Abstract
Eps8, a bi-functional actin cytoskeleton remodeler, is a positive regulator of cell proliferation and motility. Here, we describe an unrecognized mechanism regulating Eps8 that is required for proper mitotic progression: While Eps8 is stable in G1 and S phase, its half-life drops sharply in G2. This requires G2-specific proteasomal degradation mediated by the Ubiquitin E3 ligase SCFFbxw5. Consistent with a short window of degradation, Eps8 disappears from the cell cortex early in mitosis, but reappears at the midzone of dividing cells. Failure to reduce Eps8 levels in G2 prolongs its localization at the cell cortex and markedly delays cell rounding and prometaphase duration. However, during late stages of mitosis and cytokinesis, Eps8 capping activity is required to prevent membrane blebbing and cell shape deformations. Our findings identify SCFFbxw5-driven fluctuation of Eps8 levels as an important mechanism that contributes to cell-shape changes during entry into – and exit from - mitosis.
Epidermal growth factor receptor pathway substrate 8 (Eps8) is a signalling adaptor that controls various cellular protrusions by regulating actin cytoskeleton dynamics and architecture1-4. Depending on its association with other signal transducers, Eps8 can regulate the Rac GTPase5, 6 or directly control actin dynamics by binding actin filaments and exerting either actin bundling or actin barbed-end capping activity3, 4, 7. Eps8 has been implicated in the regulation of processes such as axonal filopodia growth, stereocilia length, dendritic cell migration, cancer cell migration and invasion8-11, and was shown to contribute to cell transformation in response to growth factor treatment2. Consistent with this, increased Eps8 levels have been linked to human tumor development and progression9, 12-16. Together, these findings indicate that Eps8 levels need to be tightly regulated. When overexpressed at high levels (e.g. in some pancreatic cancers), Eps8 is subject to chaperone-mediated autophagy17, but whether regulated degradation contributes to normal Eps8 biology has so far remained elusive.
Regulated protein degradation is a key mechanism to control cellular processes. A major player in regulated degradation is the Ubiquitin system, which marks proteins for proteasomal or lysosomal degradation18. Target specificity within the Ubiquitin system is conferred by Ubiquitin E3 ligases, which bind substrates and catalyze the transfer of Ubiquitin from an Ubiquitin E2 enzyme to a specific substrate19.
Amongst several hundred E3 ligases, Cullin-RING-based E3 ligases (CRLs) comprise the largest family. They are composed of a modular E3 core containing a cullin, a RING domain protein (Rbx1 or Rbx2), and a substrate specificity module usually composed of a linker protein and interchangeable substrate receptors (SRs)20. Human cells express six closely related cullin proteins that nucleate different subfamilies of CRLs (CRL1-CRL5). In combination with dedicated substrate specificity modules these CRLs bind and ubiquitylate distinct sets of substrates21. The best-characterized subfamily of CRLs, Skp1-Cul1-F-box (SCF) complexes, uses interchangeable F-box proteins as SRs22, 23.
Here, we demonstrate that Eps8 is subject to regulated degradation specifically in the G2 phase of the cell cycle. This requires the E3 ligase SCFFbxw5 and proteasomal degradation. Failure to transiently degrade Eps8 prior to mitosis results in prolonged presence of Eps8 at the cell cortex, a delay in cell rounding, and prolonged prometaphase duration. On the other hand, insufficient Eps8 capping activity during anaphase and telophase induces membrane blebbing and cell shape deformations. Together, these findings implicate SCFFbxw5-mediated regulation of Eps8 levels as a crucial mechanism to regulate cell-shape changes required for mitotic progression.
Results
Eps8 is an interaction partner of the F-box protein Fbxw5
During a project aimed at identifying binding partners for the F-box protein Fbxw5, a substrate receptor of SCF-type Ubiquitin E3 ligases24, 25, we identified Eps8 peptides in an IP / mass spectrometry-based screen (Supplementary Fig. 1a,b, online). We verified the Fbxw5-Eps8 interaction both with anti-Flag IPs from HEK293T cells stably expressing flag-Fbxw5 (Supplementary Fig. 1c), and with anti-Fbxw5 IPs (using affinity purified polyclonal antibodies) from untransfected cells (Supplementary Fig. 1d). To exclude that the observed interaction is mediated via Skp1, Cul1, or Rbx1, we repeated the anti-Flag IP upon transfection of an Fbxw5 derivative (flag-Fbxw5ΔF-box) that cannot be integrated into SCF complexes. The interaction between Eps8 and Fbxw5 remained unchanged (Supplementary Fig. 1e), suggesting that it may reflect a bona fide E3 ligase - substrate interaction. These findings inspired us to follow up on a possible regulation of Eps8 by the ubiquitin/proteasome system.
Eps8 is subject to Fbxw5-dependent proteasomal degradation during G2 phase of the cell cycle
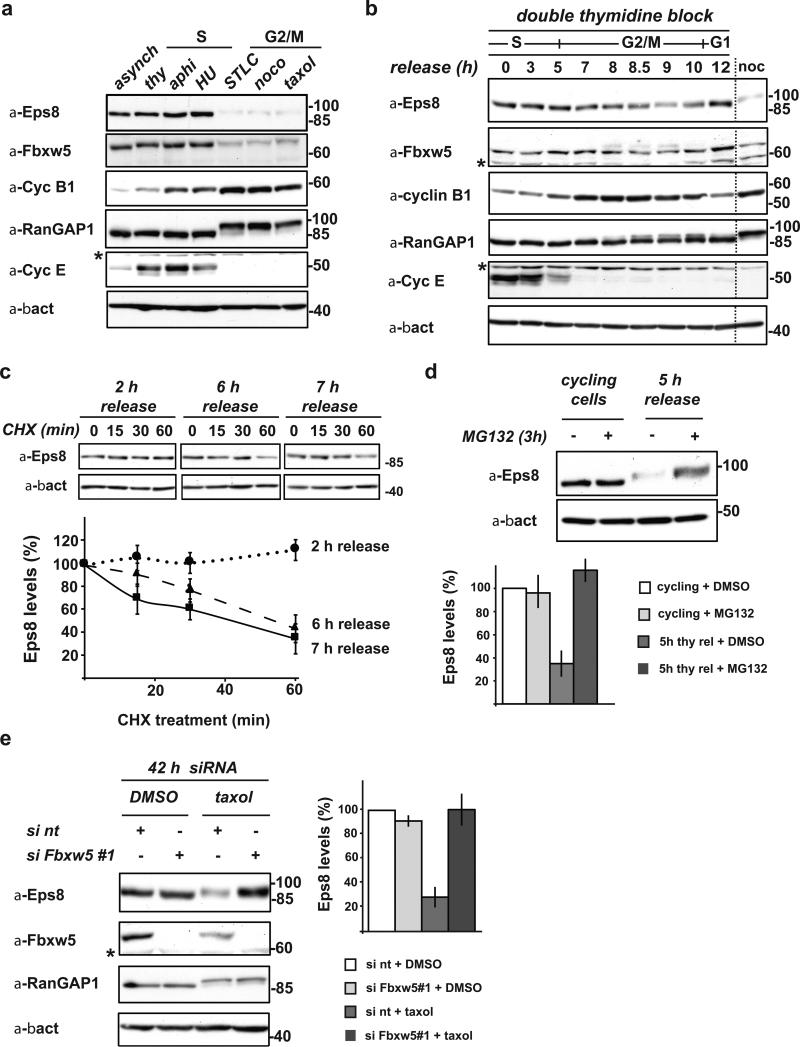
Initial experiments with asynchronous cells indicated that Eps8 is a stable protein (see below). However, targets for SCF E3 ligases are frequently degraded at a specific time in the cell cycle. To explore this possibility, we compared cycling versus S-phase (aphidocholine, thymidine, or hydroxyurea) or G2/M-phase (nocodazole, taxol, or STLC) arrested HeLa cells. Indeed, immunoblotting revealed a striking reduction of Eps8 levels in G2/M-arrested cells (Fig. 1a). To test whether this reduction is also observed during normal cell cycle progression, we synchronized Hela cells using a double thymidine block/release protocol (Fig. 1b). Cyclin E26, Cyclin B127 and phosphorylated RanGAP128 served as S, G2/M, and M-Phase markers, respectively. Indeed, Eps8 levels started to decline approximately 7 h after release (predominantly late G2: Cyclin B high, RanGAP1 not yet phosphorylated) and reached S phase levels again at the 12 h time point (predominantly G1 phase). Eps8 levels inversely correlated with Cyclin B1 levels, and minimal Eps8 levels were observed when RanGAP1 phosphorylation was maximal (8.5-9 h release). Hence, we concluded that Eps8 is a cell cycle-regulated protein.
