Abstract
Immune cells are thoroughbreds, moving farther and faster and surveying more diverse tissue space than their non-hematopoietic brethren. Intravital 2-photon microscopy has provided insights into the movements and interactions of many immune cell types in diverse tissues, but much more information is needed to link such analyses of dynamic cell behavior to function. Here we describe additional methods whose application promises to extend our vision, allowing more complete, multiscale dissection of how immune cell positioning and movement are linked to system state, host defense, and disease.
Keywords: 2-photon, dynamic, imaging, in vivo, multicolor
The immune system is like a fine mechanical watch – there are a large number of parts that must work together to achieve the right result. For the watch, the goal is keeping perfect time, for the immune system it is optimally protecting the host. If the pieces are not machined and assembled properly the watch (immune system) can run too slowly (immunodeficiency) or too fast (autoimmunity / inflammatory disease). There is a delicate balance in the interaction of the watch parts – they must move properly, engage for just the right amount of time, then disengage and move again. Likewise the cells of the immune system must circulate and migrate, find the right cellular partner at the right time, engage for the proper duration, signal effectively, change gene expression, then move once again. Dynamics and positioning are crucial aspects of immune function that need to be described and understood if we are to have an accurate picture of the system and how it carries out its functions.
Like a watchmaker, who uses magnifying lenses to peer at the minute parts of a complex timepiece to check their function, investigators have turned to optical imaging to gain knowledge about the dynamic properties of immune cells in their in vivo environments. Over the past decade in particular, intravital 2-photon imaging has provided a wealth of insights into what has come to be called ‘immunodynamics.”[1–3] We have seen how naïve T and B cells move within secondary lymphoid tissues and acquire antigenic information [4–14], the intricate dance of T and B cells at the T/B-border and within germinal centers [15–20] and of developing thymocytes in that organ [21, 22], the reactivation of memory T cells [23–25], osteoclast, platelet and neutrophil mobilization in the bone marrow [26–28], as well as the movement of innate and adaptive effectors in tissues such as skin [29–31], liver [32–34], central nervous system [35–37], lung [38, 39], and tumors [40, 41] among others. Migration and local probing behavior of dendritic cells in diverse sites has been examined [42–45]. The role of stromal elements in guiding immune cell migration has been discovered [46], the key contribution of adequate cell-cell adhesion in overcoming the dispersive migratory properties of lymphocytes and permitting effective inter-cellular co-operation has become clear [47], the restricted anatomical domains in secondary lymphoid organs within which some innate and adaptive immune cells migrate while awaiting evidence of host invasion have been revealed [48], and the in vivo operation of chemokines with regards to facilitating the encounter between rare cell populations delineated [49, 50].
While these discoveries have ‘animated’ the field for years, we are still far from where we need to be to link information on molecules, signaling pathways, and gene regulatory events to these descriptive dynamics. New tools and techniques are required to see more for longer in larger volumes. Methods that allow simultaneous tracking of receptor signaling events and cell movement, of cytokine production and the response to these key mediators and of gene activation are essential for connecting dynamic behavior with function and differentiation. Regions of tissues currently inaccessible to our imaging platforms need to be made visible, tracking needs to occur in larger volumes to avoid loss of cells over the time span involved in their progression from resting cells to a differentiated state, many more cell types (including stromal elements such as mesenchymal cells, nerves, and vessels) need to be distinctively labeled for visualization at one time, and resolution must be increased to permit intracellular elements to be monitored. New computational tools must be developed to cope with the vast amount of data that will be generated from imaging more colors, with greater resolution, for longer times, and in larger volumes, including visualization methods that make such complex data understandable to the experimentalist. In this Viewpoint, we briefly describe the evolving methods (Figure 1A–E) that will contribute to overcoming these limitations and how their implementation will provide essential insights into immune function in health and disease.
Figure 1. Overview of improved imaging modalities.
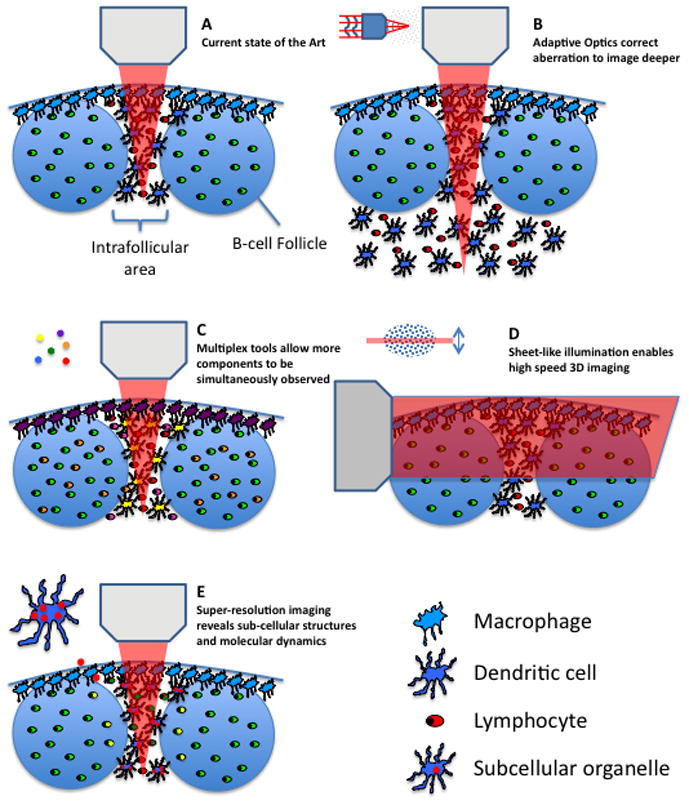
(A) The current state of the art in imaging the interfollicular area and B-cell follicles of a lymph node is shown. (B) Improved imaging depth by means of adaptive optics is shown. (C) Detection of multiple different signals by means of multiplexing tools such as multiple lasers or new chromophores is shown. (D) High-speed 3D imaging using sheet-illumination is shown. (E) The visualization of sub-cellular compartments using super-resolution imaging techniques is shown.
