Abstract
A decrease in genomic methylation commonly occurs in aging cells; however, whether this epigenetic modification leads to age-related phenotypes has not been evaluated. Alu elements are the major interspersed repetitive DNA elements in humans that lose DNA methylation in aging individuals. Alu demethylation in blood cells starts at approximately 40 years of age, and the degree of Alu hypomethylation increases with age. Bone mass is lost with aging, particularly in menopausal women with lower body mass. Consequently, osteoporosis is commonly found in thin postmenopausal women. Here, we correlated the Alu methylation level of blood cells with bone density in 323 postmenopausal women. Alu hypomethylation was associated with advanced age and lower bone mass density, (P<0.05). The association between the Alu methylation level and bone mass was independent of age, body mass, and body fat, with an odds ratio [1] = 0.4316 (0.2087–0.8927). Individuals of the same age with osteopenia, osteoporosis, and a high body mass index have lower Alu methylation levels (P = 0.0005, 0.003, and ≤0.0001, respectively). Finally, when comparing individuals with the same age and body mass, Alu hypomethylation was observed in individuals with lower bone mass (P<0.0001). In conclusion, there are positive correlations between Alu hypomethylation in blood cells and several age-related phenotypes in bone and body fat. Therefore, reduced global methylation may play a role in the systemic senescence process. Further evaluation of Alu hypomethylation may clarify the epigenetic regulation of osteoporosis in post-menopausal women.
Introduction
Global hypomethylation, the reduction of genome methylation at interspersed repetitive sequences, is an epigenomic alteration that occurs with aging and human diseases, such as cancer and autoimmune diseases. The global hypomethylation pattern in aging is distinct from that observed in some diseases. Methylation levels of Alu elements and long interspersed element-1 s (LINE-1 s) have been evaluated in cancer, autoimmune disease, and aging. Alu and LINE-1 hypomethylation are commonly reported in cancer [2]–[4].(Kitkumthorn N, 2011 #2) Hypomethylation of LINE-1 but not Alu has been reported in systemic lupus erythematosus patients [5]. By contrast, hypomethylation of Alu but not LINE-1 was reported as a global hypomethylation event in aging cells [6]. In this study, we evaluated the association of Alu hypomethylation with age-related phenotypes, focusing on osteoporosis.
Osteoporosis is a common age-related disease. Among individuals older than 50 years of age, half of all women and a quarter of all men break a bone due to osteoporosis [7], [8]. This is most notable in postmenopausal women. The World Health Organization (WHO) characterizes osteoporosis by low bone mass and micro architectural deterioration of bone tissue, which results in an excessively fragile skeleton that is susceptible to fractures [9]–[11]. The most common clinical outcomes are fractures of the spine, hip, and wrist [11]. The prevalence of femoral neck and lumbar spine osteoporosis in 40–80-year-old Thai women in 2000–2001 was 13.6% and 19.8%, respectively [12], [13]. Skeletal size and density develop from early embryogenesis through the intrauterine period, infancy, childhood, and adult life [10]. Genetics and environmental conditions, such as nutrition (calcium and vitamin D), hormones, and lifestyle components, during these all periods contribute to disease burden by impacting low bone mass density (BMD) and modulating on the determination of osteoporotic fractures besides BMD [11], [14]–[16]. BMD is under strong genetic control [11]. Some specific genes involved in all periods of bone development have been shown to be involved in osteoporosis, and the impacts of these genes on osteoporosis are likely to be different among populations with different ethnic backgrounds [10]. Single nucleotide polymorphisms (SNPs) and copy number variations (CNVs) are responsible for a fraction of the genetic component of osteoporosis [10]. Gene-environment interactions are involved in fetal bone development and may cause osteoporosis later in life [1], [10], [17]–[19]. Lifestyle factors associated with low BMD and osteoporosis include smoking, alcohol intake, low calcium intake, vitamin D insufficiency, delayed menarche, low body weight, and physical inactivity [11].
DNA methylation is a possible mechanism of epigenetic regulation of bone development and osteoporosis and plays a significant role in osteogenic cell differentiation and bone metabolism [20]–[22]. Alu is the most abundant of the short-interspersed nuclear elements (SINE), with over one million copies per genome. Alu elements comprise approximately 11% of the human genome and contain 30% of its methylation sites [23], [24]. Methylation of repetitive elements such as Alu is correlated with the total genomic methylation content [23]–[25]. Recently, we demonstrated that a direct correlation between age and a reduction of Alu methylation levels becomes apparent at 34–68 years and that Alu methylation is progressively lost with increased age [6]. Loss of global methylation is associated with genomic instability [26]. Genomic instability is also proposed to be related to the aging process [27]. In addition to overall methylation levels, we recently reported the methylation statuses of LINE-1 loci by classifying the LINE-1 loci into 4 classes: hypermethylated, hypomethylated, and 2 types of partially methylated loci [28]–[31]. We observed that in the presence of certain conditions (smoking), changes in the LINE-1 methylation pattern occurred while the overall LINE-1 methylation level was not significantly changed [30]. Moreover, in some conditions, such as cancer, hypomethylated loci were more sensitive and specific than overall methylation levels [28], [29], [31]. Because osteoporosis is a disease of aging and epigenetic changes are related to age, we were interested in correlating age-related epigenetic variation with bone density. Our aim was to examine whether methylation of Alu is associated with osteoporosis.
Results
Age-related phenotypes in correlation with Alu methylation level
In this study, in addition to overall methylation levels, we also classified Alu loci based on the methylation statuses of 2 CpG dinucleotides as hypermethylated loci, mCmC, hypomethylated loci, uCuC, and 2 types of partially methylated, mCuC and uCmC (Fig. S1). The mean and median characteristics of 323 postmenopausal women are described in Table 1 and Table 2. There was a negative correlation between Alu methylation level (%mC) and age and Alu methylation level (%mC) and waist/hip (W/H) ratio and a positive correlation between %mC and the bone mass density (BMD) of many bone regions (Table 1). Using BMD indicator; age, %uCuC and %uCmC were higher and body mass (weight, body mass index (BMI), W/H ratio and %total body fat), systolic blood pressure (SBP), diastolic blood pressure (DBP), spine BMD, hip BMD, radius BMD, bone total BMD, %mC level and %mCmC loci were lower in osteopenia and osteoporosis than normal (Table S1).
