CONSPECTUS
In living organisms, biological molecules often organize into multi-component complexes. Such assemblies consist of various proteins and carry out essential functions, ranging from cell division, transport, and energy transduction to catalysis, signaling, and viral infectivity. To understand the biological functions of these assemblies, in both healthy and disease states, researchers need to study their three-dimensional architecture and molecular dynamics. To date, the large size, the lack of inherent long-range order, and insolubility have made atomic-resolution studies of many protein assemblies challenging or impractical using traditional structural biology methods such as X-ray diffraction and solution NMR spectroscopy. In the past ten years, we have focused our work on the development and application of magic angle spinning solid-state NMR (MAS NMR) methods to characterize large protein assemblies at atomic-level resolution.
In this Account, we discuss the rapid progress in the field of MAS NMR spectroscopy, citing work from our laboratory and others on methodological developments that have facilitated the in-depth analysis of biologically important protein assemblies. We emphasize techniques that yield enhanced sensitivity and resolution, such as fast MAS (spinning frequencies of 40 kHz and above) and non-uniform sampling protocols for data acquisition and processing. We also discuss the experiments for gaining distance restraints and for recoupling anisotropic tensorial interactions under fast MAS conditions. We give an overview of sample preparation approaches when working with protein assemblies.
Following the overview of contemporary MAS NMR methods, we present case studies into the structure and dynamics of two classes of biological systems under investigation in our laboratory. We will first turn our attention to cytoskeletal microtubule motor proteins including mammalian dynactin and dynein light chain 8. We will then discuss protein assemblies from the HIV-1 retrovirus.
Keywords: protein assemblies, solid-state NMR, SSNMR, magic angle spinning, MAS, HIV-1, HIV-AIDS, CA, capsid protein, microtubule-associated proteins, dynactin, CAP-Gly
1. INTRODUCTION
The cell is a crowded environment. The most essential cellular functions are performed by assemblies of biological molecules (e.g. proteins, nucleic acids and small biomolecules) working in concert. Such assemblies consisting of two or more interacting partners often undergo changes in conformation and/or molecular motions upon binding to each other, and knowledge of their molecular architectures and mobility at atomic resolution is critical to our understanding of their biological functions in the healthy cell/organism and in the disease-associated states. While several structural biology techniques yield atomic-level structural information (X-ray diffraction and solution NMR spectroscopy), these methods are limited to systems that can be crystallized or are soluble. Solid-state NMR (SSNMR) spectroscopy does not have limitations with respect to molecular size, solubility, and long-range order, and is thus an excellent technique for studying large macromolecular assemblies.
A wide range of sample conditions amenable to SSNMR characterization and the uniquely high information content of SSNMR experiments make the method particularly attractive. 3D architectures, molecular motions on timescales ranging from picoseconds to seconds and longer, interactions between macromolecules and a wide range of their binding partners including small molecules, can be inferred from the measurements, all with unprecedented site specificity and atomic-level resolution. Until recently, SSNMR studies of large biomolecules have been impaired by limited sensitivity and resolution. The rapid advances in all aspects of the technique- from hardware (e.g., high magnetic fields of 17.6 T and above, magic angle spinning (MAS) probes delivering rotation frequencies of 40–110 kHz, as well as the newest generation of digital radio frequency (rf) consoles), to novel pulse sequences and data acquisition and processing protocols, to advanced sample preparation approaches, have resulted in a surge of exciting investigations by multiple researchers into large macromolecular assemblies.
2. RECENT METHODOLOGICAL ADVANCES FOR MAS NMR STUDIES OF PROTEIN ASSEMBLIES
Contemporary SSNMR spectroscopy is a powerful tool for studying molecular structure and dynamics of protein assemblies, at atomic resolution. In large biological systems, sensitivity and resolution are often a challenge, and many SSNMR laboratories including ours focused major efforts in development of sensitivity and resolution enhancement techniques. High magnetic fields (17.6 T and above) have proven necessary in studies of protein assemblies, to achieve the required sensitivity and resolution. Often, additional signal enhancement is needed, and it can be attained (without compromising the resolution) by non-uniform sampling (NUS),1–3 paramagnetic-relaxation-assisted condensed data collection (PACC),4,5 and a combination of these two methods (NUS-PACC).6
At high magnetic fields and in conjunction with MAS frequencies of 10–60 kHz now routinely available, multidimensional dipolar-based (through space) and scalar-based (through bond) correlation experiments remain the cornerstone of experimental protocols for resonance assignments and distance restraints in protein assemblies.7 Anisotropic tensorial interactions (such as chemical shift and dipolar) contain indispensable structural and dynamics information, and to re-introduce those under MAS, a variety of rf field pulse schemes have been developed recently.7 In this section, we review the current MAS NMR methods for studying protein assemblies that work under a wide range of spinning frequencies.
2.1 Through-Space Correlation Spectroscopy
Dipolar correlations are necessary for gaining distance restraints that form the basis for resonance assignments and 3D protein structure determination. In the past two decades, many approaches for dipolar correlation spectroscopy in solids have been developed that rely both on direct polarization transfer between heteronuclei (13C, 15N) and on proton-mediated or proton-driven polarization transfer (reviewed in7). Most of these methods were established for MAS frequencies of 10–30 kHz. In the recent years, another promising approach has emerged- the application of fast MAS frequencies (ωr = 40–110 kHz) to enhance spectral resolution and sensitivity (for 1H and, in some cases, heteronuclei detection).8–10 Under these conditions, the conventional spin diffusion experiments (such as DARR and PDSD) and many of the sequences for dipolar and chemical shift recoupling do not work. Therefore, we pursued the development of methods that overcome the limitations of the existing techniques. We introduced a family of rotor-synchronized R2nv symmetry sequences for spin diffusion (RDSD).8 The R11, R21, and R22 recoupling efficiency at ωr of 40 kHz is comparable with or better than that of DARR at ωr of 10–20 kHz. One drawback of this approach is the dependence of the polarization transfer efficiency on the isotropic chemical shift difference, yielding efficient recoupling for only subsets correlations in the spectra, which are different for each of the four R2nv sequences.8 Subsequently, we have demonstrated that combined supercycled R2nv–driven (CORD) sequences are the best suited for 13C-13C spin diffusion experiments under fast MAS as they yield efficient broadband polarization transfer and uniform distribution of cross peak intensities across the entire correlation spectrum11 (see Figure 1). CORD sequences are useful for both resonance assignments and acquisition of long-range distance restraints for protein structure determination.
Figure 1.
