Abstract
Photoactivation of caged fluorescent tubulin was used mark the microtubule (MT) lattice and monitor MT behavior in interphase cells. A broadening of the photoactivated region occurred as MTs moved bidirectionally. MT movement was not inhibited when MT assembly was suppressed with nocodazole or Taxol; MT movement was suppressed by inhibition of myosin light chain kinase with ML7 or by a peptide inhibitor. Conversely, MT movement was increased after inhibition of cytoplasmic dynein with the antibody 70.1. In addition, the half-time for MT turnover was decreased in cells treated with ML7. These results demonstrate that myosin II and cytoplasmic dynein contribute to a balance of forces that regulates MT organization, movement, and turnover in interphase cells.
Keywords: microtubule motion
The capacity for microtubules (MTs) to undergo dynamic rearrangement is critical to the various vital cellular activities to which they contribute, including the assembly and function of the mitotic spindle, intracellular transport of organelles and vesicles, and the establishment and maintenance of cell shape and polarity. MTs are nucleated at the centrosome and maintain a generally radial organization, with plus-ends at the cell periphery. Direct observations show that peripheral plus-ends undergo dynamic instability behavior, characterized by the stochastic switching between phases of elongation and rapid shortening (1). Dynamic instability is tempered, such that turnover is more rapid in the peripheral region of the cell (2). MT turnover in the more central cytoplasmic domain is thought to occur by subunit turnover at both plus- and minus-ends (3, 4).
MTs modulate the behavior of the actin cytoskeleton in a temporally and spatially regulated manner. For example, MTs are required to establish the site of contractile ring formation in cytokinesis and to restrict ruffling behavior to the leading edge of motile fibroblasts (5). The nature of the cross-talk between the MT and actin cytoskeletons is not well understood, but recent work indicates that a feedback mechanism involving Rho family members may contribute to these regulatory interactions (6). In addition, molecular motors may integrate the MT and actin systems. Actomyosin moves MTs rearward at the cell periphery (3), and myosin-generated forces are responsible for MT transport in motile cells and for axon retraction in cultured neurons (7, 8). Cytoplasmic dynein contributes to MT organization in interphase cells, to the assembly and function of the mitotic spindle, and to MT transport in neurons (9, 10). These observations led us to test the hypothesis that molecular motors modulate the organization, rearrangement, and turnover of MTs in interphase cells.
Materials and Methods
Materials.
All materials for cell culture were obtained from Life Technologies (Gaithersburg, MD), with the exception of FCS, which was obtained from Atlanta Biologicals (Norcross, GA). Unless otherwise noted, all other chemicals were obtained from Sigma.
Cell Culture, Cell Staining, and Microinjection.
Cell culture, labeling of fluorescent tubulins, and microinjection were performed exactly as described (11). Tubulin labeled with a reactive fluorescent dye, Cy3, was microinjected at a needle concentration of 2.0–2.7 mg/ml; tubulin labeled with a caged derivative of carboxyfluorescein called C2CF (1) was microinjected at a needle concentration of 4.1–4.7 mg/ml. Cells that were to be double injected with fluorescent tubulin and antibodies or inhibitory peptide were injected with tubulin and allowed to recover at 37°C for 60 min. The same cells were relocated and injected with anti-dynein antibody (Sigma clone 70.1) at a needle concentration of 30–50 mg/ml, anti-kinesin HD antibody (gift of Fatima Gyoeva) at a needle concentration of 9.5 mg/ml, anti-HSET antibody at a needle concentration of 10 mg/ml, anti-Eg5 antibody (both gifts of Duane Compton) at a needle concentration of 5.7 mg/ml, nonimmune IgM (Sigma) at a needle concentration of 43 mg/ml, or myosin light chain kinase (MLCK) pseudosubstrate inhibitory peptide (Peptides International) at a needle concentration of 1 mM. The peptide, sequence AKKLSKDRMKKYMARRKWQKTG, specifically inhibits myosin II by acting both as a pseudosubstrate for the autoinhibitory domain of MLCK and by binding to calmodulin and inhibiting Ca2+/calmodulin-dependent activation of MLCK (12, 13).
Pharmacological inhibitors were used as follows. Cells were pretreated for 1 h with 100 nM Taxol or nocodazole, and were photoactivated and imaged for an additional 75 min. Cells were treated with 100 μM ML7 or 20 mM 2,3-butanedionemonoxime (BDM), and were photoactivated and imaged within 75 min of drug application. Cytokinesis was blocked in mitotic cells incubated with BDM or ML7 under these conditions.