Challenge #1 – Imaging depth
Even in mice, the main experimental animal used for immune system dynamic imaging, many events that need to be visualized are hundreds of microns to millimeters from the surface of a tissue. Current fluorescence-based intravital imaging techniques can only probe the region near the surface (up to 200–300 microns) in dense lymphoid and other tissues; this is because of the dispersive optical properties of these biological structures. To gain information deep in live tissue by multi-photon imaging, photons carrying input information (excitation) have to be efficiently delivered to the location of interest (focus), and the output information (fluorescence) has to be transmitted back via emitted photons that can be collected and detected by light sensors. Within deeper areas of tissues, scattering and absorption processes will dramatically attenuate the excitation laser intensity that reaches the focal point and the heterogeneous structure of the tissue will distort the light propagation wave front, so the excitation light cannot be tightly focused. Such optical aberration is a major factor leading to degradation of image quality.
Recognizing this limitation, many groups have been working on solutions. Near term, the simplest approach is the use of far-red or near-infrared fluorochromes and fluorescent proteins [51–54]. The longer wavelength of the excitation and emission photons involved in imaging such fluorochromes and proteins allow for better tissue penetration with less scattering or absorption than shorter wavelength photons. A penetration depth up to 1.6 mm in mouse cortex has been demonstrated with the laser wavelength tuned at 1.28 μm and emission in the near infrared (IR) range [55].
A very promising method that is not limited by wavelength in this way involves a technique that is termed adaptive optics (AO) (Figure 1B). Such a method aims to pre-compensate the light wavefront distortion inside the live tissue and allow a maximum amount of coherent laser light to reach the focal point. This approach can dramatically improve imaging contrast [56, 57]. While the speed of the wave front optimization process has been increased significantly, compatible with typical dynamic multiphoton intravital imaging methods, the improvement of signal quality using AO approaches comes with the price of a smaller field of view. This is because the wavefront distortion has to be measured and compensated at different locations inside the heterogeneous tissue sample, a process that takes time both due to the physical properties and software control of the adaptive mirrors employed to control the beam. These limitations will eventually be overcome through development of faster hardware and software. Even with current AO technology, one can now perform highly resolved imaging in small fields deep within tissues that are structurally stable (i.e., bone marrow, skin).
Other new optical approaches that extend the penetration depth even further, to beyond millimeter scale, are also in development. One such technique is ultrasound-guided optical imaging [58, 59], which takes advantage of an acoustic wave. The latter is less sensitive to the tissue medium that scatters the light wave and can be used to guide excitation and emission photons. This method can currently achieve imaging resolution up to 12 μm at a depth of 2 mm [59], making it potentially useful not only for deep animal imaging but for application to the dermis in humans, as one example.
These methods not only promise to allow visualization of objects deeper in tissues than presently accessible, but also the imaging over time of larger volumes than can be presently examined. Because of cell motility, capture of information from a larger volume not only reduces sampling error (more events can be tracked), but also permits cells to be followed over longer time intervals (because they stay within the imaging volume). In combination with hardware improvements that permit faster scanning, more sensitive photon detection, and faster movement of the stage holding the specimen, these advances will provide an enhanced ability to link early and late events during an ongoing response on a per-cell basis throughout a larger range of organ compartments. Ultimately, this will allow for continuous tracking of cells and their fates after asymmetric division and during differentiation [60].
Challenge #2 – Multiplex detection
Immunity is a highly concerted process involving many different cellular and molecular players. For this reason, the present imaging methods that typically involve 3–4 colors per experiment are incapable of revealing many of the elements that play an important role in immune processes. This limitation applies at the macro scale (insufficient diversity of cell types and stromal structures visualized), so that the impact of tissue organization and the many different types of cell-cell contacts is not well appreciated. It is also true at the nano-scale – without an ability to multiplex more extensively, we cannot track both cell behavior and the molecules involved in cell interactions and signaling or gene activity. Yet such co-measurements are critical if we are to link bulk dynamics to the processes of cellular activation and response crucial to immune function. Achieving these goals requires new experimental capability to observe multiple different cellular and molecular players at the same time, in the same sample, and with the same imaging configuration.
