Abstract
Celiac disease is characterized by intestinal inflammation caused by gluten, proteins which are widely contained in the Western diet. Mammalian digestive enzymes are only partly capable of cleaving gluten, and fragments remain that induce toxic responses in celiac patients. We found that the oral microbiome is a novel and rich source of gluten degrading enzymes. Here we report on the isolation and characterization of the cultivable resident oral microbes that are capable of cleaving gluten, with special emphasis on its immunogenic domains. Bacteria were obtained by a selective culturing approach and enzyme activities were characterised by: 1) Hydrolysis of paranitroanilide-derivatised gliadin-derived tripeptide substrates; 2) Gliadin degradation in-gel (gliadin zymography); 3) Gliadin degradation in solution; 4) Proteolysis of the highly immunogenic α-gliadin-derived 33-mer. For select strains pH activity profiles were determined. The culturing strategy yielded 87 aerobic and 63 anaerobic strains. Species with activity in at least two of the four assays were typed as: Rothia mucilaginosa HOT-681, Rothia aeria HOT-188, Actinomyces odontolyticus HOT-701, Streptococcus mitis HOT-677, Streptococcus sp. HOT-071, Neisseria mucosa HOT-682 and Capnocytophaga sputigena HOT-775, with Rothia species being active in all four assays. Cleavage specificities and substrate preferences differed among the strains identified. The approximate molecular weights of the enzymes were ~75 kD (Rothia spp.), ~60 kD (A. odontolyticus) and ~150 kD (Streptococcus spp.). In conclusion, this study identified new gluten-degrading microorganisms in the upper gastro-intestinal tract. A cocktail of the most active oral bacteria, or their isolated enzymes, may offer promising new treatment modalities for celiac disease.
Keywords: celiac disease, gliadin, oral bacteria, proteases, degradation
Introduction
Celiac disease (CD) is an inflammatory disorder with auto-immune features and a primary manifestation in the human small intestine. The disease is triggered upon ingestion of wheat gluten or similar proteins found in barley and rye (mainly hordeins and secalins, respectively). Gluten, comprising glutenins and gliadins, contain a relatively high concentration of glutamine and proline residues within their structures [1, 2], endowing certain gluten-derived domains with high resistance to degradation by human gastrointestinal proteases. None of the major human gastrointestinal proteases, such as pepsin, trypsin, chymotrypsin, carboxypeptidases A and B, elastases and brush-border membrane enzymes of the small intestine contain the necessary proteolytic capabilities to effectively cleave certain immunogenic gluten peptides, due to a lack of post-proline cleavage-site specificity. An example of a gliadin-derived, human intestinal protease resistant peptide is the 33-mer peptide derived from α-gliadins (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) [3]. Apart from its resistance to proteolysis, it contains multiple, in part overlapping, immunogenic epitopes which stimulate CD4+ T-cells in the lamina propria of HLA-DQ2-positive CD patients [4].
In most patients adherence to a gluten-free diet reverses the immune-mediated small intestinal damage and fully restores absorptive functions, whereas reintroduction of dietary gluten leads to renewed disease [5]. However, to completely exclude even traces gluten from the daily diet is a difficult task, since gluten is an abundant constituent of most refined foods. In addition, there are other complicating factors such as cross-contamination of foods and inadequate food labeling. Finally, the necessity for leading a strict gluten-free life (as opposed to a diet that only excludes obvious and major sources of gluten such as bread, pizza or pasta) means a socio-economic burden to the patient. Therefore, alternate or adjunctive treatments beyond the strict gluten free diet are considered an area of therapeutic need [6].
To achieve a more complete digestion of gluten, and specifically its immunogenic domains, proline and glutamine-specific endopeptidases from bacteria, fungi and barley are currently being explored, and such therapies are in phase I–II clinical studies [7–13]. Notably, research in our laboratory has recently shown that gluten-degrading proteases are naturally present in the upper human gastro-intestinal tract. These enzymes are not from human origin, but produced by the wealth of bacteria that colonise the oral cavity and duodenum [14, 15]. Here we performed a large scale screening of the cultivable portion of the human oral microbiome for bacteria with proteolytic enzymes cleaving any of five selected gliadin-derived synthetic tripeptide enzyme substrates, intact gliadins, and the immunodominant α-gliadin 33-mer peptide. Such bacteria could possibly fulfill physiological roles in the digestion of gluten in vivo. Moreover, they may serve as a safe source of enzyme(s) with clinical potential in the treatment of gluten sensitivity and CD.
Materials and Methods
Collection of dental plaque and whole saliva
Protocols for the collection of human dental plaque and whole saliva (WS) were approved by the Institutional Review Board at Boston University. Written informed consent was obtained from the participating subject prior to sample collection. Samples were collected after the donor refrained from oral hygiene for 24 hr. Plaque was collected using a sterile dental scaler and suspended in a buffer mimicking the ion composition of saliva, containing 50 mM KCl, 1.5 mM potassium phosphate, 1 mM CaCl2 and 0.1 mM MgCl2, pH 7.0. Bacterial aggregates were resuspended with a pipet. Whole saliva (WS) was collected under conditions of masticatory stimulation as described [15].
