Abstract
Ferrets are widely used as a small animal model for a number of viral infections, including influenza A virus and SARS coronavirus. To further analyze the microbiological status of ferrets, their fecal viral flora was studied using a metagenomics approach. Novel viruses from the families Picorna-, Papilloma-, and Anelloviridae as well as known viruses from the families Astro-, Corona-, Parvo-, and Hepeviridae were identified in different ferret cohorts. Ferret kobu- and hepatitis E virus were mainly present in human household ferrets, whereas coronaviruses were found both in household as well as farm ferrets. Our studies illuminate the viral diversity found in ferrets and provide tools to prescreen for newly identified viruses that potentially could influence disease outcome of experimental virus infections in ferrets.
Introduction
Infectious viral diseases, both emerging and re-emerging, pose a continuous health threat and disease burden to humans. Many important human pathogens are zoonotic or originated as zoonoses before adapting to humans [1]–[4]. This is exemplified by recently emerged diseases in which mortality ranged from a few hundred people due to infection with H5N1 avian influenza A virus to millions of HIV-infected people from acquired immunodeficiency syndrome [5]–[8]. Severe acute respiratory syndrome (SARS) coronavirus and the pandemic influenza A/H1N1(2009) virus in humans were linked to transmission from animal to human hosts as well and have highlighted this problem [9]–[11]. An ongoing systematic global effort to monitor for emerging and re-emerging pathogens in animals, especially those in key reservoir species that have previously shown to represent an imminent health threat to humans, is crucial in countering the potential public health threat caused by these viruses.
Relatively few studies have been conducted on diseases of non-domestic carnivores, especially regarding diseases of small carnivores (e.g. mustelids). Ferrets (Mustela putorius furo) can carry bacteria and parasites such as Campylobacter, Giardia, and Cryptosporidium in their intestinal tract and potentially spread them to people [12], [13]. In addition, they can transmit influenza A virus to humans and possibly on rare occasions rabies virus as well [14]–[16]. Because of their susceptibility to several human respiratory viruses, including human and avian influenza viruses, SARS coronavirus, nipah virus, and morbilliviruses [17]–[21], ferrets have been used as small animal model for these viruses. To further characterize this important animal model and to obtain epidemiological baseline information about pathogens in ferrets, the fecal viral flora of ferrets was studied using a metagenomics approach. Both known and new viruses were identified.
Materials and Methods
Clinical specimens
Rectal swabs were collected from 39 ferrets (Mustela putorius furo) in the Netherlands and Sweden and stored at −80°C (Table 1) from 2010–2012. The first cohort consisted of rectal swabs from ferrets from a farm in The Netherlands in 2010 (n = 12). The second cohort consisted of rectal swabs from ferrets (Sweden) that were used in an experimental procedure that consisted of 3 weeks of immunosuppression [22] gathered in March 2012 (n = 12). The third cohort were rectal swabs from household ferrets (the Netherlands) with (n = 3) or without (n = 12) diarrhea gathered in 2010. This study was carried out in strict accordance with European guidelines (EU directive on animal testing 86/609/EEC) and Dutch legislation (Experiments on Animals Act, 1997). In compliance with relevant laws and institutional guidelines, no ethical approval is required for most of the samples in this study, because live animals were not primarily and purposefully sampled to obtain material for scientific purposes, but for health care purposes, such as surveillance of viruses that could cause underlying ferret disease. Some of the samples from the ferrets in this study were obtained under ethical approval by the independent animal experimentation ethical review committee of the Netherlands Vaccine Institute (permit number 20110286) in compliance with relevant laws and institutional guidelines. All efforts were made to minimize animal suffering and the owners of the ferrets gave permission for their animals to be used in this study.
Table 1. Summary of mammalian viruses found in ferret fecal material.
| Ferret | Housingc | Total no. readsa | Eukaryotic viruses (No. of reads) | Virus in-depthd | Taqmand |
| 1 | Farm NL | 3981 | Picobirnavirus (2) | FRCoV | |
| 2 | Farm NL | 10149 | Papillomavirus (3) | FRCoV | |
| 3 | Farm NL | 5186 | Coronavirus (17) | FRCoV | |
| Picobirnavirus (10) | |||||
| Kobuvirus (75) | |||||
| 4 | Farm NL | 8919 | Torque teno virus (3) | ||
| 5 | Farm NL | 71706 | |||
| 6 | Farm NL | 7710 | |||
| 7 | Farm NL | 7344 | |||
| 8 | Farm NL | 7686 | Aleutian mink disease virus (2) | FRCoV | |
| 9 | Farm NL | 9576 | |||
| 10 | Farm NL | 7702 | Aleutian mink disease virus (2) | FRCoV | |
| 11 | Farm NL | 1149 | |||
| 12 | Farm NL | 4351 | |||
| 13 | Farm SE | 4947 | |||
| 14 | Farm SE | 6560 | |||
| 15 | Farm SE | 4632 | Coronavirus (116) | FRCoV | |
| 16 | Farm SE | 5985 | |||
| 17 | Farm SE | 4766 | Papillomavirus (92) | MpPV1 | MpPV1 |
| 18 | Farm SE | 4728 | |||
| 19 | Farm SE | 3641 | Coronavirus (19) | FRCoV | |
| Kobuvirus (90) | MpKoV | ||||
| 20 | Farm SE | 5627 | Kobuvirus (11) | ||
| 21 | Farm SE | 6091 | Coronavirus (43) | FRCoV | |
| Torque teno virus (48) | MpfTTV1 | ||||
| 22 | Farm SE | 3422 | |||
| 23 | Farm SE | 706 | |||
| 24 | Farm SE | 3099 | Coronavirus (22) | FRCoV | |
| 25 | Household | 6249 | FRCoV | ||
| 26 | Household | 7745 | Astrovirus (636) | ||
| Coronavirus (13) | FRCoV | ||||
| Picobirnavirus (3) | |||||
| 27 | Household | 10002 | |||
| 28 | Household | 13634 | |||
| 29 | Household | 4298 | |||
| 30 | Household | 4886 | Picobirnavirus (9) | ||
| Polyomavirus (3) | |||||
| 31 | Household | 8959 | |||
| 32 | Household | 8369 | Kobuvirus (4280) | MpKoV32 | MpKoV |
| Polyomavirus (2) | |||||
| 33 | Household | 8459 | Herpesvirus (2) | ||
| Kobuvirus (2039) | MpKoV | ||||
| Hepevirus (562) | FRHEV | ||||
| Coronavirus (35) | FRCoV | ||||
| 34 | Household | 7116 | FRCoV | ||
| 35 | Household | 1167 | Hepevirus (898) | FRHEV | |
| 36 | Household | 9539 | Kobuvirus (50) | MpKoV | |
| Parechovirus (71) | MpPeV1 | MpPeV1 | |||
| 37 | Householdb | 5069 | Astrovirus (1464) | ||
| Hepevirus (516) | FRHEV | ||||
| 38 | Householdb | 13387 | Coronavirus (19) | MpKoV38 | FRCoV |
| Kobuvirus (6160) | MpKoV | ||||
| 39 | Householdb | 9117 | Kobuvirus (3582) | MpKoV39 | MpKoV |
Total no. of trimmed reads that were analyzed.