The current spectral range of fluorescence detection is from near ultraviolet (UV) to near IR. Considering the intrinsic fluorescent bandwidth of typical chromophores, the number of different fluorophores that can be distinguished within this spectral range is limited using conventional filtration methods, even assuming that an ideal set of labels with well-separated fluorescent spectra can be employed experimentally. Four methods promise to overcome these present limitations by facilitating the simultaneous detection of multiple components (Figure 1C):
The use of multiple lasers tuned to different excitation wavelengths. Employed in the proper way, this allows optimized excitation of a variety of fluorochromes rather than the severe compromise typical of single laser instruments, permitting substantial improvement in detection of labels expressed at low levels and the use of more distinct fluorochromes that in combination would not be well excited with single laser systems [61, 62].
The development of new chromophores (synthetic dyes or fluorescent proteins) with fluorescence spectra beyond the current range. For example, more near-IR or IR fluorescent proteins have already been developed in the past few years [51, 63–66], extending the palette available for imaging studies with current microscope systems.
-
The application of spectral unmixing strategies. Rather than using filters to isolate distinct (non-overlapping) regions of emission spectra to identify targets, one can “deconvolve” the entire emitted fluorescent spectrum to identify the target chromophore by its specific emission spectral profile [67]. This requires the chromophore to have a signal strong enough to be split into the multiple spectral windows, each of which captures fewer total emitted photons. One has to find a balance between the spectral precision (more windows) and sensitivity (signal/noise = larger windows), setting a limit to the use of this approach when the cell or molecule of interest can only have a limited overall fluorescent output.
A related approach is to label each target component with multiple chromophores and modulate the ratio between different chromophores to identify various biological components (spectral painting). This approach has been successfully demonstrated in the application of single molecule mRNA FISH [68] and in Brainbow [69] and similar transgenic animals. Here, the complex emission spectrum from each different color combination provides a signature for that target. This approach can be further enhanced by careful choice of chromophores with distinct absorption spectra permitting selective excitation at different laser wavelengths when using multi-line instruments.
The combination of standard wavelength detection methods with other strategies for fluorochrome identification. One well-established approach in single cell imaging is fluorescence lifetime measurement (FLIM) [70, 71] and this has recently been incorporated into multiphoton intravital imaging [72]. In this method, special detectors and software statistically characterize the time interval (on a nanosecond scale) between the excitation and emission events for each chromophore molecule. Many spectrally-overlapped chromophores have very distinct fluorescence lifetimes, so this technique can be used to add another dimension to the “identity space” of these labels. With the newest generation of high sensitivity detectors on commercial microscopes, implementation of FLIM is becoming an available tool for intravital multiphoton imaging.
A final area in which substantial progress is needed in the area of multiplex detection is in the creation of optimized probes for the analysis of intracellular signaling, molecular localization, or gene expression. A few intravital studies have characterized such events, most involving dye-based calcium sensors [13, 73], in vivo staining [74], fluorescent chimeric proteins [75, 76], and fluorescent gene reporters [77–80]. The calcium studies are limited by the leak rate of the sensor dyes to just an hour or two after cell transfer, the chimeric proteins are both hard to detect and the analysis suffers from artifacts of optical resolution limitations and signal intensity differences in the axial dimension, and the gene reporters produce cytoplasmic proteins whose lifetime greatly exceeds that of cytokine transcripts, thus failing to provide a properly time-resolved record of gene activity that can be linked to cell dynamics [81]. To make progress in these areas, these limitations must be overcome. New fluorescent proteins have been developed [82] for the generation of optimized FRET sensors that can detect signaling events such as calcium elevation, MAPK pathway activation, and the like and should allow creation of genetically-labeled cells whose signaling can be tracked without the present post-transfer time limitation. The improved sensitivity of newer instruments will permit better detection of chimeric proteins expressed at close to physiologic levels and some of the methods described below and by Sixt in this issue will enhance axial resolution that presently limits such analyses. The creation of a new generation of genetic reporters producing destabilized fluorescent reporters, or secreted versions of such proteins, will improve the temporal connectivity between appearance of these labeled proteins and the underlying genetic activities of the cell of interest [83]. Together, such tools will allow investigators to link molecular events with cell behavior.
Challenge #3 - Faster imaging
The complexity of biological systems not only exists in the space domain, but also in the time domain. Cellular and molecular dynamics have multiplex temporal features ranging from femtoseconds to hours. Both the sampling speed and length of image collection of present instruments and their linked computers dictate the time resolution that can be achieved.
To date, the predominant scheme for sample illumination is point scanning, which limits information collection rate due to the time needed to move the beam over the entire x-y dimensions of the imaging field. A major breakthrough in the past few years allows for parallel illumination of biological samples, sometimes in three dimensions. In these methods, a modified optical system illuminates a selected plane rather than a single point inside the biological sample, with the fluorescence from the whole plane detected by a wide field imager (Figure 1D). For example, light sheet microscopy [84–86] uses one lens to create a narrow sheet of laser focus inside the tissue, and another objective is used to acquire fluorescent images at the perpendicular direction. Bessel beam-based light sheet microscopy [87] further improves the spatial resolution especially in the axial dimension. Another temporal focusing scheme [88, 89] modulates the excitation laser pulse width so that the multiphoton excitation is confined in a sheet-like region. Although the temporal focusing method is limited to non-linear excitation, it brings the convenience of using only a single objective lens with less spatial restriction for sample arrangement.
