Abstract
Background
Cell‐free DNA (cfDNA) circulating in blood is currently used for noninvasive diagnostic and prognostic tests. Minimizing background DNA is vital for detection of low abundance cfDNA. We investigated whether a new blood collection device could reduce background levels of genomic DNA (gDNA) in plasma compared to K 3 EDTA tubes, when subjected to conditions that may occur during sample storage and shipping.
Methods
Blood samples were drawn from healthy donors into K 3 EDTA and Cell‐Free DNA™ BCT (BCT). To simulate shipping, samples were shaken or left unshaken. In a shipping study, samples were shipped or not shipped. To assess temperature variations, samples were incubated at 6°C, 22°C, and 37°C. In all cases, plasma was harvested by centrifugation and total plasma DNA (pDNA) assayed by quantitative real‐time polymerase chain reaction (qPCR).
Results
Shaking and shipping blood in K 3 EDTA tubes showed significant increases in pDNA, whereas no change was seen in BCTs. Blood in K 3 EDTA tubes incubated at 6°C, 22°C, and 37°C showed increases in pDNA while pDNA from BCTs remained stable.
Conclusions
BCTs prevent increases in gDNA levels that can occur during sample storage and shipping. This new device permits low abundance DNA target detection and allows accurate cfDNA concentrations.
Keywords: hematology, investigative techniques, clinical laboratory techniques, blood preservation, real‐time polymerase chain reaction
INTRODUCTION
Cell‐free DNA (cfDNA) naturally occurs in blood and has been largely attributed to apoptotic and necrotic processes 1, 2. While the presence of cfDNA in blood was discovered in 1948 3, its implications in clinical medicine were not realized for more than two decades. Later, it was demonstrated that cfDNA was present in the serum of patients with systemic lupus erythematosus 4 and that cfDNA levels were elevated in the serum of cancer patients 5. These findings sparked an interest in the potential use of cfDNA in disease diagnosis and prognosis. Investigations carried out with rheumatoid arthritis 6, colorectal, breast, pancreatic, head, and neck cancer patients have all shown a marked rise in cfDNA concentrations 7, 8, 9, 10, 11. The cfDNA extracted from plasma or serum of cancer patients has shown characteristics typical of tumor DNA and may serve as noninvasive biomarkers for cancer detection and management 12, 13.
There is also growing interest in the potential use of cell‐free fetal nucleic acids in noninvasive prenatal diagnosis. Lo et al. 14 were the first to show that pregnant donor blood samples have elevated maternal cfDNA concentrations and also demonstrated the presence of fetal cfDNA in maternal plasma. Clinical applications involving fetal cfDNA analysis include sex determination, single‐gene disorders, pregnancy‐related disorders, and aneuploidy detection 15.
Due to the low abundance of the cfDNA biomarkers of cancer or fetal origin, it is recommended that genomic DNA (gDNA) background levels be minimized to provide accurate measurements cfDNA levels 16. Therefore, it is necessary to address several preanalytical issues that arise during the time between blood drawn and DNA isolation. These include delays in blood processing, blood storage temperature, agitation of the sample during transport, and shipment of blood. Such conditions may alter plasma DNA (pDNA) levels by causing gDNA release from lysed nucleated blood cells and obfuscate true cfDNA. As a result, it is important to consider the type of blood collection device and postphlebotomy conditions while working with cfDNA samples 16.
In this study, we compared two blood collection devices, K3EDTA and Cell‐Free DNA BCTTM (BCT), under conditions that can occur during sample storage, shipping, and processing. We assessed the ability of these devices to prevent gDNA release.
MATERIALS AND METHODS
Recruitment of Blood Donors
All blood donors were anonymous volunteers who were recruited from Streck, Inc. in Omaha, NE. Donors were both male and female and presumed to be healthy. All donor samples were drawn by venipuncture.