To stain for the 58-kDa Golgi protein after imaging, cells were fixed in 3.7% formaldehyde and incubated in a monoclonal antibody (Sigma, clone 58K-9; 1:100 dilution) followed by incubation in FITC-conjugated goat anti-mouse secondary antibody (Organon Teknika-Cappel; 1:13 dilution). Staining for MTs and F-actin was performed exactly as described (11). Some cells that were injected with Eg5 antibody were followed into mitosis, and monopolar spindles were observed in phase contrast to verify the inhibitory activity of the antibody. Vital staining with rhodamine 123 (Molecular Probes) was performed by incubating cells in 10 μg/ml dye for 5 min, and rinsing twice in dye-free medium.
Image Collection.
Cells that were injected with a mixture of caged fluorescein tubulin and Cy3-labeled tubulin were imaged on a Bio-Rad 600 Confocal microscope with a 60 × 1.4 numerical aperture lens. Photoactivation was performed by using a 4-sec exposure to unattenuated 360-nm light generated by a 100-watt mercury arc lamp and a 4′,6-diamidino-2-phenylindole filter cube (Nikon). A 25-μm × 3-mm slit (Coherent Ealing, Auburn, CA) was placed on a slider in a conjugate image plane. Oxyrase (Oxyrase Inc., Mansfield, OH) was added to the medium at 0.3 unit/ml to reduce photodamage to cells (14). Images were printed by using ADOBE PHOTOSHOP software (Adobe Systems, Mountain View, CA) running on a Power Macintosh G3 with an Epson Stylus 850N printer.
For analysis of individual MT dynamics and observation of Rhodadmine 123-stained cells, a Nikon Eclipse TE 300 inverted microscope equipped with a 100 × 1.3 numerical aperture objective lens was used. Images were acquired by using a Princeton Instruments micromax interline transfer-cooled CCD camera and METAMORPH software (Universal Imaging, Media, PA).
Data Analysis.
The behavior of individual MTs was measured and analyzed as described (15). To measure turnover of the MT array, fluorescence intensity values, within a rectangle of sufficient dimensions to ensure that the photoactivated region was within the box for the entire sequence, were measured and corrected for background fluorescence (determined by measuring fluorescence in a rectangle of the same dimensions placed in a region of the cell that was not photoactivated). Several cells were measured, and the values for each time point were averaged; percentage of fluorescence (± SD) versus time, in min, was plotted by using ORIGIN (Microcal Software, Northampton, MA) analytical software and fitted to a single exponential decay. We measured the fluorescence intensity in the original photoactivated region just after photoactivation, and at 14 min after photoactivation. The 14-min value was corrected for the dynamic turnover of MTs, by using the rate constant for turnover generated for each cell, and the resulting fluorescence intensity, which represents the percentage of MTs remaining in the original photoactivated region, was expressed as a percentage of the initial intensity. The difference between 100 and the percentage left in the original region is the percentage of MTs that have moved. Decay of fluorescent signal because of photobleaching was determined to be negligible.
Results and Discussion
Bidirectional Movement of MTs in Interphase Cells.
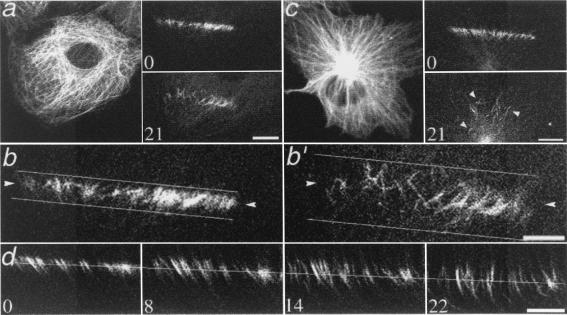
We have used photoactivation of a caged fluorescent derivative of tubulin (16) to observe and quantify MT behavior in nonperipheral regions of interphase PtK2 cells. Bidirectional movement, either toward the nucleus or toward the cell periphery, of individual and small bundles of photoactivated MTs is shown in Fig. 1. The rate of movement was measured from sequences of images collected at 2-min intervals for 16 min. The average rate of movement (0.21 ± 0.09 μm/min, range 0.11–0.45, n = 28 measurements in 12 cells) was the same for MTs moving toward or away from the nucleus. Individual MTs behaved independently: adjacent MTs could move in opposite directions, indicating that the mechanism for movement is highly local (Fig. 1d) (17). The apparent lengthening of the fluorescent mark on some MTs was likely to be the result of closely aligned MTs moving at different rates or in opposite directions (Fig. 1d).
Figure 1.