Table 1. Mean and median of characteristic data and the correlation (r) between %mC and age-related phenotypes among all menopausal subjects.
| Characteristics | mean ± SE | median (min-max) | r | N |
| age (yrs) | 57.23±0.39 | 57.00 (36–75) | −0.13* | 323 |
| weight (kg) | 56.74±0.53 | 55.50 (38.1–95) | NS | 323 |
| BMI (kg/m2) | 23.58±0.20 | 22.97 (15.46–39.54) | NS | 321 |
| waist (cm) | 78.70±0.51 | 77.75 (26.0–111) | NS | 318 |
| W/H ratio | 0.82±0.01 | 0.81 (0.68–1.05) | −0.12* | 336 |
| SBP (mmHg) | 124.81±0.77 | 122.00 (100–168) | NS | 253 |
| DBP ((mmHg) | 76.55±0.53 | 80.00 (50–98) | NS | 253 |
| total body fat (%) | 36.63±0.42 | 36.90 (19.7–57.90) | NS | 236 |
| L1 BMD (g/cm2) | 0.93±0.01 | 0.91 (0.42–1.4) | NS | 249 |
| L2 BMD (g/cm2) | 0.98±0.01 | 0.95 (0.5–1.49) | NS | 258 |
| L3 BMD (g/cm2) | 1.06±0.01 | 1.03 (0.69–1.57) | NS | 258 |
| L4 BMD (g/cm2) | 1.05±0.01 | 1.02 (0.71–1.66) | NS | 258 |
| L12 BMD (g/cm2) | 0.96±0.01 | 0.94 (0.64–1.44) | 0.14* | 218 |
| L13 BMD (g/cm2) | 1.02±0.01 | 0.99 (0.68–1.47) | 0.15* | 217 |
| L14 BMD (g/cm2) | 1.02±0.01 | 0.99 (0.70–1.78) | 0.15* | 217 |
| L23 BMD (g/cm2) | 1.03±0.01 | 1.00 (0.69–1.65) | 0.16** | 217 |
| L24 BMD (g/cm2) | 1.04±0.01 | 1.01 (0.7–1.56) | 0.15* | 218 |
| L34 BMD (g/cm2) | 1.06±0.01 | 1.04 (0.64–1.60) | 0.16* | 217 |
| femur neck BMD (g/cm2) | 0.81±0.01 | 0.79 (0.29–1.67) | NS | 252 |
| hip ward BMD (g/cm2) | 0.65±0.01 | 0.64 (0.29–1.24) | 0.14* | 252 |
| femur trochanteric BMD (g/cm2) | 0.69±0.01 | 0.68 (0.41–1.07) | NS | 252 |
| hip total BMD (g/cm2) | 0.88±0.01 | 0.87 (0.23–1.32) | NS | 251 |
| radius ud BMD (g/cm2) | 0.32±0.00 | 0.31 (0.18–0.52) | NS | 284 |
| radius 33 BMD (g/cm2) | 0.62±0.01 | 0.63 (0.32–0.82) | NS | 285 |
| radius total BMD (g/cm2) | 0.49±0.00 | 0.48 (0.26–0.96) | NS | 284 |
| bone total BMD (g/cm2) | 1.08±0.01 | 1.07 (0.82–1.38) | NS | 190 |
L, spine region lumbar; ± SE, standard error; r, correlation coefficient; n, number of cases;
P<0.05,
P<0.001.
Table 2. Mean and median of %mC level, %mCmC loci, %mCuC loci, %uCmC loci, %uCuC loci and %mCuC loci+%uCmC loci among all menopausal subjects.
| Alu methylation | mean ± SE | median (min-max) | N |
| %mC level | 30.13±0.30 | 30.78 (16.71–43.61) | 323 |
| %mCmC loci | 8.58±0.26 | 8.06 (0.32–24.22) | 323 |
| %mCuC loci | 18.00±0.14 | 17.89 (3.76–25.93) | 323 |
| %uCmC loci | 25.29±0.24 | 26.00 (11.76–45.03) | 323 |
| %uCuC loci | 47.48±0.44 | 45.93 (21.28–70.85) | 323 |
| %mCuC loci+%uCmC loci | 43.08±0.27 | 43.20 (15.98–67.53) | 323 |
± SE, standard error; n, number of cases; %mCmC, %hypermethylated loci;
%uCuC, %hypomethylated loci; %uCmC and %mCuC, %partially methylated loci.
Age-adjusted correlation between Alu hypomethylation and osteoporosis
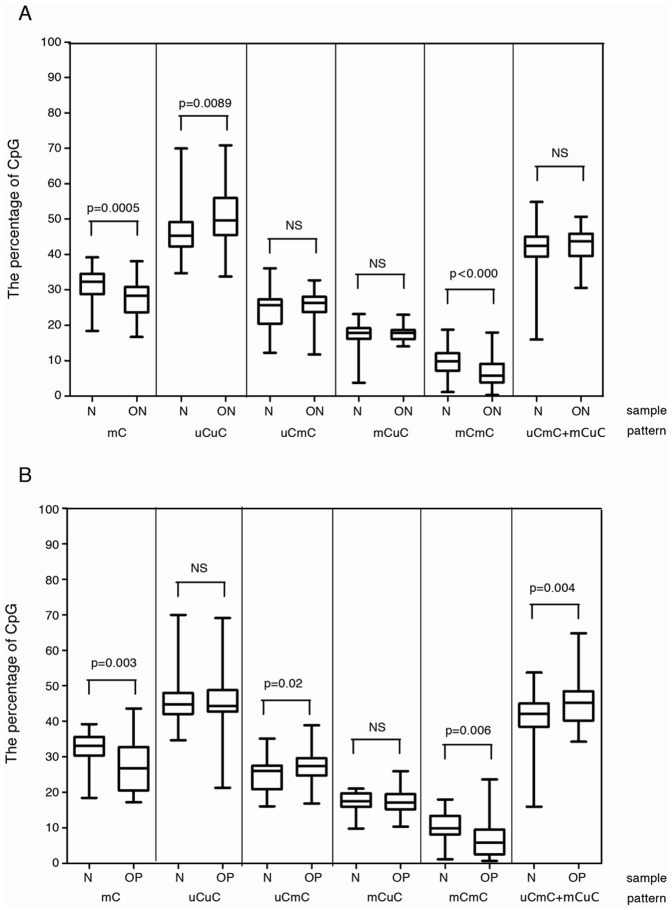
To exclude the influence of age on Alu methylation, we paired each osteopenia and osteoporosis individual with a healthy person of the same age. Individuals with osteopenia or osteoporosis had significantly lower %mC than healthy individuals (P≤0.001 and P≤0.001) (Figure 1A&Figure 1B). The hypomethylation level was correlated with a reduction in %mCmC and an increase in %uCuC (Figure 1A&Figure 1B). These data confirmed the correlation between Alu hypomethylation and lower BMD phenotypes; this correlation was not due to age.