FIGURE 1. Eps8 is a cell cycle regulated protein that undergoes Fbxw5-mediated proteasomal degradation specifically during G2/M.
(a) Immunoblot analysis of exponentially growing (cycling), thymidine-, aphidocolin-, or hydroxurea-arrested (S-Phase), and STLC, nocodazole, or taxol-arrested HeLa cells (G2/M) with indicated antibodies.
(b) HeLa cells were synchronized by a standard double thymidine block/release protocol. Samples were taken after indicated time periods and lysates analyzed by immunoblotting. For comparison, lysate of nocodazole-arrested cells (as in (a)) were loaded (noc).
(c) HeLa cells were synchronized by a standard double thymidine block and released. After indicated time periods, cycloheximide (CHX) was added. Samples were taken at indicated time points after CHX addition, analyzed by immunoblotting. Eps8 levels were quantified relative to β-actin levels and plotted against time after addition of CHX. Eps8 levels at 0 min CHX were set to 100% (mean, +/- s.e.m. of three independent experiments).
(d) Exponentially growing HeLa cells and cells released from a double thymidine block for 5 h were incubated with 20 μM MG132 or mock-treated with DMSO for 3 h. Eps8 levels were anaylzed by immunoblotting (top panel) and quantified relative to β-actin levels (bottom panel); relative Eps8 levels of mock-treated cells set to 100 (mean, +/- s.e.m. of three independent experiments).
(e) Immunoblot analysis of HeLa cells transfected with non-targeting siRNA (si nt) or siFbxw5#1 for 42 h with indicated antibodies. 24 h after siRNA treatment, cells were treated with taxol or mock-treated with DMSO for 18 h. Eps8 levels were quantified relative to β-actin levels; relative Eps8 levels of control cells (si nt + DMSO) were set to 100 % (mean, +/- s.e.m. of three independent experiments). * cross-reactive band
To investigate whether fluctuating Eps8 levels are due to regulated degradation, we treated synchronized HeLa cells 2, 6, or 7 hours after thymidine release with cycloheximide. While Eps8 levels were stable in S-Phase (2 h after thymidine release), they rapidly decreased in cells that had been released from thymidine for 6 or 7 h (Fig. 1c). This reduction of Eps8 half-life to approximately 45 min indicates that Eps8 is significantly reduced but not completely degraded during G2 and M phase. Consistent with G2-specific Eps8 regulation, treatment of HeLa cells with MG132 for 3 hours increased Eps8 levels in G2 cells (released from thymidine-arrest for 5 hours), but not in unsynchronized cells (Fig. 1d). Thus, Eps8 undergoes specific degradation by the ubiquitin/proteasome pathway during G2. To address whether Fbxw5 contributes to G2-specific degradation of Eps8, we treated HeLa cells with Fbxw5 siRNA for 24 h, added taxol for 18 h, and investigated Eps8 levels by immunoblotting. While depletion of Fbxw5 for 42 h had no visible effect on asynchronous cells (Fig. 1e, Supplementary Fig. 2a, online), it efficiently blocked Eps8 degradation in taxol-arrested cells. Thus, Fbxw5 is required for Eps8 degradation in G2.
Eps8 is a substrate for SCFFbxw5- dependent ubiquitylation
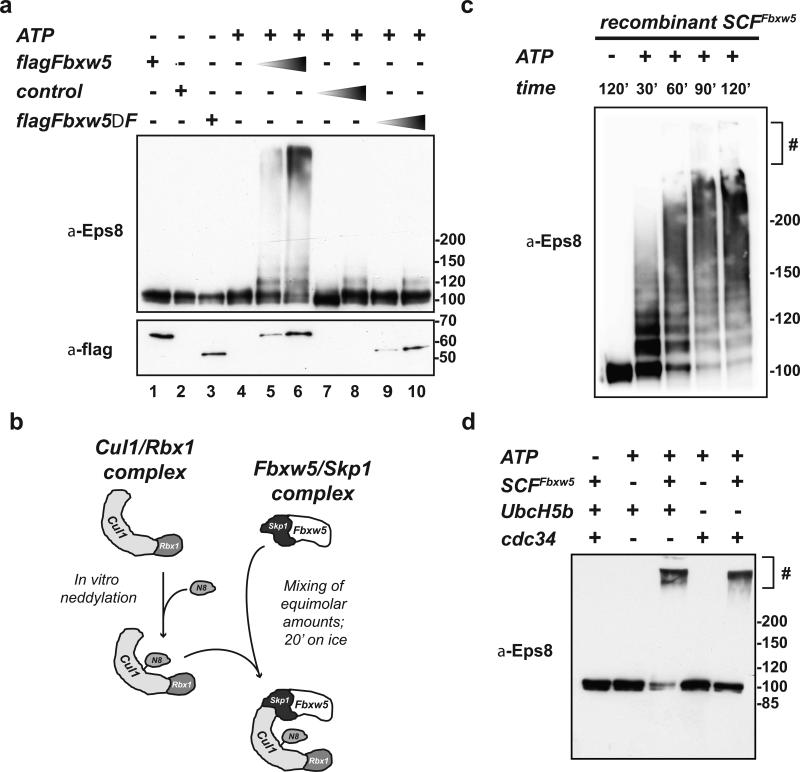
To test whether Eps8 is a target for Fbxw5-dependent ubiquitylation, we subjected purified His-Eps8 to in vitro ubiquitylation assays. First, we used mammalian Fbxw5 complexes as a source of E3 ligases. Increasing amounts of flag-Fbxw5 IP fractions (Fig. 2a, lanes 5-6) induced Eps8 ubiquitylation, while control IP fractions from mock-transfected cells did not (Fig 2a, lanes 7-8). Although the most likely interpretation of these findings was that Eps8 is ubiquitylated by SCFFbxw5 E3 ligase complexes, previous studies had suggested that Fbxw5 functions not only in the context of SCF complexes24, but also in the context of CRL4 complexes29,30. To determine which of these E3 ligases mediates Eps8 ubiquitylation, we included flag-Fbxw5ΔF-box in our analyses. This Fbxw5 variant is deficient in binding of Skp1, the linker of SCF E3s, but still capable of binding DDB1, the linker of CRL4 E3s (Supplementary Figure 3a,b). As shown in Figure 2a (lanes 9-10) flag-Fbxw5ΔF-box IPs showed no activity, indicating that Fbxw5 requires the SCF core for its activity towards Eps8.
