Abstract
The growing amount of data in operational electronic health record (EHR) systems provides unprecedented opportunity for its re-use for many tasks, including comparative effectiveness research (CER). However, there are many caveats to the use of such data. EHR data from clinical settings may be inaccurate, incomplete, transformed in ways that undermine their meaning, unrecoverable for research, of unknown provenance, of insufficient granularity, and incompatible with research protocols. However, the quantity and real-world nature of these data provide impetus for their use, and we develop a list of caveats to inform would-be users of such data as well as provide an informatics roadmap that aims to insure this opportunity to augment CER can be best leveraged.
Introduction
The increasing adoption of electronic health records (EHRs) and their “meaningful use” offer great promise to improve the quality, safety, and cost of healthcare [1]. EHR adoption also has the potential to enhance our collective ability to advance biomedical and healthcare science and practice through the re-use of clinical data [2–4]. This investment sets the foundation for a “learning” healthcare system that facilitates clinical research, quality improvement, and other data-driven efforts to improve health [5, 6].
At the same time, there has also been substantial federal investment in comparative effectiveness research (CER) that aims to study populations and clinical outcomes of maximal pertinence to real-world clinical practice [7]. These efforts are facilitated by other investments in research infrastructure, such as the Clinical and Translational Research Award (CTSA) program of the US National Institutes of Health [8]. Many institutions funded by CTSA awards are developing research data warehouses of data derived from operational systems [9]. Additional federal investment has been provided by the Office of the National Coordinator for Health Information Technology (ONC) through the Strategic Health IT Advanced Research Projects (SHARP) Program, with one of the four major research areas focusing on re-use of clinical data [10].
A number of successes have already been achieved. Probably the most concentrated success has come from the Electronic Medical Records and Genomics (eMERGE) Network [11], which has demonstrated the ability to validate existing research results and generate new findings mainly in the area of genome-wide association studies (GWAS) that associate specific findings from the EHR (the “phenotype”) with the growing amount of genomic and related data (the “genotype”) [12]. Using these methods, researchers have been able to identify genomic variants associated with atrioventricular conduction abnormalities [13], red blood cell traits [14], while blood cell count abnormalities [15], and thyroid disorders [16].
Other researchers have also been able to use EHR data to replicate the results of randomized controlled trials (RCTs). One large-scale effort has come from the Health Maintenance Organization Research Network’s Virtual Data Warehouse (VDW) Project [17]. Using the VDW, for example, researchers were able to demonstrate a link between childhood obesity and hyperglycemia in pregnancy [18]. Another demonstration of this ability has come from the longitudinal records of general practitioners in the UK. Using this data, Tannen et al. were able to demonstrate the ability to replicate the findings of the Women’s Health Initiative [19] [20] and RCTs of other cardiovascular diseases [21, 22]. Likewise, Danaei et al. were able to combine subject-matter expertise, complete data, and statistical methods emulating clinical trials to replicate RCTs demonstrating the value of statin drugs in primary prevention of coronary heart disease. In addition, the Observational Medical Outcomes Partnership (OMOP) has been able to apply risk-identification methods to records from ten different large healthcare institutions in the US, although with a moderately high sensitivity vs. specificity tradeoff [23].
However, routine practice data are collected for clinical and billing uses, not research. The reuse of these data to advance clinical research can be challenging. The timing, quality and comprehensiveness of clinical data are often not consistent with research standards [3]. Research assessing information retrieval (search) systems to identify candidates for clinical studies from clinical records has shown many reasons not only why appropriate records are not retrieved but also why inappropriate ones are retrieved [24].
A number of authors have explored the challenges associated with use of EHR data for clinical research. A review of the literature of studies evaluating the data quality of EHRs for clinical research identified five dimensions of data quality assessed: completeness, correctness, concordance, plausibility, and currency [25]. The authors identified many studies with a wide variety of techniques to assess these dimensions and, similar to previous reviews, a wide divergence of results. Another analyses have highlighted the potential value but also the cautions of using EHR for research purposes [4, 26].
In this paper, we describe the caveats of using operational EHR data for CER and provide recommendations for moving forward. We discuss a number of specific caveats for use of EHR data for clinical research generally, with the goal of helping CER and other clinical researchers address the limitations of EHR data. We then provide an informatics framework that provides a context for better understanding of these caveats and providing a path forward toward improving data in clinical systems and their effective use.
Caveats
The intuitive appeal of re-using large volumes of existing operational clinical data for clinical research, quality measurement and improvement, and other purposes for improving health care is great. While the successes described above are noteworthy, a growing body of literature and our own analysis remind us that there are many informatics challenges and caveats associated with such approaches. Biases may be introduced at several steps along the process of the patient receiving care, including having it documented, billed for, and processed by insurers [27].
Under the following headings, we describe some specific caveats that have been identified either from research or our own observations about clinical data. In the last caveat, we further delineate issues of “data idiosyncrasies” for special attention. We view coded data as part of the EHR and, as such, include the clinical portions of administrative databases within our notion of using the EHR for clinical research, since many of the same caveats apply to that sort of data. We are viewing the entire collection of data on the patient as the EHR, and recognizing caveats for its use for CER.
Caveat #1: EHRs may contain inaccurate (or incorrect) data
Accuracy (correctness) of data relies on correct and careful documentation, which is not always a top priority for busy clinicians [28]. Errors in EHR records can be produced at any point. For example, data entry errors were demonstrated in a recent analysis in the English National Health Service (NHS), where yearly hospital statistics showed approximately 20,000 adults attending pediatric outpatient services, approximately 17,000 males admitted to obstetrical inpatient services, and about 8,000 males admitted to gynecology inpatient services [29]. Although the admission of males admitted to obstetrical units was explained by male newborns, the other data remain more problematic and difficult to explain [30]. In addition, a large sample of United States records showed 27% of patients who were emergently intubated in an emergency department were dispositioned either as discharged or admitted to non-critical care units, a highly unlikely outcome [31]. One systematic review identified 35 studies assessing data quality for reliability and validity of quality measures from EHR data [32]. These studies were found to have tremendous diversity in data elements, study settings, populations, clinical conditions, and EHR systems. The authors called for further research to focus on the quality of data from specific components in the EHR and to pay attention to the granularity, timeliness, and comparability of data. A more recent analysis assessed how the EHR systems of four known national leaders in EHR implementation would be able to use their data for CER studies on the treatment of hypertension. Researchers at each institution determined which data elements were necessary and whether and how they might be extracted from their EHR. The analysis found five categories of reasons why the data were problematic. These included data there were missing, erroneous, un-interpretable, inconsistent, and/or inaccessible in text notes [33].