Blood Collection
For each experiment, donor samples were drawn into two different blood collection tubes. Control samples (10 ml) were drawn into K3EDTA tubes (BD Vacutainer®, Becton Dickinson, Franklin Lakes, NJ) and compared to samples (10 ml) drawn into Cell‐Free DNA™ BCT (Part Number 218962, Streck, Inc., Omaha, NE). Cell‐Free DNA BCTTM is a new collection device that contains a novel chemical cocktail. This cocktail stabilizes nucleated blood cells and contains K3EDTA as an anticoagulant. Blood was mixed immediately after the draw by inverting tubes ten times.
Sample Processing
After phlebotomy, blood samples were stored at room temperature (22°C), except where otherwise noted, and plasma was separated at noted time points. For the separation of plasma, blood samples were centrifuged at 22°C. An initial low speed centrifugation at 300 × g for 20 min was used to prevent damage to nucleated blood cells. The plasma layer was carefully removed without disturbing the buffy coat, transferred to a new vial, and then centrifuged at 22°C at 5,000 × g for 10 min to remove residual cells.
cfDNA Isolation From Plasma
The QIAamp® Circulating Nucleic Acid Kit (Qiagen, Santa Clarita, CA) was used for the extraction of pDNA. The manufacturer's recommended protocol was modified slightly by increasing the duration of the Proteinase K treatment from 30 min to 1 hr at 60°C. Samples were eluted in 50 μl sterile nuclease‐free water and stored at −80°C until analysis by Quantitative Real‐Time Polymerase Chain Reaction (qPCR).
Quantitative Real‐Time PCR
β‐Actin is a relatively abundant gene, we chose to measure endogenous plasma cfDNA and increases in pDNA due to gDNA release. The Y‐chromosomal sex‐determining region (SRY) gene served as a marker for nonnative DNA, that is, a low abundance target, since it was spiked into nonpregnant female plasma. Human β‐actin primers and probe and primers for the human SRY were prepared as previously described 17, 18. The following probe was designed for the quantification of SRY sequence: 6FAM‐ATG GCT CTA GAG AAT CCC AGA ATG CGA AAC TCA GAG A‐TAMRA. For each assay, tenfold dilutions (300,000–30 copies) of plasmid DNA constructs were used to establish qPCR standard curves. Constructs were prepared by cloning a single copy of β‐actin or SRY DNA sequence, which produced amplicons of 136 bp or 148 bp, respectively. β‐Actin genomic equivalents were calculated from qPCR copy number and then converted to nanograms of cfDNA per milliliter plasma (ng/ml) as previously described 19. The final SRY DNA concentration was expressed as DNA copies recovered per milliliter (copies/ml) plasma. All primers (500 nM final concentration) were purchased from Integrated DNA Technologies (Coralville, IA). The qPCR hydrolysis probes (50 nM final concentration) and TaqMan® Universal PCR Master Mix II were purchased from Applied Biosystems (Foster City, CA). All qPCR reaction volumes were 25 μl.
Effect of Chemical Cocktail Present in BCTs on pDNA Amplification
Blood was drawn from five donors into K3EDTA tubes (10 ml of blood from each donor), centrifuged, and plasma separated. Then plasma from all donors was pooled and mixed well. This pooled plasma sample (20 ml) was divided into two 10 ml aliquots and one aliquot was treated with BCT chemical cocktail to obtain a final concentration equal to the concentration present in a standard 10 ml blood drawn. The untreated plasma aliquot was used as control. Both treated and untreated plasma samples were incubated at 22°C and five 0.5 ml portions of plasma were removed from both aliquots at four different time points. Two short duration time points (3 and 6 hr) and two long duration time points (7 and 14 days) were taken. Plasma collected at each time point was stored at −80°C until pDNA was extracted and analyzed by qPCR using the β‐actin assay.