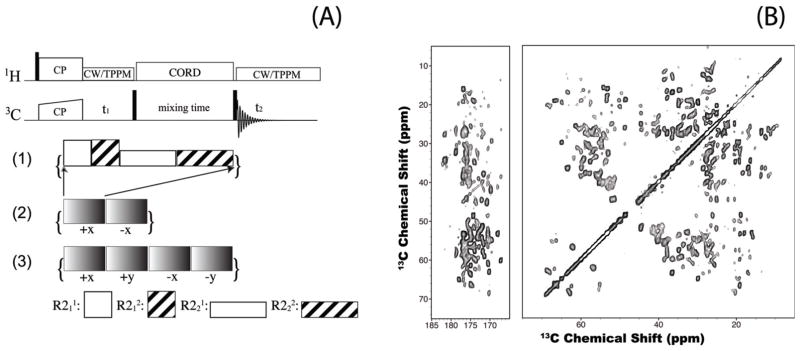
(A) 2D 13C-13C CORD sequences: (1) basic CORD; (2) CORDxix; (3) CORDxy4. These irradiation schemes are composites of rotor-synchronized R2nv- symmetry sequences, R211 (ωrf= ωr), R212 (ωrf= ωr), R221 (ωrf= ½ ωr), and R222 (ωrf= ½ ωr). Each R2nv element consists of two π pulses per n rotor periods. (B) 2D 13C-13C CORDxy4 spectra of U-13C,15N-LC8 (ωr=40 kHz). Polarization transfer is uniformly efficient in both aliphatic and carbonyl regions of the spectrum.11
In protein assemblies, knowledge of structure and dynamics of intermolecular interfaces formed by the binding partners is also essential, and our laboratory employs differential isotopic labeling that permits to selectively illuminate through-interface contacts. Using 1-73(U-13C,15N)/74-108(U-15N) E. coli thioredoxin reassembly, we introduced a family of 2D experiments to detect through-interface heteronuclear 13C/15N or 13C/1H dipolar couplings.12 These experiments are based on heteronuclear 13C/15N or 13C/1H dipolar dephasing of the signals belonging to the U-13C,15N-enriched molecule followed or preceded by either 15N-13C long-range magnetization transfer across the intermolecular interfaces or by 1H-15N or 15N-15N magnetization transfer within the 15N-enriched molecule.
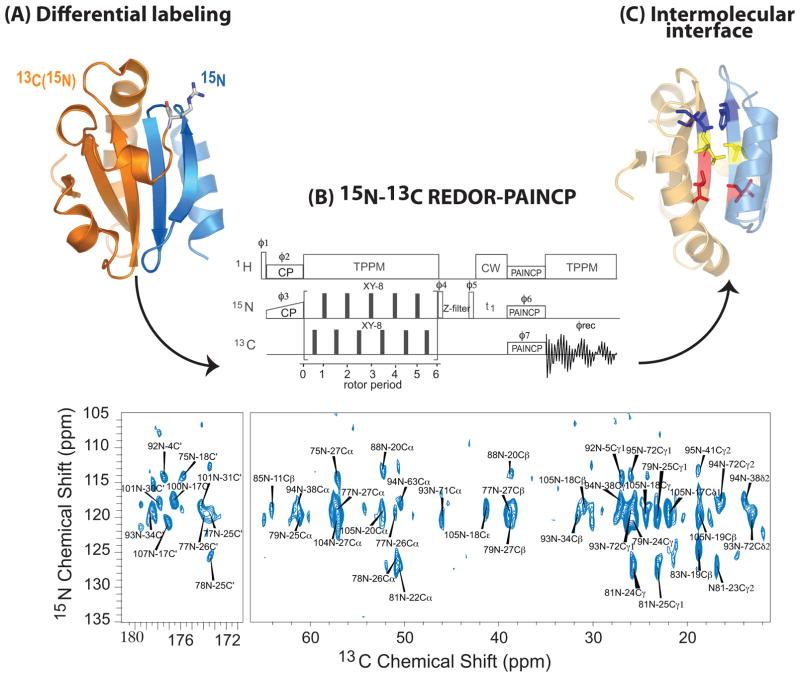
As illustrated in Figure 2, the 15N-13C REDOR-PAINCP experiment yields long-range 15N-13C correlations arising exclusively from the interfaces formed by the pair of differentially enriched complementary fragments of thioredoxin. In 15N-15N PDSD-REDOR and 1H-15N HETCOR-REDOR experiments, correlations are observed corresponding solely to the 74-108(U-15N) thioredoxin fragment, while those associated with the 1-73(U-13C,15N) fragment are eliminated by 13C/15N REDOR filter.12 The 1H-(13C)-15N REDOR-HETCOR experiment additionally highlights the residues situated at the interfaces between the two complementary fragments of reassembled thioredoxin.12 This family of experiments is applicable to a broad range of macromolecular assemblies including (and particularly beneficial to) large systems.
Figure 2.
An experimental procedure for studying intermolecular interfaces in protein assemblies as illustrated with 1–73-(U-13C,15N)/74–108-(U-15N) thioredoxin reassembly. (A) Differential labeling: the N-terminal fragment of thioredoxin (residues 1–73) is U-13C,15N labeled; the C-terminal fragment (residues 74-108) contains U-15N labels. These two fragments assemble spontaneously in solution to form a non-covalent complex with the 3D structure of intact thioredoxin. In (B), the 15N-13C REDOR-PAINCP sequence (top) and the corresponding 2D 15N-13C correlation spectrum (bottom) are shown. The cross-peaks are correlations between residues comprising the intermolecular interface, as illustrated in (C). Adapted with permission from 12.
2.2 Through-Bond Correlation Spectroscopy
Through-bond, scalar-coupling-driven correlation spectroscopy is another promising approach for structural investigations of protein assemblies.13 Scalar correlations offer information complementary to dipolar couplings. Scalar coupling-based experiments are less sensitive to the molecular motions than their dipolar-based counterparts. Several approaches exist for scalar-based spectroscopy in solids.14–17 For resonance assignments, constant-time J-based spectroscopy yields outstanding resolution, which is essential when working with large proteins and protein assemblies.18 The sensitivity of these experiments is on par with or better than that in the dipolar-based transfers. J-based experiments are compatible with in-phase/anti-phase (IPAP) selection in the direct dimension, yielding pure-phase spectra and further resolution enhancements.15 The performance of scalar-based polarization transfers is dramatically enhanced as MAS frequencies and decoupling field strengths are increased, and we anticipate that this approach will become widespread under fast-MAS conditions.
2.3 Recoupling of Anisotropic Spin Interactions
Magic angle spinning is essential for recording high-resolution SSNMR spectra. At the same time, MAS suppresses valuable information about molecular structure and dynamics, contained in the orientational dependence of the anisotropic spin interactions. Therefore, re-introduction of such interactions under MAS by suitable rf pulse sequences (termed recoupling) has been an active area of research since the 1990’s.7 An elegant approach for the design of efficient recoupling sequences was established by Levitt and co-workers, who discovered symmetry theorems defining the relationships between the various degrees of freedom (spin, space, rotation, and radiofrequency),19 which in turn serve as a guide in selecting the desired terms of the Hamiltonian while discarding or minimizing the unwanted interactions. This approach has been used by multiple groups including ours for recoupling of chemical shift and dipolar interactions.