Caveat #2: EHRs often do not tell a complete patient story
EHRs, whether those of a single institution or aggregated across institutions, do not always tell the whole story; i.e., patients may get care in different healthcare organizations or are otherwise lost to follow-up. Some estimate of the potential consequences of this incomplete picture can be gleaned from recent studies that have assessed data availability for health information exchange (HIE). One study of 3.7 million patients in Massachusetts found 31% visited two or more hospitals over five years (57% of all visits) and 1% visited five or more hospitals (10% of all visits) [34]. Another analysis of 2.8 million emergency department (ED) patients in Indiana found that 40% of patients had data at multiple institutions, with all 81 EDs sharing patients in common to create a completely linked network [35]. In addition, a study aiming to identify patients with Type 2 diabetes mellitus (DM) found that searching data from two medical centers in Minnesota had better predictive power than from a single center alone [36]. These same researchers also found that the ability to identify DM successively increased as the time frame of assessing records was increased from one through ten years of analysis [37].
Other studies have shown that data recorded in a patient's record at a single institution may be incomplete. Two systematic reviews have been performed that assess the quantity of data that were needed for clinical research but were unavailable from EHR sources. The first assessed studies on the scope and quality of data through 2001 [38]. A second review focused on the use of EHR data for outcomes research through 2007 [39] and identified 98 studies. In 55% of the studies found, additional non-EHR sources of data were also used, suggesting that an EHR data alone were not sufficient to answer the investigator's questions. About 40% of the studies supplemented EHR data with patient-reported data.
As examples of other studies on data completeness, at a New York academic medical center, 48.9% of patients with ICD-9-CM code for pancreatic cancers did not have corresponding disease documentation in pathology reports, with many data elements incompletely documented [40]. In another study comparing data from a specialty-specific EHR from community oncology clinics against data from the Surveillance Epidemiology and End Results (SEER) cancer registry and two claims databases (Medicare and commercial claims), significant proportions of data were missing from the EHR for race (40%) and tumor stage (63%) [41].
Evidence exists that there is significant variability in the quality of EHR data. One study, for example, found that relying solely on discrete EHR data, as opposed to data manually abstracted from the electronic record including text fields, led to persistent under-capture of clinical quality measures in a New York outpatient setting, with great variation in the amount of under-capture based on variation in clinical workflow and documentation practices [42].
Additional studies have evaluated EHR data for quality measurement purposes. For example, different ways of calculating adverse drug event rates from a single institution’s EHR were associated with significantly different results [43]. Likewise, quality metrics using EHR data required substantial validation to ensure accuracy [44]. An additional analysis compared a manually abstracted observational study of community-acquired pneumonia to a fully EHR-based study without manual data abstraction [45]. In the pure EHR study, mortality in the healthiest subjects appeared to exceed mortality in the sicker subjects due to several biases, including incomplete EHR data entry on patients who died quickly in the emergency department.
Determining the timing of a diagnosis from clinical data is also challenging. Not every diagnosis is recorded at every visit, and the absence of evidence is not always evidence of absence. This is just one example of a concern known by statisticians as censoring [46]. Left censoring is the statistical property where events prior to the start of an observation are missed, or their timing is not known with certainty. The result is that the first appearance of a diagnosis in an electronic record may not be the incident occurrence of the disease. Related to the notion of left censoring is right censoring, which refers to missing the occurrence of events that appear after the end of the interval under observation. While many clinical data warehouses may have millions of patients and cover many years of activity, patient turnover can be very high and individual patients in the warehouse may only have a few years’ worth of longitudinal data. The implication of patient turnover is that, for exposure-outcome pairs that take years to develop, groups of individual patients may not have a sufficiently long observation period to ascertain the degree of association. Even one of the most well-defined outcomes – death – may not be recorded in an EHR if the fatal episode occurred outside the institution.
Caveat #3: Many of the data have been transformed/coded for purposes other than research and clinical care
The most commonly known problematic transformation of data occurs when data are coded, often for billing purposes. While the underlying data may not be missing from the medical record, they are often inaccessible, either because they are paper-based or the electronic data are, for whatever reason, not available to researchers. This leads many researchers to rely solely on administrative (i.e., “claims”) data, of which a great deal of research has found to be problematic. Errors can be introduced in the clinical coding process for many reasons along the pathway of a patient’s hospitalization from admission to discharge [47]. These include inadequate or incomplete documentation, lack of access to information by clinicians and/or coders, illegible writing, suboptimal training and experience of the coder, upcoding for various reasons, inadequate review by the clinician, and errors made by anyone involved in the process.
In the early 1990s, one study reported that claims data lacked important diagnostic and prognostic information on patients admitted for cardiac catheterization; this information was contained in the medical record [48]. Later in the decade, another investigator assessed administrative data for quality measurement, finding that coding based on ICD-9-CM did not provide any clinical description beyond each code itself, including any prognostic indicators, or capture problems typical to outpatient settings, such as functional, socioeconomic, or psychosocial factors [49]. More recent studies have documented disparities between claims and EHR data in surgical conditions [50], children with pharyngitis [51], pediatric EDs [52], and patients with DM [53]. One recent study of patients with hospital-acquired, catheter-associated urinary tract infection, a complication denied payment from Medicare, found that claims data vastly under-reported the condition [54].