MT movement in interphase cells. (a) PtK2 epithelial cell. (Left) Rhodamine tubulin fluorescence of the entire MT array. (Right) Photoactivated fluorescence immediately (Upper) and 21 min after (Lower) photoactivation. (Bar, 10 μm.) (b and b′) Enlargement of the photoactivated region shown in a. Horizontal lines illustrate the increased width of the mark over time. Arrowheads at the same pixel location in both images show the bidirectionality of the movement. (c) Photoactivation in a COS-7 fibroblast. The more rapid dissipation of fluorescence, compared with cell in a, is due to faster dynamic turnover of MTs in fibroblasts. Arrowheads highlight the widespread position of remaining MTs. (Bar, 5 μm.) (d) Additional example of photoactivated region of a PtK2 cell, which shows bidirectional MT movement and apparent lengthening of photoactivated MTs as marks on adjacent MTs are moved; time, in min, after photoactivation is given in the corner of each panel. Fiduciary line indicates the approximate center of the original photoactivated region. (Bar, 5 μm.) Note that the example shown in d contains an unusually uniform MT arrangement; the arrangement shown in b is more typical.
The extent of movement was measured by visually marking the boundaries of the photoactivated region at the initial and final time points (Fig. 1 b and b′) and measuring the fold increase in the distance between the boundaries. Note that the photoactivated mark was made perpendicular to the radial orientation of the MTs; therefore, only MT movement in the y axis is measured by our assay, and measurements of the extent of movement are likely to be underestimations. For control PtK2 cells, the photoactivated mark spread an average of 2.45 times its original width in 28 min after photoactivation (Table 1). The direction of mark widening in control cells was biased toward the cell center; 64% of the increase in width was rearward and 36% was forward. Movement of MTs was observed regardless of whether photoactivation was performed nearer to the nucleus or nearer to the cell periphery, and was also observed in BSC1 epithelial cells and COS-7 fibroblasts (Fig. 1c), both of which are characterized by a more centrosomal microtubule array than PtK2 cells. The extent of movement was greater in both BSC1 and COS-7 cells than in PtK2 cells, which may result from differences in the distribution or activity of motor proteins in these cells.
Table 1.
Quantitative analysis of MT movement in PtK2 epithelial cells
| Cell type | Extent of movement (28 min) | Percentage transported (14 min) | Turnover half-time, min |
|---|---|---|---|
| Control PtK2 | 2.45 ± 0.61 (14) | 24.9 ± 9.8 (8) | 10.2 (12) |
| Nocodazole treated | 3.05 ± 1.10 (11) | — | — |
| Kinesin inhibited (HSET) | 2.23 ± 0.80 (7) | — | — |
| Kinesin inhibited (Eg5) | 2.21 ± 0.36 (5) | — | — |
| Kinesin inhibited (HD) | 2.87 ± 0.89 (5) | — | — |
| Dynein inhibited (70.1) | 4.18 ± 0.92* (13) | 29.6 ± 12.7 (6) | 10.2 (10) |
| Myosin inhibited (peptide) | 1.81 ± 0.37† (6) | — | — |
| Myosin inhibited (ML7) | 1.87 ± 0.38† (12) | 2.0 ± 15.2* (5) | 15.6 (8) |
| Myosin inhibited (BDM) | 1.85 ± 0.61† (13) | — | — |
| Myosin and dynein inhibited | 1.78 ± 0.56† (10) | — | — |
The extent of movement is the fold increase in the width of the photoactivated mark between the initial and final time points (t = 0 to t = 28 min); units are arbitrary. Values are average ± SD (number of cells). Statistically different from control:
, P = 0.001;
, P ≤ 0.05.
MT Movement Does Not Depend on MT Dynamics or on Kinesin-Related Motors.
We tested the role of MT assembly in MT movement by treating cells with nocodazole or Taxol, at concentrations that have little effect on polymer level but greatly suppress subunit addition and loss from the ends of MTs (18, 19). Treatment with nocodazole (100 nM for 1 h) did not inhibit bidirectional MT movement (Fig. 2b, Table 1), nor did treatment with Taxol (100 nM for 1 h); however, Taxol-induced rearrangements of the MT array precluded meaningful quantitative analysis of movement. It should be noted that these subtle rearrangements occur even at the low levels of Taxol that inhibit dynamic instability without affecting polymer level. The results of these experiments indicate that dynamic instability of MTs is not required for MT movement in these cells, and is likely to be an independent process.