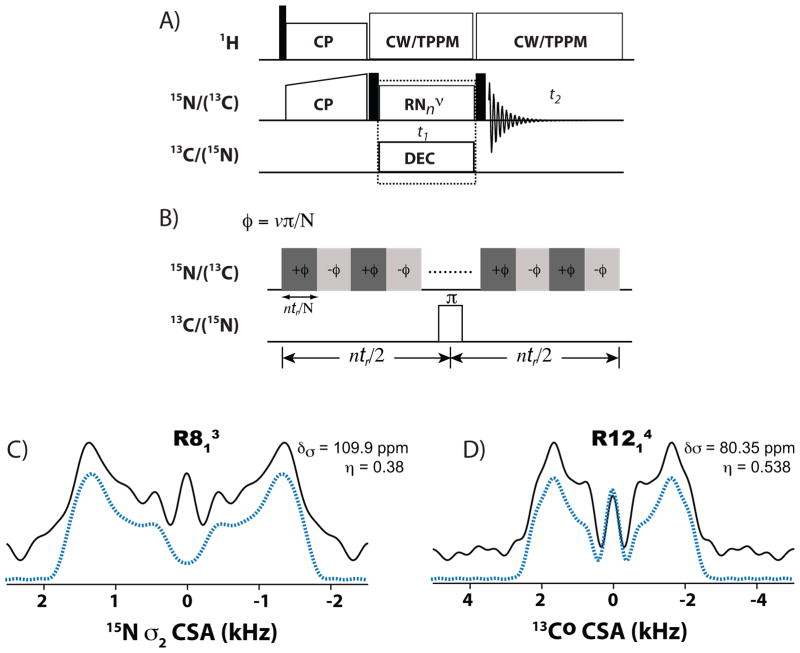
We explored the applications of RNnv-symmetry sequences for the recoupling of 13C/15N chemical shift20,21 and 1H-13C(15N) dipolar22,23 anisotropies. In proteins, the CSA and dipolar tensors yield insights into structure and dynamics unavailable from the isotropic parameters. For the CSA recoupling, we have demonstrated that with the appropriately designed γ-encoded RNnv symmetry sequences either the first-rank or the second-rank spatial components of 13C/15N CSA interaction can be efficiently reintroduced.20 These γ-encoded RNCSA symmetry schemes are suitable for CSA recoupling under a wide range of MAS frequencies, including 40 kHz and above, and perform well in both isotopically-enriched and natural-abundance systems20 (see Figure 3). RNnv-symmetry based CSA recoupling sequences are advantageous due to their tunability to experimental conditions (rf field and MAS frequencies), tolerance to experimental imperfections (rf mis-set), and ability to recouple large CSA’s (such as for carbonyl and aromatic carbons).
Figure 3.
(A) General pulse scheme for R-symmetry-based recoupling of CSA interactions. (B) Rotor-synchronized RNnv-symmetry rf pulses are applied on heteronuclei during t1 evolution time to reintroduce CSA interactions under MAS conditions. RNnv sequences consist of N π-pulses over n rotor periods; ωrf=(N/2n)*ωr. (C) Experimental (black solid line) and simulated (blue dotted line) 15N σ1-CSA R813 lineshapes of Y169 residue in U-[13C, 15N]-Tyr HIV-1 C-terminal domain (CTD) of CA protein. (D) Experimental (black solid line) and simulated (blue dotted line) 13C° σ2-CSA R1214 lineshapes of Y169 in U-[13C, 15N]-Tyr HIV-1 CTD of CA protein. Adapted with permission from 20.
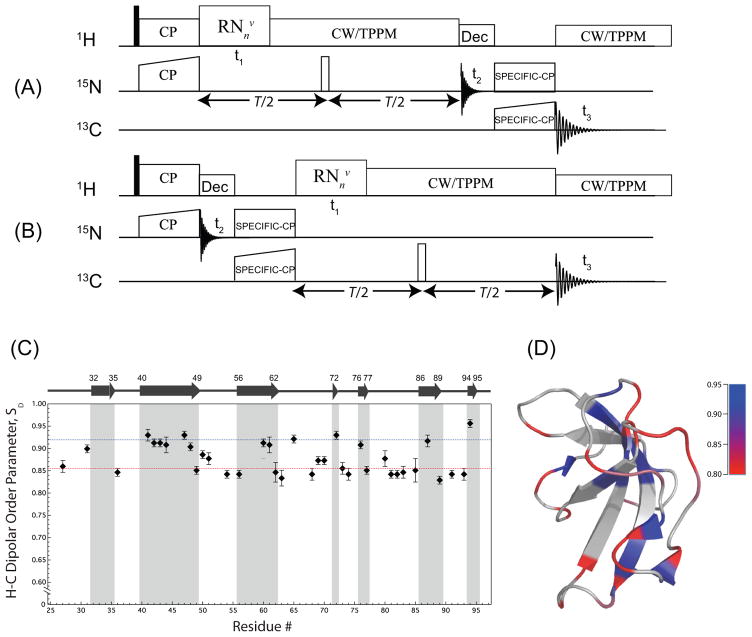
We have examined the performance of RNnv-symmetry sequences for the recoupling of heteronuclear (1H-13C and 1H-15N) dipolar interaction. As shown in Figure 4, these sequences display excellent selectivity for the recoupling of the first-order heteronuclear dipolar average Hamiltonian terms while the homonuclear dipolar Hamiltonian terms are suppressed.22 Most importantly, these experiments work very well under MAS frequencies of 40 kHz and above, and the high spectral resolution attained under these conditions makes them particularly well suited for studies of uniformly, extensively and sparsely enriched proteins and protein assemblies, as we have demonstrated for the CAP-Gly domain of mammalian dynactin22 (see Figure 4).
Figure 4.
(A and B) Three-dimensional DIPSHIFT sequences for 1H-15N and 1H-13C heteronuclear dipolar recoupling experiments under MAS frequencies of 40 kHz and above. (C) Experimental H-Cα dipolar order parameters SD (R1632-based DIPSHIFT) plotted as a function of the residue number for the sparsely-13C/U-15N enriched CAP-Gly. The blue and red dotted lines correspond to SD of 0.919 and 0.854, which are the average values for the residues comprising β-sheets and loops/turns, respectively. (D) The nanosecond-to-microsecond timescale backbone dynamics of CAP-Gly mapped onto its tertiary structure, as captured by SD. Blue: SD=0.95; red: SD=0.80; the values between these limiting order parameters were interpolated continuously. Reproduced with permission from22.
Finally, by correlating two anisotropic interactions one can obtain the relative orientation between the two corresponding tensors, and this in turn informs on the bond and dihedral angles.7 We have demonstrated that using γ-encoded R-symmetry sequences in the context of a 3D experiment, where the combined CSA and heteronuclear dipolar recoupling are recorded, permits determination of the relative orientations between the 1H–15N dipolar and 15N CSA tensors.21 These experiments yield the relative tensor orientations for multiple sites and work well in U-13C,15N-labeled proteins and assemblies.