Researchers have also looked at coding completeness of patient assessments or their outcomes for clinical research purposes. In one Texas academic practice, billing data alone only identified 22.7% and 52.2% respectively of patients with endometrial and breast cancer, although this increased to 59.1% and 88.6% respectively with use of other data and a machine learning algorithm [55]. A similar study found somewhat better results for identifying patients with myocardial infarction (MI), ischemic stroke, and severe upper gastrointestinal (UGI) bleed events, with improvements also seen upon refinement of the selection algorithm [56]. Another study attempted to identify patients with metastatic cancer of the breast, lung, colon, and prostate using algorithmic models from claims data, finding that acceptable positive predictive value and specificity could be obtained but was only possible as a tradeoff with sensitivity [57]. An additional study found that coding tended to be better at identifying variance in utilization whereas patient reporting was better for disease burden and emotional symptoms [58].
Sometimes changes in coding practices inadvertently imply clinical differences where they may not exist. For example, in a national sample of coded data, it was noted that hospitalization and inpatient mortality rates for patients with a diagnosis of pneumonia decreased while hospitalizations with a diagnosis of sepsis or respiratory failure along with a secondary diagnosis of pneumonia increased and mortality declined. This analysis found, however, that when the three pneumonia diagnoses were combined, the decline in the hospitalization rate was much smaller and inpatient mortality was barely changed, suggesting the temporal trends were due more to differences in diagnostic coding than care factors [59]. Another study in community health clinics in Oregon found variations between claims and EHR data for a variety of services used in quality measures, such as cholesterol screenings, influenza vaccinations, diabetic nephropathy screenings, and tests for Hemoglobin A1c. While some measures were found with claims data only, a much larger proportion were found with EHR data only, especially in patients who were older, male, Spanish-speaking, above the federal poverty level, or who had discontinuous insurance [60].
Sometimes even improvements in coding can alter reporting, e.g., a transition to the more comprehensive ICD-10 coding system in the Centers for Disease Control WONDER database was determined to be the explanation for an increased rate of death from falls in the US between 1999–2007 [61]. Furthermore, changes in the coding system itself over time can impede comparability of data, as codes undergo “semantic drift” [62]. This drift has been shown to go unrecognized by researchers who were analyzing EHR data collected across multiple years [63].
Caveat #4: Data captured in clinical notes (text) may not be recoverable for CER
Many clinical data are “locked” in narrative text reports [64]. This includes information-rich sources of data from the initial history and physical report through radiology, pathology, and operative and procedural reports to discharge summaries. It may also include the increasingly used summaries of care [65]. One promising approach for recovering these data for research is natural language processing (NLP) [66]. This approach has been most successful when applied to the determination of specific data elements, such as the presence of a diagnosis or treatment. For example, the eMERGE studies described above used NLP to identify the presence of specific phenotype characteristics of the patient [12]. However, while the state of the art for performance of NLP has improved dramatically over the last couple decades, it is still far from perfect [67]. Furthermore, we really do not know how good is “good enough” for NLP in data re-use for clinical research, quality measurement and other purposes [68].
Caveat #5: EHRs may present multiple sources of data that affect data provenance
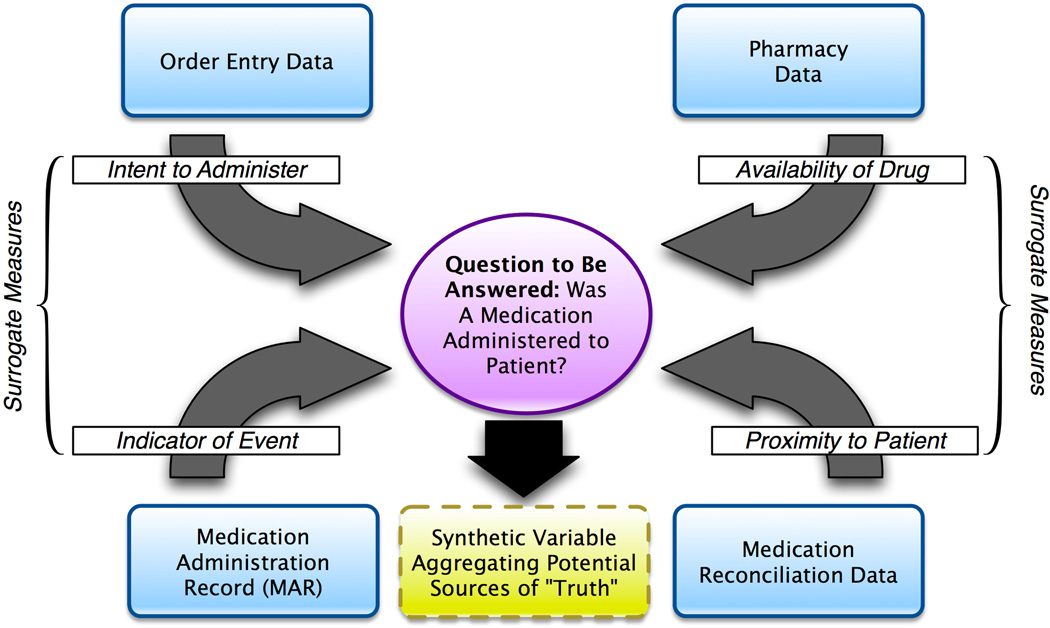
Another critical issue that contributes to the difficulty of re-using operational data for research purposes is data provenance, which is the understanding of the authoritative or definitive source(s) of a given measure or indicator of interest, given the existence of multiple potential sources for such a variable (i.e., “knowing where your data comes from”). Data provenance is concerned with establishing and systematically using a data management strategy that ensures that definitive findings are derived from multiple potential source data elements in a logical and reproducible manner [69]. For example, given a scenario where we would like to determine if a patient has received a given medication, there may be multiple possible data sources, namely: 1) order entry data, which may indicate an intent to give a medication to a patient; 2) pharmacy data, which may indicate the availability of the given medication for administration to a patient; 3) the medication administration record; and 4) medication reconciliation data, which aims to reconcile what a patient is supposed to receive and actually receives. Unfortunately, none of these elements indicate the ground truth of medication administration, but rather serve as surrogate measures for such ground truth (e.g., there is not a single variable that directly measures or otherwise indicates the physical administration of the medication in question) [70]. An example of this is illustrated in Figure 1.