Effect of Elevated Background gDNA in Plasma on Detection of Rare cfDNA Sequences
For this study, blood was collected from five nonpregnant female donors. Blood was drawn from each donor into one 10 ml K3EDTA tube and one 10 ml BCT. Blood in each tube type was divided into three aliquots and stored at 22°C. Blood aliquots were removed on days 0, 7, and 14, plasma separated and stored at −80°C until extraction and analysis. All plasma samples (1 ml) were thawed and spiked with 5 μl of a stock reagent containing a plasmid DNA construct with a sequence fragment from the Y‐chromosomal SRY region (600,000 copies/ml). This resulted in a final SRY plasmid concentration of 3000 copies/ml plasma and served to simulate a low abundance cfDNA target. Following extraction, qPCR was used to measure pDNA concentration and Y‐chromosomal sequence copy number, using β‐actin and SRY gene targets, respectively.
Effect of Storage Temperature on pDNA Concentration in Blood Samples
This study was conducted using blood obtained from six donors. Blood was drawn from each donor into three 10 ml K3EDTA tubes and three 10 ml BCTs. One K3EDTA tube and one BCT from each donor were taken and each tube was divided into three aliquots and stored at 6°C. The second K3EDTA tube and BCT from each donor was taken and each tube divided into three aliquots and stored at 22°C. The final set of tubes from each donor were divided into three aliquots and stored at 37°C. Blood samples were taken on days 0, 7, and 14 and plasma separated by centrifugation and stored at −80°C until pDNA was extracted and analyzed by qPCR using the β‐actin assay.
Effect of Shaking and Shipping on pDNA Concentration in Blood Samples
To simulate shipping, blood from eight donors was drawn into K3EDTA tubes and BCTs. Tubes were secured onto the platform of an orbital shaker and shaken at 150 rpm at 22°C. At time 0, 3, 6, and 24 hr, plasma from one K3EDTA and one BCT was harvested and frozen at −80°C until extraction and analysis. A control experiment was done where the blood was not shaken while all other experimental parameters remained the same. In a separate shipping study, blood was drawn from ten donors. Blood was drawn from each donor into three 10 ml K3EDTA tubes and three 10 ml BCTs. One K3EDTA tube and one BCT from each donor were processed within 2 hr of blood drawn. Another K3EDTA tube and BCT from each donor were shipped from Streck in Omaha, NE, to a laboratory in Springfield, MA, and back during the course of 4 days in an insulated foam cooler in December. Ambient temperatures in both locations were around 4°C. The remaining K3EDTA tube and BCT from each donor were kept at 22°C for 4 days and processed with the returned shipped blood tubes. For shaken, shipped, and control samples, pDNA was measured by qPCR using the β‐actin assay.
Statistical Analysis
Statistical analysis was carried out using Microsoft Excel for Office 2007. Analysis was performed using paired, two‐tailed Student's t‐test and P < 0.05 was considered statistically significant.
RESULTS
Effect of the Chemical Cocktail Present in BCTs on pDNA Amplification
We investigated the effect of the chemical cocktail used in the new device on pDNA amplification by qPCR. In Figure 1, comparison of pDNA concentrations in chemically treated and untreated plasma samples showed no decrease in amplification after 3 and 6 hr of incubation at 22°C (Fig. 1A, P = 0.0958 and 0.1226, respectively); samples incubated for 7 and 14 days showed similar results (Fig. 1B, P = 0.0002 and 0.0290, respectively.
Figure 1.
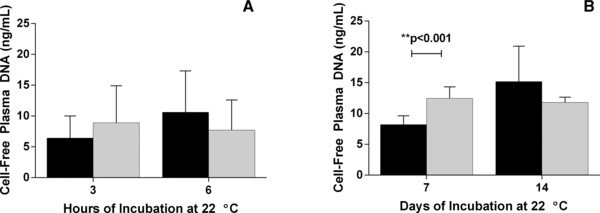
Effect of the novel chemical cocktail on plasma DNA (pDNA) detection by qPCR. Blood was drawn into K3EDTA tubes and plasma was separated by centrifugation. The plasma supernatant was aliquoted and divided into two treatment groups. To one group of plasma samples (gray bars), the chemical cocktail from a BCT was added at a final concentration equal to the amount present in a standard 10 ml blood drawn. The other group of plasma samples (black bars) was untreated and served as a control. Samples from each group were processed at 3 and 6 hr (panel A) and at 7 and 14 days (panel B). The pDNA concentration was determined using the β‐actin qPCR assay. When compared to untreated samples, samples treated with the chemicals showed no decrease in either DNA extraction from plasma or its amplification by qPCR. Error bars indicate SD, n = 5 for both panels.