Figure 1. Alu methylation of osteopenia and osteoporosis.
(A) paired age normal (n = 65) and osteopenia (n = 65); (B) paired age normal (n = 35) and osteoporosis (n = 35). N, normal control. ON, osteopenia. OP, osteoporosis. NS, not significant.
Age-adjusted correlation between Alu hypomethylation and body mass
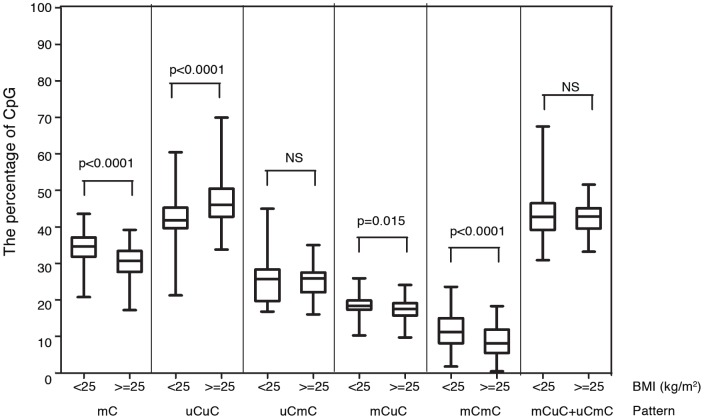
Subjects with lower body mass (weight, BMI, waist circumference (WC), and %body fat) had significantly lower BMD values (Tables S2 and S3). We examined the correlation between Alu methylation and BMI. High (BMI≥25 kg/m2) and low BMI (BMI<25 kg/m2) individuals were matched on age.
Subjects with a high BMI had significantly lower %mC, %mCmC, and %mCuC than subjects with low BMI (P≤0.001, P≤0.001, and P≤0.05, respectively) but had significantly higher %uCuC (P≤0.001) (Figure 2). Therefore, whereas lower BMI is associated with lower BMD, Alu hypomethylation is directly associated with lower BMD and higher BMI.
Figure 2. Alu methylation of osteopenia and osteoporosis.
Paired age between cases with body mass index (BMI)<25 kg/m2 (n = 80) and cases with BMI≥25 kg/m2 (n = 80). NS, not significant.
Age and BMI adjusted correlation between Alu hypomethylation and osteoporosis
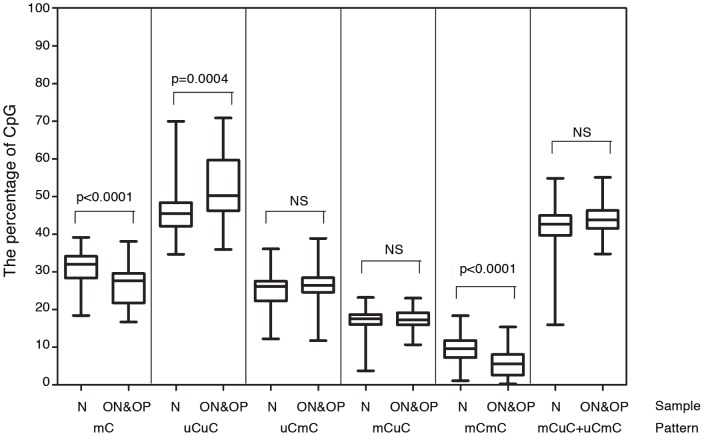
We further analyzed the association between Alu hypomethylation and osteoporosis by excluding the influences of age and BMI. We matched osteopenia and osteoporosis subjects with controls based on age and BMI. We observed that %mC and %mCmC were significantly lower and %uCuC was significantly higher in the osteopenia and osteoporosis subjects than the controls (P<0.0001 and P<0.0001; and P = 0.0004, respectively) (Figure 3).
Figure 3. Alu methylation of osteopenia and osteoporosis.
Normal (n = 52) and combined osteopenia and osteoporosis (n = 63) were paired by age and BMI.
The association between osteoporosis and Alu methylation, age, BMI, and %body fat using logistic regression models
Logistic regression models were used to test the association between several parameters and osteoporosis. Significant associations with age, BMI, %body fat and Alu methylation were confirmed. The %mC (Odds Ratio (OR) = 0.43, P = 0.02), %mCmC (OR = 1.62, P = 0.05), and %uCuC (OR = 0.71, P = 0.04) were considered risk factors for osteoporosis (Table 3). The Alu associations with osteopenia/osteoporosis are independent of age and BMI. These results confirmed the association between Alu hypomethylation in osteoporosis in postmenopausal women.
Table 3. Logistic regression analysis when bone mineral density by DEXA (T-score) cut-offs of normal and osteoporosis was the dependent variable and %mC, %mCmC loci, %uCmC loci, %mCuC loci, %uCuC loci, age, BMI, and %body fat were independent variables.
| Independent variables | Odds ratio | P-value | 95% confidence interval |
| %mC | 0.4316 | 0.02 | 0.2087–0.8927 |
| %mCmC loci | 1.6172 | 0.05 | 0.9988–2.6186 |
| %uCmC loci | 1.1121 | NS | 0.9353–1.3223 |
| %mCuC loci | 1.0887 | NS | 0.8325–1.4236 |
| %uCuC loci | 0.7115 | 0.04 | 0.5124–0.9879 |
| age | 1.2589 | 0.000 | 1.1486–1.3797 |
| BMI | 0.7030 | 0.000 | 0.5922–0.8345 |
| %body fat | 0.9998 | 0.000 | 0.9997–0.9999 |
Different levels of significance at P≤0.05.