Figure 1.
Example model of data provenance challenges associated with the identification of data related to medication administration.
Caveat #6: Data granularity in EHRs may not match the needs of CER
Data granularity is the level of detail or specificity used to encode or otherwise describe a measure or indicator of interest (i.e., “knowing what your data mean”). At a base level, this issue is important due to the wide variation of data granularity that results from the various reasons for data capture. For example, diagnostic codes assigned for billing purposes may, due to regulatory and documentation requirements, be generalized to a broad class of diagnosis (e.g., a patient with a set of complex cytogenetic and morphologic indicators of a pre-leukemic state would be described as having “myelodysplastic syndromes (MDS)” for billing purposes – an indicator of a broad set of such conditions, rather than a specific sub-set). In contrast, data collected for the purposes of clinical sub-specialties, intended to elucidate the etiology or contributing factors surrounding the initial diagnosis or treatment planning for a disease state may be highly granular and provides detailed descriptions of data types and sub-types that contribute to an overall class of diagnosis (e.g., extending the previous example, a research study might include specific variables corresponding to the cytogenetic and morphologic indicators underlying an overall diagnosis of MDS).
Caveat #7: There are differences between research protocols and clinical care
The research and theoretical concerns cited above show that there are many documented challenges to re-use of clinical data for research that are related to the data themselves. There are differences in methods and purposes between clinical care and research. Research protocols tend to be highly structured with strict definitions of inclusion and exclusion criteria, data collection that is thorough and rigorous, treatment assignment is often randomized, follow-up visits are scheduled at pre-specified intervals, and medication use is closely monitored. Clinical care, on the other hand, is geared toward patient needs. Treatments are assigned based on clinical impression of benefit balanced with patient wishes, data collection is limited to what the clinician believes is necessary, follow-up visits are scheduled at different intervals depending on clinical and non-clinical factors, and assessments of patient preferences are inconsistent at best. There are many common “idiosyncrasies” in clinical data that are enumerated and described in Table 1.
Table 1.
Data idiosyncrasies for use of operational electronic health record data in comparative effectiveness research.
| Type | Description | Example |
|---|---|---|
| Diagnostic uncertainty | - Diagnosis may be recorded when there is only a suspicion of disease | - Patient with suspected diabetes mellitus before diagnosis confirmed by laboratory testing |
| - Some overlapping clinical conditions are difficult to distinguish reliably | - Various forms of upper respiratory and related infections, e.g., sinusitis, pharyngitis, bronchitis, rhinitis, etc. | |
| - Patients may only partially fit diagnostic criteria | - Patients with non-diagnostic gastrointestinal symptoms may partially fit diagnostic criteria for one or multiple diseases | |
| - Patients in whom diagnostic testing is done but is negative are still more likely to have disease | - Patients undergoing echocardiography for shortness of breath and edema who are found to have normal left ventricular function are different from asymptomatic patients with normal left ventricular function | |
| Diagnostic timing | - Repeated diagnosis codes over time may represent a new event or a follow up to an prior event | - Two hospitalizations with a primary diagnosis of MI are likely two events, but a code for myocardial infarction in outpatient setting is more likely a follow up to an inpatient MI |
| -First diagnosis in a database is not necessarily an incident case of disease | - A new patient in the system with diabetes may have had diabetes for many years prior to presentation | |
| - Chronic diseases may vary in severity over time | - Patient with congestive heart failure with waxing and waning of symptoms | |
| Treatment choice and timing | - Many conditions do not require immediate drug or other treatment | - Hyperlipidemia or hypertension may have a trial of lifestyle changes before initiation of drug therapy |
| - Patient co-morbidities may effect timing and choice of treatment | - Patient with hypertension may have related diagnoses that were not recorded before initiation of treatment, but may be recorded later to indicate the compelling reason for a treatment choice, such as the use of ACE inhibitors in hypertensive patients with heart failure | |
| Treatment decisions | - Treatment decisions not randomized | - Physician choosing treatment based on personal views or biases regarding efficacy |
| - Some treatment decisions are remote to the patient-provider interaction | - Restrictions by patient insurance or institutional drug formulary | |
| - Some treatments not reliably recorded | - Medications available over the counter and not requiring a prescription may not be recorded, e.g., aspirin, proton pump inhibitors | |