Culturing of bacteria
Dental plaque and whole saliva suspensions were diluted 103, 104 and 105 times in saliva ion buffer. Aliquots of 50 µl were plated either on Brucella agar (BA; Hardy Diagnostics, Santa Maria, CA) or on gluten-limited agar (GA) which was prepared in-house [15]. Plates were incubated aerobically or anaerobically in a jar using GasPak pouches (BD Diagnostics, Sparks, MD) at 37°C for 24–72 hr. Individual colonies were sub-cultured on segmented gluten agar or Brucella agar, respectively, with up to 20 strains per plate. Colonies were then subcultured repeatedly on Brucella agar (1 strain per plate) to obtain macroscopically pure cultures. The final pure strains were plated on gluten agar to confirm their growth on this selective medium. The bacterial strains were maintained frozen in a mixture of BHI/glycerol (80/20% v/v).
16S rDNA identification of bacteria
DNA extraction from colonies grown on Brucella agar was performed using the UltraClean Microbial DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA) followed by sequencing using an ABI prism cycle-sequencing kit (BigDye® Terminator Cycle Sequencing kit) as previously described [16].
Hydrolysis of gliadin-derived enzyme substrates
Cells were harvested with a sterile cotton swab from Brucella agar plates and resuspended in saliva ion buffer to an OD620 1.2. Enzymatic activity was examined towards five gliadin-derived synthetic enzyme substrates: Z-YPQ-pNA, Z-QQP-pNA, Z-PPF-pNA, Z-PFP-pNA and Z-LPY-pNA. The tripeptide substrates represent sequences that are common in the immunogenic gliadin domains. Substrates were mixed with 200 µl bacterial suspensions (OD620 1.2) to a final concentration of 200 µM and incubated at 37°C. For initial screening of bacteria for substrate hydrolysis, data were interpreted macroscopically over a 48 hr time interval with assessment time points of 1 hr, 16 hr, 24 hr, 40 hr and 48 hr. Kinetic enzyme assays were carried out for selected strains, measuring substrate hydrolysis at 405 nm using a Genios microtiter plate reader (Tecan, Männedorf, Switzerland).
Degradation of gliadin in-gel (gliadin zymography)
Cells were harvested with a cotton swab from Brucella agar plates and suspended in saliva ion buffer to an OD620 5.0. Cells contained in a 150 µl aliquot were centrifuged and suspended in zymogram sample buffer containing 0.25 M Tris/HCl, 10% glycerol, 2% SDS and 0.0025% bromophenol blue. Gliadin zymography (6%) was carried out as described previously [15]. Gels were processed in renaturing and developing buffers (InVitrogen) according to the manufacturer’s instructions. After 48 hr incubation at 37°C, gels were stained in 0.2% Coomassie Brilliant blue dissolved in 40% methanol and destained without the dye.
Degradation of gliadin in solution
A mixture of gliadins obtained from Sigma (St. Louis, MO) was dissolved to 5 mg/ml in 60% (v/v) ethanol. The solution was diluted in bacterial cell suspensions in saliva ion buffer (OD620 1.2) to 250 µg/ml. After incubation time points of 0, 2 and 5 hr at 37°C, 100 µl aliquots were removed and boiled for 5 minutes to inactivate enzyme activity. For some strains different time points and cell densities were used (noted in the Figure legends). To each sample aliquot EDTA was added to a final concentration of 2.5 mM to complex calcium ions in the saliva ion buffer, and samples were dried using a Speedvac. Pellets were suspended in 1x SDS sample buffer and analyzed on pre-cast 12% Bis-Tris-PAGE (InVitrogen, Carlsbad, CA).
Degradation of the 33-mer gliadin peptide
A synthetic highly immunogenic 33-mer peptide derived from α2-gliadin (LQLQPFPQPQLPYPQPQLPYPQPQLPYPQPQPF) [3] was synthesized at 21st Century Biochemicals (Marlboro, MA) at a purity of 95%. The peptide was dissolved in milliQ water at 10 mg/ml and diluted to 250 µg/ml in bacterial cell suspensions (OD620 1.2). Incubations, sample aliquot removal and heat-inactivation of the enzymes were carried out as in the gliadin degradation experiment. The 100 µl aliquots were mixed with 900 µl 0.1% TFA and subjected to RP-HPLC using a C-18 column (TSK-GEL 5 µm, ODS-120T, 4.6×250 mm, TOSOHaas, Montgomeryville, PA). Peptides were eluted using a multi-step gradient as described previously [14].
Enzyme activities at various pH values
To assess the bacterial enzyme activities at various pH, three select strains were suspended to an OD620 1.2 in 20 mM Tris solutions at pH 3.0, 5.0, 7.0 or 9.0. Activities were assessed towards Z-YPQ-pNA as the enzyme substrate (200 µM) with a Genios microtiter plate reader in the kinetic mode over an 8 hour time interval.
In another experiment, equal volumes of suspensions of the same three strains, each at an OD620 1.2, were mixed. The mixture was then divided into three portions. The pH of the first two aliquots was adjusted to pH 3.0 with HCl, the third aliquot was adjusted to pH 7.0 with concentrated sodium bicarbonate. The pH of the second aliquot at pH 3.0 was adjusted to pH 7.0 after a 1.5 h incubation time interval at 37°C. Enzyme activities in all three aliquots were determined spectrophotometrically using Z-YPQ-pNA.