Bisulfite pyrosequencing analysis
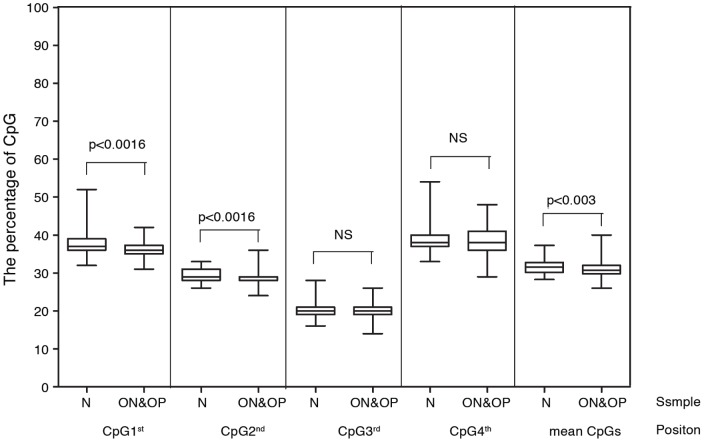
To study CpG methylation, pyrosequencing analysis was performed on 61 normal, 54 osteopenia, and 31 osteoporosis subjects. The Alu amplicon contains four CpGs, which permits variation in the percentages of methylation (Table S4). Only the 1st and 2nd positions demonstrated lower methylation levels in combined osteopenia and osteoporosis cases (P = 0.0016 and P = 0.0016, respectively) (Figure 4). Overall methylation levels were also lower (P = 0.03) (Figure 4).
Figure 4. Alu methylation levels by pyrosequencing.
4 CpG and combined methylation levels are observed. N, normal control. ON, osteopenia. OP, osteoporosis.
Discussion
Osteoporosis is a common age-related disease that causes low bone mass and micro architectural deterioration of bone tissue, resulting in a fragile skeleton. In this study, we observed lower Alu methylation in peripheral blood cells in subjects with decreased bone mass, increased age, and increased BMI. The data supported that lower Alu methylation levels are directly correlated with aging. This study demonstrates an association but not necessarily causation. Elderly individuals are likely to have lower bone density and consequently develop osteoporosis [32]. They also tend to have higher BMIs [32]. However, individuals with a high BMI are less likely to have osteoporosis because the skeleton responds to mechanical stress, such as body weight, with a stimulation of osteoblast activity and secretion of the bone-protective hormone estrogen from fat tissue [32], [33]. Here, we demonstrated an association between Alu hypomethylation and 2 aging phenotypes (osteoporosis and high BMI), even though the 2 aging phenotypes are inversely correlated.
Analysis of the Alu methylation patterns revealed decreased Alu methylation level, increased hypomethylation loci, decreased hypermethylation loci, and unchanged partial methylation loci. This finding supports the hypothesis that DNA methylation changes in aging may occur in one direction from hypermethylation to hypomethylation or hypermethylation to partial methylation to hypomethylation. This result is in contrast with our previous reports on LINE-1 methylation pattern changes caused by smoking (which are bidirectional) [30].
The mechanism responsible for Alu hypomethylation in the aging process remains unknown. However, our observation suggests that the epigenomic changes may be systemic. While we observed Alu hypomethylation in blood cells in subjects with osteoporosis and increased BMI are phenotypes of bone and fat cells, respectively. Further evaluation of Alu hypomethylation in other cell types will facilitate the elucidation of the mechanisms involved in Alu hypomethylation. Methylation variation can be induced by adverse environmental conditions such as age, gender, race/ethnicity, hormone imbalance, nutritional imbalance, oxidative stress, obese and lifestyle choices [1], [4], [25], [34]–[37]. Only two cellular environments that cause genome-wide hypomethylation have been characterized: malnutrition and increased oxidative stress. Dietary methyl donors such as folic acid, choline, betaine, zinc, vitamin B12, vitamin B6, and alcohol are precursors of DNA methylation.33 In a previous study, methylation donor supplementation resulted in an increase in the number of methylation-variable loci distributed throughout the genome [34]. The precise mechanism for the reduction of genomic methylation by oxidative stress has not yet been described. The generation of hydroxyl radicals causes a wide range of DNA lesions, including base modifications, deletions, strand breakages, and chromosomal rearrangements, that interfere with the ability of DNA to function as a substrate for DNA methyl transferases (DNMTs), resulting in global hypomethylation [38]. ROS production is associated with increased DNA damage and chromosomal degradation, with changes in both hypermethylation and hypomethylation of the DNA [38]. However, an association between global hypomethylation and oxidative stress was recently reported [31], [38], [39]. Interestingly, an association between oxidative stress and osteoporosis as well as age has also been reported [40]–[42]. Therefore, it is reasonable to further explore the connection among aging phenotypes, oxidative stress, and Alu methylation levels.
The significance of Alu hypomethylation in the promotion of aging phenotypes is still unknown. Unlike LINE-1 s, which is important in the regulation of gene expression [43], the role of Alu methylation in controlling gene expression remains to be fully characterized. There are several indications that Alu hypomethylation influences the aging process via genomic instability. First, the loss of DNA methylation in the mouse or human genome leads to genomic instability or increases in mutation rates; by contrast, genomic instability caused by, for example, oxidative stress can lead to global hypomethylation [44]. Second, we reported that histone acetylation, which is associated with DNA hypomethylation [45], spontaneously reduces delayed repair of replication-independent DNA double-stranded breaks and increases γ-H2AX bound DNA [46]. Finally, endogenous γ-H2AX foci are reported to be associated with age-related diseases [47]. Therefore, genomic instability may be one of the underlying mechanisms for the promotion of age-related phenotypes by Alu hypomethylation.