FIGURE 2. Eps8 is a substrate of SCFFbxw5 in vitro.
(a) In vitro ubiquitylation reaction of His-Eps8 (0.2 μM) with 75 μM His-Ubiquitin, 170 nM Ube1, 1 μM UbcH5b, and 5 mM ATP in the absence or presence of different amounts of control (= flag-IPs from non-transfected cells), flag-Fbxw5, and flag-Fbxw5ΔFbox immunoprecipitates at 30°C for 120 min.
(b) For SCFFbxw5 reconstitution, mouse Fbxw5/Skp1 complexes were purified from bacteria (see material and methods) and Cul1/Rbx1 complexes were purified from E.coli using a ”split and co-express“ approach and in vitro neddylated as previously described31, 32. For complex formation, both sub-complexes were mixed in equimolar amounts and incubated on ice for 20 min.
(c) Time course of SCFFbxw5-dependent Eps8 ubiquitylation. 0.2 μM His-Eps8 was incubated with 75 μM His-Ubiquitin, 170 nM Ube1, 0.5 μM UbcH5b, 50 nM SCFFbxw5, and 5 mM ATP at 30°C. Reactions were stopped at indicated time points, and analyzed by immunoblotting. # stacking gel
(d) In vitro ubiquitylation of His-Eps8 (0.2 μM) with 75 μM His-Ubiquitin, 170 nM Ube1, 1 μM UbcH5b or Cdc34, 5 mM ATP in the absence and presence of 150 nM reconstituted SCFFbxw5 at 30°C for 90 min.
To exclude contribution of other proteins that might co-IP with flag-Fbxw5, we reconstituted SCFFbxw5 complexes. For this, we used an approach described for SCFSkp2 and SCFβTRCP1 (Fig. 2b)31, 32. First, we produced in vitro neddylated Cul1/Rbx1 (Nedd8*Cul1/Rbx1) complexes using proteins obtained exclusively from bacterial expression. For SCF complex formation, we mixed Nedd8*Cul1/Rbx1 in equimolar amounts with Fbxw5/Skp1 complexes, which we had separately purified from bacteria. These recombinant SCFFbxw5 complexes efficiently stimulated ubiquitylation of His-Eps8 (Fig. 2c), either in the presence of UbcH5b or Cdc34 (Fig. 2d), two E2 enzymes known to work with SCF complexes33, 34. In conclusion, SCFFbxw5 complexes are necessary and sufficient to ubiquitylate Eps8 in vitro.
Fbxw5 contributes to timely displacement of Eps8 from the cell cortex at the onset of mitosis
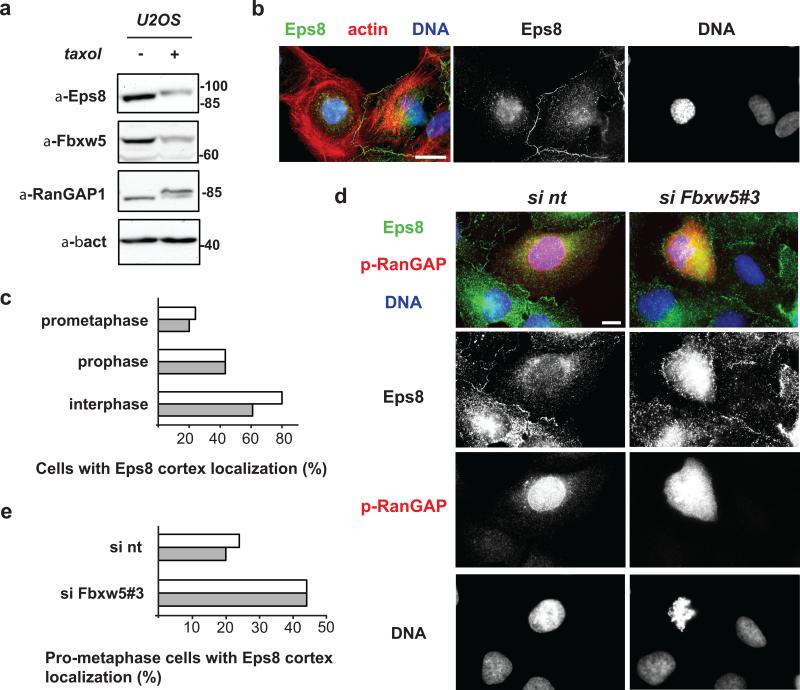
To add information on Eps8 localisation, we turned to U2OS cells; these cells express more Eps8 than HeLa cells and allow detection of endogenous Eps8 by immunofluorescence analysis. After confirmation that Eps8 is subject to cell cycle dependent degradation also in these cells (Fig. 3a, anti-Eps8 immunoblotting +/- taxol treatment), we analysed the localization of endogenous Eps8 in asynchronously growing U2OS cells. Consistent with previous studies3, 35, Eps8 was enriched at the cell cortex in the vast majority of interphase cells (Fig. 3b). Strikingly however, we found that more than 50% of prophase cells and more than 70% of pro-metaphase cells showed no prominent cell cortex association of Eps8 (Fig. 3c), revealing that - concomitant with its Fbxw5-mediated degradation at G2/M (Fig. 1, Fig. 3a) - Eps8 displaces from the cell cortex at the onset of mitosis. Hence, it was conceivable that Fbxw5 contributes to the removal of Eps8 from the cell cortex. To test this hypothesis, we analysed pro-metaphase cells of mock-transfected and Fbxw5-depleted U2OS cell population by immunofluorescence microscopy. As shown in Figure 3d (representative images) and Figure 3e (quantification), depletion of Fbxw5 resulted in almost twice as many pro-metaphase cells exhibiting Eps8 at the cell cortex compared to the control. Thus, Fbxw5-mediated degradation contributes to timely displacement of Eps8 from the cell cortex during the onset of mitosis.
FIGURE 3. Fbxw5 contributes to timely displacement of Eps8 from the cell cortex at the onset of mitosis.