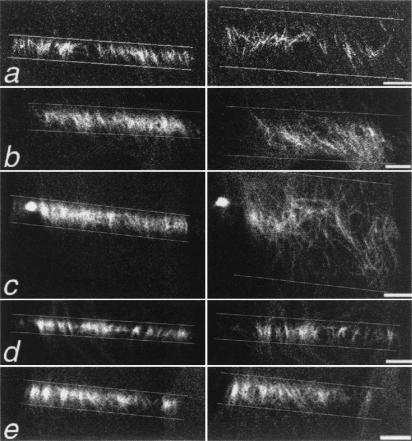
Figure 2.
MT movement requires myosin II activity and is restricted by cytoplasmic dynein/dynactin. Cells were treated as described and photoactivated as in Fig. 1. Only the photoactivated region is shown. In all panels except b, the cell center is toward the top of the figure; the cell center is toward the bottom of the figure in b. (Left) MTs immediately after photoactivation. (Right) MTs 21 min later. (a) Control PtK2 cell. The rearward movement of MTs in this cell was 89%, as compared with the average of 64%. (b) Cell treated with nocodazole; MT dynamic instability is not required for MT movement. (c) Cells injected with a function blocking dynein antibody; MT movement is extensive. Individual MTs are often observed at sites distant from their initial location. (d) Cells treated with the MLCK inhibitor ML7; MT movement is decreased. (e) Cell microinjected with a function-blocking dynein antibody and treated with ML7. The extensive movement of MTs observed on cytoplasmic dynein inhibition (c) is suppressed when myosin II is inhibited. (Bars, 5 μm.)
We wished to determine whether MT movement in interphase cells is mechanistically similar to flux in the mitotic spindle (16). In spindle flux, transport is coupled to addition and loss of subunits from the MT ends, and is inhibited by adenosine 5′-[β,γ−imido]triphosphate (20) and by monastrol, a specific inhibitor of the plus-end directed kinesin-related protein Eg5 (21, ¶). We did not observe inhibition of movement after nocodazole or Taxol treatment, indicating that subunit addition and loss were not required for MT movement. In addition, the movement that we observed is distinct from spindle flux because it is bidirectional and the behavior of the MTs is uncoordinated. We also tested the possible contribution of Eg5, and of the minus-end-directed mitotic kinesin-related protein HSET, to MT movement. Neither an antibody to HSET that inhibits meiotic spindle formation and Taxol-induced aster formation (22), nor an antibody to Eg5 that blocks bipolar spindle formation (see Materials and Methods) had any detectable effect on MT movement in interphase cells (Table 1).
The contribution of other members of the kinesin family to MT movement was also tested by using an antibody to a conserved region of the conventional kinesin heavy chain, HD, which has been previously demonstrated to block MT-dependent processes (23). Microinjection of HD into cells induced mitochondria clustering to the perinuclear region, as assessed by rhodamine 123 staining (23), and had no detectable effect on the movement of marked MTs (Table 1).
MT Movement Requires Actomyosin and Is Restricted by Cytoplasmic Dynein.
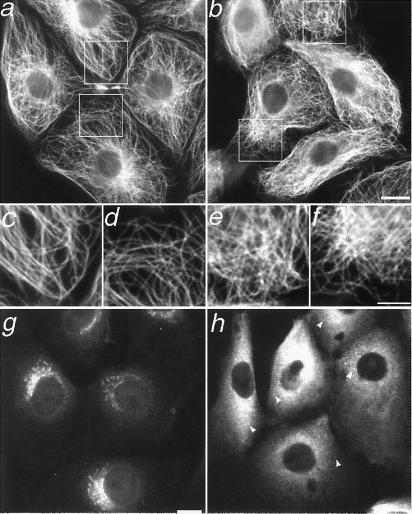
Overexpression of various components of the cytoplasmic dynein-activating complex, dynactin, disrupts MT organization in fibroblasts (9). To test the role of dynein/dynactin in MT movement, an antibody to a cytoplasmic dynein intermediate chain, clone 70.1, that blocks cytoplasmic dynein function by inhibiting its association with intact dynactin complex (24) was microinjected into cells. Our results show that marked MTs were moved farther from their original location in the absence of cytoplasmic dynein function (Fig. 2c). The movement was extensive; the photoactivated mark spread an average of 4.18 times its original width (Table 1). There was a slight rearward bias to the direction of MT movement, but this difference was not statistically significant from a 50/50 division, as it was in control cells. We hypothesize that retrograde flow of F-actin is responsible for the rearward bias in control cells, and that the inhibition of cytoplasmic dynein untethers MTs, randomizing their movement. The overall arrangement of MTs in cells injected with 70.1 was also assayed by immunofluorescence microscopy of fixed cells; consistent with recent results in fibroblasts (9), we observed a general disorganization of the MT array in epithelial cells (Fig. 3 a–f).