In conclusion, we demonstrated that Alu hypomethylation is associated with aging-associated phenotypes and diseases, increased BMI, decreased bone mass, and osteoporosis. The age-related Alu hypomethylation may be systemic, and Alu methylation levels can be an indicator of age-related phenotypes. Understanding the underlying mechanisms of the correlation between Alu hypomethylation and age-related phenotypes may clarify the roles of epigenetic modifications in age-related disabilities and diseases.
Materials and Methods
Subjects
The present analysis is based on 323 menopausal women enrolled in the Menopausal Clinic, Outpatient Department, General Practice Section of the Department of Obstetrics and Gynecology, Ramathibodi Hospital, Bangkok. Subjects were recruited according to the following criteria: each individual 1) agreed to participate and 2) had no history of chronic and/or genetic diseases and/or disorders of bone metabolism. Bone mass density (BMD), which is used as an indicator for osteoporosis diagnosis, was measured for all subjects. The DEXA T-score method of BMD was used: >−1 SD was diagnosed normal, −1 to −2.5 SD was diagnosed with osteopenia, and <−2.5 SD was diagnosed with at least one site of osteoporosis. BMD (g/cm2) was assessed at the total body, lumbar spine sites, femur sites, and radius sites, using dual-energy x-ray absorptiometry (Lunar Prodigy®, GE Healthcare, WI, USA). All menopausal women included in this analysis agreed to an assessment of socio-demographic variables and lifestyle risk factors such as smoking and alcohol consumption. Height, weight, and waist circumference (WC) measurements were obtained using standard protocols. The BMI was calculated using the height and weight measurements (weight/[height m2]). Percentage of body fat (BF) was measured with an OMRON-bioelectrical impedance analysis instrument.
Blood collection and DNA preparation for COBRA Alu
Heparinized blood was collected, and DNA was extracted from whole blood using QiAmp DNA blood kits (Qiagen, Hilden, Germany). A total of 200 ng DNA was used for bisulfite treatment. The combined bisulfite restriction analysis (COBRA) consisted of a standard sodium bisulfite PCR treatment followed by restriction digestion and quantitation. Bisulfite modification of genomic DNA was performed using previously published methods [48]. In brief, 200 ng DNA was dissolved in 50 µl distilled water and then denatured in 5.5 µl 2 M NaOH for 10–30 min at 37°C. Next, 30 µl freshly prepared 10 mM hydroquinone and 520 µl 3 M bisulfite (pH 5.0) was added and mixed. The samples were incubated at 50°C for 16 h. The bisulfite-treated DNA was isolated with the Wizard DNA Clean up System. The DNA was eluted with 50 µl of warm water and desulfonated with 5.5 µl of 3 M NaOH for 5 min. The DNA was precipitated (NH4OAC-EtOH) using glycogen as a carrier and resuspended in 20 µl water. Bisulfite-treated DNA was stored at −20°C until use.
COBRA Alu
COBRA is a standard technique for measuring methylation levels of Alu [6], [49]. The primer sequences corresponding to the nucleotides in the regulatory region of the Alu sequence (the Alu Sx subfamily) [23] were F, 5′-GGT GGT TTA MGT TTG TAA TTT TAG TAT TT-3′ and R, 5′-ATT TCA CCA TAT TAA CCA AAC TAA TC-3′. The PCR reactions were performed with the following conditions: 35 cycles of 95°C for 45 sec, 63°C for 45 sec, and 72°C for 45 sec. The PCR products were subsequently digested with 2 U TaqI (MBI Fermentas) in TE buffer 3 (Biolab) at 65°C overnight, followed by separation on an 8% nondenaturing polyacrylamide gel. The gel was stained with SyBr Green, and the band intensities were measured with a PhosphoImager using Image Quant software (Molecular Dynamics).
Genomic Alu methylation distribution pattern and methylation analysis
A schematic representation and example of the COBRA of the Alu distribution patterns is shown in Figure 1. For COBRA Alu, the Alu loci of each individual were divided into 4 groups depending on the methylation status of the 2 CpG sites as follows: 1) hypermethylated loci: mCmC, complete methylation at both CpG sites, yielded three fragments of 37, 32, and 29 bp; 2) hypomethylated loci: uCuC, fully unmethylated sequence, yielded 1 uncut amplicon with a size of 98 bp; and 3) and 4) partially methylated loci: uCmC and mCuC. The unmethylated 37 bp CpG and methylated 69 bp CpG sequence yielded 2 fragments of 69 and 29 bp. By contrast, the methylated 37 bp CpG and unmethylated 69 bp CpG sequence yielded 2 fragments of 37 and 61 bp (Figure S1A). To improve and expand the applications of Alu methylation, we determined the Alu methylation profile using the Alu methylation distribution pattern combined with the percentage of methylation. Each product was analyzed as a percentage of product intensity divided by the double strand length. The % number of hypermethylated loci (%mCmC) was %32/30, of hypomethylated loci (%uCuC) was %98/98, of partially methylated loci (%uCmC) was %69/68, and of partially methylated loci (%mCuC) was %61/60.
Bisulfite pyrosequencing Alu analysis
For the pyrosequencing Alu analysis, bisulfite modification of genomic DNA was prepared using the previously mentioned COBRA method. Alu methylation was quantitated by pyrosequencing with the following primers and conditions: 20 µM forward primer 5′-GGT GGT TTA MGT TTG TAA TTT TAG TAT TT-3′ and 20 µM reverse biotinylated primer 5′-ATT TCA CCA TAT TAA CCA AAC TAA TC-3′. The purified, single-stranded biotinylated PCR products acted as a template for annealing of the first pyrosequencing primer, 5′GTTTGTAATTTTAGTATTTTGG-3′, to detect CpG in the first 3 positions of the Alu amplicon and of the second pyrosequencing primer, 5′GATTATTTGAGGTTAGGAGT-3′, to detect CpG in the last 4 positions of the Alu amplicon (0.3 µM final concentration). The PCR products were then subjected to sequencing using an automatically generated nucleotide dispensation order for the sequences to be analyzed. The Pyrograms were analyzed using CpG mode to determine the proportion of C/T and thus the proportion of methylated and unmethylated cytosines at the targeted positions. The degree of methylation was evaluated at 4 CpG methylation sites.