Effect of Elevated Background gDNA in Plasma on Detection of Rare cfDNA Sequences
We simulated the effect of increased gDNA concentration, measured by the β‐actin assay, on the detection of low abundance cfDNA targets by introduction of a male‐specific DNA (SRY) into nonpregnant female blood. Figure 2A shows the DNA concentration of both gene targets in harvested plasma from K3EDTA samples stored at 22°C for 0, 7, and 14 days. Over time, increases in gDNA (β‐actin) levels were observed, which became statistically significant on day 14, as compared to the initial concentration on day 0 (P = 0.0581 and 0.0091). As gDNA (β‐actin) levels increased, detection of the cfDNA target (SRY fragment) significantly decreased (day 7, P = 0.0055 and day 14, P = 0.0006).
Figure 2.
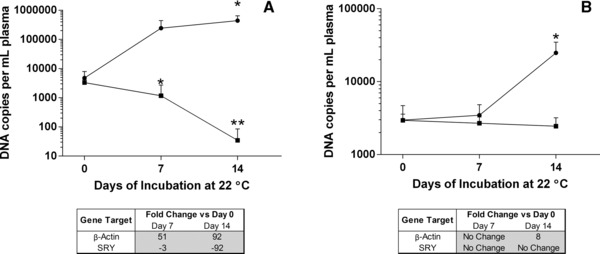
Effect of elevated background genomic DNA (gDNA) in plasma on the detection of rare DNA sequences. Blood was drawn from nonpregnant female donors into K 3 EDTA or BCT and stored at 22°C for 0, 7, or 14 days. At each time point, plasma was separated by centrifugation. A plasmid DNA construct containing a Y‐chromosomal sex‐determining region (SRY) sequence fragment (3,000 copies) was then spiked into all plasma samples and plasma DNA (pDNA) was extracted. Total pDNA concentration was determined using the β‐actin qPCR assay (●) and Y‐chromosomal sequence copy numbers were determined by the SRY qPCR assay (▪). For K 3 EDTA samples (Fig. 2A), as β‐actin pDNA concentrations increased in K 3 EDTA samples (*P < 0.05), there were statistically significant decreases in SRY sequence copy number detection (**P < 0.001). For BCT samples (Fig. 2B), pDNA concentrations remained close to the initial day 0 value (*P < 0.05) and the SRY sequence was detectible throughout the 14‐day time course. A table inset in each figure shows the fold change when comparing day 0 β‐actin and SRY levels to those on days 7 and 14. Error bars indicate SD, n = 5 for both panels.
Figure 2B shows the pDNA concentration of BCT plasma samples placed under the same storage conditions as above. Plasma harvested from BCT samples showed no significant change in the β‐actin concentration at 7 days (P = 0.4947) but did show a statistically significant change at the 14‐day time point (P = 0.0090). In BCTs, gDNA (β‐actin) levels remained close to the day 0 value and cfDNA (SRY fragment) levels did not change and remained detectible throughout the entire time course (day 7, P = 0.0749 and day 14, P = 0.0867).
In order to more clearly demonstrate how changes in gDNA levels effect cfDNA target detection, fold change calculations were performed (Fig. 2C; 20, 21). As K3EDTA gDNA levels increased (β‐actin, day 7 = 51‐fold change, day 14 = 92‐fold change), cfDNA detection decreased (SRY, day 7 = −3‐fold change, day 14 = −95‐fold change). In the BCT, gDNA levels remained relatively steady (β‐actin, day 7 = no change, day 14 = eightfold change) as did the cfDNA target detection (SRY, day 7 = no change, day 14 = no change).