Figure 3.
Disruption of cytoplasmic dynein induces a reorganization of the MT array in interphase cells. MTs in untreated cells (a) are arranged radially, whereas those in 70.1-injected cells (b) are disorganized. (Bar, 10 μm.) (c and d) Magnification of boxed regions indicated in a; (e and f) magnification of boxed regions indicated in b. (Bar, 5 μm.) Immunostaining for the 58-kDa Golgi marker protein shows compact Golgi stacks in control cells (g) and dispersed Golgi vesicles, indicated by arrowheads, in 70.1-injected cells (h). The background in h is high because the anti-mouse secondary antibody that recognizes the 58-kDa primary antibody also binds to the injected 70.1 antibody. (Bar, 10 μm.)
Cells injected with 70.1 were fixed and stained for the 58-kDa Golgi marker protein; all cells were positive for a disrupted Golgi complex, verifying the abolition of cytoplasmic dynein function (Fig. 3 g and h) (25). As an additional control, the photoactivation experiment was also performed on cells microinjected with nonimmune IgM rather than 70.1. The degree of spreading of the photoactivated mark was determined to be statistically the same as in control cells, demonstrating that the effect of greater MT movement seen with 70.1 was specific (data not shown).
Previous work has shown that MTs in the extreme cell periphery are moved by actin-dependent retrograde flow, and numerous studies have shown that coordination between MTs and actin is required for various cellular processes (5, 6). We tested whether myosin II contributes to the bidirectional movement of MTs in nonmotile cells by microinjecting an inhibitory peptide of MLCK (see Materials and Methods), and also by treating cells with the MLCK inhibitor ML7 (12, 13, 26). The results show that MT movement was, in fact, suppressed when myosin II was inhibited (Fig. 2 d and e). After 28 min, the photoactivated marks in peptide-injected and ML7-treated cells had widened to only 1.81 and 1.87 times their original width, respectively, compared with the 2.45-fold increase seen in control cells (Table 1).
Photoactivation experiments were also performed following inhibition of myosin with 20 mM BDM. BDM interferes with the ATPase cycle of myosin and is reported to inhibit myosin II and myosin V (27), although it could potentially interfere with other myosins as well, because the ATP-binding region is highly conserved among family members. The average increase in width of photoactivated marks in cells treated with BDM was 1.85 ± 0.61, similar to the peptide-injected and ML7-treated cells, and statistically different from controls (Table 1).
As an additional test, we first inhibited cytoplasmic dynein/dynactin by injection of 70.1 to allow for more extensive MT movement, and subsequently added ML7 to determine whether movement was partially or entirely dependent on myosin II. MT movement was also inhibited in these cells, to an approximately equal extent as in cells treated with ML7 alone; the average extent of mark widening was 1.78 ± 0.56 (Fig. 2e, Table 1).
To ascertain whether myosin acts to move MTs through its interactions with F-actin or through a mechanism independent of actin, we used 300 nM latrunculin B to disassemble the F-actin (28). Quantitative analysis of these experiments was difficult because of the global changes in cell morphology that occur when the actin cytoskeleton is disrupted. However, qualitative assessment indicates that F-actin is necessary for the role of myosin II in MT movement (data not shown). Our results are consistent with recent work demonstrating a role for actomyosin in spindle flux in spermatocytes (29).
To test whether Taxol-induced MT rearrangements were dependent on myosin II activity, we incubated cells simultaneously in 1 μM Taxol and 100 μM ML7 for 1 h. This treatment resulted in extensive cell rounding, a result not seen with either drug treatment alone. That either MT-dynamic behavior can be altered or myosin function can be inhibited without causing significant cell rounding, but the combination of both results in rounded cells, further emphasizes the antagonistic nature of the MT–actomyosin relationship.
We determined the percentage of photoactivated MTs that moved from the original photoactivated region at the half-time of the observation period (Materials and Methods; Table 1). MT movement was measured at the halfway point of observation because there was enough fluorescent signal remaining to provide an accurate measurement for a greater number of cells. In addition, because the fluorescence measurements must be corrected for turnover, only cells for which turnover could be accurately determined were included in the measurement. In control cells, 24.9% of the fluorescence moved to a region of the cell outside of the original photoactivated area; that number was 29.6% in 70.1-injected cells and only 2% in ML7-treated cells (Table 1). Thus, inhibition of cytoplasmic dynein-induced MTs to move further than in control cells, but increased the percentage of MTs that moved by only 5%, indicating that movement is limited, perhaps by interaction with or interference by other cellular elements. Conversely, inhibition of myosin activity dramatically reduced the percentage of MTs that moved. The lack of complete inhibition of MT movement by ML7 could result from an incomplete inhibition of MLCK and/or from movement mediated by myosins that are insensitive to ML7.