These animals had diarrhea.
NL, Netherlands; SE, Sweden.
FRCoV, ferret coronavirus; MpPV1, ferret papillomavirus; MpKoV, ferret kobuvirus; FRHEV, ferret hepevirus; MpPeV, ferret parechovirus; MpfTTV, ferret anellovirus.
Sequence-independent RNA and DNA virus screening of ferret rectal swabs
Large scale molecular virus screening, based on host nucleic acid depletion, viral nucleic acid isolation, sequence-independent amplification and next generation sequencing with a 454 GS Junior Instrument (Roche) was performed as described previously and by the manufacturer [23]–[26] on 39 rectal swabs from ferrets. More than 253,000 trimmed reads were assembled using de novo assembly in CLC Genomics Workbench 4.5.1 (CLC Bio; [27]) and analyzed according to nucleotide (contigs and singletons) and translated nucleotide BLAST searches (contigs) [28]. Sequences were classified into eukaryotic viruses, phages, bacteria, and eukaryotes based on the taxonomic origin of the best-hit sequence using MEGAN 4.40 [29], [30]. E values of 0.001 and 10−10 were used as the cutoff value of significant virus hits for BLASTn and BLASTx, respectively.
PCR amplification and Sanger sequencing
Sequence information obtained from the large scale molecular virus screening using next-generation sequencing was used to design specific primers for amplification of the papillomavirus genome MpPV1 using AmpliTaq Gold DNA polymerase (Roche), according to instructions of the manufacturer to confirm and/or extend 454-sequence reads. Primer sequences are available on request. Products were sequenced as described previously [26]. A diagnostic MpPV1 real time PCR was developed, using primers and probe VS736 (5′-ATGTGTGACATTGGCTTTGGA- 3′), VS738 (5′-CTGCAATATGACTGCTCTCGC- 3′), and VS737 (5′-FAM-CTAGAGCTGAGGTTCCTATAG-TAMRA-3′) that target the L1 genome region (KF006988) and TaqMan® Fast Virus 1-Step Master Mix (Applied Biosystems) according to instructions of the manufacturer. In addition, diagnostic ferret kobu- and parechovirus real time RT-PCRs were developed, using primers and probes VS730 (5′-CCTCCAGCTCCGCTGGCTCAA-3′), VS732 (5′-CAATCCTAGGGTGCAGTCCTT- 3′), and VS731 (5′-FAM- CCACTCTCTCTGTGGCACTTT-TAMRA- 3′) that target the 3D polymerase/3′UTR genome region of ferret kobuvirus (KF006985) and primers and probe VS733 (5′-CTGCTCCTCAATTAACAGGCT-3′), VS735 (5′-CACACTCCTGCATCCATTAGT- 3′), and VS734 (5′-FAM-TGAGATCATTGCATGACAATGT-TAMRA- 3′) that target the 3D genome region of ferret parechovirus (KF006989) with TaqMan® Fast Virus 1-Step Master Mix (Applied Biosystems) according to instructions of the manufacturer. Similarly, a diagnostic ferret hepatitis E virus real time RT-PCR was used, using primers and probe VS739 (5′- AAGATGCGTTTTGTTCTCTTGCT-3′), VS740 (5′- CCGGACGCCCTCCTGYA- 3′), and VS741 (5′-FAM- TGCTCGCCGCCCCAATGTACC-TAMRA- 3′) that target the ORF2/3 genome region of ferret hepatitis E virus (JN998606). A diagnostic ferret coronavirus real time RT-PCR was used with primers and probe as described previously (22).
Phylogenetic analysis
Multiple alignments were created using ClustalX (2.0.10) [31]. Phylogenetic analyses were carried out with Molecular Evolutionary Genetics Analysis (MEGA), version 5 [32]. Picornavirus cleavage sites were predicted by NetPicoRNA 1.0 server (http://www.cbs.dtu.dk/services/NetPicoRNA/) or predicted based on alignments with other picornaviruses. The similarity between different picornaviruses was measured using Simplot version 3.5.1 with the Kimura-2 parameter, a transition/transversion (Ts/Tv) ratio of 3.0 and window and step sizes of 600 and 20 nucleotides, respectively [33].
Genbank accession numbers
The genomes of identified viruses described in detail here were deposited in GenBank under the following accession numbers: ferret kobuviruses MpKoV38, KF006985; MpKoV32, KF006987; MpKoV39, KF006986; ferret parechovirus (MpPeV1), KF006989; ferret papillomavirus (MpPV1), KF006988; ferret anellovirus (MpfTTV1), KF006990.
Results
Large scale molecular virus screening was performed on 39 rectal swabs from ferrets (Mustela putorius furo). More than 253,000 trimmed sequence reads were generated and the sequences from each animal were assembled de novo. Sequences were classified into eukaryotic viruses, phages, bacteria, and eukaryotes based on the taxonomic origin of the best-hit sequence. Many of the identified sequences were of eukaryotic or bacterial origin, and a large proportion of the total reads did not have any significant hits for nucleotide or amino acid sequences in GenBank in agreement with viral metagenomic studies of feces from bats, turkeys, rodents, pine marten, and humans [25], [34]–[36]. Fecal samples of ferrets revealed a large degree of microbial diversity.
Virome overview
Almost all samples showed evidence for the presence of bacteriophages from the order Caudovirales and/or family Microviridae. In seven rectal swabs, evidence for quite a number of different avian viruses, among which chicken anemia virus, chicken astrovirus, chicken parvovirus, turkey hepatitis virus, fowl adenovirus, and turkey parvovirus. Six of these animals were bred on a farm and one animal was a household pet. These avian viruses may originate from the ferrets'diet (they were fed chicken) and do not represent genuine enteric infections in the ferrets. Eukaryotic viruses with relatively high homology (85–100% on the amino acid level) to ferret coronavirus ([37]; family Coronaviridae), ferret hepatitis E virus ([38]; family Hepeviridae), and Aleutian mink disease virus (family Parvoviridae), were detected (Table 1). The species demarcation criteria are not strongly defined for Corona- and Hepeviridae, but based on host range and genetic similarity they should qualify as the same species as ferret coronavirus and ferret hepatitis E virus. The presence of corona- and hepeviruses was confirmed by real time PCR assays (Table 1). Of note, ferret hepatitis E virus sequences were only detected in household ferrets, whereas coronaviruses were found both in household as well as farm ferrets, (Fig. 1; Table 1). A near complete and a partial genome of an astrovirus (family Astroviridae) were obtained from ferrets 37 and 26 respectively with ∼95% identity on the nucleotide level to murine astrovirus STL1 that was recently described (Table 1) [39], [40]. In addition, a number of ferret viruses with homology to torque teno virus (family Anelloviridae), picobirnavirus (family Picobirnaviridae), kobu- and parechovirus (family Picornaviridae), and papillomavirus (family Papillomaviridae) were identified (Table 1). We further characterized some of these novel mammalian viral sequences by full or near full genome sequencing and compared them to their closest relatives by phylogenetic analyses.