2.4 Non-Uniform Sampling
We and others have explored the utility of non-uniform sampling for efficient data collection in multidimensional SSNMR spectroscopy.1,2 Rigorous analysis of the time-domain signals by Rovnyak established that inherent gains of up to 2.5-fold are possible in each indirect dimension with appropriate NUS schedules, provided exponentially decaying signals,3 and that the resolution is maximized by sampling out to evolution times of π*T2*. SSNMR can take particular advantage of these sensitivity gains without compromising the resolution due to relatively fast spin-spin relaxation permitting sampling of data points to the required evolution times. Indeed, as we have demonstrated recently, sensitivity gains of 1.5–2.0-fold in each indirect dimension can be readily attained in multidimensional NUS-NMR experiments, compared to the uniformly sampled datasets acquired within the same experiment time.1
To assess the inherent sensitivity gains of the frequency-domain signals in NUS experiments, we have introduced Maximum Entropy INterpolation (MINT) that relies on the Maximum Entropy algorithm in the limit where the reconstructed spectra are tightly constrained to the raw data. This protocol ensures highly linear transformation between the time- and the frequency-domain signals allowing for the direct comparison of spectral features with those in the datasets acquired by uniform sampling and processed by Fast Fourier Transform (FFT).1
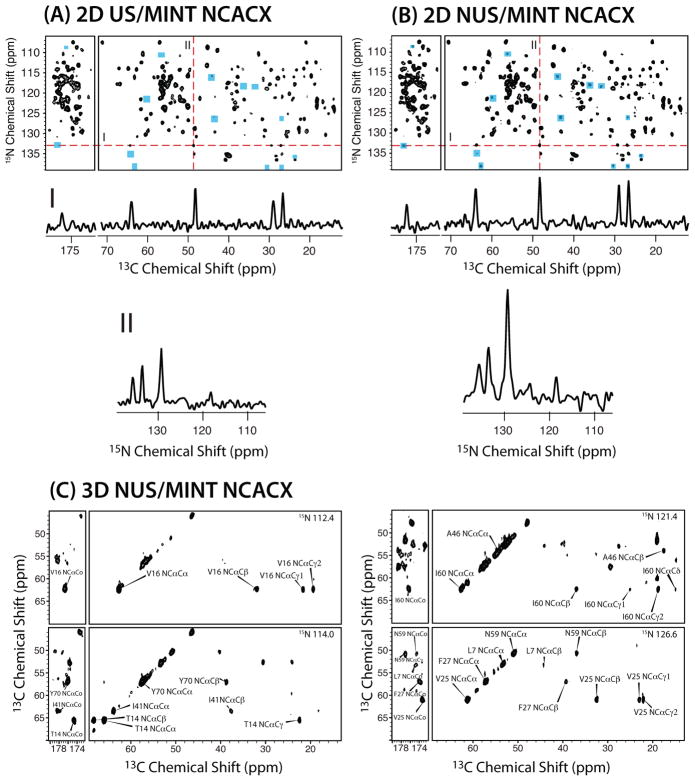
In Figure 5, we illustrate the application of NUS/MINT to 1–73-(U-13C,15N)/74–108-(U-15N) thioredoxin reassembly. The sensitivity enhancements from NUS are illustrated in the one-dimensional traces of the pair of 2D NCACX spectra (NUS and US) acquired with the same experiment time. A 3D NUS/MINT NCACX spectrum is also shown. The experiment time was 4 days, and the corresponding US/FFT spectrum would require 16 days of experiment time on this sample and would be out of reach for less sensitive samples. NUS/MINT appears to be a promising venue for SSNMR studies of large protein assemblies.
Figure 5.
2D NCACX spectra of 1-73-(U-13C,15N)/74-108-(U-15N) thioredoxin reassembly: (A) US/MINT, (B) NUS/MINT. I and II: lineshapes extracted from the direct and indirect dimensions. Significant increase in sensitivity is observed for the NUS data set. New peaks in NUS spectra are indicated with blue boxes. (C) Representative 2D planes from a 3D NCACX NUS/MINT experiment. The indirect-dimensions data points were sampled nonuniformly (25% points collected), and the experiment time is 4 days. The corresponding US experiment would be time prohibitive. Adapted with permission from1.
2.5 Sample Preparation and Characterization
Sample preparation is an important practical aspect in studies of large biomolecular assemblies. While SSNMR spectroscopy does not have inherent limitations with respect to molecular size or sample conditions (long-range order, crystallinity, solubility, etc.), preparation of high-quality samples is the most essential determinant of their successful characterization.
Generating pure isotopically labeled protein in amounts sufficient for SSNMR is often a challenge, but rapid progress has been made recently in recombinant protein expression (using E.coli or mammalian or insect cells) as well as cell-free protein production.24 While uniform labeling with 13C and 15N remains the most ubiquitous and information-rich protocol, other schemes have emerged as well, such as sparse and amino-acid-specific labeling. The various labeling protocols for biomolecular SSNMR have been reviewed recently.25
We have employed differential labeling to study interfaces formed by interacting proteins in protein assemblies.12,26,27 We have demonstrated that using a pair of protein molecules to prepare an assembly, one U-13C,15N and another- U-15N, one can gain detailed structural information on the U-13C,15N molecule and at the same time define the intermolecular interface by examining through-interface correlations in specially designed experiments (see section 2.1 and Figure 2). These experiments were established on a 108-residue E. coli thioredoxin reassembly formed by reconstitution of the complementary 1-73/74-108 fragments; these assemble spontaneously to form native thioredoxin structure. Using this procedure, we have generated a pair of thioredoxin reassemblies, 1-73(U-15N)/74-108(U-13C,15N) and 1-73(U-13C,15N)/74-108(U-15N).12,23,26,27
Once the suitable isotopic labeling protocols are identified, and the protein or protein assembly of interest is purified, functional assays need to be conducted and/or sample morphology needs to be examined. In our work with microtubule-associated proteins and HIV-1 CA assemblies, we use a number of biophysical techniques to ensure correct sample morphology and high conformational homogeneity.28–31 Among other methods, we employ transmission electron microscopy (TEM), cryo-scanning electron microscopy (cryoSEM), and confocal microscopy to confirm that desired sample morphology is formed. It is beneficial to perform the morphology characterization both before and after the MAS experiments, to ensure that the samples are intact under magic angle spinning.30
2.6 Emerging Techniques
With the ever-increasing size and complexity of biological systems under investigation, sensitivity and resolution remain the two cornerstones of biological SSNMR. In addition to the approaches outlined above which are based on high magnetic fields, fast MAS frequencies, and non-uniform sampling, there are several recent promising developments that lie at the intersection of two or more fields and deserve attention.