Results
Hydrolysis of gliadin-derived enzyme substrates
Using the selective plating approach, 87 aerobic strains and 63 anaerobic strains were obtained. Among the 87 aerobic strains 16 were isolated from human whole saliva (referred to as WSA-#) and 71 from human dental plaque (referred to as PA-#). Among the 63 anaerobic strains 18 were derived from whole saliva and 45 were from dental plaque. To evaluate enzymatic cleavage specificities, the 87 aerobic strains were incubated with gliadin-derived synthetic enzyme substrates and hydrolysis was monitored macroscopically. Since this assay was intended for screening purposes only, experiments were performed once, and repeated for select strains of interest. Of the 87 aerobic strains, 75 cleaved Z-YPQ-pNA, 7 cleaved Z-QQP-pNA, 5 cleaved PPF-pNA, 8 cleaved Z-PFP-pNA and 52 cleaved Z-LPY-pNA (Supplemental Table 1). Of the 75 strains hydrolyzing Z-YPQ-pNA, 12 hydrolyzed this substrate within 1 hr incubation. All strains that cleaved Z-LPY-pNA also cleaved Z-YPQ-pNA, but not the reverse. Lastly, 5 strains rapidly hydrolyzed all five substrates within 1 hr incubation. The anaerobic strains tested (35 out of 63 strains) did not cleave any of the five tripeptide substrates and were not further considered.
Zymography results
To determine gliadin degradation in gel, the 87 aerobic bacteria were subjected to gliadin zymography. A total of 15 strains showed the presence of gliadin-degrading enzyme(s), either in the ~60 kDa, ~75 kDa, or ~150 kDa region. The remainder of the strains did not show evidence of gliadin degrading activity in gel (supplemental Table 2). Taking the tripeptide hydrolysis and the zymogram results together, 21 aerobic strains were deemed of interest and categorised into five groups based on their patterns of activity. Group I (n=5): YPQ↓ and LPY↓ cleavage and an enzyme with a molecular weight of ~75 kDa; group II (n=1): YPQ↓ and LPY↓ cleavage and an enzyme at ~60 kDa; group III (n=9): YPQ↓ cleavage and a weak enzyme band at ~150 kDa; group IV (n=2): YPQ↓, QQP↓, and LPY↓ cleavage but no evidence for activity in the gliadin zymogram, and group V (n=5): cleaveage of YPQ↓, QQP↓, PPF↓, PFP↓ and LPY↓ and no band in the gliadin zymogram gel (Table 1).
Table 1.
Summary Evaluation of Aerobic Strainsa
| pNA-derivatized tripeptide substrates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain name | Strain ID# | YPQ | QQP | PPF | PFP | LPY | Gliadin zymo band (MW) | Gliadin degr in solution | 33-mer degradation | |
| Rothia mucilaginosa HOT-681 | WSA-2B | + 1 hr | - | - | - | + 16 hrs | ~70/75 kD | Yes | Yes |
GROUP I |
| Rothia sp. HOT-188 (Rothia aeria) | WSA-8 | + 1 hr | - | - | - | + 16 hrs | ~75 kD | Yes | Yes | |
| Rothia mucilaginosa HOT-681 | WSA-21B | + 1 hr | - | - | - | + 16 hrs | ~70/75 kD | Yes | +/− | |
| Rothia mucilaginosa HOT-681 | WSA-26 | + 1 hr | - | - | - | + 16 hrs | ~70/75 kD | Yes | Yes | |
| Actinomyces odontolyticus HOT-701 | PA-20b | + 1 hr | - | - | - | + 16 hrs | ~60 kD | No | Yes | GROUP II |
| Streptococcus mitis HOT-677 | WSA-7A | + 16 hrs | - | - | - | - | ~150 kD | n.d. | n.d. |
GROUP III |
| Streptococcus mitis HOT-677 | PA-8 | + 16 hrs | - | - | - | - | ~150 kD | n.d. | n.d. | |
| Streptococcus sp HOT-071 | PA-9 | + 16 hrs | - | - | - | - | ~150 kD | n.d. | n.d. | |
| Streptococcus mitis HOT-677 | PA-12 | + 16 hrs | - | - | - | - | ~150 kD | No | No | |
| Streptococcus sp HOT-071 | PA-21 | + 24 hrs | - | - | - | - | ~150 kD | No | No | |
| Streptococcus sp HOT-071 | PA-3b | + 16 hrs | - | - | - | - | ~150 kD | No | No | |