Statistical analysis
Data were analyzed with SPSS statistical software version 11.5. The averages and distributions of the characteristic data of menopausal women are shown as the mean±SD and median. Pearson's correlation between the Alu methylation pattern and the characteristic data of the population were used to determine 95% confidence intervals of association. The DEXA T-score was used to divide subjects into 3 groups: normal, osteopenia, and osteoporosis; and the average of the characteristic data of each group are shown as the mean±SD. A T-test was used to determine the differences at a P-value≤0.05 between the groups in the matched cases based on age. A T-test was used to determine the differences of BMD at each site, and Alu methylation pattern at a P-value≤0.05 between the groups in the matched cases divided by indicator BMI (≥25 vs <25 kg/m2). Logistic regression models were used to evaluate the association between indicator BMD by DEXA T-score as a dependent variable specified as a binary outcome, normal and osteoporosis, and %mC, %mCmC loci, %uCmC loci, %mCuC loci, %uCuC loci, age, BMI, and %body fat as independent predictors. All the independent variables entered into the model at the same time.
Ethics statement
The study protocol and procedures were approved by the Ethical Committee of the Faculty of Tropical Medicine, Mahidol University and the faculty of Medicine, Ramathibodi Hospital. All study participants gave written informed consent.
Supporting Information
Combined bisulfite restriction analysis: Alu sequence of sodium bisulfite treated whole blood DNA of menopausal subjects. (A) Alu methylation pattern by COBRA PCR. The Alu amplicons were 98 bp and contained 2 CpG dinucleotides. After digestion with the enzyme Taq1, which targets the methylated cytosine site, the Alu sequences were divided into 4 product fragments depending on the methylation status of the 2 CpG sites as follows: 1) hypermethylated loci: mCmC; 2) hypomethylated loci: uCuC; and 3) and 4) partially methylated loci: uCmC and mCuC. (B) COBRA Alu in menopausal subjects DNA treated Taq 1 enzyme, Alu sequences with complete methylation at both CpG sites yielded 3 fragments of 37, 32, 29 bp, while the fully unmethylated sequence yielded 1 fragment, 98 bp uncut amplicon. For the partially methylated sequences, unmethylated 37 bp CpG and methylated 69 bp CpG yielded 2 fragments, 69 and 29 bp. By contrast, methylated 37 bp CpG and unmethylated 69 bp CpG yielded 2 fragments, 37 and 61 bp.
(RAR)
Mean and median of age, BMI, waist, SBP, DBP, %total 1 body fat, BMD and Alu methylation pattern among normal, osteopenia, and osteoporosis diagnosed by DEXA T-score (>−1.0 SD = normal, −1.0 to −2.5 SD = osteopenia and <−2.5 SD = osteoporosis).
(PDF)
Mean difference (X ± SE) of weight, BMI, waist, systolic 1 blood pressure (SBP), diastolic blood pressure (DBP), and %total body fat between normal and osteopenia and between normal and osteoporosis as diagnosed by DEXA T-score (>−1.0 SD = normal, −1.0 to −2.5 SD = osteopenia and <−2.5 SD = osteoporosis) in the matched cases. Normal and osteopenia and normal and osteoporosis were paired by age.
(PDF)
Mean difference (X ± SE) of bone mass density (BMD) at each 1 region: L1, L2, L3, L4, L1–2, L1–3, L1–4, L2–3, L2–4, L3–4, femur neck, hip ward, femur trochanteric, hip total, radius ud, radius 33, radius total, and bone total between matched cases with body mass index (BMI)<25 kg/m2 and BMI>25 kg/m2. Paired age between cases with body mass index (BMI)<25 kg/m2 and cases with BMI>25 kg/m2.
(PDF)
The percentages of Alu methylation levels in each position 1 of CpG and mean of all CpG in normal, osteopenia, and osteoporosis cases.
(PDF)
Acknowledgments
We thank all personnel in the Department of Tropical Nutrition and Food Science, the Faculty of Tropical Medicine, Mahidol University and the Center for Excellence in Molecular Genetics of Cancer and Human Diseases, Department of Anatomy, Faculty of Medicine, Chulalongkorn University; Dr. Chaowanee Chupeerat for sample collection; and Mr. Supalarp Puangsa-art for suggestion about logistic regression analysis.
Funding Statement
Pornrutsami Jintaridth is supported by Thailand Research Fund Grant MRG 5480131 and the Faculty of Tropical Medicine Grants 2011 and 2012. Apiwat Mutirangura is supported by the Research Chair Grant 2011, the National Science and Technology Development Agency, Thailand.
References
- 1. Antoniades L, MacGregor AJ, Andrew T, Spector TD (2003) Association of birth weight with osteoporosis and osteoarthritis in adult twins. 42: 791–796. [DOI] [PubMed] [Google Scholar]
- 2. Kitkumthorn N, Keelawat S, Rattanatanyong P, Mutirangura A (2012) LINE-1 and Alu methylation patterns in lymph node metastases of head and neck cancers. Asian Pac J Cancer Prev 13: 4469–4475. [DOI] [PubMed] [Google Scholar]
- 3. Kitkumthorn N, Mutirangura A (2011) Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenetics 2: 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christensen BC, Houseman EA, Marcit CJ, Zheng S, Wrensch MR, et al. (2009) Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PloS Genetics 5: e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakkuntod J, Avihingsanon Y, Mutirangura A, Hirankan N (2011) Hypomethylation of LINE-1 but not Alu in lymphocyte subjects of systemic lupus erythematosus patients. Clin Chim Acta 15: 1457–1461. [DOI] [PubMed] [Google Scholar]
- 6. Jintaridth P, Mutirangura A (2010) Distinctive patterns of age-dependent hypomethylation in interspersed repetitive sequences. Physiol Genomics 41: 194–200. [DOI] [PubMed] [Google Scholar]
- 7. De Laet CE, van Hout BA, Burger H, Hofman A, Pols HA (1997) Bone density and risk of hip fracture in men and women: cross sectional analysis. Br Med J 315: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang QY, Kung AW (2006) Genetics of osteoporosis. Mol Genet Metab 88: 295–306. [DOI] [PubMed] [Google Scholar]
- 9. Bocheva G, Boyadjieva N (2011) Epigenetic regulation of fetal bone development and placental transfer of nutrients: progress for osteoporosis. Interdiscip Toxicol 4: 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey N, Dennison E, Cooper C (2008) Epidemiology of osteoporotic fracture. In: MJ F, editor. In: Primer on the metabolic diseases and disorders of mineral metabolism 7th ed. ASMBR, Washington. pp. 198–203.