Inhibition of Myosin Reduces Dynamic Turnover of MTs.
The half-time of MT turnover was measured for control, 70.1-injected and ML7-treated cells by measuring the dissipation of fluorescence after photoactivation. A half-time of 10.2 min was measured for both control and cytoplasmic dynein-inhibited cells; however, the half-time for myosin-inhibited cells was increased by 53% to 15.6 min, indicating a substantial reduction in MT dynamic turnover (Table 1). To determine the mechanism responsible for the increased half-time of turnover, we measured the dynamic instability behavior of the plus-ends of individual MTs in control, ML7-treated, and 70.1-injected cells (15, 19). As shown in Table 2, the dynamic instability parameters are similar for all three groups. Specifically, no statistically significant differences were detected between the control and 70.1-injected cells. In cells treated with ML7, the rate of growth was 15% slower and the rate of shortening was 36% slower than in control cells; these differences were statistically significant (Table 2). In addition, rescue was 34% less frequent in ML7-treated cells than in controls. Other parameters, including dynamicity, which is a measure of the overall growth and shortening, were similar to controls (Table 2), indicating that the reduction in turnover is not solely due to an alteration in plus-end dynamic behavior. MT minus-ends, generated by release from the centrosome and by breaking of long MTs, also contribute to MT turnover (4, 6, 30). In epithelial cells, the frequency of release from the centrosome is low (≈1.5 MTs per min), suggesting that MT breakage is the more physiologically relevant pathway for generating MT minus ends (4, 30). Our data suggest that, in the absence of myosin contractility, MT bending and breaking is reduced, and fewer free minus ends are generated, resulting in slower overall turnover. These results provide direct evidence for myosin-generated forces modulating the kinetic behavior of MTs (7, 17).
Table 2.
Dynamic instability parameters at the MT plus ends
| Parameter | Control | 70.1-injected | ML7-treated |
|---|---|---|---|
| Shortening rate, μm/min | 12.9 ± 9.2 (78) | 10.7 ± 7.3 (68) | 8.3 ± 4.5† (63) |
| Growth rate, μm/min | 6.6 ± 3.7 (121) | 7.3 ± 4.5 (92) | 5.6 ± 2.9† (108) |
| Pause duration, sec | 18.5 ± 17.5 (124) | 21.7 ± 22.5 (115) | 21.9 ± 22.7 (122) |
| % time in pause | 41.8 | 49.3 | 40.9 |
| % time in growth | 18.4 | 21.8 | 19.8 |
| % time in shortening | 39.8 | 28.9 | 39.3 |
| Rescue frequency, sec−1 | 0.090 ± 0.055 (36) | 0.105 ± 0.083 (32) | 0.059 ± 0.050* (35) |
| Catastrophe frequency, sec−1 | 0.019 ± 0.015 (40) | 0.030 ± 0.062 (39) | 0.017 ± 0.024 (49) |
| Dynamicity, μm/min | 4.81 ± 3.07 (40) | 3.99 ± 2.67 (40) | 3.76 ± 2.44 (49) |
Values are average ± SD (number of events). Number of MTs analyzed is 41 in control cells, 40 in 70.1-injected cells, and 50 in ML7-treated cells. Statistically different from control:
, P = 0.02;
, P = 0.05.
Model for the Regulation of MT Movement by Cytoplasmic Dynein and Myosin II.
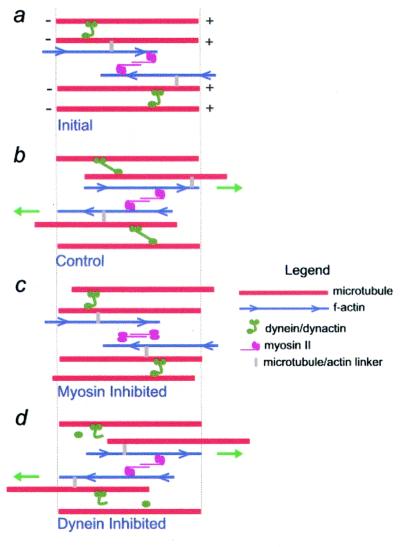
Our results can be explained by a model in which cytoplasmic dynein and myosin generate antagonistic forces that contribute to the organization of the MT array (Fig. 4). Our data support a role for cytoplasmic dynein in tethering MTs and resisting actomyosin-generated forces. One possible mechanism for tethering is that the cytoplasmic dynein heavy chain and the p150 subunit of dynactin, both of which contain MT binding domains, could cross-link adjacent MTs (31). Alternatively, dynein/dynactin could link MTs to other cellular components such as the centrosome (9), membrane systems (10), or sites of cell–substrate adhesion (5). Note that inhibition of dynein activity could enhance MT movement indirectly, by increasing MT release from the centrosome, or directly, by preventing dynein-dependent force generation on MTs bound to membrane organelles or other MTs. Further experiments are necessary to distinguish between these possibilities.
