC, Liu H, Shi Y. 2009. Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol Cell Biol 29:6245–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap LS, Hayashi K, Surani MA. 2009. ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin 2:12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Han J, Guo G, Orlov YL, Huss M, Loh YH, Yaw LP, Robson P, Lim B, Ng HH. 2009. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev 23:2507–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, Frants RR, Ball AR, Jr, Lock LF, Donovan PJ, van der Maarel SM, Yokomori K. 2009. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 5:e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]