Figure 1. Prevalence of ferret viruses.
Percentage coronavirus (A), hepatitis E virus (B) and kobuvirus (C) positive farm versus household ferrets by real time PCR assay. Significant differences (unpaired t-test P<0.05) are indicated by an asterisk.
Ferret kobuvirus
Kobuvirus is a viral genus belonging to the family Picornaviridae. The genus is composed of three species, called Aichivirus A, B, and C. Aichiviruses A, B, and C have been found in humans [41], cows [42], and pigs [43], respectively. More recently, kobuviruses from Canyon mouse (Peromyscus crinitus) [44], dogs (Canis lupus familiaris) [45], [46] and sheep [47] have been identified. This virus may be associated with diarrhea [48]. Kobuviruses are small, nonenveloped viruses with a single stranded positive sense RNA genome of ∼8 kb. The genome encodes a single polyprotein that is cleaved by virus-encoded proteases into the structural and nonstructural proteins [48].
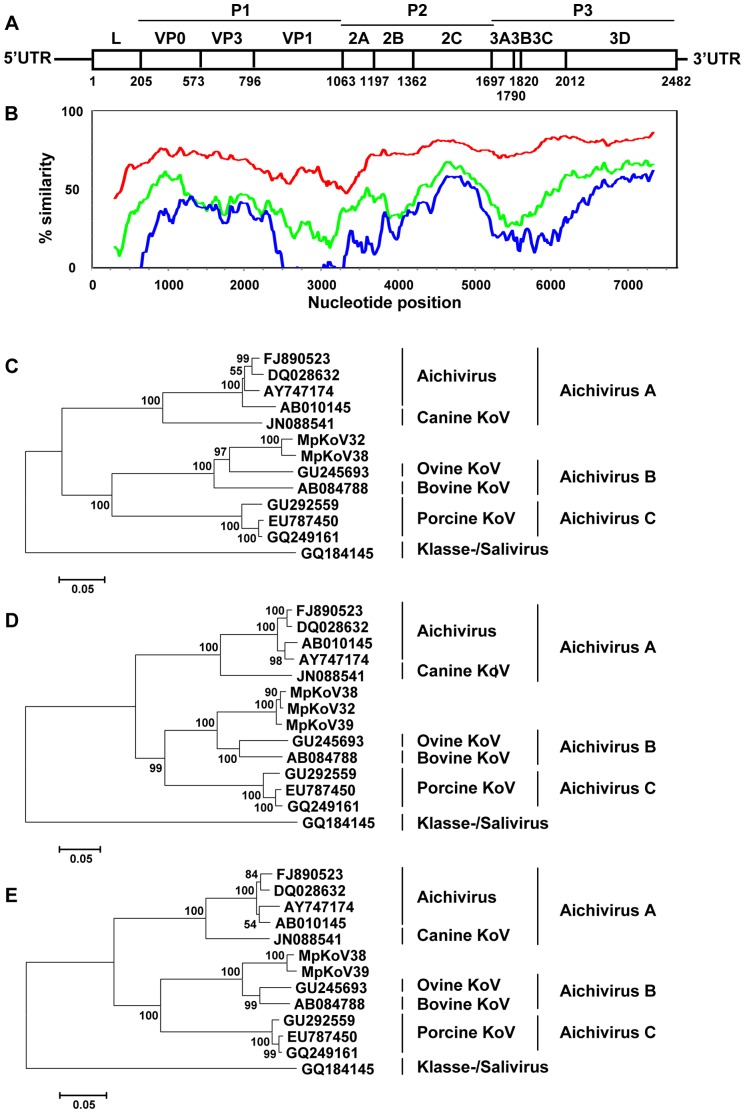
A near-complete kobuvirus genome containing the complete coding region was obtained by 454-sequencing from a rectal swab from ferret 38. In addition, two partial polyprotein sequences were obtained from rectal swabs of household ferrets 32 and 39. The ferret kobuvirus (MpKoV) from ferret 38 encodes a 2,482-amino-acid polyprotein flanked on each side by untranslated regions (UTRs) (Fig. 2A). Polyprotein cleavage sites were predicted by NetPicoRNA analysis and based on kobuvirus alignments (Fig. 2A). NetPicoRNA predictions were strong for the L/VP0, VP1/2A, 2A/2B, 2B/2C, and 2C/3A, all containing Q/G residues on either side of the predicted site. A Q/S cleavage site between 3C and 3D equivalent to that of aichiviruses was predicted at position 2012–2013 of MpKoV38. Q/H and Q/A cleavage sites in the P1 region were predicted by comparison with aichivirus sites. The 3A/3B and 3B/3C sites were tentatively placed at position 1790 (DQAQ/AAYT) and at position 1820 (VVRQ/SGPS) based on alignments with other kobuviruses.
Figure 2. Genome organization of ferret kobuvirus and amino acid sequence divergence from other aichiviruses.
(A) Predicted genome organization of ferret kobuvirus showing amino acid positions of predicted cleavage sites in the polyprotein (numbering based on the ferret kobuvirus polyprotein sequence). Sites were predicted by NetPicoRNA analysis and by alignment with known cleavage sites in aichiviruses. (B) Mean similarity of bovine kobuvirus (AB084788) to ferret kobuvirus (red), porcine kobuvirus (GU292559, green), and human aichivirus (FJ890523; blue) polyprotein-coding nucleotide sequences scaled to the genome diagram in A. (C–E) Phylogenetic trees of the amino acid sequences of ferret kobuvirus (MpKoV32, 38, and 39) with other aichiviruses in the P1 (C), P2 (D), and P3 (E) gene regions were generated using MEGA5, with the neighbor-joining method with p-distance and 1,000 bootstrap replicates. Significant bootstrap values are shown.