| Streptococcus sp HOT-071 | PA-4b | + 16 hrs | - | - | - | - | ~150 kD | n.d. | n.d. | |
| Streptococcus mitis HOT-677 | PA-19b | + 24 hrs | - | - | - | - | ~150 kD | No | No | |
| Streptococcus sp HOT-071 | PA-23b | + 24 hrs | - | - | - | - | ~150 kD | n.d. | n.d. | |
| Neisseria mucosa HOT-682 | PA-1b | + 1 hr | +/− 24hrs | - | - | + 16 hrs | - | n.d. | n.d. |
GROUP IV |
| Neisseria mucosa HOT-682 | PA-2b | + 1 hr | +/− 24hrs | - | - | + 16 hrs | - | No | No | |
| Capnocytophaga sputigena HOT-775 | PA-33B | + 1 hr | + 1 hr | + 1 hr | + 1 hr | + 1 hr | - | n.d. | n.d. |
GROUP V |
| Capnocytophaga sputigena HOT-775 | PA-33R | + 1 hr | + 1 hr | + 1 hr | + 1 hr | + 1 hr | - | No | Yes | |
| Capnocytophaga sputigena HOT-775 | PA-90B | + 1 hr | + 1 hr | + 1 hr | + 1 hr | + 1 hr | - | n.d. | n.d. | |
| Capnocytophaga sputigena HOT-775 | PA-90R | + 1 hr | + 1 hr | + 1 hr | + 1 hr | + 1 hr | - | No | Yes | |
| Capnocytophaga sputigena HOT-775 | PA-90 | + 1 hr | + 1 hr | + 1 hr | + 1 hr | + 1 hr | - | n.d. | n.d. | |
Total number of aerobic bacteria screened: 87; total number of bacteria cleaving one or more of the five tripeptide-pNA substrates: 78 +color change due to substrate hydrolysis, occuring after the indicated incubation time (macroscopic assessment) Gliadin and 33-mer degradation in solution: 250 ug/ml protein, proteolysis assessed by SDS PAGE and RP-HPLC, resp
16S rDNA sequencing results
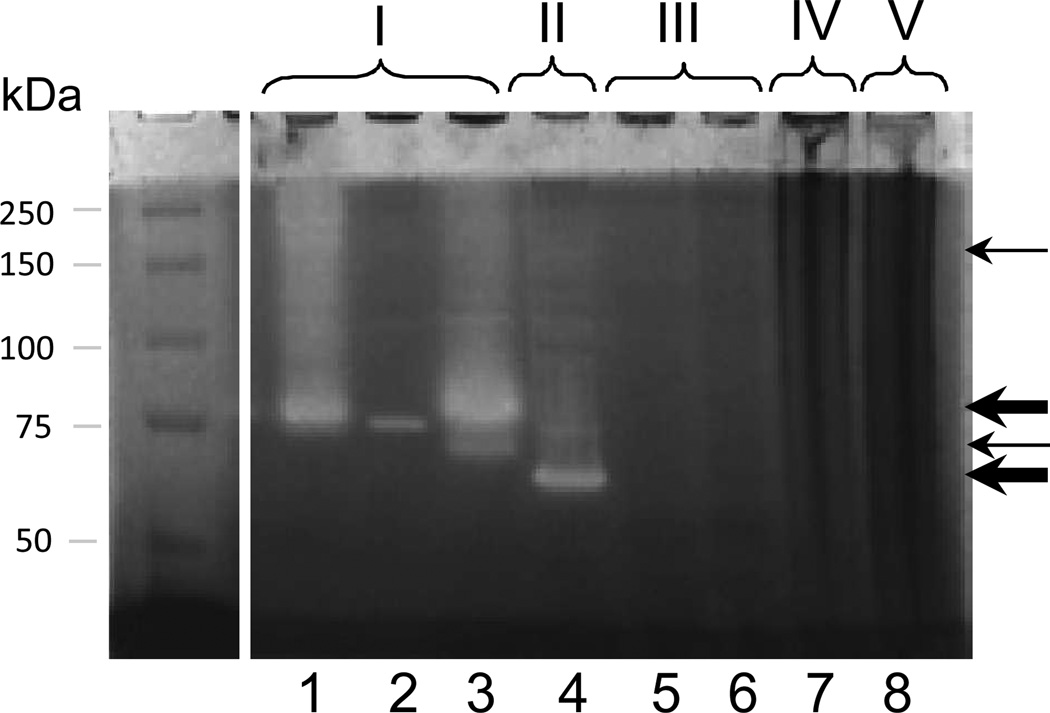
The 21 strains of interest were speciated by 16S rDNA analysis. As expected, in some cases, strains within one group represented the same species. A clear picture emerged as to the differences in species in the five groups. The taxa identified in group I were: Rothia mucilaginosa HOT-681 and Rothia aeria HOT-188; in group II: Actinomyces odontolyticus HOT-701; in group III: Streptococcus mitis HOT-677 and Streptococcus sp. HOT-071; in group IV: Neisseria mucosa HOT-682; and in group V: Capnocytophaga sputigena HOT-775 (Table 1). A representative zymogram with a selection of strains from each group is shown in Figure 1.
Figure 1.
Gliadin zymography of selected oral strains. An aliquot of 150 µl cells (OD620 5.0) was applied per lane. Lane 1, WSA (whole saliva)-2B (R. mucilaginosa); lanes 2: WSA-8 (R. aeria), lane 3: WSA-21B (R. mucilaginosa); lane 4: PA (dental plaque)-20b (A. odontolyticus); lane 5: PA-12 (Streptococcus mitis); lane 6: PA-21 (Streptococcus sp. HOT-071); lane 7: PA-2b (Neisseria mucosa); lane 8: PA-90R (Capnocytophaga sputigena).
Degradation of gliadin in Solution
A subset of strains was selected to study their capacity to degrade gliadin in solution. These were three strains from Group I (WSA-2B, WSA-21B, WSA-26), one strain from group II (PA-20b), four strains from group III (PA-12, PA-21, PA-3b, and PA-19b), two strains from group IV (PA-1b, PA-2b), and two strains from group V (PA-33R, PA-90R). Results obtained with Rothia aeria (strain WSA-8) were reported previously [15]. The final OD620 and incubation times for strains in groups I, II and III were OD620 1.2 and 0, 2 hr and 5 hr, respectively, and for strains in group IV and V the OD620 was 0.2 and the incubation times were 0, 4 hr and 8 hr, respectively. The OD and time adjustment were made based on the apparent high protein content in these strains (see e.g. Figure 1). Only bacteria in group I cleaved gliadin in solution (Supplemental Figure 1). Results for strains WSA-2B (group I), PA-20b (group II), and PA-33R (group V) are shown in Figure 2, left panels.