Challenge #4 - Higher resolution
In principle, the resolution of an optical microscope is limited by the optical diffraction, in the range of a few hundred nanometers with a lens of the highest possible numerical aperture (NA). In practice one has to find a balance between the working distance of the lens and NA, preventing use of the best resolving lenses for most intravital imaging purposes. In addition, the optical aberration inside tissues also degrades resolution, so methods to correct such aberrations, like the AO approaches discussed above, are needed to achieve near-diffraction-limit resolution deep inside tissues.
During the past decade, however, there have been a number of exciting breakthroughs that allow biological samples to be imaged at a resolution far beyond the optical diffraction limit (Figure 1E). These novel approaches can be generalized into two types: one type takes advantage of single molecule location measurement, such as PALM (Photoactivated localization microscopy), FPALM (Fluorescence photoactivated localization microscopy), and STORM (Stochastic optical reconstruction microscopy) [90–92]; the other creates sub-diffraction excitation patterns, such as STED (Stimulated emission depletion) [93] and Structured Illumination [94]. These new techniques are poised to reveal detailed molecular and structural information inside live tissues. Although to date most applications are with relatively flat cells in culture systems, some exciting recent developments have begun to incorporate such schemes into intravital imaging scenarios [95], permitting the detection of subcellular events with improved precision, at least close to the surface of various tissues [96].
Challenge #5 – Data capture and processing
All the above advances will help collect more information on the immune system in situ, but without the proper means for data analysis, we will make little progress in understanding things. A new generation of software tools is needed to better analyze the image data, preferably allowing more to be done automatically and in an unbiased manner than is presently possible [97]. Some progress has been made in this arena over the past several years, both by commercial software vendors and academic centers, but much more is needed. Optimized algorithms are required to handle the much larger number of objects to be tracked when the imaging volume is increased and more cell types are visualized in more colors, for both analytic purposes and for display of the underlying cell movements; embryologists have made substantial progress in this direction [86, 98] and their methods need to be adopted (and adapted) by immunological imaging experts. Progress is also needed to address specific issues in molecular imaging, which is highly sensitive to depth-related image intensity artifacts. Some schemes to deal with such problems have already appeared [75, 79] but as the field embraces molecular imaging going forward, further improvements will be crucial. Other analytic tools that have been introduced to handle the analysis of cell movements into and out of defined volumes such as germinal centers or to measure synaptic dimensions[99, 100], provide a display in two dimensions out of four or more parameters such as time, speed, directionality, and distance from a defined site. These parameters greatly aids in understanding chemosensing behavior (Lämmermann and Angermann, unpublished), differentiate random walk from other migratory behavior [101, 102], and evaluate contact times between two cell types [103]. These data sets may then be used to derive mathematical models of complex cellular behavior, which by iterative processes can be refined and may allow for prediction of biological outcomes that can be tested in in vivo models [102, 104–110]. Finally, the field will need to move away from the maximum projection the two-dimensional movies that are currently used to three-dimensional displays that allow a better appreciation of depth in the full imaging volume.
Conclusion
Here we have very briefly surveyed some of the emerging techniques that will aid our probing of the dynamic behavior of the immune system going forward. They will allow a larger portion of an organ to be examined, many more elements to be tracked simultaneously, cell analysis to be combined with molecular imaging, and cell function linked to dynamic behavior. An essential point to emphasize, however, is that while such imaging can be revealing in its own right, it is most valuable as a part of the larger fabric of immune investigation; dynamic imaging data need to be properly associated with information gathered by other means, such as static tissue imaging [111, 112], flow cytometry, ex vivo assessment of lymphocyte activity and polarization, genetic and epigenetic studies, and overall measurements of systemic immunity and host resistance. Only through integration of information garnered using the full range of methods available to the field can we develop a comprehensive model of immune system behavior, in which events on the micro and macro scale are linked to the meso-scale dynamics of individual cells that are the present focus of imaging analysis. With the increased depth and breadth of analysis we anticipate from rigorous application of the methods reviewed here, we are confident the future is, not to put too fine a point on it, “bright.”
Acknowledgments
This work was supported by the Intramural Research Program of NIAID, NIH. We apologize to all colleagues whose primary work could not be directly cited due to editorial length and reference limitations.
Footnotes
Conflict of interest:
The authors declare no financial or commercial conflict of interest.