The genetic relationship of MpKoV38 with other known kobuviruses was assessed by determining the pairwise distance to the corresponding genome regions of other picornaviruses (Table 2; Fig. 2B–E). MpKoV38 showed the greatest sequence identity with bovine and ovine kobuvirus throughout the genome (Table 2; Fig. 2B–E). According to the 9th ICTV Report [49], members of kobuvirus species share greater than 70% amino acid identity in the polyprotein, greater than 70% amino acid identity in P1 genome region, greater than 80% amino acid identity in 2C and 3CD genome regions, and share a common genome organization, suggesting that ferret kobuvirus belongs to aichivirus species B. The diversity of ferret kobuvirus from bovine and ovine kobuviruses, however, was consistently greater than that observed within different isolates of human aichiviruses or different ferret kobuvirus isolates, confirming its identity as a separate type (Fig. 2C–E). No evidence of genetic recombination among ferret KoV or other kobuviruses was observed (Table 2, Fig. 2B–E).
Table 2. Pairwise amino acid identity in the P1, P2, and P3 regions between ferret kobuvirus 38 (MpKoV38; KF006985) and bovine kobuvirus (BKoV; AB084788), human aichivirus (AIV; FJ890523), porcine kobuvirus (PKoV; GU292559), and klassevirus (Klasse; GQ184145).
| Region and virus | BKoV | AIV | PKoV | Klasse |
| P1 | ||||
| MpKoV38 | 80.8 | 52.1 | 63.2 | 41.7 |
| BKoV | 52.2 | 63.1 | 41.7 | |
| AIV | 56.2 | 44.2 | ||
| PKoV | 41.9 | |||
| P2 | ||||
| MpKoV38 | 83.3 | 62.7 | 70.4 | 36.0 |
| BKoV | 62.4 | 72.0 | 35.5 | |
| AIV | 63.7 | 34.2 | ||
| PKoV | 35.4 | |||
| P3 | ||||
| MpKoV38 | 89.5 | 63.8 | 72.3 | 42.8 |
| BKoV | 64.5 | 74.0 | 42.5 | |
| AIV | 66.0 | 45.0 | ||
| PKoV | 43.3 | |||
To obtain insight in the prevalence of ferret kobuvirus in ferrets, a ferret kobuvirus-specific real time PCR targeting the 3D/3′NTR region of the kobuvirus genome was performed on the total set of 39 rectal samples. Besides samples 32, 38, and 39, in which ferret kobuvirus was identified, three other rectal samples were positive as well, resulting in an overall prevalence of ∼15% (Table 1; Fig. 1). As was observed for ferret hepatitis E virus, ferret kobuvirus is significantly more often detected in household ferrets than in farm ferrets using a real time PCR assay (Fig. 1).
Ferret parechovirus
Parechovirus is a viral genus in the family Picornaviridae. The genus is composed of two species, human parechovirus and Ljungan virus from bank voles. Parechoviruses are small, non-enveloped viruses with a single stranded positive sense RNA genome of ∼7.5 kb. The genome encodes a single polyprotein that is cleaved by virus-encoded proteases into the structural and nonstructural proteins. Human parechoviruses are widely spread infectious pathogens that are associated with mild gastroenteritis, respiratory disease, flaccid paralysis, encephalitis, and myocarditis in young children [50]–[53].
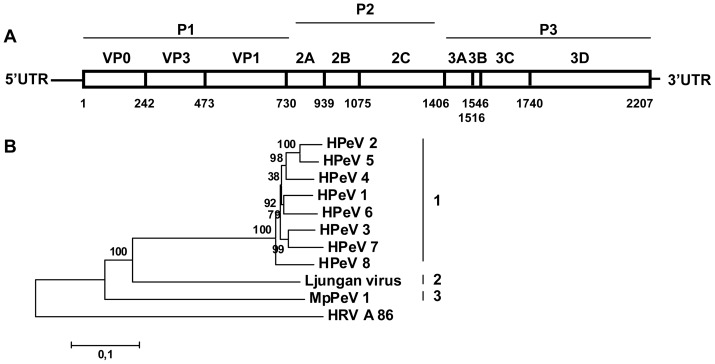
A near-complete parechovirus genome containing the complete coding region was obtained by 454-sequencing from a rectal swab from household ferret 36 without diarrhea. The ferret parechovirus from ferret 36 (MpPeV1) encodes a 2,207-amino-acid polyprotein flanked on each side by untranslated regions (UTRs) (Fig. 3A). Cleavage sites were predicted by NetPicoRNA analysis and alignments with other parechoviruses (Fig. 3A). The pairwise identity of the MpPeV1 polyprotein with the corresponding genome regions of other parechoviruses was assessed (Table 3; Fig. 3B). The Picornavirus Study Group of ICTV determined that members of a species of the parechovirus genus share greater than 70% amino acid identity in the polyprotein, and share a natural host range and genome organization. Members of a picornavirus genus should normally share phylogenetically related P1, P2 and P3 genome regions, each sharing >40%, >40% and >50% amino acid identity, respectively [49]. MpPeV1 has a typical parechovirus genome organization (Fig. 2A), but shares less than 43% identity on the amino acid level in the entire polyprotein compared to human parechoviruses and Ljungan virus (Table 3) and has been identified in a host different from bank vole and human and thus MpPeV1 constitutes a new species in the genus parechovirus, and may even represent the first species in a new genus in the Picornaviridae family.
Figure 3. Genome organization and phylogenetic analysis of ferret parechovirus.
(A) Predicted genome organization of ferret parechovirus showing amino acid positions of predicted cleavage sites in the polyprotein (numbering based on the ferret parechovirus polyprotein sequence). Sites were predicted by NetPicoRNA analysis and by alignment with other parechoviruses. (B) A phylogenetic tree of the polyprotein sequence of ferret parechovirus (MpPeV1) and representative human (HPeV1-8) and bank vole parechoviruses (Ljungan virus) was generated using MEGA5, with the neighbor-joining method with p-distance and 1,000 bootstrap replicates and human rhinovirus A 86 as an outgroup (HRV-A 86). Significant bootstrap values are shown. Genbank accession numbers are shown in Table S1.
Table 3. Pairwise amino acid identity in the polyprotein P1, P2, and P3 regions between different parechovirus species.
| Region and virus | HPeV1 | HPeV2 | Ljungan |
| P1 | |||
| MpPeV1 | 40.1 | 40.3 | 43.1 |
| HPeV1 | 79.3 | 50.5 | |
| HPeV2 | 50.4 | ||
| P2 | |||
| MpPeV1 | 40.6 | 40.1 | 46.2 |
| HPeV1 | 94.9 | 47.7 | |
| HPeV2 | 48.8 | ||
| P3 | |||
| MpPeV1 | 37.5 | 37.5 | 40.0 |
| HPeV1 | 93.6 | 46.3 | |
| HPeV2 | 46.6 | ||
To obtain insight in the prevalence of MpPeV1 in ferrets, a MpPeV1-specific real time PCR targeting the 3D region of the parechovirus genome was performed on the total set of rectal samples from 39 ferrets. Besides the sample in which MpPeV1 was identified, no samples were positive (Table 1).