| Treatment follow-up | - Some treatments confounded by clinical factors unrelated to condition being treated | - Patient with multiple co-morbidities may be seen more frequently and have conditions treated faster, e.g., hyperlipidemia in otherwise healthy person versus patient with diabetes and its complications |
| - Non-clinical factors impact availability of data | - Patient access to resources in order to follow treatment recommendations may be limited due to travel, payor systems, or other non-clinical factors | |
Informatics framework for addressing caveats
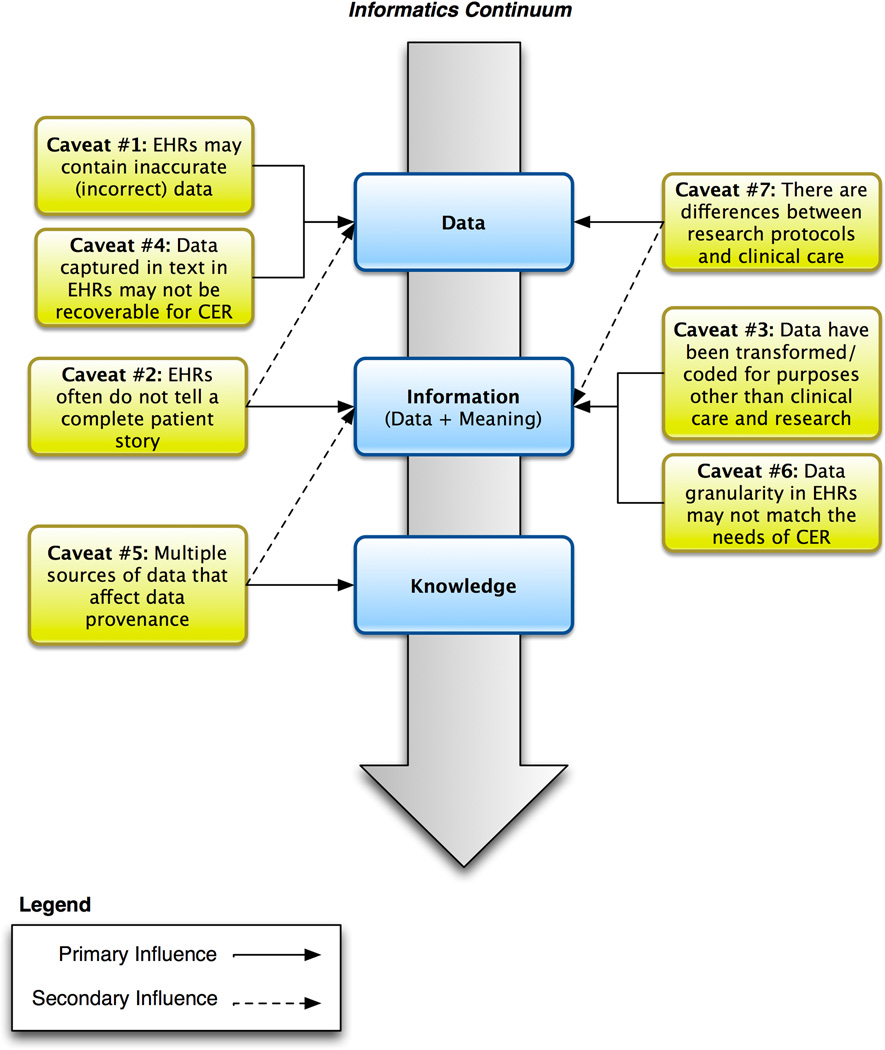
In order to address the full gamut of caveats related to the reuse of clinical data for CER and other types of research, it is helpful to have a framework to categorize the major issues at hand. One way to organize these findings is to think along a continuum historically employed in biomedical informatics that comprises data, information, and knowledge. Fundamentally, discovery requires the collection of observations (data), making sense of these observations (making the data meaningful, or transforming data into information) and deriving justified true belief (knowledge) based on this information. For example, we can collect data regarding smoking and lung cancer across institutions, map these data to a common standard to ensure that they are compatible (information) and look for correlations to determine whether smoking is associated with lung cancer (knowledge). This provides us a structure for understanding the challenges we face in reusing clinical data. Figure 2 shows the caveats and their influences on data, information, and knowledge.
Figure 2.
Relationship between central tenets of biomedical informatics and data-centric caveats pertaining to the design and conduct of comparative effectiveness research. Solid lines indicate the primary influencers of the caveats, while the secondary influencers are indicated with dashed lines.
Probably most critical to the success of using EHR data for CER and other types of research is the promotion of policies calling for, mandating, or providing incentives for the universal adoption of standards-based, interoperable healthcare data, captured seamlessly across the diverse sites where patients receive care. Other sources of data should be accessible as well, such as those in the public health system. Data use may be further enhanced by integrated personal health records and other sources of patient-collected data (e.g., sensors). All of these sources, when use is allowed by appropriate patient consent, will allow us to compare and learn what is truly effective for optimal health and treating disease.
The opportunities for using operational EHR and other clinical data for CER and other types of clinical and translational research are immense, as demonstrated by the studies cited in the introduction to this paper. If used carefully, with assessment for completeness and appropriate statistical transformation, these data can inform not only the health of individuals, but also the function of the larger health care system. However, attention must be paid to the caveats about such data that are raised in this paper. We also hope that the caveats described in this paper will lead the healthcare system to strive to improve the quality of data, through attention to standards, appropriate health information exchange, and usability of systems that will lead to improved data capture and its use for analysis. Development of a clinical research workforce trained to understand nuances of clinical data and its analytical techniques, and development of guidelines and practices for optimal data entry, structure and extraction should be part of a national research agenda to identify and implement optimal approaches in the use of EHR data for CER.
Acknowledgements
This paper was commissioned and funded by a grant from AcademyHealth. Additional resources for author time were provided by NIH National Center for Advancing Translational Science (NCATS) Grants 5UL1RR024140 (Hersh), 8UL1TR000090 (Embi and Payne), 1UL1RR024148 (Bernstam), 5UL1RR025005 (Lehmann), 2UL1RR024156 (Hripcsak), and 5UL1RR025008 (Saltz) as well as intramural research funding from the NIH Clinical Center and National Library of Medicine (Cimino). We acknowledge the support of the NIH NCATS Clinical and Translational Science Award Consortium and its CER Key Function Committee as well as the feedback and suggestions of Rosemarie Filart, MD of NCATS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
William R. Hersh, Oregon Heath & Science University, hersh@ohsu.edu.
Mark G. Weiner, AstraZeneca, mark.weiner@astrazeneca.com.
Peter J. Embi, The Ohio State University Wexner Medical Center, Peter.Embi@osumc.edu.
Judith R. Logan, Oregon Heath & Science University, loganju@ohsu.edu.
Philip R.O. Payne, The Ohio State University Wexner Medical Center, Philip.Payne@osumc.edu.
Elmer V. Bernstam, The University of Texas Health Science Center at Houston, Elmer.V.Bernstam@uth.tmc.edu.
Harold P. Lehmann, Johns Hopkins University, lehmann@jhmi.edu.
George Hripcsak, Columbia University, hripcsak@columbia.edu.
Timothy H. Hartzog, Medical University of South Carolina, hartzogt@musc.edu.