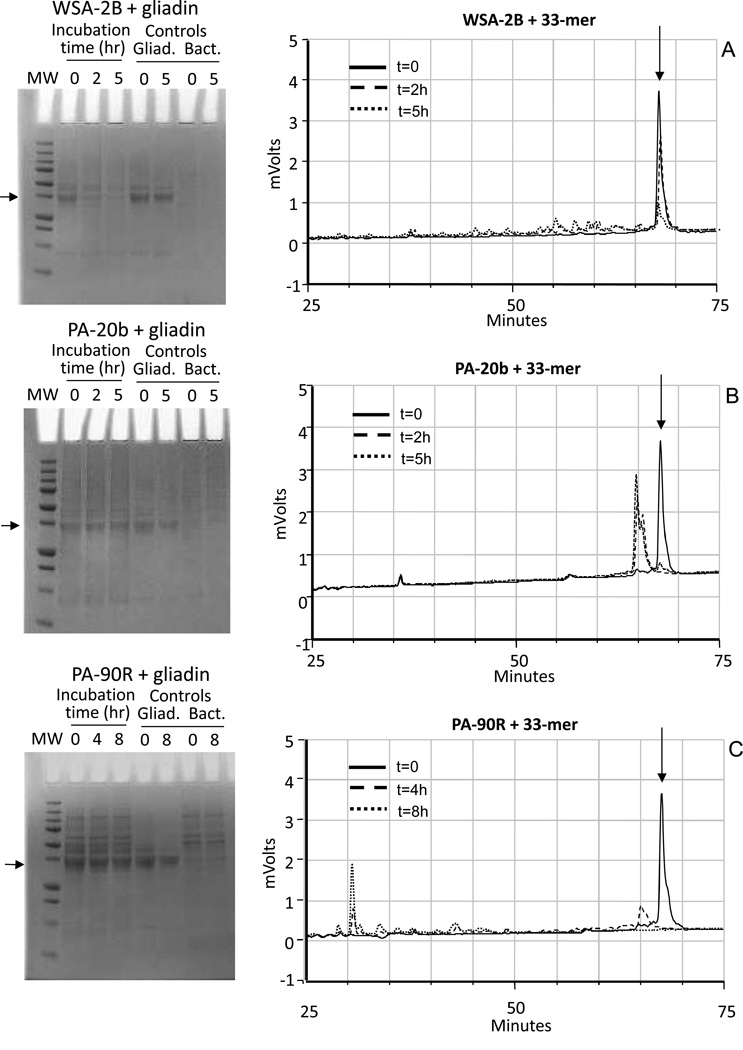
Figure 2.
Kinetics of gliadin degradation and 33-mer degradation by WSA-2B, PA-20b and PA-90R. Arrows point to the major protein component in the gliadin preparation at ~37 kD (SDS gels, left panels) and the 33-mer (RP-HPLC chromatograms, right panels). The cell densities used for the incubations were OD620 1.2 for WSA-2B and PA-20b and OD620 0.2 for PA-90R. The final concentration of mixed gliadins and 33-mer peptide were 250 µg/ml. Degradation mixtures were analyzed at t=0, 2h and 5h (strains WSA-2B and PA-20b) and t=0, 4h and 8h (strain PA90R).
Degradation of the 33-mer Gliadin Peptide
The same 12 strains were also investigated for degrading the gliadin-derived 33-mer peptide. The cell densities and incubation times used were the same as for the gliadin degradation experiments. Supplemental Figures 2A, B, C show the chromatograms obtained. Strains in group I (R. mucilaginosa HOT-681) cleaved the 33-mer partly in the 5 hr time span and buffer conditions examined. The strain in group II (A. odontolyticus HOT-701) cleaved the 33-mer completely within a 2 hr time interval, with proteolysis already noticeable in the t=0 sample (taken immediately after mixing of the 33-mer peptide with PA-20b suspension). The degradation patterns at t=2 hr and t= 5 hr were comparable, and remained stable over a 24 hr time span (not shown). Strains in groups III (S. mitis HOT-677 and Streptococcus HOT-071) and IV (N. mucosa HOT-682) were unable to cleave the 33-mer peptide. In contrast, group V bacteria (C. sputigena HOT-775) cleaved the 33-mer rapidly without any visible evidence for surviving major degradation fragments. Results for strains WSA-2B, PA-20b and PA-90R are shown in Figure 2 (right panels). Table 1 summarizes all the results obtained.