References
- 1.Germain RN, Robey EA, Cahalan MD. A decade of imaging cellular motility and interaction dynamics in the immune system. Science. 2012;336:1676–1681. doi: 10.1126/science.1221063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 3.Pittet MJ, Weissleder R. Intravital imaging. Cell. 2011;147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296:1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 5.Stoll S, Delon J, Brotz TM, Germain RN. Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science. 2002;296:1873–1876. doi: 10.1126/science.1071065. [DOI] [PubMed] [Google Scholar]
- 6.Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, et al. Visualizing dendritic cell networks in vivo. Nat Immunol. 2004;5:1243–1250. doi: 10.1038/ni1139. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki K, Grigorova I, Phan TG, Kelly LM, Cyster JG. Visualizing B cell capture of cognate antigen from follicular dendritic cells. J Exp Med. 2009;206:1485–1493. doi: 10.1084/jem.20090209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan TG, Grigorova I, Okada T, Cyster JG. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat Immunol. 2007;8:992–1000. doi: 10.1038/ni1494. [DOI] [PubMed] [Google Scholar]
- 9.Carrasco YR, Batista FD. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity. 2007;27:160–171. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Junt T, Moseman EA, Iannacone M, Massberg S, Lang PA, Boes M, Fink K, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 11.Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427:154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 12.Hickman HD, Takeda K, Skon CN, Murray FR, Hensley SE, Loomis J, Barber GN, et al. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat Immunol. 2008;9:155–165. doi: 10.1038/ni1557. [DOI] [PubMed] [Google Scholar]
- 13.Qi H, Egen JG, Huang AY, Germain RN. Extrafollicular activation of lymph node B cells by antigen-bearing dendritic cells. Science. 2006;312:1672–1676. doi: 10.1126/science.1125703. [DOI] [PubMed] [Google Scholar]
- 14.Bousso P, Robey E. Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nat Immunol. 2003;4:579–585. doi: 10.1038/ni928. [DOI] [PubMed] [Google Scholar]
- 15.Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada T, Miller MJ, Parker I, Krummel MF, Neighbors M, Hartley SB, O'Garra A, et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:1047–1061. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 19.Schwickert TA, Lindquist RL, Shakhar G, Livshits G, Skokos D, Kosco-Vilbois MH, Dustin, et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 20.Gunzer M, Weishaupt C, Hillmer A, Basoglu Y, Friedl P, Dittmar KE, Kolanus W, et al. A spectrum of biophysical interaction modes between T cells and different antigen-presenting cells during priming in 3-D collagen and in vivo. Blood. 2004;104:2801–2809. doi: 10.1182/blood-2004-03-1193. [DOI] [PubMed] [Google Scholar]
- 21.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science. 2002;296:1876–1880. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 22.Le Borgne M, Ladi E, Dzhagalov I, Herzmark P, Liao YF, Chakraborty AK, et al. 2009;10:823–830. doi: 10.1038/ni.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavanagh LL, Bonasio R, Mazo IB, Halin C, Cheng G, van der Velden AW, Cariappa A, et al. Activation of bone marrow-resident memory T cells by circulating, antigen-bearing dendritic cells. Nat Immunol. 2005;6:1029–1037. doi: 10.1038/ni1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung JH, Zhang H, Moseman EA, Alvarez D, Iannacone M, Henrickson SE, de la Torre JC, et al. Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell. 2012;150:1249–1263. doi: 10.1016/j.cell.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kastenmuller W, Brandes M, Wang Z, Herz J, Egen JG, Germain RN. Peripheral Prepositioning and Local CXCL9 Chemokine-Mediated Guidance Orchestrate Rapid Memory CD8(+) T Cell Responses in the Lymph Node. Immunity. 2013 doi: 10.1016/j.immuni.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohler A, De Filippo K, Hasenberg M, van den Brandt C, Nye E, Hosking MP, Lane, et al. G-CSF-mediated thrombopoietin release triggers neutrophil motility and mobilization from bone marrow via induction of Cxcr2 ligands. Blood. 2011;117:4349–4357. doi: 10.1182/blood-2010-09-308387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii M, Egen JG, Klauschen F, Meier-Schellersheim M, Saeki Y, Vacher J, Proia RL, et al. Sphingosine-1-phosphate mobilizes osteoclast precursors and regulates bone homeostasis. Nature. 2009;458:524–528. doi: 10.1038/nature07713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Junt T, Schulze H, Chen Z, Massberg S, Goerge T, Krueger A, Wagner DD, et al. Dynamic visualization of thrombopoiesis within bone marrow. Science. 2007;317:1767–1770. doi: 10.1126/science.1146304. [DOI] [PubMed] [Google Scholar]
- 29.Celli S, Albert ML, Bousso P. Visualizing the innate and adaptive immune responses underlying allograft rejection by two-photon microscopy. Nat Med. 2011;17:744–749. doi: 10.1038/nm.2376. [DOI] [PubMed] [Google Scholar]