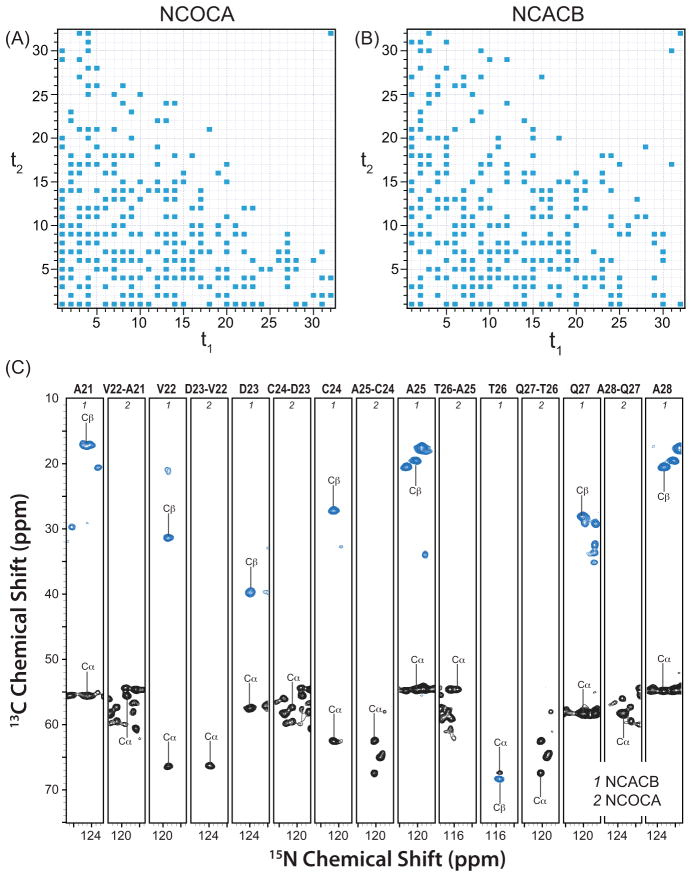
One example is the FROSTY protocol32 and its subsequent application to MAS NMR of sedimented proteins.33 These methods are based on transient protein sedimentation from solution to the MAS rotor walls upon spinning and permit characterization of large-size proteins and protein complexes without the need to precipitate them out of solution. Another direction is the use of paramagnetic probes34 for assessing long-range distance information in proteins (20–30 Å), through paramagnetic relaxation enhancements35 and paramagnetic pseudocontact shifts,36 and speeding up SSNMR data collection by taking advantage of dramatically decreased spin-lattice relaxation times in the presence of paramagnetic dopant under fast-MAS (PACC).4,5 We have recently demonstrated that combining PACC with NUS yields further dramatic time savings in multidimensional NMR experiments permitting data collection in a fraction of time it takes to acquire a conventional data set,6 as illustrated in Figure 6.
Figure 6.
(A and B) Sampling schedule for NUS-PACC 3D (A) NCOCA, and (B) NCACB experiments conducted on the U-13C,15N-LC8 protein doped with 5 mM Cu(II)-EDTA. (C) Backbone walk for the A21-A28 stretch of residues in LC8 using three-dimensional NUS-PACC NCACB and NCOCA experiments. The total experiment times are 27 and 9 hours for the NCACB and NCOCA experiments, respectively. Reproduced with permission from 6.
3. STRUCTURE AND DYNAMICS OF PROTEIN ASSEMBLIES BY SSNMR: CASE STUDIES
From the above discussion, the reader can appreciate the power and potential that contemporary SSNMR offers for atomic-resolution characterization of large biologically important protein assemblies. In this section, we provide an account of the recent studies where SSNMR offered unique insights into the structural biology of two classes of systems, cytoskeleton-associated and HIV-1 protein assemblies.
3.1 Cytoskeleton-Associated Protein Assemblies
Cytoskeletal structures and their associated proteins play fundamental roles in eukaryotic cells.37 However, the interactions between the cytoskeletal structures (microtubules, actin, and intermediate filaments) and their associated proteins still remain poorly understood at atomic level, due to the insolubility and lack of long-range order of cytoskeleton assemblies.
The focus of our laboratory is investigation of microtubule-associated motor proteins, dynein, dynactin, and kinesin, and their assemblies with microtubules, by MAS NMR spectroscopy. Microtubules (MTs) and microtubule-associated proteins (MAPs) perform essential physiological functions, including cell migration, mitosis, polarization and differentiation, and vesicle and organelle transport.38 The malfunctions of MAPs are associated with neurological disorders.39 Understanding of the disease pathology requires atomic-level knowledge of the structure and interactions between MAPs and MTs.
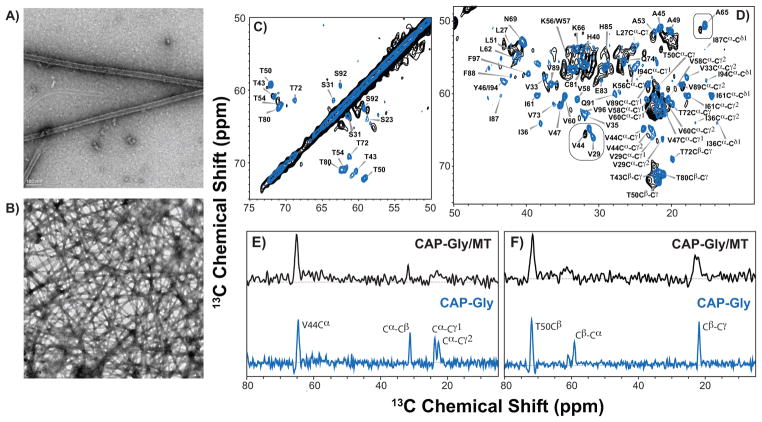
We investigated the CAP-Gly microtubule-binding domain of the mammalian dynactin in its healthy state and in pathogenic (Perry’s Syndrome-associated) mutations, by a combination of SSNMR, solution NMR, and biophysical techniques.22,29,40 We discovered that in the free state, CAP-Gly displays unusual dynamic properties associated with the high loop content, as illustrated in Figure 4.22 In the Perry’s Syndrome mutants, CAP-Gly exhibits reduced stability and a significant number of chemical shift perturbations while being well folded. Most strikingly and contrary to the original hypotheses in the literature, the binding affinity to the MTs is not decreased in the Perry’s mutants with respect to the wild type protein; at the same time, binding to an MT plus-end tracking protein EB1 is abrogated.40 In line with these observations is our discovery that when assembled on the microtubules, CAP-Gly exhibits multiple chemical shift perturbations, indicating that its conformation changes considerably upon binding.29 The results from these studies are summarized in Figure 7.
Figure 7.
(A, B) Negatively stained TEM images of CAP-Gly/microtubule complex at different magnification levels. (C, D) 21.1 T DARR spectra of CAP-Gly/MT complex (black) overlaid with the spectra of the free CAP-Gly (blue): expansions of the aliphatic regions illustrating the chemical shift differences in CAP-Gly upon binding to microtubules. (E, F) Representative 1D traces drawn through the aliphatic regions of DARR spectra and illustrating the cross peaks for V44 (E) and T50 (F). Blue: free CAP-Gly; black: CAP-Gly/MT complex. (C-F) are adapted with permission from 29.
We are studying another microtubule-associated protein, dynein light chain 8, LC8.41,42 LC8, an 89-residue protein, is an integral subunit of cytoplasmic dynein, a multisubunit microtubule-based retrograde motor. It is found in many other protein complexes and, through its interactions with diverse binding partners, LC8 has a variety of dynein-dependent and dynein-independent functions. Using a combination of NMR spectroscopy, X-ray diffraction, and other biophysical techniques, the binding interface between LC8 and PAK1 peptide was characterized and a novel sequence identified that expands the LC8 interaction repertoire.42 We have assigned the resonances of LC8 in the solid state,41 and demonstrated using LC8 that NUS-PACC approach yields dramatic time savings that will enable rapid data collection in LC8 assemblies with its binding partners and in a wide variety of protein assemblies,6 as shown in Figure 6.