James J. Cimino, NIH Clinical Center, ciminoj@cc.nih.gov.
Joel H. Saltz, Emory University, jhsaltz@emory.edu.
References
- 1.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. New England Journal of Medicine. 2010;363:501–504. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 2.Safran C, Bloomrosen M, Hammond WE, Labkoff SE, Markel-Fox S, Tang P, et al. Toward a national framework for the secondary use of health data: an American Medical Informatics Association white paper. Journal of the American Medical Informatics Association. 2007;14:1–9. doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiner MG, Embi PJ. Toward reuse of clinical data for research and quality improvement: the end of the beginning. Annals of Internal Medicine. 2009;151:359–360. doi: 10.7326/0003-4819-151-5-200909010-00141. [DOI] [PubMed] [Google Scholar]
- 4.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association. 2012;20:117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Science Translational Medicine. 2010;2(57):57cm29. doi: 10.1126/scitranslmed.3001456. http://stm.sciencemag.org/content/2/57/57cm29.full. [DOI] [PubMed] [Google Scholar]
- 6.Smith M, Saunders R, Stuckhardt L, McGinnis JM. Best Care at Lower Cost: The Path to Continuously Learning Health Care in America. Washington, DC: National Academies Press; 2012. [PubMed] [Google Scholar]
- 7.Sox HC, Goodman SN. The methods of comparative effectiveness research. Annual Review of Public Health. 2012;33:425–445. doi: 10.1146/annurev-publhealth-031811-124610. [DOI] [PubMed] [Google Scholar]
- 8.Collins FS. Reengineering translational science: the time is right. Science Translational Medicine. 2011;3:90cm17. doi: 10.1126/scitranslmed.3002747. http://stm.sciencemag.org/content/3/90/90cm17.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacKenzie SL, Wyatt MC, Schuff R, Tenenbaum JD, Anderson N. Practices and perspectives on building integrated data repositories: results from a 2010 CTSA survey. Journal of the American Medical Informatics Association. 2012;19(e1):e119–e124. doi: 10.1136/amiajnl-2011-000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rea S, Pathak J, Savova G, Oniki TA, Westberg L, Beebe CE, Building a robust, et al. scalable and standards-driven infrastructure for secondary use of EHR data: The SHARPn project. Journal of Biomedical Informatics. 2012;45:763–771. doi: 10.1016/j.jbi.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Genomics. 2010;4(1):13. doi: 10.1186/1755-8794-4-13. http://www.biomedcentral.com/1755-8794/4/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny JC. In: Mining Electronic Health Records in the Genomics Era, in PLOS Computational Biology: Translational Bioinformatics. Kann M, Lewitter F, editors. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kullo LJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS ONE. 2010;5(9):e13011. doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosslin DR, McDavid A, Weston N, Nelson SC, Zheng X, Hart E, et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Human Genetics. 2012;131:639–652. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. American Journal of Human Genetics. 2011;89:529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hornbrook MC, Hart G, Ellis JL, Bachman DJ, Ansell G, Greene SM, et al. Building a virtual cancer research organization. Journal of the National Cancer Institute Monographs. 2005;35:12–25. doi: 10.1093/jncimonographs/lgi033. [DOI] [PubMed] [Google Scholar]
- 18.Hillier TA, Pedula KL, Schmidt MM, Mullen JA, Charles MA, Pettitt DJ. Childhood obesity and metabolic imprinting: the ongoing effects of maternal hyperglycemia. Diabetes Care. 2007;30:2287–2292. doi: 10.2337/dc06-2361. [DOI] [PubMed] [Google Scholar]
- 19.Tannen RL, Weiner MG, Xie D, Barnhart K. A simulation using data from a primary care practice database closely replicated the Women's Health Initiative trial. Journal of Clinical Epidemiology. 2007;60:686–695. doi: 10.1016/j.jclinepi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Weiner MG, Barnhart K, Xie D, Tannen RL. Hormone therapy and coronary heart disease in young women. Menopause. 2008;15:86–93. doi: 10.1097/gme.0b013e3180413e45. [DOI] [PubMed] [Google Scholar]
- 21.Tannen RL, Weiner MG, Xie D. Replicated studies of two randomized trials of angiotensin-converting enzyme inhibitors: further empiric validation of the 'prior event rate ratio' to adjust for unmeasured confounding by indication. Pharmacoepidemiology and Drug Safety. 2008;17:671–685. doi: 10.1002/pds.1584. [DOI] [PubMed] [Google Scholar]
- 22.Tannen RL, Weiner MG, Xie D. Use of primary care electronic medical record database in drug efficacy research on cardiovascular outcomes: comparison of database and randomised controlled trial findings. British Medical Journal. 2009;338:b81. doi: 10.1136/bmj.b81. http://www.bmj.com/cgi/content/full/338/jan27_1/b81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan PB, Madigan D, Stang SE, Overhage JM, Racoosin JA, Hartzema AG. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Statistics in Medicine. 2012;31:4401–4415. doi: 10.1002/sim.5620. [DOI] [PubMed] [Google Scholar]
- 24.Edinger T, Cohen AM, Bedrick S, Ambert K, Hersh W. AMIA 2012 Annual Symposium. Chicago, IL: 2012. Barriers to retrieving patient information from electronic health record data: failure analysis from the TREC Medical Records Track. [PMC free article] [PubMed] [Google Scholar]
- 25.Weiskopf NG, Weng C. Methods and dimensions of electronic health record data quality assessment: enabling reuse for clinical research. Journal of the American Medical Informatics Association. 2012;20:144–151. doi: 10.1136/amiajnl-2011-000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overhage JM, Overhage LM. Sensible use of observational clinical data. Statistical Methods in Medical Research. 2011;22:7–13. doi: 10.1177/0962280211403598. [DOI] [PubMed] [Google Scholar]