pH activity profiles
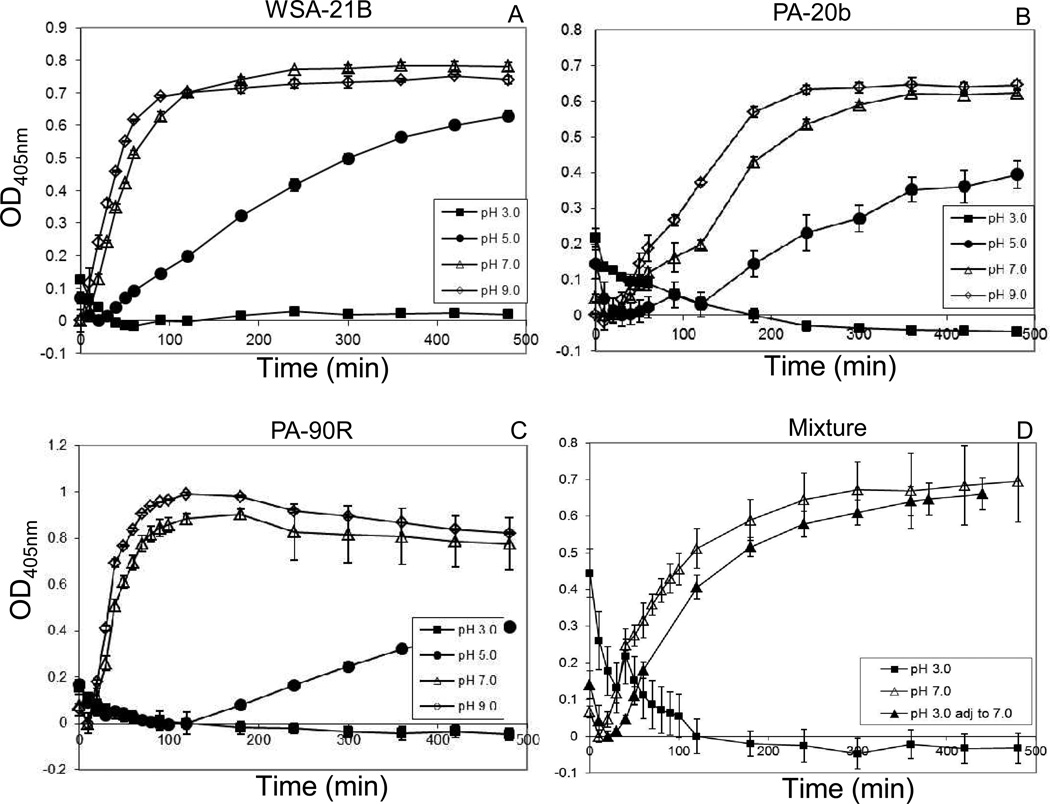
Three strains with prominent activities were selected for pH activity determinations: WSA-21B (R. mucilaginosa HOT-681), PA-20b (A. odontolyticus HOT-701) and PA-90R (C. sputigena HOT-775). Enzymatic activities for all three strains were optimal at neutral to basic pH conditions. At pH 5.0, rates of hydrolysis were reduced for all strains and at pH 3.0 none of the strains were active over the 8 hr time interval examined (Figure 3). The latter would suggest that the strains are not active at the pH conditions encountered in the fasting stomach. To investigate if the strains could withstand low pH conditions for a time interval consistent with gastric passage, we performed an experiment in which a mixture of the three strains were incubated at pH 3.0 for 1.5 hr, after which the pH was adjusted to pH 7.0. Enzyme activity assessment confirmed activity of the bacterial mixture at pH 7.0 and inactivity at pH 3.0. Importantly, bacteria preincubated at pH 3.0 were not irreversibly inactivated, since their activity could be fully restored when the pH was adjusted to pH 7.0.
Figure 3.
pH gluten peptide degrading activity profiles of WSA-21B, PA-20b and PA-90R. Strains were suspended to a final OD620 1.2 in Tris solutions with pH values adjusted to pH 3.0, 5.0, 7.0 and 9.0, and hydrolysis of Z-YPQ-pNA (final concentration 200 µM) was monitored spectrophotometrically at 405 nm over a 0–480 min time interval. A, WSA-21B; B, PA-20b; C, PA-90R. D, equal volumes of the three strains were mixed at pH 3.0, 7.0, or at pH 3.0 followed by adjustment of the pH after 1.5 h to 7.0. Z-YPQ-pNA hydrolysis in the three suspensions is shown.
Discussion
This study was aimed at the discovery and broad characterisation of natural resident microbes in the most upper part of the gastro-intestinal tract that can degrade immunogenic gliadins implicated in celiac disease. The approach consisted of a culture-based initial enrichment step for such bacteria from the mixed bacterial oral samples. Enzyme activities associated with the isolates were determined in four independent assays. Bacteria could be separated into five groups after analyzing degradation patterns and bacterial IDs. The most active microbes were tested at various pH, showing that their enzymes were not active at acidic pH, but that a temporary exposure to a low pH consistent with gastric transfer is permissible. Additional information on the identified strains’ rank of abundance in the oral cavity, and the number of known and putative peptidases, are given in Table 2.
Table 2.
Oral abundance and number of proteases in selected species
| Strain name | Group | Approximate Rank abundance in the oral cavity (#)a |
Number of known and putative peptidasesb |
|---|---|---|---|
| Rothia mucilaginosa HOT-681 | I | 170 | 13 |
| Rothia aeria HOT-188 | I | 221 | unavailable |
| Actinomyces odontolyticus HOT-701 | II | 326 | 41 |
| Streptococcus mitis HOT-677 | III | 2 | 19 |
| Streptococcus sp. HOT-071 | III | 37 | unavailable |
| Neisseria mucosa HOT-682 | IV | 64 | 8 |
| Capnocytophaga sputigena HOT-775 | V | 166 | 22 |
Information from Dewhirst et al 2010.
Information from www.merops.ac.uk
Bacteria in Group I comprised Rothia mucilaginosa HOT-681 and R. aeria HOT-188. Both strains were active in all four assays and therefore are of high interest. Rothia species are harmless residents of the oral cavity and the oral pharynx [16], and have also been identified in duodenal biopsies [17]. There are case reports of infections caused by R. mucilaginosa in immunocompromised patients, but overall it is considered a harmless coloniser of the oral cavity [18] They hydrolyzed the Z-YPQ-pNA and Z-LPY-pNA substrates, and cleaved the highly immunogenic α-gliadin 33-mer peptide after QPQ↓ and LPY↓ [14]. The LPY↓ specificity is unique and adds to the post-proline and post-glutamine activities currently being the focus of luminal enzyme therapy [19].