- 30.Peters NC, Egen JG, Secundino N, Debrabant A, Kimblin N, Kamhawi S, Lawyer P, et al. In vivo imaging reveals an essential role for neutrophils in leishmaniasis transmitted by sand flies. Science. 2008;321:970–974. doi: 10.1126/science.1159194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, Carbone FR, et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature. 2011;477:216–219. doi: 10.1038/nature10339. [DOI] [PubMed] [Google Scholar]
- 32.Egen JG, Rothfuchs AG, Feng CG, Horwitz MA, Sher A, Germain RN. Intravital imaging reveals limited antigen presentation and T cell effector function in mycobacterial granulomas. Immunity. 2011;34:807–819. doi: 10.1016/j.immuni.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CC, Beck PL, et al. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 34.Geissmann F, Cameron TO, Sidobre S, Manlongat N, Kronenberg M, Briskin MJ, Dustin ML, et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson EH, Harris TH, Mrass P, John B, Tait ED, Wu GF, Pepper M, et al. Behavior of parasite-specific effector CD8+ T cells in the brain and visualization of a kinesis-associated system of reticular fibers. Immunity. 2009;30:300–311. doi: 10.1016/j.immuni.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawakami N, Nagerl UV, Odoardi F, Bonhoeffer T, Wekerle H, Flugel A. Live imaging of effector cell trafficking and autoantigen recognition within the unfolding autoimmune encephalomyelitis lesion. J Exp Med. 2005;201:1805–1814. doi: 10.1084/jem.20050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JV, Kang SS, Dustin ML, McGavern DB. Myelomonocytic cell recruitment causes fatal CNS vascular injury during acute viral meningitis. Nature. 2009;457:191–195. doi: 10.1038/nature07591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreisel D, Nava RG, Li W, Zinselmeyer BH, Wang B, Lai J, Pless R, et al. In vivo two-photon imaging reveals monocyte-dependent neutrophil extravasation during pulmonary inflammation. Proc Natl Acad Sci U S A. 2010;107:18073–18078. doi: 10.1073/pnas.1008737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruns S, Kniemeyer O, Hasenberg M, Aimanianda V, Nietzsche S, Thywissen A, Jeron A, et al. Production of extracellular traps against Aspergillus fumigatus in vitro and in infected lung tissue is dependent on invading neutrophils and influenced by hydrophobin RodA. PLoS Pathog. 2010;6:e1000873. doi: 10.1371/journal.ppat.1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breart B, Lemaitre F, Celli S, Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J Clin Invest. 2008;118:1390–1397. doi: 10.1172/JCI34388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boissonnas A, Fetler L, Zeelenberg IS, Hugues S, Amigorena S. In vivo imaging of cytotoxic T cell infiltration and elimination of a solid tumor. J Exp Med. 2007;204:345–356. doi: 10.1084/jem.20061890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–2852. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, Knoop KA, Newberry RD, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–349. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thornton EE, Looney MR, Bose O, Sen D, Sheppard D, Locksley R, Huang X, et al. Spatiotemporally separated antigen uptake by alveolar dendritic cells and airway presentation to T cells in the lung. J Exp Med. 2012;209:1183–1199. doi: 10.1084/jem.20112667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller MJ, Hejazi AS, Wei SH, Cahalan MD, Parker I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc Natl Acad Sci U S A. 2004;101:998–1003. doi: 10.1073/pnas.0306407101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bajenoff M, Glaichenhaus N, Germain RN. Fibroblastic reticular cells guide T lymphocyte entry into and migration within the splenic T cell zone. J Immunol. 2008;181:3947–3954. doi: 10.4049/jimmunol.181.6.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider H, Downey J, Smith A, Zinselmeyer BH, Rush C, Brewer JM, Wei B, et al. Reversal of the TCR stop signal by CTLA-4. Science. 2006;313:1972–1975. doi: 10.1126/science.1131078. [DOI] [PubMed] [Google Scholar]
- 48.Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012;150:1235–1248. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 50.Hugues S, Scholer A, Boissonnas A, Nussbaum A, Combadiere C, Amigorena S, Fetler L. Dynamic imaging of chemokine-dependent CD8+ T cell help for CD8+ T cell responses. Nat Immunol. 2007;8:921–930. doi: 10.1038/ni1495. [DOI] [PubMed] [Google Scholar]
- 51.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin MZ. Beyond the rainbow: new fluorescent proteins brighten the infrared scene. Nat Methods. 2011;8:726–728. doi: 10.1038/nmeth.1678. [DOI] [PubMed] [Google Scholar]
- 53.Mojzisova H, Vermot J. When multiphoton microscopy sees near infrared. Curr Opin Genet Dev. 2011;21:549–557. doi: 10.1016/j.gde.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Shcherbo D, Shemiakina II, Ryabova AV, Luker KE, Schmidt BT, Souslova EA, Gorodnicheva TV, et al. Nat Methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobat D, Horton NG, Xu C. In vivo two-photon microscopy to 1. 6-mm depth in mouse cortex. J Biomed Opt. 2011;16:106014. doi: 10.1117/1.3646209. [DOI] [PubMed] [Google Scholar]