Effect of Shaking and Shipping on pDNA Concentration in Blood Samples
Agitation conditions present during sample transport were simulated with an orbital shaker. A control experiment was also performed using both types of tubes without agitation. Figure 3A shows that shaking blood drawn into K3EDTA tubes caused a statistically significant increase in the pDNA concentration at 6 and 24 hr postblood drawn (P = 0.0015 and 0.0046), whereas shaking blood drawn into the new collection tube showed no change in the pDNA concentration except for the 3 hr time point (P = 0.0182, 0.1717, and 0.2592). Nonshaken control samples drawn into K3EDTA only showed statistically significant change at the 3 hr time point (P = 0.0029) but not at 12 or 24 hr (P = 0.1461 and 0.1646). The nonshaken BCTs samples show no change in pDNA concentration (P = 0.0815, 0.3544, and 0.5430) for all three time points.
Figure 3.
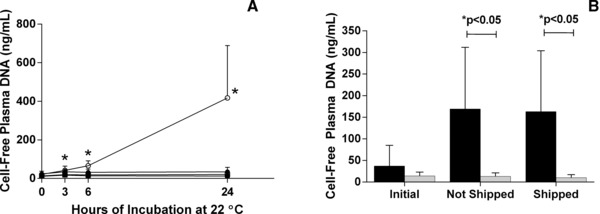
Effect of shaking and shipping on plasma DNA (pDNA) concentration in blood samples. Blood was drawn into K 3 EDTA tubes and the new device and then agitated at 22°C on an orbital shaker. As shown in panel A, at times 0, 3, 6, and 24 hr, one K 3 EDTA tube (○) and one BCT (□) was removed and pDNA was isolated. A control experiment was also performed with blood drawn from the same donors into K 3 EDTA tubes (●) and BCTs (▪) over the same time period without shaking. The pDNA concentration was determined using the β‐actin qPCR assay. Samples drawn into K 3 EDTA tubes that were subjected to shaking showed statistically significant increases in pDNA concentration at 6 and 24 hr compared to time 0 (*P < 0.05). Total plasma DNA concentration remained stable in shaken BCTs through those same time periods. Error bars indicate SD, n = 8 in panel A. In a shipping study, as shown in panel B, blood was drawn into K 3 EDTA tubes (black bars) and BCTs (gray bars) and either shipped round trip to a laboratory in Springfield, MA, or not shipped. Upon return, plasma from shipped and nonshipped samples was isolated and the pDNA concentration was determined using the β‐actin qPCR assay. Samples drawn into K 3 EDTA tubes that were shipped showed a statistically significant increase in pDNA when compared to samples shipped in the new tube (*P < 0.05). The pDNA concentration in shipped and nonshipped BCT samples showed no statistically significant change when compared to time zero. Error bars indicate SD, n = 10 in panel B.
After shipping was mimicked with a shaking experiment, new samples were drawn from ten donors into both K3EDTA and BCT. Tubes were either shipped to a laboratory in Springfield, MA, and back over the course of 4 days or not shipped and left at 22°C. Figure 3B illustrates a significant increase in pDNA concentration between shipped K3EDTA and shipped BCT (P = 0.0076) after they had returned. There was also a statistically significant increase in pDNA between initial and shipped K3EDTA (P = 0.0155) and between initial and nonshipped K3EDTA (P = 0.0284). In the BCT samples, however, there was no change in pDNA concentration between the initial and nonshipped or the nonshipped and shipped tubes (P = 0.7561 and 0.1355).