Figure 4.
Model for regulation of MT movement by cytoplasmic dynein and myosin II. MT position immediately after (a) and 21 min after (b–d) photoactivation is shown. (a) Dynein/dynactin complexes cross-link adjacent MTs, restricting MT movement. It is assumed that the majority of MTs in the lamella are of uniform polarity. MTs are linked to F-actin by multifunctional cross-linking molecules, shown at different positions along the filaments to emphasize their dynamic nature. (b) Bipolar myosin II molecules interact with neighboring microfilaments of opposite polarity, resulting in bidirectional MT movement. When myosin II moves actin filaments in the direction(s) indicated by the green arrows, adjacent MTs are also moved in the same direction. Some MTs may not be moved if they do not have a dynamic link to actin. The extent of MT movement is limited by dynein/dynactin. (c) Actin filaments are not moved when myosin II is inactivated, reducing MT movement. (d) Inhibition of cytoplasmic dynein relieves the restraint of MT movement, and MTs are moved further with actin movement. Note that the MTs and microfilaments shown in the diagram are meant to represent only a portion of the polymer lattice; assembly/disassembly behavior at polymer ends is not included on the diagram. In addition, although dynein is shown linking adjacent MTs, it could also function by linking MTs to the centrosome or to membranous organelles in the cytoplasm.
In our model, myosin moves MTs because actin filaments and MTs are linked, directly or indirectly. One possibility is that multifunctional proteins link the two filament systems in cells; movement of myosin on actin would then indirectly move MTs. This possibility is supported by the recent identification of several molecules that contain both MT and actin binding domains (32) and by recent in vitro work, which reports MT-dependent F-actin motility and implicates myosin V in linking the two systems (33). Alternatively, myosin might act directly on MTs, independently of actin, for example, through interactions with MT-associated proteins or other motor proteins (5). The observation that F-actin is necessary for the role of myosin in MT movement supports the former, rather than the latter, possibility. Additional experiments are necessary to elucidate the precise mechanism by which actomyosin mediates MT movement, and how dynein/dynactin functions to restrict MT movement.
Summary.
Our results provide evidence for the view that a dynamic feedback system exists between the actin and MT cytoskeletal systems. Myosin-generated forces are highly localized, as indicated by the pattern of MT movements, and myosin-generated tension modulates the turnover half-time of MTs. Our data also demonstrate that forces generated by molecular motors play an important and previously unrecognized role in mediating interactions between cytoskeletal polymers: tension generated by actomyosin is responsible for moving MTs in cells and this tension is resisted by cytoplasmic dynein/dynactin complexes (6, 7). Temporal and spatial regulation of both the myosin- and cytoplasmic dynein/dynactin-generated forces is likely to play a significant role in diverse cellular processes including cell locomotion, cell division, and the establishment and maintenance of asymmetric cell morphologies.
Acknowledgments
We thank Drs. A. Desai and T. Mitchison for the C2CF and the Salmon laboratory for C2CF-labeled tubulin, Dr. Aline Valster for assistance with preparation of tubulin, Mr. Dale Callaham for assistance with confocal microscopy, and the members of the Wadsworth laboratory for their support. This work was supported by a grant from the National Institutes of Health (to P.W.).
Abbreviations
- BDM
2,3 butanedione monoxime
- MLCK
myosin light chain kinase
- MT
microtubule
Footnotes
Kapoor, T. M., Mayer, T. U., Desai, A., Maddox, P., Salmon, E. D., Schreiber, S. L. & Mitchison, T. J. (1999) Mol. Biol. Cell 10, 128 (abstr).