(a) Immunoblot analysis of exponentially growing or taxol-arrested U2OS cells with indicated antibodies.
(b) U2OS cells were fixed and stained with anti-Eps8 antibodies, phalloidin, and Hoechst. Scale bar = 20 μm
(c) U2OS cells were fixed and stained with anti-Eps8 and phospho-RanGAP1 antibodies and Hoechst. Random pictures were taken and interphase, prophase, and prometaphase cells were counted and classified according to Eps8 localization. The percentage of cells with Eps8 cortex localization is depicted (two independent experiments, 25 cells were analyzed per cell cycle state and experiment).
(d) U2OS cells were treated with non-targeting siRNA (si nt) or siRNA targeting Fbxw5 (si Fbxw5#3) for 72 hours. Cells were fixed and stained with anti-Eps8 and anti-phospho-RanGAP1 antibodies and HOECHST. Scale bar = 10 μM.
(e) U2OS cells were treated as in (c), random pictures were taken, and cells in prometaphase were counted and classified with respect to Eps8 localization. The percentage of cells showing cortex localization is shown (two independent experiments, 25 cells were analyzed per cell cycle state and experiment)
SCFFbxw5-mediated Eps8 degradation in G2 is required for timely progression into metaphase
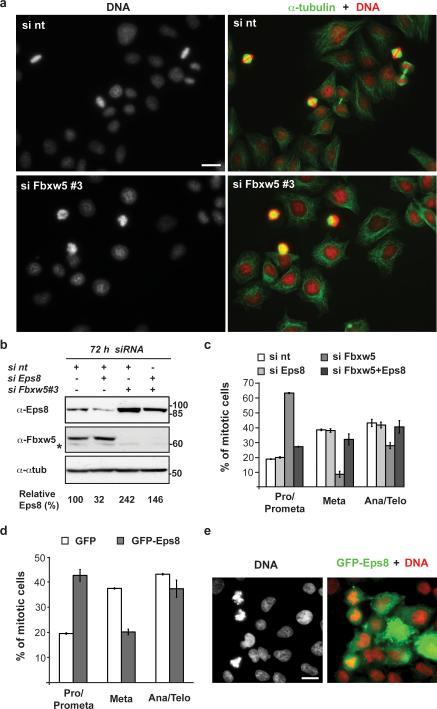
To begin to address whether SCFFbxw5-dependent Eps8 degradation contributes to mitotic progression, and to test for possible changes in tubulin and actin organisation, we transfected two different stable HeLa cell lines expressing either α-tubulin-GFP and H2B-RFP36 or Lifeact-GFP and H2B-RFP37 with non-targeting siRNA or siRNA targeting Fbxw5 and monitored cells undergoing mitosis in fluorescence microscopy time-lapses. Bipolar spindle formation (α-tubulin-GFP cells) and overall actin distribution (Lifeact-GFP cells) appeared normal at this level of resolution (Supplementary Fig. S4a and Supplementary Videos S1-S3, online). However, both cell lines exerted a striking delay in prometaphase upon Fbxw5 depletion (Supplementary information, Fig 4. b-c, video 3, online). Consistent with this, regular HeLa cells fixed 72 hours after treatment with Fbxw5 siRNA showed a significant accumulation of prometaphase cells when compared with control populations (Fig. 4a; corresponding Eps8 levels +/- siRNA are shown in Fig. 4b and Supplementary Fig. 2b, online). Quantification revealed an increase of cells in pro- and prometaphase from 19 % in control to 63 % in Fbxw5-depleted cells (Fig. 4c). This was accompanied by a decrease of cells in metaphase (from 39 % to 9 %) and ana-/telophase (from 43 % in control to 28 % in Fbxw5-depleted cells). Of note, the overall mitotic index of wt and Fbxw5-depleted HeLa cells remained the same (Supplementary Fig S5a,b, online).
FIGURE 4. Fbxw5-mediated Eps8 degradation contributes to progression into metaphase.
(a) HeLa cells were treated with non-targeting siRNA (si nt) or siRNAs targeting Fbxw5 (si Fbxw5 #3) for 72 h. Cells were fixed, stained with Hoechst and anti-α-tubulin antibodies. Scale bar = 20 μm.
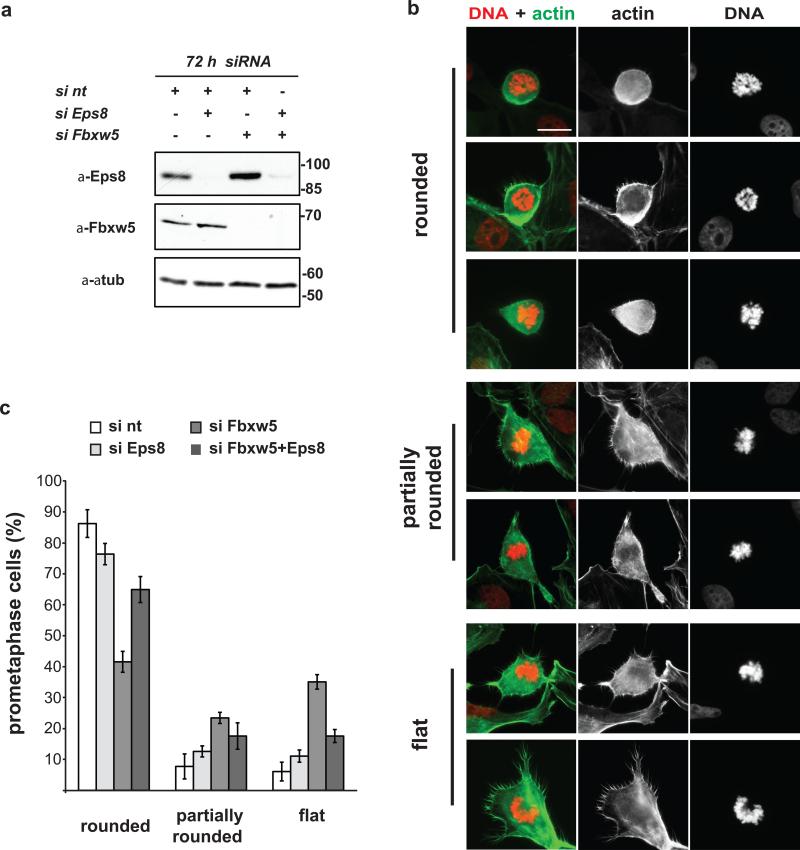
(b) Immunoblot analysis of HeLa cells transfected with different combinations of non-targeting siRNA (si nt) or siRNAs targeting Eps8 or Fbxw5#3 for 72 h with indicated antibodies. For each condition, final concentration of siRNAs was 10 nM.
(c) HeLa cells were treated with siRNA as in (a), fixed, and stained with anti-α-tubulin and anti-γ-tubulin antibodies plus Hoechst. Random pictures were taken and mitotic cells were counted and classified into different mitotic stages. For pictures exemplifying mitotic stages refer to Supplementary Information Fig 5a. The percentage of different mitotic stages is shown (mean, +/- s.e.m. of three independent experiments, ~50 mitotic cells were counted per condition and experiment).