Effect of Storage Temperature on pDNA Concentration in Blood Samples
To demonstrate the effect of temperature on pDNA levels in blood drawn into K3EDTA tubes and BCTs, samples were stored at 6°C, 22°C, or 37°C for up to 14 days. Figure 4A shows that samples stored at 6°C had large increases in pDNA concentration (78‐fold on day 14) but these changes were not statistically significant (P = 0.0547 on day 7 and P = 0.0908 on day 14). BCTs stored in the same fashion showed statistically significant change on day 7 (P = 0.0096) and day 14 (P = 0.0277), however, when looking at fold changes, these increases amounted to only a threefold change over all 14 days of storage. When samples were incubated at 22°C (Fig. 4B), there were statistically significant changes seen in the pDNA concentration in K3EDTA samples (P = 0.0932 and 0.0028), but no change was seen in BCTs (P = 0.4953 and 0.0757). The fold change calculations showed dramatic increases in K3EDTA (233‐fold on 14 days) with minor increases in BCTs (sevenfold on day 14). The samples incubated at 37°C (Fig. 4C) showed a similar trend, with statistically significant increases occurring in the K3EDTA samples (P = 0.0104 and 0.0009) as well as in fold changes (53 on day 14). No significant changes were observed in the BCTs (P = 0.2235 and 0.1137), which had a fivefold change at day 14.
Figure 4.
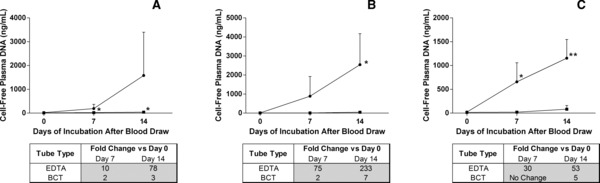
Effect of storage temperature on plasma DNA (pDNA) concentration in blood samples. Blood was drawn from six donors into three 10 ml K 3 EDTA tubes and three 10 ml BCTs per donor. One BCT (▪) and one K 3 EDTA (●) tube from each donor was stored (as aliquots) at either 6°C (A), 22°C (B), or 37°C (C) and these aliquots were harvested after 0, 7, and 14 days of storage at the previously mentioned temperatures. The pDNA concentration in the samples was determined using the β‐actin qPCR assay. Samples drawn into K 3 EDTA tubes and stored at all temperatures showed statistically significant increases in pDNA concentration as compared to the day 0 value except for the samples stored at 6°C, where fold change calculations showed an increase of 78‐folds after 14 days. The only statistically significant change in pDNA isolated from the new blood collection device was when samples were stored at 6°C. In this case the fold change indicated an increase of only threefold after 14 days (*P < 0.05, **P < 0.001). Tables below each graph show the fold change. Error bars indicate SD, n = 6 for both panels.
DISCUSSION
Transportation of blood samples from the site of phlebotomy to another facility is commonly required for molecular diagnostic testing. In this regard, several studies have focused on preanalytical variables that might compromise the accuracy cfDNA measurements, including the selection of blood collection devices, sample storage, and shipping conditions 16, 22, 23. Each of these parameters affects the amount of nucleated blood cell lysis that occurs postphlebotomy. Nucleated cell lysis leads to release of gDNA, elevating the background in pDNA and contaminating true cfDNA measurement. Here, we have minimized background gDNA increases by developing a novel chemical cocktail contained within a blood collection tube with the ability to stabilize nucleated blood cells.
While traditional chemicals used in cell stabilization, such as formaldehyde and glutaraldehyde, are known to damage DNA and RNA by causing nucleic acid–protein cross‐links and make extraction of nucleic acids difficult 24, the proprietary stabilization cocktail in BCTs has been shown by 13C‐NMR to be formaldehyde free 25. Other internal studies (Das et al., unpublished) have shown that incubation with the cocktail for 14 days at RT had no detectible effect on both qPCR and end point PCR. We have also assessed the effect of the cell stabilizing agents present in BCTs on pDNA extraction and amplification (Fig. 1) and found no decrease when compared to K3EDTA alone.
During transportation, shaking may disrupt nucleated blood cell integrity and compromise accuracy, as described above. We simulated shipping conditions over a 24‐hr period by placing blood in the control and the experimental collection devices on an orbital shaker. An increase in pDNA concentration was observed at 6 and 24 hr in K3EDTA blood samples. BCT stabilized blood cells showed a statistically significant increase in pDNA after 3 hr of shaking, but not at 6 or 24 hr, which was the same as shaken K3EDTA samples. Nonshaken BCTs showed no change at all time point.