- 27.Schneeweiss S, Avorn J. A review of uses of health care utilization databases for epidemiologic research on therapeutics. Journal of Clinical Epidemiology. 2005;58:323–337. doi: 10.1016/j.jclinepi.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 28.de Lusignan S, vanWeel C. The use of routinely collected computer data for research in primary care: opportunities and challenges. Family Practice. 2005;23:253–263. doi: 10.1093/fampra/cmi106. [DOI] [PubMed] [Google Scholar]
- 29.Brennan L, Watson M, Klaber R, Charles T. The importance of knowing context of hospital episode statistics when reconfiguring the NHS. British Medical Journal. 2012;344:e2432. doi: 10.1136/bmj.e2432. http://www.bmj.com/content/344/bmj.e2432. [DOI] [PubMed] [Google Scholar]
- 30.Roebuck C. The importance of knowing context of hospital episode statistics when reconfiguring the NHS. British Medical Journal. 2012 doi: 10.1136/bmj.e2432. Rapid Response. http://www.bmj.com/content/344/bmj.e2432/rr/578977. [DOI] [PubMed]
- 31.Green SM. Congruence of disposition after emergency department intubation in the National Hospital Ambulatory Medical Care Survey. Annals of Emergency Medicine. 2013;61:423–426. doi: 10.1016/j.annemergmed.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Chan KS, Fowles JB, Weiner JP. Electronic health records and reliability and validity of quality measures: a review of the literature. Medical Care Research and Review. 2010;67:503–527. doi: 10.1177/1077558709359007. [DOI] [PubMed] [Google Scholar]
- 33.Savitz L, Bayley KB, Masica A, Shah N. Challenges in using electronic health record data for CER: experience of four learning organizations. Journal of the American Medical Informatics Association. 2012 doi: 10.1097/MLR.0b013e31829b1d48. In review. [DOI] [PubMed] [Google Scholar]
- 34.Bourgeois FC, Olson KL, Mandl KD. Patients treated at multiple acute health care facilities: quantifying information fragmentation. Archives of Internal Medicine. 2010;170:1989–1995. doi: 10.1001/archinternmed.2010.439. [DOI] [PubMed] [Google Scholar]
- 35.Finnell JT, Overhage JM, Grannis S. AMIA Annual Symposium Proceedings. Washington, DC: 2011. All health care is not local: an evaluation of the distribution of emergency department care delivered in Indiana; pp. 409–416. [PMC free article] [PubMed] [Google Scholar]
- 36.Wei WQ, Leibson CL, Ransom JE, Kho AN, Caraballo PJ, Chai HS, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. Journal of the American Medical Informatics Association. 2012;19:219–224. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei WQ, Leibson CL, Ransom JE, Kho AN, Chute CG. The absence of longitudinal data limits the accuracy of high-throughput clinical phenotyping for identifying type 2 diabetes mellitus subjects. International Journal of Medical Informatics. 2013;82:239–247. doi: 10.1016/j.ijmedinf.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiru K, Hassey A, Sullivan F. Systematic review of scope and quality of electronic patient record data in primary care. British Medical Journal. 2003;326:1070. doi: 10.1136/bmj.326.7398.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dean BB, Lam J, Natoli JL, Butler Q, Aguilar D, Nordyke RJ. Review: use of electronic medical records for health outcomes research: a literature review. Medical Care Research and Review. 2009;66:611–638. doi: 10.1177/1077558709332440. [DOI] [PubMed] [Google Scholar]
- 40.Botsis T, Hartvigsen G, Chen F, Weng C. AMIA Summits on Translational Science Proceedings. San Francisco, CA: 2010. Secondary use of EHR: data quality issues and informatics opportunities. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3041534/ [PMC free article] [PubMed] [Google Scholar]
- 41.Lau EC, Mowat FS, Kelsh MA, Legg JC, Engel-Nitz NM, Watson HN, et al. Use of electronic medical records (EMR) for oncology outcomes research: assessing the comparability of EMR information to patient registry and health claims data. Clinical Epidemiology. 2011;3:259–272. doi: 10.2147/CLEP.S23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parsons A, McCullough C, Wang J, Shih S. Validity of electronic health record-derived quality measurement for performance monitoring. Journal of the American Medical Informatics Association. 2012;19:604–609. doi: 10.1136/amiajnl-2011-000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kahn MG, Ranade D. The impact of electronic medical records data sources on an adverse drug event quality measure. Journal of the American Medical Informatics Association. 2010;17:185–191. doi: 10.1136/jamia.2009.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benin AL, Fenick A, Herrin J, Vitkauskas G, Chen J, Brandt C. How good are the data? Feasible approach to validation of metrics of quality derived from an outpatient electronic health record. American Journal of Medical Quality. 2011;26:441–451. doi: 10.1177/1062860611403136. [DOI] [PubMed] [Google Scholar]
- 45.Hripcsak G, Knirsch C, Zhou L, Wilcox A, Melton G. Bias associated with mining electronic health records. Journal of Biomedical Discovery and Collaboration. 2011;6:48–52. doi: 10.5210/disco.v6i0.3581. http://www.uic.edu/htbin/cgiwrap/bin/ojs/index.php/jbdc/article/view/3581/2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z, Sun J. Interval censoring. Statistical Methods in Medical Research. 2010;19:53–70. doi: 10.1177/0962280209105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: ICD code accuracy. Health Services Research. 2005;40:1620–1639. doi: 10.1111/j.1475-6773.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jollis JG, Ancukiewicz M, DeLong ER, Pryor DB, Muhlbaier MH, Mark DB. Discordance of databases designed for claims payment versus clinical information systems: implications for outcomes research. Annals of Internal Medicine. 1993;119:844–850. doi: 10.7326/0003-4819-119-8-199310150-00011. [DOI] [PubMed] [Google Scholar]