- 56.Ji N, Milkie DE, Betzig E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat Methods. 2010;7:141–147. doi: 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- 57.Tang J, Germain RN, Cui M. Superpenetration optical microscopy by iterative multiphoton adaptive compensation technique. Proc Natl Acad Sci U S A. 2012;109:8434–8439. doi: 10.1073/pnas.1119590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang YM, Judkewitz B, Dimarzio CA, Yang C. Deep-tissue focal fluorescence imaging with digitally time-reversed ultrasound-encoded light. Nat Commun. 2012;3:928. doi: 10.1038/ncomms1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Si K, Fiolka R, Cui M. Breaking the spatial resolution barrier via iterative sound-light interaction in deep tissue microscopy. Sci Rep. 2012;2:748. doi: 10.1038/srep00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 61.Mahou P, Zimmerley M, Loulier K, Matho KS, Labroille G, Morin X, Supatto W, et al. Multicolor two-photon tissue imaging by wavelength mixing. Nat Methods. 2012;9:815–818. doi: 10.1038/nmeth.2098. [DOI] [PubMed] [Google Scholar]
- 62.Entenberg D, Wyckoff J, Gligorijevic B, Roussos ET, Verkhusha VV, Pollard JW, Condeelis J. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat Protoc. 2011;6:1500–1520. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hilderbrand SA, Weissleder R. Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol. 2010;14:71–79. doi: 10.1016/j.cbpa.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 64.Luo S, Zhang E, Su Y, Cheng T, Shi C. A review of NIR dyes in cancer targeting and imaging. Biomaterials. 2011;32:7127–7138. doi: 10.1016/j.biomaterials.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 65.Andresen V, Alexander S, Heupel WM, Hirschberg M, Hoffman RM, Friedl P. Infrared multiphoton microscopy: subcellular-resolved deep tissue imaging. Curr Opin Biotechnol. 2009;20:54–62. doi: 10.1016/j.copbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 66.Subach FV, Piatkevich KD, Verkhusha VV. Directed molecular evolution to design advanced red fluorescent proteins. Nat Methods. 2011;8:1019–1026. doi: 10.1038/nmeth.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimmermann T. Spectral imaging and linear unmixing in light microscopy. Adv Biochem Eng Biotechnol. 2005;95:245–265. doi: 10.1007/b102216. [DOI] [PubMed] [Google Scholar]
- 68.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 69.Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 70.Dong CY, French T, So PT, Buehler C, Berland KM, Gratton E. Fluorescence-lifetime imaging techniques for microscopy. Methods Cell Biol. 2003;72:431–464. doi: 10.1016/s0091-679x(03)72021-4. [DOI] [PubMed] [Google Scholar]
- 71.French T, So PT, Dong CY, Berland KM, Gratton E. Fluorescence lifetime imaging techniques for microscopy. Methods Cell Biol. 1998;56:277–304. doi: 10.1016/s0091-679x(08)60431-8. [DOI] [PubMed] [Google Scholar]
- 72.Fruhwirth GO, Matthews DR, Brock A, Keppler M, Vojnovic B, Ng T, Ameer-Beg S. Deep-tissue multiphoton fluorescence lifetime microscopy for intravital imaging of protein-protein interactions. 2009:71830L-71830L. [Google Scholar]
- 73.Bhakta NR, Oh DY, Lewis RS. Calcium oscillations regulate thymocyte motility during positive selection in the three-dimensional thymic environment. Nat Immunol. 2005;6:143–151. doi: 10.1038/ni1161. [DOI] [PubMed] [Google Scholar]
- 74.Schwendele B, Brawek B, Hermes M, Garaschuk O. High-resolution in vivo imaging of microglia using a versatile nongenetically encoded marker. Eur J Immunol. 2012;42:2193–2196. doi: 10.1002/eji.201242436. [DOI] [PubMed] [Google Scholar]
- 75.Friedman RS, Beemiller P, Sorensen CM, Jacobelli J, Krummel MF. Real-time analysis of T cell receptors in naive cells in vitro and in vivo reveals flexibility in synapse and signaling dynamics. J Exp Med. 2010;207:2733–2749. doi: 10.1084/jem.20091201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCombs JE, Palmer AE. Measuring calcium dynamics in living cells with genetically encodable calcium indicators. Methods. 2008;46:152–159. doi: 10.1016/j.ymeth.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoffman RM. Cellular and subcellular imaging in live mice using fluorescent proteins. Curr Pharm Biotechnol. 2012;13:537–544. doi: 10.2174/138920112799436311. [DOI] [PubMed] [Google Scholar]
- 78.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 79.Melichar HJ, Li O, Herzmark P, Padmanabhan RK, Oliaro J, Ludford-Menting MJ, Bousso P, et al. Quantifying subcellular distribution of fluorescent fusion proteins in cells migrating within tissues. Immunol Cell Biol. 2011;89:549–557. doi: 10.1038/icb.2010.122. [DOI] [PubMed] [Google Scholar]
- 80.Croxford AL, Buch T. Cytokine reporter mice in immunological research: perspectives and lessons learned. Immunology. 2011;132:1–8. [Google Scholar]