When samples were shipped, similar trends were observed as to when samples were shaken. We shipped blood in both K3EDTA and the new collection devices and compared the resulting pDNA concentration between the two tube types. The BCT showed stable pDNA concentrations before and after shipping, whereas K3EDTA showed increases in pDNA under shipping conditions. This suggests nucleated cell disruption occurred in blood samples that were shipped in K3EDTA, which led to gDNA release, but this did not occur in stabilized samples.
Variation in sample storage temperature is another postphlebotomy condition that can cause changes in gDNA concentration. Here, we studied the effect of three different storage temperatures on the pDNA concentration of blood drawn into K3EDTA and BCT. Following blood drawn, samples were incubated at 6°C, 22°C, or 37°C for 14 days. For all storage temperatures, we observed either large changes in terms of fold change, or statistically significant increases in K3EDTA blood sample pDNA concentrations. Blood drawn into the new device showed statistically significant increases only for days 7 and 14 during 6°C incubation; however, the corresponding fold changes showed only minor increases in pDNA concentration. This data demonstrates the ability of the new blood collection device to stabilize pDNA levels and prevent gDNA release across a broad temperature range for an extended period of time.
Recent studies have shown plasma to contain low abundance cfDNA targets, such as circulating tumor cell‐derived DNA or fetal DNA present in maternal blood 26, 27. PCR detection of these cell‐free targets within a high gDNA background is challenging, requiring specialized protocols and/or large volumes of starting material 28. As a result, minimizing the release of gDNA from nucleated cells is essential for accurate analysis of true cfDNA. Using the new tube, we had previously shown that it is possible to preserve the original proportion of fetal cfDNA by minimizing maternal gDNA background 29. Here, we have shown that increased background gDNA adversely affects the detection of low abundance cfDNA targets (Fig. 2). As pDNA concentration increased in K3EDTA samples due to gDNA release, detection of the introduced SRY fragments decreased (Fig. 2A). In contrast, there was no statistically significant change in rare target (SRY) detection when blood was stored in the new collection device (Fig. 2B).
In current practice, it is recommended that blood samples be centrifuged to isolate plasma and frozen to prevent gDNA contamination of cfDNA during sample processing, transportation, and storage 21. We have shown that the novel stabilizing chemical cocktail prevents the release of gDNA into plasma postphlebotomy up to 14 days, avoiding these labor‐intensive requirements. Using this new blood collection device, ex vivo storage at room temperature becomes possible, allowing flexibility for offsite blood drawn to be sent to centralized laboratories for downstream analysis of the cfDNA without preliminary centrifugations or cryopreservation.
ACKNOWLEDGMENT
We gratefully acknowledge the technical assistance provided by Gary Krzyzanowski.
REFERENCES
- 1. Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139–142. [DOI] [PubMed] [Google Scholar]
- 2. Bischoff F, Lewis D, Simpson, J . Cell‐free fetal DNA in maternal blood: Kinetics, source and structure. Hum Reprod Update 2005;11:59–67. [DOI] [PubMed] [Google Scholar]
- 3. Mandel P, Métais P. Les acides nucléiques du plasma sanguine chez l'homme. C R Acad Sci Paris 1948;142:241–243. [PubMed] [Google Scholar]
- 4. Tan EM, Schur PH, Carr RI, et al. Deoxyribonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest 1966;45:1732–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646–650. [PubMed] [Google Scholar]
- 6. Leon SA, Ehrlich GE, Shapiro B, et al. Free DNA in the serum of rheumatoid arthritis patients. J Rheumatol 1977;4:139–143. [PubMed] [Google Scholar]
- 7. Fournié GJ, Courtin JP, Laval F, et al. Plasma DNA as a marker of cancerous cell death. Investigations in patients suffering from lung cancer and in nude mice bearing human tumours. Cancer Lett 1995;91:221–227. [DOI] [PubMed] [Google Scholar]