3.2 HIV-1 and Other Viruses
Viruses and assemblies of viral proteins and nucleic acids are of enormous biological importance and interest. Two systems have been under intense scrutiny during the past several years: the major coat protein of intact Pf1 filamentous phage whose structure and dynamics were examined by MAS NMR43,44 and the HIV-1 viral capsid.30,31,45
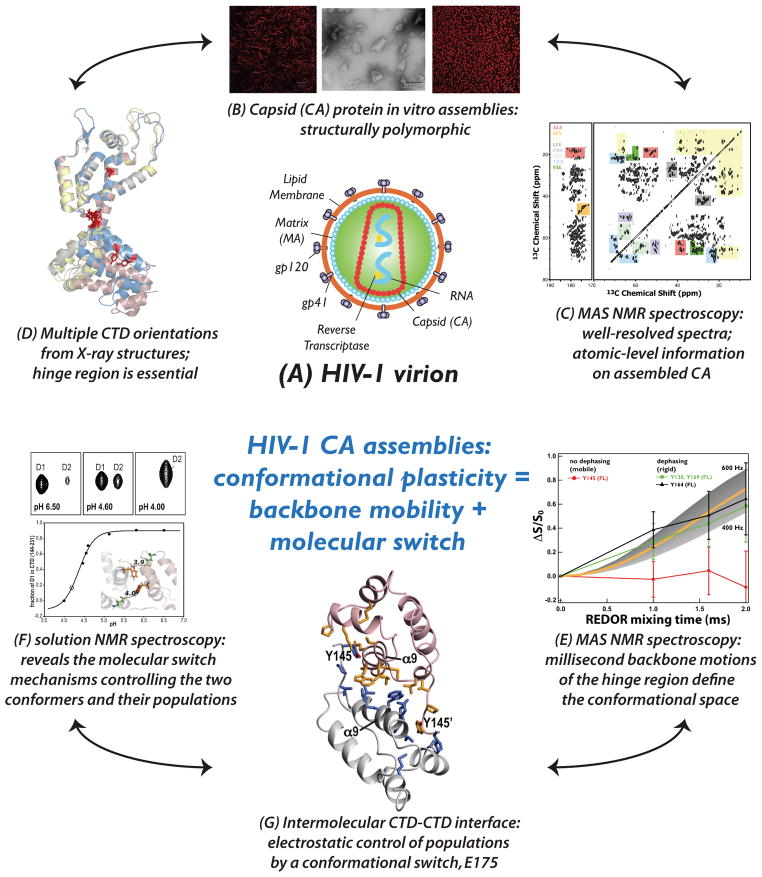
Our group investigates structure and dynamics of HIV-1 CA protein assemblies.8,20,30,31 HIV-1 capsid is a fullerene cone-like structure comprised of 1500 copies of a 231-residue CA protein, which encloses the viral RNA and a small complement of viral proteins.46 The early stages of the HIV-1 infection are accompanied by the capsid disassembly (uncoating), a tightly spatially and temporally controlled process, to release its contents into the cytoplasm of the host cell for subsequent virus replication. Formation of the conical structure requires pleiomorphic capsid with pentameric and hexameric oligomers co-existing in the same assembly (for recent review, see46). In vivo and in vitro, CA protein assembles into various morphologies, such as cones, spheres, and tubes. We examined the molecular mechanism of the polymorphism and structural plasticity responsible for the pleiomorphic capsid assemblies. Assignment of two-thirds of the residues in the CA assemblies of conical morphology was possible from the high-quality 21.1 T SSNMR spectra, as illustrated in Figure 8.30 Through the measurements of the 13C-15N and 1H-15N dipolar couplings in the U-13C,15N-Tyr-labeled CA conical assembly, we have identified the presence of slow, millisecond-timescale motions in the flexible hinge region linking the N- and the C-terminal domains of CA, which open up the conformational space and permit formation of multiple, varied conformers, as shown in Figure 8.31 Solution NMR spectroscopy revealed a molecular switch mechanism controlling the populations of the two conformers (one major, one minor) that co-exist in solution and are available for the capsid assembly into cones.31 Studies are under way to determine the full 3D structure and dynamics of the CA assemblies and assemblies of Gag maturation intermediates, and to elucidate their interactions with various HIV-1 inhibitors.
Figure 8.
Summary of structural and dynamics studies HIV-1 CA assemblies of conical morphologies. (A) Schematic representation of the HIV-1 virion. (B) Morphologies of the in vitro assemblies of HIV-1 CA protein: tubes (left, confocal), cones (center, negatively stained TEM), and spheres (right, confocal). (C) 21.1 T DARR spectra of conical assemblies of U-13C,15N CA. (D) Superposition of several X-ray structures of unassembled CA revealing the different orientations of the C-terminal domain (CTD). (E) 13C-15N REDOR dephasing curves reveal millisecond timescale motions in the hinge region (Y145) connecting the N- and C-terminal domains of CA, as probed through the tyrosine residues in U-13C,15N-Tyr-CA assembled into cones. These millisecond motions provide the conformational flexibility required for the assembly of CA into hexamers and pentamers and into the capsid cone surface. (F) pH dependence of the W184 Hε1N resonances of the two predominant conformers probed through HSQC spectra of the CA CTD dimer. (G) The intermolecular CTD-CTD interface structure. The solution NMR results reveal that the actual number of conformers accessible for CA assembly into varied morphologies is finite, and their relative populations in solution appear associated with an electrostatic interaction between the W184 and E175 side chains. Parts of the figure are reproduced with permission from30,31.
4. CONCLUSIONS AND FUTURE OUTLOOK
Solid-state MAS NMR spectroscopy has emerged as an essential technique for structure and dynamics characterization of protein assemblies. We hope that we conveyed to the reader our excitement and the tremendous potential of the field to provide atomic-resolution information inaccessible from other structural biology and biophysics methods currently available to the research community. We anticipate that synergistic collaborations between SSNMR spectroscopists and scientists in the fields of cell biology and virology as well as the use of hybrid methodologies will open new opportunities and new systems leading to exciting discoveries into the biological functions and mechanisms of complex cellular machineries.
Acknowledgments
The various aspects of this work were supported by the National Science Foundation (NSF-CAREER CHE-0237612) and the National Institutes of Health (R01GM085396, P50GM082251 P30GM103519, and P30RR031160). The 21.1 T spectra were acquired at the Environmental Molecular Sciences Laboratory. We thank our coworkers and collaborators who have contributed to the studies presented here.
Biographies
Si Yan received her B.S. degree from the University of Science and Technology of China in 2008. She is a Ph.D. candidate at the University of Delaware. Her research focuses on microtubule-associated protein assemblies.
Christopher L. Suiter received his B.A. degree from West Virginia University in 2009. He is a Ph.D. candidate at the University of Delaware. His research focuses on HIV-1 protein assemblies.
Guangjin Hou is an NMR Spectroscopist at the University of Delaware. He received his M.S. degree from Huazhong University of Science and Technology in 2004, and his Ph.D. degree from the Chinese Academy of Sciences in 2007. After the postdoctoral appointments at the Max-Planck Institute for Polymer Research (Mainz, Germany) and University of Delaware, he joined the staff at the University of Delaware in 2012. His research focuses on developments of SSNMR methods and on HIV-1 protein assemblies.
Huilan Zhang received her B.S. degree from Central China Normal University in 2001 and Ph.D. from Huazhong University of Science and Technology in 2007. After a postdoctoral appointment at Leibniz-Institute for Polymer Research (Dresden, Germany) she joined the University of Delaware as a volunteer scientist. Her research focuses on HIV-1 protein assemblies.