The bacterial species identified in Group II was A. odontolyticus HOT-701. Its enzymes also cleaved the substrates Z-YPQ-pNA and Z-LPY-pNA, mixed gliadins and the 33-mer peptide. The molecular weight of the enzyme was distinctly lower than that of the Rothia bacteria. A. odontolyticus HOT-701 is typically isolated from patients with advanced dental caries [20], and has rarely been implicated in bacteremia [21]. It is described as an anaerobic microorganism, but we could culture it both under aerobic and under anaerobic conditions. The typing of A. odontolyticus was made based on 16S rDNA analysis. Since the level of homology in the 490 bp sequence after removal of any ambiguous base calls was 98%, it cannot be excluded that strain PA-20b represents a closely related Actinomyces subspecies. Therefore we obtained ATCC strain 17929 (A. odontolyticus Batty) for validation of the results with isolate PA-20b. The zymogram results of the ATCC strain were indistinguishable from PA-20b, confirming the gluten-degrading capacity of A. odontolyticus (data not shown). Notably, strain PA-20b degraded gliadin in-gel but not in solution, suggesting that substrate conformation is an important determinant for recognition. PA-20b quickly degraded the 33-mer, faster than the Rothia bacteria. However, the degradation fragments were stable and of substantial length, given their high retention times in the HPLC chromatograms. Therefore, PA-20b enzymes would work optimally in combination with other enzymes to achieve the full fragmentation of immunogenic gliadin peptides.
The bacteria in group III comprised Streptococcus mitis HOT-677 and Streptococcus sp. HOT-071. Streptococcus mitis HOT-677 is the second most abundant oral microorganism. It has been detected on the buccal epithelium, maxillary anterior vestibule, tongue dorsum, hard and palate, tonsils, tooth surfaces and subgingival plaque [22]. The selection of S. mitis following the plating approach might have been due to its high abundance, rather than it being a truly gliadin-degrading organism. Indeed, it exhibited low gliadin-degrading activities in gel, and negligible or undetectable activities in solution. Neither of the streptococcal species cleaved the 33-mer peptide. Nevertheless, because of its high abundance, S. mitis could still contribute substantially to the overall gliadin-degrading capacity in the human mouth.
The bacterium in Group IV was typed as N. mucosa HOT-682. This is a gram-negative aerobic bacterium that is a nonpathogenic resident of the oropharynx of humans. As for the other strains identified, rare cases of systemic infections with Neisseria mucosa have been reported [23, 24]. Interestingly, Neisseria mucosa strains hydrolyzed three of the five gliadin-related tripeptide substrates, but not any of the larger substrates (33-mer, or intact gliadin, either in gel or in solution). Given this limited activity, Neisseria mucosa is deemed of less interest for enzyme isolation and potential exploitation in CD.
Lastly, group V comprised the bacterium C. sputigena HOT-775. This species stood out from the other identified microorganisms in being able to cleave all five gliadin-based tripeptide substrates. This result demonstrates that a wide variety of protease specificities are contained within this single microorganism. C. sputigena HOT-775 is a gram negative oral coloniser. It has been implicated in a number of infectious diseases, e.g. septicaemia, osteomyelitis, abscesses and keratitis [25], perhaps, owing to its high and diverse protease activities (Table 2). C. sputigena HOT-775, like A. odontolyticus HOT-701, rapidly cleaved the immunogenic 33-mer gliadin peptide, grouping this bacterium automatically into the category of high interest. In contrast to A. odontolyticus HOT-701, no larger protease-resistant peptides were observed, indicating extensive fragmentation of the 33-mer peptide by C. sputigena HOT-775. It can be assumed, but needs further proof, that immunogenic epitopes would thus be neutralised. N. mucosa HOT-682 and C. sputigena HOT-775 did not cleave intact gliadin, neither in a partly denatured state in the gel matrix, nor in a native conformation in solution, suggesting that larger substrates are not recognised by their proteases.
While ingested gluten is a necessary trigger for CD development, gluten reactivity alone cannot explain the absence of disease in most subjects with HLA-DQ2 and −DQ8 phenotypes. A prominent role for the gastrointestinal microbiome in the switch from tolerance to an inflammatory immune response to gluten is becoming evident [26]. Recently, this concept was further supported by studies showing a difference between the developing gut microbiome in HLA-DQ2+/DQ8+ children of patients with biopsy-proven CD [27] and children from healthy mothers [28]. Besides modulating the immune response, our study provides evidence that the microbiota of the (upper) GI tract may also play a more direct role by degrading toxic gluten peptides, and perhaps aiding to gluten detoxification in vivo.
How far the gluten-degrading microorganisms contribute to gluten digestion in vivo remains to be established. Apart from their potential physiological effects as natural colonizers of the upper GI tract, the bacteria - and their enzymes- could also be exploited therapeutically as digestive aids. Thus gluten-degrading enzymes from other sources are already being pursued as adjunctive therapeutics for celiac disease [7–13]. Similarly, the gluten-degrading enzymes from the oral bacteria could be isolated and further pursued as a pharmaceutical drug, or dietary supplement. The advantage is that the sources of these enzymes are natural human body-associated microbes, and most oral bacteria are indeed harmless residents. Given their tolerated natural colonization of the upper GI tract an alternative approach would be to develop the gluten-degrading bacteria themselves, as probiotic agents, to achieve gluten digestion. Successful application would require the probiotic to be effective along the gastrointestinal canal and to be safe when consumed in larger quantities, aspects that are currently being addressed.