Ferret papillomavirus
Papillomaviruses (PVs) are a highly diverse family of viruses with double stranded circular DNA genomes of ∼8 kb in size. They infect a wide variety of mammals, as well as birds and reptiles, and are highly species specific. Some papillomaviruses cause benign or malignant epithelial tumors of the skin and mucous membranes in their natural hosts [54].
A full-length papillomavirus genome sequence was amplified and sequenced from a rectal swab of ferret 17, to complete and/or confirm the findings by next-generation sequencing. The circular genome organization of the ferret papillomavirus (MpPV1) was typical for papillomaviruses, consisting of a long control region (LCR), and early (E6, E7, E1, E2, E4) and late regions (L1, L2) (Fig. 4A). Whole genome sequence alignments revealed that the most closely related papillomavirus to MpPV1 was canine papillomavirus 2 (60% identity) from the genus Taupapillomavirus (Fig. 4B). Papillomaviruses are classified based on nucleotide sequence identity in the L1 ORF and their biological and pathological properties [54], [55]. A papillomavirus strain is a new type if the complete genome has been sequenced and the L1 ORF shares less than 90% homology with the closest known papillomavirus type [54], [55]. L1 is the most conserved region among papillomaviruses and according to the current genus classification, most papillomavirus types within a genus share more than 60% nucleotide identity in L1. Based on these criteria, MpPV1 could be designated as a new papillomavirus type in the genus Taupapillomavirus (Fig. 4C), as MpPV1 L1 shares 68.2% nucleotide identity to canine papillomavirus 2.
Figure 4. Genome organization of ferret papillomavirus and nucleotide sequence divergence from other papillomaviruses.
(A) Predicted genome organization of ferret papillomavirus with early (E) and late (L) genes indicated. (B) A phylogenetic tree of the complete ferret papillomavirus (MpPV1) genome and representative human and animal papillomaviruses was generated using MEGA5, with the maximum-likelihood method with Kimura-2 parameter and 1,000 bootstrap replicates. Significant bootstrap values are shown. (C) A phylogenetic tree of the L1 genome region of ferret papillomavirus (MpPV1) and representative human and animal papillomaviruses was generated using MEGA5, with the maximum-likelihood method with Kimura-2 parameter and 1,000 bootstrap replicates. Significant bootstrap values are shown. Genbank accession numbers are shown in Table S1.
To obtain insight in the prevalence of MpPV1 in ferrets, a MpPV1-specific real time PCR targeting the L1 region of the papillomavirus genome was performed on the total set of 39 rectal samples. Besides the sample in which MpPV1 was identified, no samples were positive for MpPV1 (Table 1).
Ferret anellovirus
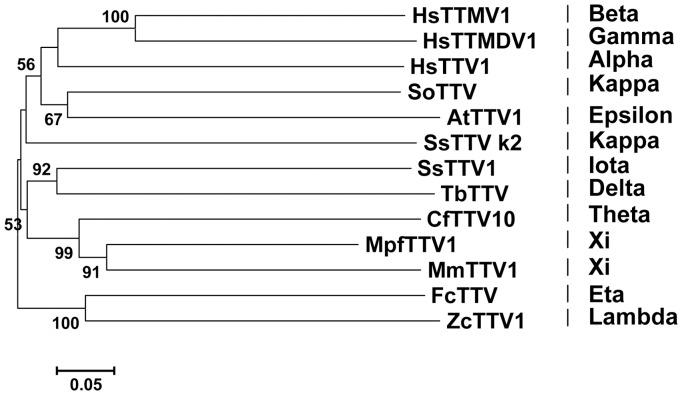
The partial anellovirus genome sequence from sample 21 was designated MpfTTV1 for Mustela putorius furo torque teno virus 1. This sequence encodes for a partial open reading frame 1 (ORF1) as determined by alignment with ORF1 amino acid sequences from representative torque teno viruses. A taxonomic proposal submitted to the International Committee on the Taxonomy of Viruses (ICTV) by the Anelloviridae-Circoviridae Study Group proposes that genera in the family Anelloviridae are defined as having >56% divergence in the nucleotide sequence of ORF1 [56]. Divergence analysis using p-distance calculated with MEGA5 [32] demonstrated that the partial ORF1 nucleotide sequence of anellovirus MpfTTV1 was in general >56% divergent on the nucleotide level from anelloviruses identified in wildlife, with the exception of MmTTV1 (∼50%) (data not shown). Neighbor-joining phylogenetic trees were generated using the partial ORF1 nucleotide alignments, which underlined the nucleotide divergence analysis (Fig. 5). Our data therefore suggest that the torque teno virus species identified in ferrets belongs to the genus Xitorquevirus.
Figure 5. Phylogenetic analysis of ferret anellovirus.
A phylogenetic tree of the partial ORF1 nucleotide sequence of ferret torque teno virus (MpfTTV1) and the corresponding region of representative human and animal anelloviruses was generated using MEGA5, with the neighbor-joining method with p-distance and 1,000 bootstrap replicates. Significant bootstrap values are shown. Genbank accession numbers are shown in Table S1.
Discussion
Previously, virus discovery in animals has focused on pathogenic infections or on animals that on the basis of relevance to (re-)emerging viruses are thought to represent a key risk host for emerging virus-associated disease in humans [57]. With the recent advances in the metagenomics field, a substantial increase in studies looking at virus epidemiological baseline levels in different (wildlife) animals has been observed [25], [36], [46], [57]–[68]. Even though ferrets are a very important animal model for a number of human viral infections, not much is known about viruses that naturally occur in ferrets [17]–[21]. This study describes the viral communities in fecal material of ferrets (Mustela putorius furo).
Sequences closely related to known viral sequences were identified, with homology to ferret coronavirus, ferret hepatitis E virus, Aleutian mink disease virus, different avian viruses and murine astrovirus STL1 [37], [38], [40]. The avian viruses may be a reflection of the diet of these ferrets, which were fed chicken. It is of note that both the viral screening with random amplification and next-generation sequencing and a specific ferret hepatitis E virus taqman assay indicated that household ferrets that are kept as pets are significantly more likely to excrete ferret hepatitis E virus in their fecal material than farm animals in this study. Such a correlation was not observed for ferret coronavirus excretion for which prevalence in both cohorts seemed similar, but not as high as previously reported [37]. Sporadic cases of human hepatitis E virus infections seem to be increasing and several observations suggest that these cases are caused by zoonotic spread of infection from wild or domestic animals [69]–[71]. Follow-up studies into seroprevalence and/or virus prevalence in household ferrets, their owners, and possibly other pets may provide indications for cross species transmission.