(d) HeLa cells were transfected with GFP or GFP-Eps8, fixed, and stained with Hoechst. Random pictures were taken and GFP-positive, mitotic cells were counted and classified into different mitotic stages. The percentage of different mitotic stages is shown (mean +/- s.e.m. of three independent experiments, ~25 mitotic cells were counted per condition and experiment)
(e) Representative image of GFP–Eps8-transfected HeLa cells of the experiment in d. Scale bar, 20 μm. Uncropped images of blots are shown in Supplementary Fig. S7.
These findings suggested that Eps8 degradation is required for timely progression through prometaphase. However, E3 ligases frequently have many targets, and Fbxw5 is known to mediate degradation of tumor suppressor TSC229 and centriolar protein Sas-624. SAS-6 is required for centrosome duplication38, and both GFP-Sas-6 overexpression38 and siRNA-mediated Fbxw5 depletion24 cause centrosome overduplication and an increase of mitotic cells with multipolar spindles, which could cause a prometaphase delay. However, most cells had bipolar spindles. Consistent with this, we observed only few cells with multipolar spindles upon Fbxw5 siRNA knockdown for 48 or 72 h, and excluded them from our analyses. We employed two independent approaches to test whether Eps8 regulation is involved in prometaphase progression: first, rescue of the Fbxw5 depletion- induced prometaphase delay by co-depletion of Eps8 (Fig 4b,c) and second, investigation of mitotic stage distribution upon ectopic expression of GFP-Eps8 (Fig 4d-e). As shown in Fig. 4b, simultaneous depletion of Fbxw5 and Eps8 resulted in Eps8 levels slightly above those of control cells (siRNA against Eps8 only caused partial depletion, down to approx. 30%). Importantly, this reduction in Eps8 levels could restore a normal distribution of mitotic phases (Fig. 4c), while reduction of Eps8 alone had no effect. Moreover, ectopic expression of GFP-Eps8 (but not of GFP) was sufficient to significantly increase the percentage of mitotic cells in prometaphase (Fig 4d-e). Taken together, these data demonstrate that indeed Eps8 needs to be degraded by SCFFbxw5 for timely transition through pro-metaphase.
Failure to degrade Eps8 leads to a delay in cell rounding in early mitosis
The actin cytoskeleton undergoes dramatic changes to allow for cell shape changes required for proper transition into and out of mitosis39. Recently, the actin cross-linking protein Moesin40 and the RhoGEF Ect237 were identified as crucial regulators of mitotic cell rounding. Considering that Eps8 functions as an actin capper in interphase3, we hypothesized that increased Eps8 levels during early mitosis may interfere with cell rounding (life cell analysis provided first hints, but was not conclusive at our level of resolution). To test this idea, we transfected U2OS cells with different combinations of control, Fbxw5, and Eps8 siRNAs. As in HeLa cells (Fig. 4b), Eps8 siRNA treatment decreased - and Fbxw5 siRNA treatment increased - steady state levels of Eps8 72 h after transfection (Fig. 5a). Analysis of fixed cells stained with phalloidin and Hoechst revealed a striking accumulation of flat or only partially rounded prometaphase cells in Fbxw5-depleted compared to control populations (Fig. 5b): rounded prometaphase cells dropped from 86% in control to 42% in Fbxw5 knock down populations, while partially rounded (and flat) prometaphase cells increased from 8% (6%) in control to 23% (35%) in Fbxw5-depleted populations (Fig. 5c). Importantly, while removal of Eps8 alone had little effect, co-depletion of Fbxw5 and Eps8 partially rescued the cell-rounding defect (Fig. 5c). Together, these findings reveal that Fbxw5-mediated degradation of Eps8 at the onset of mitosis contributes to timely cell rounding.
FIGURE 5. Fbxw5-mediated Eps8 degradation contributes to timely cell rounding in early mitosis.
(a) Immunoblot analysis of U2OS cells transfected with different combinations of non-targeting siRNA (si nt) or siRNAs targeting Eps8 or Fbxw5#3 for 72 h with indicated antibodies. For each condition, final concentration of siRNAs was 10 nM.
(b) U2OS cells were treated with non-targeting siRNA (si nt) or siRNAs targeting Fbxw5 (si Fbxw5 #3) for 72 h. Cells were fixed, stained with Hoechst (DNA) and phalloidin (actin). Representative pictures of prometaphase cells are depicted. Scale bar = 20 μm
(c) U2OS cells were treated as in (a) and fixed and stained with phalloidin and Hoechst. Random pictures of prometaphase cells were taken. Prometaphase cells were counted and classified according to cell shape. The percentage of rounded, partially rounded, and flat prometaphase cells is shown (mean, +/- s.e.m. of three independent experiments, ~25 prometaphase cells were counted per condition and experiment).