- 11. Kung AW, Huang QY (2007) Genetic and environmental determinants of osteoporosis. J Musculoskelet Neuronal Interact 7: 26–32. [PubMed] [Google Scholar]
- 12. Pongchaiyakul C, Songpattanasilp T, Taechakraichana N (2008) Burden of osteoporosis in Thailand. J Med Assoc Thai 91: 261–267. [PubMed] [Google Scholar]
- 13. Ongphiphadhanakul B (2002) Osteoporosis in Thailand. Clin calcium 12: 822–826. [PubMed] [Google Scholar]
- 14. Espallargues M, Sampietro-Colomb L, Estrada MD, Sola M, Del Rio L, et al. (2001) Identifying bone-mass-related risk factors for fracture to guide bone densitometry measurements: a systematic review of the literature. Osteoporosis Int 12: 811–822. [DOI] [PubMed] [Google Scholar]
- 15. Tromp AM, Ooms ME, Popp-Snijders C, Roos JC, Lips P (2000) Predictors of fractures in elderly women. Osteoporosis Int 11: 134–140. [DOI] [PubMed] [Google Scholar]
- 16. Cummings SR, Nevitt MC, Browner WS, Stone K, Fox KM, et al. (1995) Risk factors for hip fracture in white women. New Engl J Med 332: 767–773. [DOI] [PubMed] [Google Scholar]
- 17. Wong CC, Caspi A, Williams B, Craig IW, Houts R, et al. (2010) A longitudinal study of epigenetic variation in twins. Epigenetics 5: 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burwell RG, Dangerfield PH, Moulton A, Grivas TB (2011) Adolescent idiopathic scoliosis (AIS), environment, exposome and epigenetics: a molecular perspective of postnatal normal spinal growth and the etiopathogenesis of AIS with consideration of a network approach and possible implications for medical therapy. Scoliosis [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Javaid MK, Godfrey KM, Taylor P, Shore SR, Breier B, et al. (2004) Umbilical venous IGF-1 concentration, neonatal bone mass, and body composition. J Bone Miner Res 19: 56–63. [DOI] [PubMed] [Google Scholar]
- 20. Kitazawa R, Kitazawa S (2007) Methylation status of a single CpG locus 3 bases upstream of TATA-Box of receptor activator of nuclear factor-κB ligand (RANKL) gene promoter modulates cell- and tissue-specific RANKL expression and osteoclastogenesis. Molecular Endocrinology 21: 148–158. [DOI] [PubMed] [Google Scholar]
- 21. Villagra A, Gutiérrez J, Paredes R, Sierra J, Puchi M, et al. (2002) Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J Cell Biochem 85: 112–122. [PubMed] [Google Scholar]
- 22. Lambertini E, Penolazzi L, Sollazzo V, Pezzetti F, de Mattei M, et al. (2002) Modulation of gene expression in human osteoblasts by targeting a distal promoter region of human estrogen receptor-alpha gene. J Endocrinol 172: 683–693. [DOI] [PubMed] [Google Scholar]
- 23. Batzer MA, Deininger PL (1999) Alu repeats and human genomic diversity. Nat Rev 3: 370–380. [DOI] [PubMed] [Google Scholar]
- 24. Deininger PL, Moran JV, Batzer MA, Kazazian JHH (2003) Mobile elements and mammalian genome evolution. Curr Opin Genet Dev 13: 651–658. [DOI] [PubMed] [Google Scholar]
- 25. Subramanyam MA, Diez-Roux AV, Richard Pilsner J, Villamor E, Donohue KM, et al. (2013) Social Factors and leukocyte DNA methylation of repetitive sequences: The multi-ethnic study of atherosclerosis. PLoS One 8: e54018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R (1998) DNA hypomethylation leads to elevated mutation rates. Nature 3: 89–93. [DOI] [PubMed] [Google Scholar]
- 27. Vijg J, Suh Y (2013) Genome instability and aging. Annu Rev Physiol 10: 645–668. [DOI] [PubMed] [Google Scholar]
- 28. Pobsook T, Subbalekha K, Sannikorn P, Mutirangura A (2011) Improved measurement of LINE-1 sequence methylation for cancer detection. Clin Chim Acta 30: 314–321. [DOI] [PubMed] [Google Scholar]
- 29. Kitkumthorn N, Tuangsintanakul T, Rattanatanyong P, Tiwawech D, Mutirangura A (2012) LINE-1 methylation in the peripheral blood mononuclear cells of cancer patients. Clin Chim Acta 18: 869–874. [DOI] [PubMed] [Google Scholar]
- 30. Wangsri S, Subbalekha K, Kitkumthorn N, Mutirangura A (2012) Patterns and possible roles of LINE-1 methylation changes in smoke-exposed epithelia. PLoS One 7: e45292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patchsung M, Boonla C, Amnattrakul P, Dissayabutra T, Mutirangura A, et al. (2012) Long interspersed nuclear element-1 hypomethylation and oxidative stress: correlation and bladder cancer diagnostic potential. PLoS One 7: e37009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wardlaw GM (1996) Putting body weight and osteoporosis into perspective. Am J Clin Nutr 63 Suppl: 433S–436S. [DOI] [PubMed] [Google Scholar]
- 33. Ribot C, Pouilles JM, Bonneu M, Tremollieres F (1992) Assessment of the risk of post menopausal osteoporosis using clinical factors. Clin Endoclinol 36: 225–228. [DOI] [PubMed] [Google Scholar]
- 34. Li CCY, Cropley JE, Cowley MJ, Preiss T, Martin DIK, et al. (2011) A sustained dietary change increases epigenetic variation in isogenic mice. PloS Genetics 7: e1001380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Day JK, Bauer AM, desBordes C, Zhuang Y, Kim B, et al. (2002) Genistein alters methylation patterns in mice. J Nutr 132: 2419S–2423S. [DOI] [PubMed] [Google Scholar]