We report on the first identification of a kobuvirus, parechovirus, papillomavirus and anellovirus in ferrets. The kobu- and parechovirus belong to the family Picornaviridae that currently comprises 17 genera and many more different virus species [49]. They infect a wide range of mammals and are well-known for their involvement in gastroenteritis [49]. Our phylogenetic analysis revealed that the ferret kobuvirus clustered with bovine and ovine kobuvirus in species Aichivirus B. The close relationship between bovine, ovine, and ferret kobuviruses may indicate past cross species transmission events and subsequent evolution in the separate host species resulting in the distinct types seen today. As was observed for ferret hepatitis E virus but not ferret coronavirus, ferret kobuvirus is significantly more often detected in household ferrets than in farm ferrets by a specific ferret kobuvirus taqman assay, although random amplification and next-generation sequencing suggested the presence of kobuvirus in two additional samples from farm ferrets, which could reflect the presence of another divergent kobuvirus species. Also here follow-up studies into seroprevalence and/or virus prevalence in household ferrets, their owners, and possibly other pets should be performed. An overall kobuvirus prevalence of ∼15% was observed in this study. Two out of 3 animals (67%) with diarrhea and 4 animals out of 36 (11%) without diarrhea were positive by taqman for ferret kobuvirus. Because of the small sample size and the fact that other causes for diarrhea were not excluded, further in depth studies are needed to show that these viruses can cause diarrhea in ferrets.
The ferret parechovirus is the third parechovirus species identified to date. It is highly divergent from human parechovirus and Ljungan virus and may even constitute a new genus in the family Picornaviridae based on the observed sequence diversity. Interestingly, the P3 genome region of MpPeV1, encoding the nonstructural proteins NS3A–3D, is not more conserved than the P1 genome region, encoding the structural capsid proteins, when compared to the corresponding regions of human parechoviruses 1 and 2. This has been observed for Ljungan virus as well by us and others [72] and requires more in-depth studies. Only 1 out of 39 ferrets was positive for MpPeV1, suggesting that the ferret parechovirus prevalence may be relatively low (<2.5%). In humans, parechovirus infections are most common in young children and the prevalence in adults may be low as well [73]. It is of note that the parechovirus-positive animal did not show any signs of gastroenteritis. Low virus prevalence was observed for the identified ferret papillomavirus as well. Papillomaviruses are, however, not typically detected in fecal material or associated with gastroenteritis like parechoviruses. Although recently a full-length papillomavirus genome was characterized from fecal material of a deer mouse as well [36]. Not much knowledge exists on papillomatosis in ferrets as a disease, or the virus(es) causing them. The impact of papillomavirus infections on ferrets in general and the specific role of MpPV1 thus has to be further addressed.
Like many other mammals, it seems that ferrets also harbor anelloviruses. The ferret anellovirus MpfTTV1 is most closely related to the recently identified anellovirus from pine marten [25]. Based on phylogenetic analysis, we propose that MpfTTV1 should be placed in the proposed new anellovirus genus, Xitorquevirus, in analogy to the classification of torque teno viruses in nine genera named Alpha-, Beta-, Gamma-, Delta-, Epsilon-, Eta-, Iota-, Theta-, and Zetatorquevirus, and the proposed five genera Kappa-, Lambda-, Mu-, Nu-, and Xitorquevirus [25], [56], [74]. A closely related virus to MpfTTV1 seemed to be present in one other ferret (Table 1) besides ferret 21, resulting in a virus prevalence of ∼5% in this study. Anelloviruses do not generally seem to be as abundantly present in fecal material as in serum, as evidenced by fecal virome studies in different host species [25], [35], [44], [46], [56], [58], [59], [66].
Ferrets are carnivores that are significantly affected by a number of human pathogens and hence are widely used as a small animal model for viral infections [17]–[21]. In addition, they are capable of spreading viral infections to humans, among which influenza A virus and on rare occasions rabies virus [14]–[16]. The characterization of the fecal virome of ferrets provides epidemiological baseline information about pathogens which allows the swift identification of possible sources of future zoonotic infections and their subsequent control. Especially the seemingly interesting correlation between certain viral infections in ferrets and their presence in human households as pets needs to be further addressed. In addition, this study shows that ferrets can be infected with a wide range of different viruses, some of which may influence the outcome in experimental virus infection studies, thus delivering a strong argument in favor of development of specific pathogen-free ferret colonies.
Supporting Information
GenBank accession numbers of representative parecho-, papilloma-, and anellovirus strains.
(DOC)
Acknowledgments
Dr. A.D.M.E. Osterhaus and Dr. S.L. Smits are part time chief scientific officer and senior scientist respectively of Viroclinics Biosciences B.V.