Tatyana Polenova is a Professor of Chemistry and Biochemistry at the University of Delaware. She received her undergraduate degree with honors from Moscow State University (Moscow, Russia) and Ph.D. at Columbia University. After a faculty position at the City University of New York, she joined the faculty at the University of Delaware in 2003. Professor Polenova’s work focuses on structural biology and biophysics of cytoskeleton and HIV-1 protein assemblies and on SSNMR spectroscopy.
Contributor Information
Si Yan, Email: syan@udel.edu.
Christopher L. Suiter, Email: csuiter@udel.edu.
Guangjin Hou, Email: hou@udel.edu.
Huilan Zhang, Email: zhang@udel.edu.
Tatyana Polenova, Email: tpolenov@udel.edu.
References
- 1.Paramasivam S, Suiter CL, Hou GJ, Sun SJ, Palmer M, Hoch JC, Rovnyak D, Polenova T. Enhanced Sensitivity by Nonuniform Sampling Enables Multidimensional MAS NMR Spectroscopy of Protein Assemblies. J Phys Chem B. 2012;116:7416–7427. doi: 10.1021/jp3032786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuki Y, Eddy MT, Griffin RG, Herzfeld J. Rapid Three-Dimensional MAS NMR Spectroscopy at Critical Sensitivity. Angew Chem Int Ed. 2010;49:9215–9218. doi: 10.1002/anie.201003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rovnyak D, Sarcone M, Jiang Z. Sensitivity Enhancement for Maximally Resolved Two-Dimensional NMR by Nonuniform Sampling. Magn Reson Chem. 2011;49:483–491. doi: 10.1002/mrc.2775. [DOI] [PubMed] [Google Scholar]
- 4.Wickramasinghe NP, Parthasarathy S, Jones CR, Bhardwaj C, Long F, Kotecha M, Mehboob S, Fung LWM, Past J, Samoson A, Ishii Y. Nanomole-Scale Protein Solid-State NMR by Breaking Intrinsic 1H T1 Boundaries. Nat Methods. 2009;6:215–218. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laage S, Sachleben JR, Steuernagel S, Pierattelli R, Pintacuda G, Emsley L. Fast Acquisition of Multi-Dimensional Spectra in Solid-State NMR Enabled by Ultra-Fast MAS. J Magn Reson. 2009;196:133–141. doi: 10.1016/j.jmr.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Sun S, Yan S, Guo C, Li M, Hoch JC, Williams JC, Polenova T. A Timesaving Strategy for MAS NMR Spectroscopy by Combining Non-Uniform Sampling and Paramagnetic Relaxation Assisted Condensed Data Collection. J Phys Chem B. 2012;116:13585–13596. doi: 10.1021/jp3005794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDermott A, Polenova T, editors. Solid-State NMR Studies of Biopolymers. John Wiley & Sons Ltd; 2010. p. 572. [Google Scholar]
- 8.Hou GJ, Yan S, Sun SJ, Han Y, Byeon IJL, Ahn J, Concel J, Samoson A, Gronenborn AM, Polenova T. Spin Diffusion Driven by R-Symmetry Sequences: Applications to Homonuclear Correlation Spectroscopy in MAS NMR of Biological and Organic Solids. J Am Chem Soc. 2011;133:3943–3953. doi: 10.1021/ja108650x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst M, Meier MA, Tuherm T, Samoson A, Meier BH. Low-Power High-Resolution Solid-State NMR of Peptides and Proteins. J Am Chem Soc. 2004;126:4764–4765. doi: 10.1021/ja0494510. [DOI] [PubMed] [Google Scholar]
- 10.Zhou DH, Shah G, Cormos M, Mullen C, Sandoz D, Rienstra CM. Proton-Detected Solid-State NMR Spectroscopy of Fully Protonated Proteins at 40 kHz Magic-Angle Spinning. J Am Chem Soc. 2007;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- 11.Hou G. Broadband Homonuclear Correlation Spectroscopy Driven by Combined R2nv Sequences under Fast Magic Angle Spinning for NMR Structural Analysis of Organic and Biological Solids. J Magn Reson. 2012 doi: 10.1016/j.jmr.2013.04.009. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Tasayco ML, Polenova T. Magic Angle Spinning NMR Experiments for Structural Studies of Differentially Enriched Protein Interfaces and Protein Assemblies. J Am Chem Soc. 2008;130:5798–5807. doi: 10.1021/ja711304e. [DOI] [PubMed] [Google Scholar]
- 13.Lesage A, Auger C, Caldarelli S, Emsley L. Determination of Through-Bond Carbon-Carbon Connectivities in Solid-State NMR Using the INADEQUATE Experiment. J Am Chem Soc. 1997;119:7867–7868. [Google Scholar]
- 14.Chen L, Kaiser JM, Polenova T, Yang J, Rienstra CM, Mueller LJ. Backbone Assignments in Solid-State Proteins Using J-Based 3D Heteronuclear Correlation Spectroscopy. J Am Chem Soc. 2007;129:10650–10651. doi: 10.1021/ja073498e. [DOI] [PubMed] [Google Scholar]
- 15.Chen LL, Kaiser JM, Lai JF, Polenova T, Yang J, Rienstra CM, Mueller LJ. J-based 2D Homonuclear and Heteronuclear Correlation in Solid-State Proteins. Magn Reson Chem. 2007;45:S84–S92. doi: 10.1002/mrc.2107. [DOI] [PubMed] [Google Scholar]
- 16.Tian Y, Chen LL, Niks D, Kaiser JM, Lai JF, Rienstra CM, Dunn MF, Mueller LJ. J-Based 3D Sidechain Correlation in Solid-State Proteins. Phys Chem Chem Phys. 2009;11:7078–7086. doi: 10.1039/b911570f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertini I, Emsley L, Felli IC, Laage S, Lesage A, Lewandowski JR, Marchetti A, Pierattelli R, Pintacuda G. High-Resolution and Sensitivity Through-Bond Correlations in Ultra-Fast Magic Angle Spinning (MAS) Solid-State NMR. Chem Sci. 2011;2:345–348. [Google Scholar]
- 18.Chen LL, Olsen RA, Elliott DW, Boettcher JM, Zhou DHH, Rienstra CM, Mueller LJ. Constant-Time Through-Bond 13C Correlation Spectroscopy for Assigning Protein Resonances with Solid-State NMR Spectroscopy. J Am Chem Soc. 2006;128:9992–9993. doi: 10.1021/ja062347t. [DOI] [PubMed] [Google Scholar]
- 19.Levitt MH. Symmetry-Based Pulse Sequences in Magic-Angle Spinning Solid-State NMR. Encyclopedia of Magnetic Resonance. 2007 [Google Scholar]