Capping activity of Eps8 is required for proper exit from mitosis
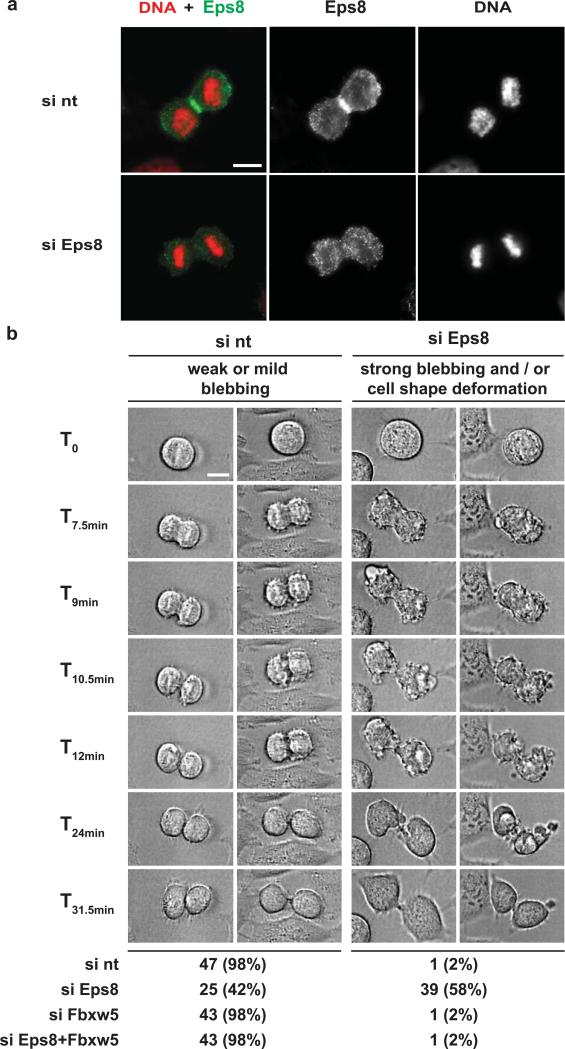
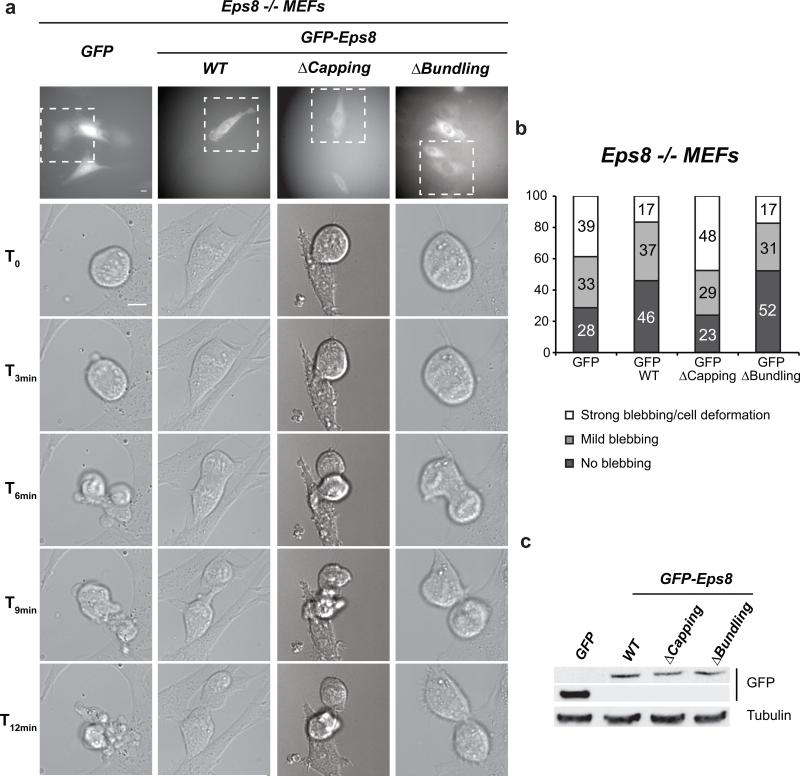
As shown in Figure 1b, Eps8 levels are restored once cyclin B levels are again low (12 hours after release from a double thymidine block). Consistent with this, a recent report revealed that Fbxw5 is inactivated via APC/C-mediated proteasomal degradation during mitosis24. We thus wondered when Eps8 would be needed again at the cell cortex - after metaphase/anaphase transition or only upon completion of M-phase? When analysing the localisation of endogenous Eps8 in U2OS cells, we could detect prominent enrichment of Eps8 at the equatorial cortex in anaphase and telophase (Fig. 6a). To test whether this pool plays a functional role in mitotic exit, we analysed mitotic HeLa cells transfected with different combinations of non-targeting, Fbxw5, and Eps8 siRNAs via live-cell imaging using differential interference contrast (DIC) microscopy. This revealed a striking blebbing phenotype: As has been observed by many labs (e.g.41, 42), control cells exhibited weak or intermediate membrane blebbing during progression through ana- and telophase and both developing daughter cells exhibited a round shape at all times (Figure 6b, Supplementary Information, video 4, online). The same was true for Fbxw5-depleted cells. In contrast, Eps8-depleted cells frequently displayed pronounced membrane blebbing and massive cell deformations after metaphase until completion of mitosis (61%, 39 out of 64 cells). The blebbing phenotype could be rescued by co-depletion of Fbxw5, indicating that it is indeed caused by lack of Eps8 (as shown in Fig. 4a, Eps8 knock down is partial in HeLa cells and Eps8 levels close to those of the control can be restored in the double knock-down). To verify these findings by an independent approach, we monitored mitotic Eps8 null mouse embryonic fibroblasts (MEF Eps8 -/-) infected with control viruses or viruses encoding for GFP-Eps8, a capping-deficient mutant of Eps8 (GFP-Eps8ΔCap7), or a bundling deficient mutant of Eps8 (GFP-Eps8ΔBund7) by DIC microscopy time lapses (Fig. 7a-c). Indeed, 39% (28 of 72) of control-infected MEFs exhibited strong blebbing and cell deformation during exit of mitosis, while only 17% Eps8-infected MEFs showed a comparable phenotype (Fig. 7b). Consistent with this, only 28% of control-infected MEFs but 46% of Eps8 expressing MEFs showed no recognizable sign of blebbing. Importantly, while the bundling mutant of Eps8 also rescued the blebbing phenotype of MEF Eps8 -/- cells, re-expression of GFP-Eps8ΔCap even aggravated blebbing and cell deformations during late mitosis. Together, these findings reveal that capping activity of Eps8 is required after metaphase for proper exit from mitosis.
FIGURE 6. Failure to restore Eps8 levels induces membrane blebbing and cell shape deformation after metaphase in HeLa cells.
(a) U2OS cells were treated with non-targeting siRNA (si nt) or siRNA targeting Eps8 (siEps8) for 72 hours. Cells were fixed and stained with anti-Eps8 antibodies and Hoechst. Scale bar = 10 μM.
(b) Still images of differential interference contrast (DIC) time lapses of mitotic HeLa cells. Cells were synchronized by a double thymidine block/release protocol and transfected with different combinations of si nt, si Eps8, and si Fbxw5 #3. 60 h after transfection, DIC time lapses were started and performed for 12-14 h (time interval 1.5 min). Representative examples of cells undergoing mitosis from control cells (left two time lapses) or Eps8-depleted cells (right two time lapses) are shown. ~40-60 cells were recorded per condition and classified as indicated. Scale bar = 10 μm
FIGURE 7. MEF Eps8 -/- cells exhibit a post-metaphase membrane blebbing phenotype that can be rescued by re-expression of Eps8 versions with actin capping activity.
(a) Still images of DIC time lapses of mitotic mouse embryo fibroblasts (MEFs). MEFs derived from Eps8 -/- mice5 and infected with either control pBABE-GFP vector (GFP) or pBABE-GFP-Eps8 WT, ΔCapping, or ΔBundling were seeded onto gelatine-coated coverslips at low confluency. The day after seeding, cells were subjected to time-lapse analysis (24 hours, time interval 2 minutes). 30 MEFs passaging through mitosis were recorded and classified per condition. Upper panels: fluorescence images taken immediately before the time-lapse. Lower panels: DIC images of the GFP-positive cells (white dashed line insets in upper panels) followed during mitosis. Bar = 10 μm.
(b) Quantification of the experiment in (a).
(c) Immunoblot analysis of expression levels of GFP-Eps8 versions in infected MEF Eps8 -/- cells. α-tubulin levels serve as loading control.