Funding Statement
The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under the project “European Management Platform for Emerging and Re-emerging Infectious disease Entities” (EMPERIE) EC grant agreement number 223498 and the Virgo consortium. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kuiken T, Leighton FA, Fouchier RA, LeDuc JW, Peiris JS, et al. (2005) Public health. Pathogen surveillance in animals. Science 309: 1680–1681 309/5741/1680 [pii];10.1126/science.1113310 [doi] [DOI] [PubMed] [Google Scholar]
- 2. Taylor LH, Latham SM, Woolhouse ME (2001) Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 356: 983–989 10.1098/rstb.2001.0888 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Woolhouse ME, Gowtage-Sequeria S (2005) Host range and emerging and reemerging pathogens. Emerg Infect Dis 11: 1842–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woolhouse ME, Haydon DT, Antia R (2005) Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 20: 238–244 S0169–5347(05)00038–8 [pii];10.1016/j.tree.2005.02.009 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, et al. (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397: 436–441 10.1038/17130 [doi] [DOI] [PubMed] [Google Scholar]
- 6. Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR (1989) An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339: 389–392 10.1038/339389a0 [doi] [DOI] [PubMed] [Google Scholar]
- 7. Osterhaus A (2001) Catastrophes after crossing species barriers. Philos Trans R Soc Lond B Biol Sci 356: 791–793 10.1098/rstb.2001.0856 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Osterholm MT, Kelley NS (2012) Mammalian-transmissible H5N1 influenza: facts and perspective. MBio 3: e00045–12 mBio.00045-12 [pii];10.1128/mBio.00045-12 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, et al. (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459: 1122–1125 nature08182 [pii];10.1038/nature08182 [doi] [DOI] [PubMed] [Google Scholar]
- 10. Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, et al. (2005) Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A 102: 2430–2435 0409608102 [pii];10.1073/pnas.0409608102 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, et al. (2006) Prevalence and genetic diversity of coronaviruses in bats from China. J Virol 80: 7481–7490 80/15/7481 [pii];10.1128/JVI.00697-06 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abe N, Read C, Thompson RC, Iseki M (2005) Zoonotic genotype of Giardia intestinalis detected in a ferret. J Parasitol 91: 179–182 10.1645/GE-3405RN [doi] [DOI] [PubMed] [Google Scholar]
- 13. Hankenson FC, Johnston NA, Weigler BJ, Di Giacomo RF (2003) Zoonoses of occupational health importance in contemporary laboratory animal research. Comp Med 53: 579–601. [PubMed] [Google Scholar]
- 14. Campagnolo ER, Moll ME, Tuhacek K, Simeone AJ, Miller WS, et al. (2013) Concurrent 2009 pandemic influenza A (H1N1) virus infection in ferrets and in a community in Pennsylvania. Zoonoses Public Health 60: 117–124 –10.1111/j.1863–2378.2012.01503.x [doi] [DOI] [PubMed] [Google Scholar]
- 15. Chomel BB (1992) Zoonoses of house pets other than dogs, cats and birds. Pediatr Infect Dis J 11: 479–487. [DOI] [PubMed] [Google Scholar]
- 16. Marini RP, Adkins JA, Fox JG (1989) Proven or potential zoonotic diseases of ferrets. J Am Vet Med Assoc 195: 990–994. [PubMed] [Google Scholar]
- 17. Bossart KN, Zhu Z, Middleton D, Klippel J, Crameri G, et al. (2009) A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog 5: e1000642 10.1371/journal.ppat.1000642 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martina BE, Haagmans BL, Kuiken T, Fouchier RA, Rimmelzwaan GF, et al. (2003) Virology: SARS virus infection of cats and ferrets. Nature 425: 915 10.1038/425915a [doi];425915a [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith H, Sweet C (1988) Lessons for human influenza from pathogenicity studies with ferrets. Rev Infect Dis 10: 56–75. [DOI] [PubMed] [Google Scholar]
- 20. Svitek N, von Messling V (2007) Early cytokine mRNA expression profiles predict Morbillivirus disease outcome in ferrets. Virology 362: 404–410 S0042-6822(07)00004-9 [pii];10.1016/j.virol.2007.01.002 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zitzow LA, Rowe T, Morken T, Shieh WJ, Zaki S, et al. (2002) Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol 76: 4420–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Vries E, Stittelaar KJ, van Amerongen G, Veldhuis Kroeze EJB, de Waal L, et al.. (2013) Prolonged Influenza Virus Shedding and Emergence of Antiviral Resistance in Immunocompromised Patients and Ferrets. PLoS Pathog. In press. [DOI] [PMC free article] [PubMed]
- 23. Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J (2001) A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci U S A 98: 11609–11614 10.1073/pnas.211424698 [doi];211424698 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, et al. (2005) Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A 102: 12891–12896 0504666102 [pii];10.1073/pnas.0504666102 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Brand JM, van Leeuwen M., Schapendonk CM, Simon JH, Haagmans BL, et al.. (2011) Metagenomic analysis of the viral flora of pine marten and European badger feces. J Virol. JVI.06373-11 [pii];10.1128/JVI.06373-11 [doi]. [DOI] [PMC free article] [PubMed]
- 26. van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, et al. (2010) Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol 48: 1787–1794 JCM.02452-09 [pii];10.1128/JCM.02452-09 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Losada L, Varga JJ, Hostetler J, Radune D, Kim M, et al. (2011) Genome sequencing and analysis of Yersina pestis KIM D27, an avirulent strain exempt from select agent regulation. PLoS One 6: e19054 10.1371/journal.pone.0019054 [doi]; PONE-D-11-01493 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al..(1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. gka562 [pii]. [DOI] [PMC free article] [PubMed]
- 29. Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17: 377–386 gr.5969107 [pii];10.1101/gr.5969107 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC (2011) Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560 gr.120618.111 [pii];10.1101/gr.120618.111 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 btm404 [pii];10.1093/bioinformatics/btm404 [doi] [DOI] [PubMed] [Google Scholar]
- 32. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 msm092 [pii];10.1093/molbev/msm092 [doi] [DOI] [PubMed] [Google Scholar]
- 33. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, et al. (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73: 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Day JM, Ballard LL, Duke MV, Scheffler BE, Zsak L (2010) Metagenomic analysis of the turkey gut RNA virus community. Virol J 7: 313 1743-422X-7-313 [pii];10.1186/1743-422X-7-313 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L, Victoria JG, Wang C, Jones M, Fellers GM, et al. (2010) Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J Virol 84: 6955–6965 JVI.00501-10 [pii];10.1128/JVI.00501-10 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, et al. (2011) The fecal viral flora of wild rodents. PLoS Pathog 7: e1002218 10.1371/journal.ppat.1002218 [doi]; PPATHOGENS-D-11-00801 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Provacia LB, Smits SL, Martina BE, Raj VS, Doel PV, et al. (2011) Enteric coronavirus in ferrets, The Netherlands. Emerg Infect Dis 17: 1570–1571 10.3201/eid1708.110115 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raj VS, Smits SL, Pas SD, Provacia LB, Moorman-Roest H, et al. (2012) Novel hepatitis E virus in ferrets, the Netherlands. Emerg Infect Dis 18: 1369–1370 10.3201/eid1808.111659 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farkas T, Fey B, Keller G, Martella V, Egyed L (2012) Molecular detection of novel astroviruses in wild and laboratory mice. Virus Genes 45: 518–525 10.1007/s11262-012-0803-0 [doi] [DOI] [PubMed] [Google Scholar]
- 40. Yokoyama CC, Loh J, Zhao G, Stappenbeck TS, Wang D, et al. (2012) Adaptive immunity restricts replication of novel murine astroviruses. J Virol 86: 12262–12270 JVI.02018-12 [pii];10.1128/JVI.02018-12 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamashita T, Sakae K, Tsuzuki H, Suzuki Y, Ishikawa N, et al. (1998) Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J Virol 72: 8408–8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, et al. (2003) Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol 84: 3069–3077. [DOI] [PubMed] [Google Scholar]
- 43. Reuter G, Boldizsar A, Pankovics P (2009) Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol 154: 101–108 10.1007/s00705-008-0288-2 [doi] [DOI] [PubMed] [Google Scholar]
- 44. Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, et al. (2011) The fecal viral flora of wild rodents. PLoS Pathog 7: e1002218 10.1371/journal.ppat.1002218 [doi]; PPATHOGENS-D-11-00801 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kapoor A, Simmonds P, Dubovi EJ, Qaisar N, Henriquez JA, et al. (2011) Characterization of a canine homolog of human Aichivirus. J Virol 85: 11520–11525 JVI.05317-11 [pii];10.1128/JVI.05317-11 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li L, Pesavento PA, Shan T, Leutenegger CM, Wang C, et al. (2011) Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J Gen Virol 92: 2534–2541 vir.0.034611-0 [pii];10.1099/vir.0.034611-0 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reuter G, Boros A, Pankovics P, Egyed L (2010) Kobuvirus in domestic sheep, Hungary. Emerg Infect Dis 16: 869–870 10.3201/eid1605.091934 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reuter G, Boros A, Pankovics P (2011) Kobuviruses – a comprehensive review. Rev Med Virol 21: 32–41 10.1002/rmv.677 [doi] [DOI] [PubMed] [Google Scholar]
- 49.Knowles NJ, Hovi T, Hyypia T, King AMQ, Lindberg AM, et al. (2012) Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier. 855–880.