- 81.Beuneu H, Lemaitre F, Deguine J, Moreau HD, Bouvier I, Garcia Z, Albert ML, et al. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity. 2010;33:412–423. doi: 10.1016/j.immuni.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 82.Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Houser JR, Ford E, Chatterjea SM, Maleri S, Elston TC, Errede B. An improved short-lived fluorescent protein transcriptional reporter for Saccharomyces cerevisiae. Yeast. 2012;29:519–530. doi: 10.1002/yea.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 85.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 86.Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 87.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Durst ME, Zhu G, Xu C. Simultaneous Spatial and Temporal Focusing in Nonlinear Microscopy. Opt Commun. 2008;281:1796–1805. doi: 10.1016/j.optcom.2007.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oron D, Tal E, Silberberg Y. Scanningless depth-resolved microscopy. Opt Express. 2005;13:1468–1476. doi: 10.1364/opex.13.001468. [DOI] [PubMed] [Google Scholar]
- 90.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 91.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 94.Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.York AG, Parekh SH, Dalle Nogare D, Fischer RS, Temprine K, Mione M, Chitnis AB, et al. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat Methods. 2012;9:749–754. doi: 10.1038/nmeth.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ludewig B, Stein JV, Sharpe J, Cervantes-Barragan L, Thiel V, Bocharov G. A global “imaging” view on systems approaches in immunology. Eur J Immunol. 2012;42:3116–3125. doi: 10.1002/eji.201242508. [DOI] [PubMed] [Google Scholar]
- 98.Tomer R, Khairy K, Amat F, Keller PJ. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods. 2012;9:755–763. doi: 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- 99.Klauschen F, Ishii M, Qi H, Bajenoff M, Egen JG, Germain RN, Meier-Schellersheim M. Quantifying cellular interaction dynamics in 3D fluorescence microscopy data. Nat Protoc. 2009;4:1305–1311. doi: 10.1038/nprot.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Klauschen F, Qi H, Egen JG, Germain RN, Meier-Schellersheim M. Computational reconstruction of cell and tissue surfaces for modeling and data analysis. Nat Protoc. 2009;4:1006–1012. doi: 10.1038/nprot.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harris TH, Banigan EJ, Christian DA, Konradt C, Tait Wojno ED, Norose K, Wilson EH, et al. Generalized Levy walks and the role of chemokines in migration of effector CD8+ T cells. Nature. 2012;486:545–548. doi: 10.1038/nature11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beltman JB, Maree AF, Lynch JN, Miller MJ, de Boer RJ. Lymph node topology dictates T cell migration behavior. J Exp Med. 2007;204:771–780. doi: 10.1084/jem.20061278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dustin ML, Bromley SK, Kan Z, Peterson DA, Unanue ER. Antigen receptor engagement delivers a stop signal to migrating T lymphocytes. Proc Natl Acad Sci U S A. 1997;94:3909–3913. doi: 10.1073/pnas.94.8.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garin A, Meyer-Hermann M, Contie M, Figge MT, Buatois V, Gunzer M, Toellner KM, et al. Toll-like receptor 4 signaling by follicular dendritic cells is pivotal for germinal center onset and affinity maturation. Immunity. 2010;33:84–95. doi: 10.1016/j.immuni.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 105.Figge MT, Garin A, Gunzer M, Kosco-Vilbois M, Toellner KM, Meyer-Hermann M. Deriving a germinal center lymphocyte migration model from two-photon data. J Exp Med. 2008;205:3019–3029. doi: 10.1084/jem.20081160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bogle G, Dunbar PR. On-lattice simulation of T cell motility, chemotaxis, and trafficking in the lymph node paracortex. PLoS One. 2012;7:e45258. doi: 10.1371/journal.pone.0045258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beltman JB, Henrickson SE, von Andrian UH, de Boer RJ, Maree AF. Towards estimating the true duration of dendritic cell interactions with T cells. J Immunol Methods. 2009;347:54–69. doi: 10.1016/j.jim.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beltman JB, Maree AF, de Boer RJ. Spatial modelling of brief and long interactions between T cells and dendritic cells. Immunol Cell Biol. 2007;85:306–314. doi: 10.1038/sj.icb.7100054. [DOI] [PubMed] [Google Scholar]
- 109.Bogle G, Dunbar PR. Agent-based simulation of T-cell activation and proliferation within a lymph node. Immunol Cell Biol. 2010;88:172–179. doi: 10.1038/icb.2009.78. [DOI] [PubMed] [Google Scholar]
- 110.Mandl JN, Liou R, Klauschen F, Vrisekoop N, Monteiro JP, Yates AJ, Huang AY, et al. Quantification of lymph node transit times reveals differences in antigen surveillance strategies of naive CD4+ and CD8+ T cells. Proc Natl Acad Sci U S A. 2012;109:18036–18041. doi: 10.1073/pnas.1211717109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37:364–376. doi: 10.1016/j.immuni.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moreau HD, Lemaitre F, Terriac E, Azar G, Piel M, Lennon-Dumenil AM, Bousso P. Dynamic in situ cytometry uncovers T cell receptor signaling during immunological synapses and kinapses in vivo. Immunity. 2012;37:351–363. doi: 10.1016/j.immuni.2012.05.014. [DOI] [PubMed] [Google Scholar]