Discussion
SCF E3 ligases have been implicated in diverse aspects of cell cycle regulation e.g. through the F-box proteins Fbxw7, Skp2, β-TRCP, and Cyclin F43, 44. Here, we show that SCFFbxw5 contributes to mitotic progression by regulating the actin-binding protein Eps8. In combination with the recently described role of SCFFbxw5 in centrosome duplication during S-Phase24, 25, our findings identify Fbxw5 as an important player in cell cycle control. An intriguing question is how recognition of Eps8 specifically in G2 is controlled: while recognition by prototypic SCF E3 ligases typically requires target phosphorylation45, 46-51, both mock- or CIP-treated Eps8 from SF9 cells (Supplementary information Fig. 6a,b, online) are efficiently ubiquitylated by SCFFbxw5 in vitro. Thus, Fbxw5 has no absolute requirement for target phosphorylation. Whether Eps8 has a degron that can be regulated by stimulatory or inhibitory modifications in vivo remains to be seen. Target recognition could also be regulated at the level of the E3 ligase52. However, SCFFbxw5 complexes efficiently ubiquitylate Eps8 in vitro, irrespective of whether they are immuno-purified from mammalian cells or reconstituted from bacterially expressed proteins (Fig. 2, Supplementary information, Fig. 6 a,b). An inhibitory phosphorylation has been described for Fbxw5, Plk4-dependent phosphorylation of Ser151, in the context of Sas-6 degradation24. However, SCFFbxw5 variants with S151A or S151D showed no differences compared to wt SCFFbxw5 in binding or ubiquitylation of Eps8 (Supplementary information, Fig. 6 c,d, online). The fact that both Eps8 and SCFFbxw5 are competent for ubiquitylation in vitro raises the intriguing question how Fbxw5-dependent Eps8 degradation is restricted to G2 phase. One possibility is spatial control: Cortical Eps8 may be released from - or SCFFbxw5 may be recruited to - the cortex specifically in G2. It is interesting to note that a fraction of Fbxw5 undergoes a mobility shift at that time (Fig. 1b) and we will follow up on this in the future.
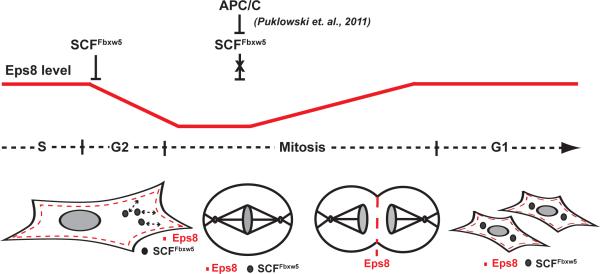
Eps8 controls actin-based protrusion events such as axonal filopodia growth, stereocilia length and cell migration by either capping or cross-linking actin filaments7,3, 4, 8, 53. Here, we discovered a so far unrecognized role for Eps8 in mitosis: it is required for cell shape stability late in mitosis and cytokinesis. On the other hand, cortical Eps8 inhibits timely cell rounding at the onset of mitosis, and its levels need to be temporally reduced by SCFFbxw5 dependent degradation in G2. These findings are intriguing in light of the emerging importance of actin cortex dynamics throughout M phase (e.g. for centrosome separation, spindle positioning, cytokinesis)37, 39, 42, 54, 55. In interphase cells, lammellipodia are composed of highly branched and short actin filaments (a situation that requires elevated levels of capping proteins56, 57), mitotic cells are characterized by long actomyosin-positive filaments under high tension39, 40. Recently, Moesin, a member of the ERM (Erzin, Radixin, Moesin) family of actin-binding proteins40 and the RacGEF Ect237 were identified as key players in driving cell rounding and spindle assembly through establishment of a stiff, rounded metaphase cortex. Given that under conditions of reduced Eps8 capping activity the actin network can become sparser10 and presumably less stiff, we speculate that displacement of Eps8 from the cortex at the onset of mitosis favours changes in actin network architecture to facilitate ERM- and Ect2-driven cell rounding and cortex stiffening. Consistent with this, Fbxw5-depleted (Fig. 5), Moesin-depleted40, and Ect2-depleted37 cells exhibit similar defects in cell rounding at the onset of mitosis. After metaphase-anaphase transition, insufficient Eps8 capping activity causes dramatic membrane blebbing and cell shape deformations (Fig. 6b, 7). Given the recently described roles of membrane blebs and actomyosin contractility in cytokinesis42, these observations suggest that Eps8 is required for the establishment of an actin-rich furrow and the generation of a branched actin network to fill blebs. Collectively, our data are consistent with the following model (Fig. 8): SCFFbxw5-mediated degradation contributes to displacement of Eps8 from the cell cortex in late G2 to facilitate cell rounding and establishment of an ERM-linked actomyosin contractile cortex network in metaphase. Before or at the onset of anaphase, SCFFbxw5 is inactivated by APC/CCdc20-mediated degradation24. This prevents further degradation of Eps8 and allows enrichment of newly synthesized and/or residual Eps8 at the equatorial cortex, which in turn contributes to changes in the actin cortex architecture favouring proper completion of mitosis. How Eps8, ERM proteins and Ect2 cooperate in regulating mitotic cell shape changes will thus be an important field for future studies. An intriguing question that arises from our study is also whether the contribution of aberrant Eps8 levels to tumor development 9, 12-16 is mediated - at least in part - by improperly executed mitosis.
FIGURE 8.
Model: SCFFbxw5 regulates Eps8 levels to control actin dynamics for proper mitotic progression
Material and Methods
Constructs
pCDNA3.1-flag-Fbxw5 was constructed by PCR amplification of Fbxw5 from HeLa cDNA and subsequent cloning of the PCR product into pCDNA3.1-flag-USP2558 using BamHI and NotI sites (thereby replacing USP25). To obtain pCDNA3.1-flag-Fbxw5ΔF-box, an Fbxw5 construct lacking the F-box domain, nucleotides corresponding to aa 81-566 were PCR amplified from pCDNA3.1-flag-Fbxw5 and were cloned into BamHI and XhoI sites of pCDNA3.1-flag-USP25. Full length Eps8 cDNA was obtained by PCR and cloned in pBABE-hygro vector59.
Protein Preparation
Purification of mouse His-Eps8 from SF9 cells and human Ube1-His, UbcH5b, Cdc34, and His-Ubiquitin from E.coli were described previously 3, 58, 60, 61. Mouse Fbxw5 was purified in complex with Skp1 from bacteria. In short, pRSFDuet-HisMBP-mFbxw5 and pGEX6P1-GST-Skp1 were co-transformed in BL21(DE3) and protein production was induced by 0.5 mM IPTG at an OD600 of 0.8. Cells were grown at 16°C over night, harvested by centrifugation and resuspended in Buffer A (50 mM Tris pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol, 10 mM imidazole, and protease inhibitors). Cells were lyzed, the lysate was cleared by centrifugation, and passed over Ni-NTA agarose (Qiagen). After washing with buffer A containing 20 mM imidazole, the His-MPB-mFbxw5/GST-Skp1 complex was eluted with buffer A containing 250 mM imidazole and further purified by molecular sieving over a preparative Superdex 200 column in Buffer B (50 mM Tris pH 8.0, 100 mM NaCl, 5 mM β-mercaptoethanol). For tag cleavage, TEV protease was added, followed by incubation at 4°C for 12 h. Separation of untagged mFbxw5/Skp1 complex from tags and TEV protease was achieved by a second run over a preparative Superdex 200 column in buffer B. mFbxw5/Skp1 fractions were concentrated, aliquoted, flash-frozen in liquid nitrogen, and stored at -80°C. For in vitro reconstitution of SCFFbxw5 complexes, mouse Fbxw5 purified in complex with Skp1 from bacteria was mixed in equimolar amounts with Nedd8*Cul1/Rbx1 complexes (generated as described previously32) and incubated on ice for 20 min.