Supplementary Material
Acknowledgments
The studies were funded by grants from the National Institutes of Health: AI087803 (EJH), DE07652 (FO), DE16937 (FED) and AI078385 (DS).
The authors thank Dr. Maram Zamakhchari for assistance with bacterial culturing.
References
- 1.Wieser H. The precipitating factor in coeliac disease. Bailliere's clinical gastroenterology. 1995;9:191–207. doi: 10.1016/0950-3528(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 2.Wieser H. Chemistry of gluten proteins. Food microbiology. 2007;24:115–119. doi: 10.1016/j.fm.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Shan L, Molberg O, Parrot I, et al. Structural basis for gluten intolerance in celiac sprue. Science (New York NY. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 4.Vader LW, de Ru A, van der Wal Y, et al. Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. The Journal of experimental medicine. 2002;195:643–649. doi: 10.1084/jem.20012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leffler D, Schuppan D, Pallav K, et al. Kinetics of the histological, serological and symptomatic responses to gluten challenge in adults with coeliac disease. Gut. 2012 doi: 10.1136/gutjnl-2012-302196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sollid LM, Khosla C. Novel therapies for coeliac disease. J Intern Med. 2011;269:604–613. doi: 10.1111/j.1365-2796.2011.02376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitea C, Havenaar R, Drijfhout JW, Edens L, Dekking L, Koning F. Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: Implications for coeliac disease. Gut. 2008;57:25–32. doi: 10.1136/gut.2006.111609. [DOI] [PubMed] [Google Scholar]
- 8.Marti T, Molberg O, Li Q, Gray GM, Khosla C, Sollid LM. Prolyl endopeptidase-mediated destruction of t cell epitopes in whole gluten: Chemical and immunological characterisation. The Journal of pharmacology and experimental therapeutics. 2005;312:19–26. doi: 10.1124/jpet.104.073312. [DOI] [PubMed] [Google Scholar]
- 9.Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: Implications for coeliac sprue. The Biochemical journal. 2004;383:311–318. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stepniak D, Spaenij-Dekking L, Mitea C, et al. Highly efficient gluten degradation with a newly identified prolyl endoprotease: Implications for celiac disease. American journal of physiology. 2006;291:G621–G629. doi: 10.1152/ajpgi.00034.2006. [DOI] [PubMed] [Google Scholar]
- 11.Siegel M, Bethune MT, Gass J, et al. Rational design of combination enzyme therapy for celiac sprue. Chemistry & biology. 2006;13:649–658. doi: 10.1016/j.chembiol.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Cerf-Bensussan N, Matysiak-Budnik T, Cellier C, Heyman M. Oral proteases: A new approach to managing coeliac disease. Gut. 2007;56:157–160. doi: 10.1136/gut.2005.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bethune MT, Khosla C. Oral enzyme therapy for celiac sprue. Methods Enzymol. 2012;502:241–271. doi: 10.1016/B978-0-12-416039-2.00013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmerhorst EJ, Zamakhchari M, Schuppan D, Oppenheim FG. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity. PloS one. 2010;5:e13264. doi: 10.1371/journal.pone.0013264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamakhchari M, Wei G, Dewhirst F, et al. Identification of rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PloS one. 2011;6:e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bact. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou G, Hedberg M, Horstedt P, et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am J Gastroenterol. 2009;104:3058–3067. doi: 10.1038/ajg.2009.524. [DOI] [PubMed] [Google Scholar]
- 18.Yamane K, Nambu T, Yamanaka T, et al. Complete genome sequence of Rothia mucilaginosa DY-18: a clinical isolate with dense meshwork-like structures from a persistent apical periodontitis lesion. Sequencing. 2010 [Google Scholar]
- 19.Caputo I, Lepretti M, Martucciello S, Esposito C. Enzymatic strategies to detoxify gluten: Implications for celiac disease. Enzyme Res. 2010;2010:174354. doi: 10.4061/2010/174354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batty I. Actinomyces odontolyticus, a new species of actinomycete regularly isolated from deep carious dentine. J Pathol Bacteriol. 1958;75:455–459. doi: 10.1002/path.1700750225. [DOI] [PubMed] [Google Scholar]
- 21.Cone LA, Leung MM, Hirschberg J. Actinomyces odontolyticus bacteremia. Emerg Infect Dis. 2003;9:1629–1632. doi: 10.3201/eid0912.020646. [DOI] [PubMed] [Google Scholar]
- 22.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacon AE, 3rd, Pal PG, Schaberg DR. Neisseria mucosa endocarditis. J Infect Dis. 1990;162:1199–1201. doi: 10.1093/infdis/162.5.1199. [DOI] [PubMed] [Google Scholar]
- 24.Davis CL, Towns M, Henrich WL, Melby K. Neisseria mucosus endocarditis following drug abuse. Case report and review of the literature. Arch Intern Med. 1983;143:583–585. [PubMed] [Google Scholar]
- 25.Frandsen EV, Poulsen K, Kononen E, Kilian M. Diversity of capnocytophaga species in children and description of capnocytophaga leadbetteri sp. Nov. And capnocytophaga genospecies ahn8471. Int J Syst Evol Microbiol. 2008;58:324–336. doi: 10.1099/ijs.0.65373-0. [DOI] [PubMed] [Google Scholar]
- 26.Cinova J, De Palma G, Stepankova R, et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: Study in germ-free rats. PloS one. 2011;6:e16169. doi: 10.1371/journal.pone.0016169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sellitto M, Bai G, Serena G, et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PloS one. 2012;7:e33387. doi: 10.1371/journal.pone.0033387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.