- 36. Piyathilake CJ, Badiga S, Alvarez RD, Partridge EE, Johanning GL (2013) Lower degree of PBMC L1 methylation is associated with excess body weight and higher HOMA-IR in the presence of lower concentrations of plasma folate. Plos One 8: e54544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Intarasunanont P, Navasumrit P, Waraprasit S, Chaisatra K, Suk WA, et al. (2012) Effects of arsenic exposure on DNA methylation in cord blood samples from newborn babies and in a human lymphoblast cell line. Environmental Health 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Donkena KV, Young CYF, Tindall DJ (2010) Oxidative stress and DNA methylation in prostate cancer. Obstetrics and Gynecology International [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Potocki L, Lewinska A, Klukowska-Rötzler J, Bugno-Poniewierska M, Koch C, et al. (2012) DNA hypomethylation and oxidative stress-mediated increase in genomic instability in equine sarcoid-derived fibroblasts. Biochimie 94: 2013–2024. [DOI] [PubMed] [Google Scholar]
- 40. Sánchez-Rodríguez MA, Ruiz-Ramos M, Correa-Muñoz E, Mendoza-Núñez VM (2007) Oxidative stress as a risk factor for osteoporosis in elderly Mexicans as characterized by antioxidant enzymes. BMC Musculoskeletal Disorders 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baek KH, Oh KW, Lee WY, Lee SS, Kim MK, et al. (2010) Association of oxidative stress with postmenopausal osteoporosis and the effects of hydrogen peroxide on osteoclast formation in human bone marrow cell cultures. Calcif Tissue Int 87: 226–235. [DOI] [PubMed] [Google Scholar]
- 42. Manolagas SC (2010) From Estrogen-Centric to Aging and Oxidative Stress: A Revised Perspective of the Pathogenesis of Osteoporosis. Endocrine Reviews 31: 266–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aporntewan C, Phokaew C, Piriyapongsa J, Ngamphiw C, Ittiwut C, et al. (2011) Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One 6: e17934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R (1998) DNA hypomethylation leads to elevated mutation rates. Nature 395: 89–93. [DOI] [PubMed] [Google Scholar]
- 45. Eden S, Hashimshony T, Keshet I, Cedar H, Thorne AW (1998) DNA methylation models histone acetylation. Nature 394: 842. [DOI] [PubMed] [Google Scholar]
- 46. Kongruttanachok N, Phuangphairoj C, Thongnak A, Ponyeam W, Rattanatanyong P, et al. (2010) Replication independent DNA double-strand break retention may prevent genomic instability. Mol Cancer 9 doi:10.1186/1476-4598-1189-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schurman SH, Dunn CA, Greaves R, Yu B, Ferrucci L, et al. (2012) Age-related disease association of endogenous γ-H2AX foci in mononuclear cells derived from leukapheresis. PLoS One 7: e45728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chalichagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, et al. (2004) Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 23: 8841–8846. [DOI] [PubMed] [Google Scholar]
- 49. Cho NY, Kin BH, Choi M, Yoo EJ, Moon KC, et al. (2007) Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol 211: 269–277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Combined bisulfite restriction analysis: Alu sequence of sodium bisulfite treated whole blood DNA of menopausal subjects. (A) Alu methylation pattern by COBRA PCR. The Alu amplicons were 98 bp and contained 2 CpG dinucleotides. After digestion with the enzyme Taq1, which targets the methylated cytosine site, the Alu sequences were divided into 4 product fragments depending on the methylation status of the 2 CpG sites as follows: 1) hypermethylated loci: mCmC; 2) hypomethylated loci: uCuC; and 3) and 4) partially methylated loci: uCmC and mCuC. (B) COBRA Alu in menopausal subjects DNA treated Taq 1 enzyme, Alu sequences with complete methylation at both CpG sites yielded 3 fragments of 37, 32, 29 bp, while the fully unmethylated sequence yielded 1 fragment, 98 bp uncut amplicon. For the partially methylated sequences, unmethylated 37 bp CpG and methylated 69 bp CpG yielded 2 fragments, 69 and 29 bp. By contrast, methylated 37 bp CpG and unmethylated 69 bp CpG yielded 2 fragments, 37 and 61 bp.
(RAR)
Mean and median of age, BMI, waist, SBP, DBP, %total 1 body fat, BMD and Alu methylation pattern among normal, osteopenia, and osteoporosis diagnosed by DEXA T-score (>−1.0 SD = normal, −1.0 to −2.5 SD = osteopenia and <−2.5 SD = osteoporosis).
(PDF)
Mean difference (X ± SE) of weight, BMI, waist, systolic 1 blood pressure (SBP), diastolic blood pressure (DBP), and %total body fat between normal and osteopenia and between normal and osteoporosis as diagnosed by DEXA T-score (>−1.0 SD = normal, −1.0 to −2.5 SD = osteopenia and <−2.5 SD = osteoporosis) in the matched cases. Normal and osteopenia and normal and osteoporosis were paired by age.
(PDF)
Mean difference (X ± SE) of bone mass density (BMD) at each 1 region: L1, L2, L3, L4, L1–2, L1–3, L1–4, L2–3, L2–4, L3–4, femur neck, hip ward, femur trochanteric, hip total, radius ud, radius 33, radius total, and bone total between matched cases with body mass index (BMI)<25 kg/m2 and BMI>25 kg/m2. Paired age between cases with body mass index (BMI)<25 kg/m2 and cases with BMI>25 kg/m2.
(PDF)
The percentages of Alu methylation levels in each position 1 of CpG and mean of all CpG in normal, osteopenia, and osteoporosis cases.
(PDF)