- 8. Anker P, Mulcahy H, Chen XQ, et al. Detection of circulating tumor DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 1999;18:65–73. [DOI] [PubMed] [Google Scholar]
- 9. Anker P, Mulcahy H, Stroun M. Circulating nucleic acids in plasma and serum as a noninvasive investigation for cancer: Time for large‐scale clinical studies? Int J Cancer 2003;103:149–152. [DOI] [PubMed] [Google Scholar]
- 10. Gal S, Fidler C, Lo YM. Quantitation of circulating DNA in the serum of breast cancer patients by real‐time PCR. Br J Cancer 2004;90:1211–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nawroz H, Koch W, Anker P, et al. Microsatellite alterations in serum DNA of head and neck cancer patients. Nat Med 1996;2:1035–1037. [DOI] [PubMed] [Google Scholar]
- 12. Schwarzenbach H, Hoon DS, Pantel K. Cell‐free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2001;11:426–437. [DOI] [PubMed] [Google Scholar]
- 13. Kohler C, Barekati Z, Radpour R, et al. Cell‐free DNA in the circulation as a potential cancer biomarker. Anticancer Res 2011;31:2623–2628. [PubMed] [Google Scholar]
- 14. Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350:485–487. [DOI] [PubMed] [Google Scholar]
- 15. Wright CF, Burton H. The use of cell‐free fetal nucleic acids in maternal blood for non‐invasive prenatal diagnosis. Hum Reprod Update 2009;15:139–151. [DOI] [PubMed] [Google Scholar]
- 16. Hung EC, Chiu RWK, Lo YM. Detection of circulating fetal nucleic acids: A review of methods and applications. J Clin Pathol 2009;62:308–313. [DOI] [PubMed] [Google Scholar]
- 17. Chan KC, Ding C, Gerovassili A, et al. Hypermethylated RASSF1A in maternal plasma: A universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin Chem 2006;52:2211–2218. [DOI] [PubMed] [Google Scholar]
- 18. Lee TH, Paglieroni T, Ohto H, et al. Survival of donor leukocyte subpopulations in immunocompetent transfusion recipients: frequent long‐term microchimerism in severe trauma patients. Blood 1999;93:3127–3139. [PubMed] [Google Scholar]
- 19. Boddy JL, Gal S, Malone PR, et al. Prospective study of quantitation of plasma DNA levels in the diagnosis of malignant versus benign prostate disease. Clin Cancer Res 2005;11:1394–1399. [DOI] [PubMed] [Google Scholar]
- 20. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001;98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Witten D, Tibshirani R. A comparison of fold‐change and the t‐statistic for microarray data analysis. Department of Statistics, Stanford University technical report; 2007. [Google Scholar]
- 22. Jung M, Klotzek S, Lewandowski M, et al. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem 2003;49:1028–1029. [DOI] [PubMed] [Google Scholar]
- 23. Dhallan R, Au WC, Mattagajasingh S, et al. Methods to increase the percentage of free fetal DNA recovered from the maternal circulation. JAMA 2004;291:1114–1119. [DOI] [PubMed] [Google Scholar]
- 24. Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 2002;161:1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Das K, Dumais J, Basiaga S, Krzyzanowski GD. Carbon‐13 nuclear magnetic resonance analyses of formaldehyde free preservatives. Acta Histochem 2012. doi: 10.1013/j.actahis.2001.11.004. (epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 26. Skvortsova T, Rykova E, Tamkovich S, et al. Cell‐free and cell‐bound circulating DNA in breast tumours: DNA quantification and analysis of tumour‐related gene methylation. Br J Cancer 2006;94:1492–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barrett A, Zimmermann B, Wang D, et al. Implementing prenatal diagnosis based on cell‐free fetal DNA: Accurate identification of factors affecting fetal DNA yield. PloS ONE 2011;6(10):e25202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu M, Stott S, Toner M, Maheswaran S, et al. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol 2011;192:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fernando MR, Chen K, Norton S, et al. A new methodology to preserve the original proportion and integrity of cell‐free fetal DNA in maternal plasma during sample processing and storage. Prenat Diagn 2010;30:418–424. [DOI] [PubMed] [Google Scholar]