- 50. Boivin G, Abed Y, Boucher FD (2005) Human parechovirus 3 and neonatal infections. Emerg Infect Dis 11: 103–105 10.3201/eid1101.040606 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Harvala H, Robertson I, McWilliam Leitch EC, Benschop K, et al. (2008) Epidemiology and clinical associations of human parechovirus respiratory infections. J Clin Microbiol 46: 3446–3453 JCM.01207-08 [pii];10.1128/JCM.01207-08 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levorson RE, Jantausch BA, Wiedermann BL, Spiegel HM, Campos JM (2009) Human parechovirus-3 infection: emerging pathogen in neonatal sepsis. Pediatr Infect Dis J 28: 545–547 10.1097/INF.0b013e318194596a [doi];00006454-200906000-00021 [pii] [DOI] [PubMed] [Google Scholar]
- 53. Verboon-Maciolek MA, Groenendaal F, Hahn CD, Hellmann J, van Loon AM, et al. (2008) Human parechovirus causes encephalitis with white matter injury in neonates. Ann Neurol 64: 266–273 10.1002/ana.21445 [doi] [DOI] [PubMed] [Google Scholar]
- 54. de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H (2004) Classification of papillomaviruses. Virology 324: 17–27 10.1016/j.virol.2004.03.033 [doi]; S004268220400220X [pii] [DOI] [PubMed] [Google Scholar]
- 55. Bernard HU, Burk RD, Chen Z, Van DK, Hausen H, et al. (2010) Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 401: 70–79 S0042-6822(10)00100-5 [pii];10.1016/j.virol.2010.02.002 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maggi F, Bendinelli M (2010) Human anelloviruses and the central nervous system. Rev Med Virol 20: 392–407 10.1002/rmv.668 [doi] [DOI] [PubMed] [Google Scholar]
- 57. Mokili JL, Rohwer F, Dutilh BE (2012) Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2: 63–77 S1879-6257(11)00190-8 [pii];10.1016/j.coviro.2011.12.004 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ge X, Li Y, Yang X, Zhang H, Zhou P, et al. (2012) Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J Virol 86: 4620–4630 JVI.06671-11 [pii];10.1128/JVI.06671-11 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Shan T, Wang C, Cote C, Kolman J, et al.. (2011) The fecal viral flora of California sea lions. J Virol. JVI.05026-11 [pii];10.1128/JVI.05026-11 [doi]. [DOI] [PMC free article] [PubMed]
- 60. Li L, Kapoor A, Slikas B, Bamidele OS, Wang C, et al. (2010) Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J Virol 84: 1674–1682 JVI.02109-09 [pii];10.1128/JVI.02109-09 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li L, Pesavento PA, Woods L, Clifford DL, Luff J, et al. (2011) Novel amdovirus in gray foxes. Emerg Infect Dis 17: 1876–1878 10.3201/eid1710.110233 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lipkin WI (2010) Microbe hunting. Microbiol Mol Biol Rev 74: 363–377 74/3/363 [pii];10.1128/MMBR.00007-10 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ng TF, Willner DL, Lim YW, Schmieder R, Chau B, et al. (2011) Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS One 6: e20579 10.1371/journal.pone.0020579 [doi]; PONE-D-11-07213 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ng TF, Wheeler E, Greig D, Waltzek TB, Gulland F, et al. (2011) Metagenomic identification of a novel anellovirus in Pacific harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J Gen Virol 92: 1318–1323 vir.0.029678-0 [pii];10.1099/vir.0.029678-0 [doi] [DOI] [PubMed] [Google Scholar]
- 65. Ng TF, Manire C, Borrowman K, Langer T, Ehrhart L, et al. (2009) Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J Virol 83: 2500–2509 JVI.01946-08 [pii];10.1128/JVI.01946-08 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shan T, Li L, Simmonds P, Wang C, Moeser A, et al. (2011) The fecal virome of pigs on a high-density farm. J Virol 85: 11697–11708 JVI.05217-11 [pii];10.1128/JVI.05217-11 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Smits SL, van Leeuwen M, Kuiken T, Hammer AS, Simon JH, et al. (2010) Identification and characterization of deer astroviruses. J Gen Virol 91: 2719–2722 vir.0.024067-0 [pii];10.1099/vir.0.024067-0 [doi] [DOI] [PubMed] [Google Scholar]
- 68. Smits SL, Poon LL, van Leeuwen M, Lau PN, Perera HK, et al. (2011) Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis 17: 2328–2330 10.3201/eid1712.110934 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Aggarwal R, Jameel S (2011) Hepatitis E. Hepatology. 54: 2218–2226 10.1002/hep.24674 [doi] [DOI] [PubMed] [Google Scholar]
- 70. Dalton HR, Bendall R, Ijaz S, Banks M (2008) Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis 8: 698–709 S1473-3099(08)70255-X [pii];10.1016/S1473-3099(08)70255-X [doi] [DOI] [PubMed] [Google Scholar]
- 71. Meng XJ (2010) Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol 140: 256–265 S0378-1135(09)00146-1 [pii];10.1016/j.vetmic.2009.03.017 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johansson S, Niklasson B, Maizel J, Gorbalenya AE, Lindberg AM (2002) Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins. J Virol 76: 8920–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harvala H, Simmonds P (2009) Human parechoviruses: biology, epidemiology and clinical significance. J Clin Virol 45: 1–9 S1386-6532(09)00123-1 [pii];10.1016/j.jcv.2009.03.009 [doi] [DOI] [PubMed] [Google Scholar]
- 74. Biagini P (2009) Classification of TTV and related viruses (anelloviruses). Curr Top Microbiol Immunol 331: 21–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenBank accession numbers of representative parecho-, papilloma-, and anellovirus strains.
(DOC)