References
- 1.Desai A, Mitchison T. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 2.Sammak P J, Gorbsky G J, Borisy G G. J Cell Biol. 1987;104:395–405. doi: 10.1083/jcb.104.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waterman-Storer C M, Salmon E D. J Cell Biol. 1997;139:417–434. doi: 10.1083/jcb.139.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keating T J, Peloquin J G, Rodionov V I, Momcilovic D, Borisy G G. Proc Natl Acad Sci USA. 1997;94:5078–5083. doi: 10.1073/pnas.94.10.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goode B L, Drubin D G, Barnes G. Curr Opin Cell Biol. 2000;12:63–71. doi: 10.1016/s0955-0674(99)00058-7. [DOI] [PubMed] [Google Scholar]
- 6.Waterman-Storer C M, Salmon E D. Curr Op Cell Biol. 1999;11:61–67. doi: 10.1016/s0955-0674(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad F J, Hughey J, Wittmann T, Hyman A, Greaser M, Baas P W. Nat Cell Biol. 2000;2:276–280. doi: 10.1038/35010544. [DOI] [PubMed] [Google Scholar]
- 8.Yvon A C, Wadsworth P. J Cell Biol. 2000;151:1003–1012. doi: 10.1083/jcb.151.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quintyne N J, Gill S R, Eckley D M, Crego C L, Compton D A, Schroer T A. J Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karki S, Holzbaur E L. Curr Opin Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 11.Yvon A C, Wadsworth P. J Cell Sci. 1997;110:2391–2401. doi: 10.1242/jcs.110.19.2391. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari M B, Ribbeck K, Hagler D J, Spitzer N C. J Cell Biol. 1998;141:1349–1356. doi: 10.1083/jcb.141.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp B, Pearson R B, Guerriero V, Bagchi I C, Means A R. J Biol Chem. 1987;262:2542–2548. [PubMed] [Google Scholar]
- 14.Waterman-Storer C M, Sanger J W, Sanger J M. Cell Motil Cytoskeleton. 1993;26:19–39. doi: 10.1002/cm.970260104. [DOI] [PubMed] [Google Scholar]
- 15.Rusan N M, Fagerstrom C J, Yvon A M, Wadsworth P. Mol Biol Cell. 2001;12:971–980. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchison T J. J Cell Biol. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heidemann S R, Kaech S, Buxbaum R E, Matus A. J Cell Biol. 1997;145:109–122. doi: 10.1083/jcb.145.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasquez R J, Howell B, Yvon A C, Wadsworth P, Cassimeris L. Mol Biol Cell. 1996;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yvon A C, Wadsworth P, Jordan M A. Mol Biol Cell. 1998;10:947–959. doi: 10.1091/mbc.10.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawin K E, Mitchison T J. J Cell Biol. 1991;112:941–954. doi: 10.1083/jcb.112.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer T U, Kapoor T M, Haggarty S J, King R W, Schreiber S L, Mitchison T J. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 22.Mountain V, Simerly C, Howard L, Ando A, Schatten G, Compton D A. J Cell Biol. 1999;147:351–365. doi: 10.1083/jcb.147.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodionov V I, Gyoeva F K, Tanaka E, Bershadsky A D, Vasiliev J M, Gelfand V I. J Cell Biol. 1993;123:1811–1820. doi: 10.1083/jcb.123.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Compton D A. J Cell Sci. 1998;111:1477–1481. doi: 10.1242/jcs.111.11.1477. [DOI] [PubMed] [Google Scholar]
- 25.Burkhardt J K, Echeverri C J, Nilsson T, Vallee R B. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh M, Ishikawa T, Matsushima S, Naka M, Hidaka H. J Biol Chem. 1987;262:7796–7801. [PubMed] [Google Scholar]
- 27.Cramer L P, Mitchison T J. J Cell Biol. 1995;131:179–189. doi: 10.1083/jcb.131.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spector I, Sochet N R, Kashman Y, Groweiss A. Science. 1983;241:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 29.Silverman-Gavrila R V, Forer A. J Cell Sci. 2000;113:597–609. doi: 10.1242/jcs.113.4.597. [DOI] [PubMed] [Google Scholar]
- 30.Odde D J, Ma L, Briggs A H, DeMarco A, Kirschner M W. J Cell Sci. 1999;112:3283–3388. doi: 10.1242/jcs.112.19.3283. [DOI] [PubMed] [Google Scholar]
- 31.Waterman-Storer C M, Karki S, Holzbaur E L. Proc Natl Acad Sci. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houseweart M K, Cleveland D W. Curr Biol. 1999;9:R864–R866. doi: 10.1016/s0960-9822(00)80048-2. [DOI] [PubMed] [Google Scholar]
- 33.Waterman-Storer C, Duey D Y, Weber K L, Keech J, Cheney R E, Salmon E D, Bement W M. J Cell Biol. 2000;150:361–376. doi: 10.1083/jcb.150.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]