- 49.Iezzoni LI. Assessing quality using administrative data. Annals of Internal Medicine. 1997;127:666–674. doi: 10.7326/0003-4819-127-8_part_2-199710151-00048. [DOI] [PubMed] [Google Scholar]
- 50.Lawson EH, Louie R, Zingmond DS, Brook RH, Hall HL, Han L, et al. A comparison of clinical registry versus administrative claims data for reporting of 30-day surgical complications. Annals of Surgery. 2012;256:973–981. doi: 10.1097/SLA.0b013e31826b4c4f. [DOI] [PubMed] [Google Scholar]
- 51.Benin AL, Vitkauskas G, Thornquist E, Shapiro ED, Concato J, Aslan M, et al. Validity of using an electronic medical record for assessing quality of care in an outpatient setting. Medical Care. 2005;43:691–698. doi: 10.1097/01.mlr.0000167185.26058.8e. [DOI] [PubMed] [Google Scholar]
- 52.Gorelick MH, Knight S, Alessandrini EA, Stanley RM, Chamberlain JM, Kuppermann N, et al. Lack of agreement in pediatric emergency department discharge diagnoses from clinical and administrative data sources. Academic Emergency Medicine. 2007;14:646–652. doi: 10.1197/j.aem.2007.03.1357. [DOI] [PubMed] [Google Scholar]
- 53.Harris SB, Glazier RH, Tompkins JW, Wilton AS, Chevendra V, Stewart MA, et al. Investigating concordance in diabetes diagnosis between primary care charts (electronic medical records) and health administrative data: a retrospective cohort study. BMC Health Services Research. 2010;10:347. doi: 10.1186/1472-6963-10-347. http://www.biomedcentral.com/1472-6963/10/347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meddings JA, Reichert H, Rogers MA, Saint S, Stephansky J, McMahon LF. Effect of nonpayment for hospital-acquired, catheter-associated urinary tract infection: a statewide analysis. Annals of Internal Medicine. 2012;157:305–312. doi: 10.7326/0003-4819-157-5-201209040-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernstam EV, Herskovic JR, Reeder P, Meric-Bernstam F. Oncology research using electronic medical record data. Journal of Clinical Oncology. 2010;28(suppl):e16501. abstr http://www.asco.org/ascov2/Meetings/Abstracts?&vmview=abst_detail_view&confID=74&abstractID=42963. [Google Scholar]
- 56.Wahl PM, Rodgers K, Schneeweiss S, Gage BF, Butler J, Wilmer C, et al. Validation of claims-based diagnostic and procedure codes for cardiovascular and gastrointestinal serious adverse events in a commercially-insured population. Pharmacoepidemiology and Drug Safety. 2010;19:596–603. doi: 10.1002/pds.1924. [DOI] [PubMed] [Google Scholar]
- 57.Nordstrom BL, Whyte JL, Stolar M, Mercaldi C, Kallich JD. Identification of metastatic cancer in claims data. Pharmacoepidemiology and Drug Safety. 2012;21(Suppl 2):21–28. doi: 10.1002/pds.3247. [DOI] [PubMed] [Google Scholar]
- 58.Bayliss EA, Ellis JL, Shoup JA, Zeng C, McQuillan DB, Steiner JF. Association of patient-centered outcomes with patient-reported and ICD-9-based morbidity measures. Annals of Family Medicine. 2012;10:126–133. doi: 10.1370/afm.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindenauer PK, Lagu T, Shieh MS, Pekow PS, Rothberg MB. Association of diagnostic coding with trends in hospitalizations and mortality of patients with pneumonia, 2003–2009. Journal of the American Medical Association. 2012;307:1405–1413. doi: 10.1001/jama.2012.384. [DOI] [PubMed] [Google Scholar]
- 60.Devoe JE, Gold R, McIntire P, Puro J, Chauvie S, Gallia CA. Electronic health records vs Medicaid claims: completeness of diabetes preventive care data in community health centers. Annals of Family Medicine. 2011;9:351–358. doi: 10.1370/afm.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu G, Baker SP. An explanation for the recent increase in the fall death rate among older Americans: a subgroup analysis. Public Health Reports. 2012;127:275–281. doi: 10.1177/003335491212700307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods of Information in Medicine. 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- 63.Yu AC, Cimino JJ. A comparison of two methods for retrieving ICD-9-CM data: the effect of using an ontology-based method for handling terminology changes. Journal of Biomedical Informatics. 2011;44:289–298. doi: 10.1016/j.jbi.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hripcsak G, Friedman C, Anderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Annals of Internal Medicine. 1995;122:681–688. doi: 10.7326/0003-4819-122-9-199505010-00007. [DOI] [PubMed] [Google Scholar]
- 65.D'Amore JD, Sittig DF, Ness RB. How the continuity of care document can advance medical research and public health. American Journal of Public Health. 2012;102:e1–e4. doi: 10.2105/AJPH.2011.300640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. Journal of the American Medical Informatics Association. 2011;18:544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanfill MH, Williams M, Fenton SH, Jenders RA, Hersh WR. A systematic literature review of automated clinical coding and classification systems. Journal of the American Medical Informatics Association. 2010;17:646–651. doi: 10.1136/jamia.2009.001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hersh W. Evaluation of biomedical text mining systems: lessons learned from information retrieval. Briefings in Bioinformatics. 2005;6:344–356. doi: 10.1093/bib/6.4.344. [DOI] [PubMed] [Google Scholar]
- 69.Seiler KP, Bodycombe NE, Hawkins T, Shell R, Lemon A, deSouza A, et al. Master data management: getting your house in order. Combinatorial Chemistry & High Throughput Screening. 2011;14:749–756. doi: 10.2174/138620711796957189. [DOI] [PubMed] [Google Scholar]
- 70.de Lusignan S, Liaw ST, Krause P, Curcin V, Vicente MT, Michalakidis G, et al. Key Concepts to Assess the Readiness of Data for International Research: Data Quality, Lineage and Provenance, Extraction and Processing Errors, Traceability, and Curation. In: Haux R, Kulikowski CA, Geissbuhler A, editors. IMIA Yearbook of Medical Informatics 2011. Stuttgart, Germany: Schattauer; 2011. pp. 112–120. [PubMed] [Google Scholar]