- 20.Hou G, Byeon IJ, Ahn J, Gronenborn AM, Polenova T. Recoupling of Chemical Shift Anisotropy by R-Symmetry Sequences in Magic Angle Spinning NMR Spectroscopy. J Chem Phys. 2012;137:134201. doi: 10.1063/1.4754149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou G, Paramasivam S, Byeon IJL, Gronenborn AM, Polenova T. Determination of Relative Tensor Orientations by Gamma-Encoded Chemical Shift Anisotropy/Heteronuclear Dipolar Coupling 3D NMR Spectroscopy in Biological Solids. Phys Chem Chem Phys. 2010;12:14873–14883. doi: 10.1039/c0cp00795a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou GJ, Byeon IJL, Ahn J, Gronenborn AM, Polenova T. 1H-13C/1H-15N Heteronuclear Dipolar Recoupling by R-Symmetry Sequences Under Fast Magic Angle Spinning for Dynamics Analysis of Biological and Organic Solids. J Am Chem Soc. 2011;133:18646–18655. doi: 10.1021/ja203771a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J, Tasayco ML, Polenova T. Dynamics of Reassembled Thioredoxin Studied by Magic Angle Spinning NMR: Snapshots from Different Time Scales. J Am Chem Soc. 2009;131:13690–13702. doi: 10.1021/ja9037802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atreya HS. Isotope Labeling in Biomolecular NMR. Vol. 992 Springer Berlin Heidelberg; New York, NY: 2012. [Google Scholar]
- 25.Renault M, Pawsey S, Bos MP, Koers EJ, Nand D, Tommassen-van Boxtel R, Rosay M, Tommassen J, Maas WE, Baldus M. Solid-State NMR Spectroscopy on Cellular Preparations Enhanced by Dynamic Nuclear Polarization. Angew Chem Int Ed. 2012;51:2998–3001. doi: 10.1002/anie.201105984. [DOI] [PubMed] [Google Scholar]
- 26.Marulanda D, Tasayco ML, McDermott A, Cataldi M, Arriaran V, Polenova T. Magic Angle Spinning Solid-State NMR Spectroscopy for Structural Studies of Protein Interfaces. Resonance Assignments of Sifferentially Enriched Escherichia coli Thioredoxin Reassembled by Fragment Complementation. J Am Chem Soc. 2004;126:16608–16620. doi: 10.1021/ja0464589. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Paramasivan S, Marulanda D, Cataidi M, Tasayco ML, Polenova T. Magic Angle Spinning NMR Spectroscopy of Thioredoxin Reassemblies. Magn Reson Chem. 2007;45:S73–S83. doi: 10.1002/mrc.2092. [DOI] [PubMed] [Google Scholar]
- 28.Renault M, Cukkemane A, Baldus M. Solid-State NMR Spectroscopy on Complex Biomolecules. Angew Chem Int Ed. 2010;49:8346–8357. doi: 10.1002/anie.201002823. [DOI] [PubMed] [Google Scholar]
- 29.Sun SJ, Siglin A, Williams JC, Polenova T. Solid-State and Solution NMR Studies of the CAP-Gly Domain of Mammalian Dynactin and Its Interaction with Microtubules. J Am Chem Soc. 2009;131:10113–10126. doi: 10.1021/ja902003u. [DOI] [PubMed] [Google Scholar]
- 30.Han Y, Ahn J, Concel J, Byeon IJL, Gronenborn AM, Yang J, Polenova T. Solid-State NMR Studies of HIV-1 Capsid Protein Assemblies. J Am Chem Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byeon IJL, Hou GJ, Han Y, Suiter CL, Ahn J, Jung J, Byeon CH, Gronenborn AM, Polenova T. Motions on the Millisecond Time Scale and Multiple Conformations of HIV-1 Capsid Protein: Implications for Structural Polymorphism of CA Assemblies. J Am Chem Soc. 2012;134:6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mainz A, Jehle S, van Rossum BJ, Oschkinat H, Reif B. Large Protein Complexes with Extreme Rotational Correlation Times Investigated in Solution by Magic-Angle-Spinning NMR Spectroscopy. J Am Chem Soc. 2009;131:15968–15969. doi: 10.1021/ja904733v. [DOI] [PubMed] [Google Scholar]
- 33.Bertini I, Luchinat C, Parigi G, Ravera E, Reif B, Turano P. Solid-State NMR of Proteins Sedimented by Ultracentrifugation. Proc Natl Acad Sci U S A. 2011;108:10396–10399. doi: 10.1073/pnas.1103854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaroniec CP. Solid-State Nuclear Magnetic Resonance Structural Studies of Proteins Using Paramagnetic Probes. Solid State Nucl Magn Reson. 2012;43–44:1–13. doi: 10.1016/j.ssnmr.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Nadaud PS, Helmus JJ, Hofer N, Jaroniec CP. Long-Range Structural Restraints in Spin-Labeled Proteins Probed by Solid-State Nuclear Magnetic Resonance Spectroscopy. J Am Chem Soc. 2007;129:7502–7503. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- 36.Knight MJ, Felli IC, Pierattelli R, Bertini I, Emsley L, Herrmann T, Pintacuda G. Rapid Measurement of Pseudocontact Shifts in Metalloproteins by Proton-Detected Solid-State NMR Spectroscopy. J Am Chem Soc. 2012;134:14730–14733. doi: 10.1021/ja306813j. [DOI] [PubMed] [Google Scholar]
- 37.Fulton A. The Cytoskeleton: Cellular Architecture and Choreography. Chapman and Hall; 1984. [Google Scholar]
- 38.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 39.Gerdes JM, Katsanis N. Small Molecule Intervention in Microtubule-Associated Human Disease. Hum Mol Genet. 2005;14:R291–R300. doi: 10.1093/hmg/ddi269. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed S, Sun S, Siglin AE, Polenova T, Williams JC. Disease-Associated Mutations in the p150(Glued) Subunit Destabilize the CAP-Gly Domain. Biochemistry. 2010;49:5083–5085. doi: 10.1021/bi100235z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun SJ, Butterworth AH, Paramasivam S, Yan S, Lightcap CM, Williams JC, Polenova T. Resonance Assignments and Secondary Structure Analysis of Dynein Light Chain 8 by Magic-Angle Spinning NMR Spectroscopy. Can J Chem. 2011;89:909–918. doi: 10.1139/v11-030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lightcap CM, Sun S, Lear JD, Rodeck U, Polenova T, Williams JC. Biochemical and Structural Characterization of the Pak1-LC8 Interaction. J Biol Chem. 2008;283:27314–27324. doi: 10.1074/jbc.M800758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldbourt A, Gross BJ, Day LA, McDermott AE. Filamentous Phage Studied by Magic-Angle Spinning NMR: Resonance Assignment and Secondary Structure of the Coat Protein in Pf1. J Am Chem Soc. 2007;129:2338–2344. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- 44.Lorieau JL, Day LA, McDermott AE. Conformational Dynamics of an Intact Virus: Order Parameters for the Coat Protein of Pf1 Bacteriophage. Proc Natl Acad Sci U S A. 2008;105:10366–10371. doi: 10.1073/pnas.0800405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen B, Tycko R. Structural and Dynamical Characterization of Tubular HIV-1 Capsid Protein Assemblies by Solid State Nuclear Magnetic Resonance and Electron Microscopy. Protein Sci. 2010;19:716–730. doi: 10.1002/pro.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganser-Pornillos BK, Yeager M, Sundquist WI. The Structural Biology of HIV Assembly. Curr Opin Struc Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]