Antibodies
Anit-Fbxw5 antibodies were raised in rabbits against GST-Fbxw5 and affinity-purified (0.12 μg/mL in IB). Anti-RanGAP1 sera (1:3,000 in IB) were previously described28. Anti-β-actin (clone AC-74, A2228 Sigma, 1:10,000 in IB), Anti-Cyclin B1 (clone V152, #4135 Cell Signaling, 1:1,000 in IB), Anti-Cyclin E (clone HE12, #4129 Cell Signaling, 1:1,000 in IB), Anti-flag (clone M2, F3165 Sigma, 1:5,000 in IB), anti-HA11 (MMS-101P, Covance, 1:5,000 in IB), anti-Eps8 (clone 15/Eps8, #610144 BD Bioscience, 1:1,000 in IB and 1:100 in IF), anti-DDB1 (A300-462A Bethyl, 1:1,000 in IB), anti-Skp1 (sc-1568 Santa Cruz, 1:250 in IB), anti−α-tubulin (clone DM1A, T6199 Sigma, 1:10,000 in IB, 1:1000 in IF), and anti-γ-tubulin (T5192 Sigma, 1:500 in IF) antibodies were commercially purchased.
Cell culture and transfections
HeLa, U2OS, and HEK293T cells were maintained in DMEM with 10% fetal bovine serum. For HeLa cells stably expressing α-tubulin-GFP and H2B-RFP (kind gift from D. Gerlich), 1mg/mL puromycin and 400 μg/mL G418 were supplemented to the medium. HeLa (CSH) suspension cells were propagated in Jokliks medium supplemented with 5 % (v/v) NCS, 5 % (v/v) FBS. Immortalized fibroblasts from Eps8 -/- embryos were generated and maintained as previously described5. For synchronization, HeLa cells or HeLa cells stably expressing α-tubulin-GFP and H2B-RFP were subjected to a double thymidine block following standard procedures (18 h 2 mM thymidine, 9 h release, 18 h 2 mM thymidine). For arrest at G2/M transition, HeLa cells were treated with 75 ng/mL nocodazole, 10 nM taxol, or 10 μM STLC for 18 h. Cycloheximide chases were performed using 40 μg/mL CHX. Plasmid transfection of HEK293T and HeLa cells was with calcium phosphate and JetPRIME™ (Peqlab), respectively. For generation of HEK293T cells stably expressing flag-Fbxw5, HEK 293T cells were transfected with pCDNA3.1-flag-Fbxw5 and single clones were selected with G418. siRNA transfection of adherent HeLa cells was with Lipofectamine RNAiMAX (Invitrogen) according to manufacturer's instructions using 10 nM siRNA. siRNA sequences were:
| name | Sense (5’→3’) | Antisense (5’→3’) | Sequence Origin |
| Non-targeting | UAGCGACUAAACACAUCAAUU | UUGAUGUGUUUAGUCGCUAUU | Ambion |
| Fbxw5 #1 | CCACAGGCGCCAAGAGCAAdTdT | UUGCUCUUGGCGCCUGUGGdTdT | Designed within this work |
| Fbxw5 #2 | CGGGAGAGGUGGAGAUGCUdTdT | AGCAUCUCCACCUCUCCCGdTdT | Designed within this work |
| Fbxw5 #3 | CCCUACAACUGGAGCUACAdTdT | UGUAGCUCCAGUUGUAGGGdTdT | Designed within this work |
| Fbxw5 #4 | GGACCACGUCAUAGACAUAtt | UAUGUCUAUCAGUGGUCCag | Pre-designed (Applied Biosystems) |
| Eps8 | GGCCCUUUAUGAACAAGGdTdT | CCUUUGUUCAUAAAGGGCCdTdT | Previously described9 |
GFP-expressing and GFP-Eps8(WT/ΔBundling/ΔCapping) re-expressing MEFs were obtained as described previously7.
Immunoprecipitations
Anti-flag immunoprecipitations (IPs) were performed from extracts of HEK293T cells transiently or stably expressing flag-Fbxw5(Δ–Fbox). Lysis was in two pellet volumes of 20 mM HEPES pH 7.3, 110 mM KOAc, 2 mM Mg(OAc)2, 5 mM EDTA, 5 mM EGTA, 0.2% Tween20, protease inhibitors, and 10 mM NEM. Lysates were cleared by centrifugation and incubated with ANTI-FLAG-M2 agarose (Sigma) for 2 h at 4 °C. After washing with lysis buffer containing no NEM, bound proteins were eluted with lysis buffer containing 0.5 mg/mL flag peptide. Anti-Fbxw5 IPs were performed from HeLa suspension cell lysates (108 cells per sample prepared as above). After incubation with Fbxw5 or control antibodies at 4°C for 1h, Protein A beads (Roche) were added for 2h. After washing, bound proteins were eluted with 2x SDS sample buffer.
In vitro ubiquitylation assays
In vitro ubiquitylation assays were performed following previously described protocols60. 170 nM Ube1, 0.5-1 μM UbcH5b or Cdc34, 75 μM His-Ubiquitin, 0.2 μM His-Eps8, and 5 mM ATP were incubated in the absence or presence of flag-Fbxw5 immunoprecipitates or 75-150 nM in vitro reconstituted SCFFbxw5 in SAB buffer (20 mM HEPES pH 7.3, 110 mM KOAc, 2 mM Mg(OAc)2, 1mM EGTA, 0.2 mg/ml ovalbumin, 0.05 % (v/v) Tween 20, 1 mM DTT, and protease inhibitors) at 30°C. Reactions were stopped by addition of 1 volume of 2x SDS sample buffer. Samples were analyzed by SDS PAGE followed by immunoblotting.
Mass Spectrometry
For mass spectrometry analysis, gel lanes of flag-Fbxw5 and control flag IP were cut into slices and each gel slice was subjected to in-gel digestion using Trypsin62. Peptides were extracted and analysed by LC-coupled MSMS (Q-Tof Ultima, Waters) under standard conditions, and proteins were identified by searching the fragment spectra against NCBInr database using Mascot as search engine. Taxonomy was human, two missed cleavages were allowed, mass deviation for MS was set to 30 ppm, and for MSMS 0.8 amu. Oxidations at M, W, H, and carbamidomethyl at C were chosen as variable modifications. Only protein hits with a score higher than 20 and absent in the control lane were considered putative Fbxw5 interactors.
Immunofluorescence microscopy
For immunofluorescence analysis, HeLa cells were seeded on converslips and transfected with various combinations of siRNAs for 72 h. Cells were fixed with methanol or PFA and stained with indicated antibodies, alexa488-labeled phalloidin (Invitrogen) or Hoechst. Coverslips were mounted and analyzed by fluorescence microscopy using an Axiovert ObserverZ.1 microscope system (Zeiss).
Life cell imaging
Live cell microscopy of HeLa cells or HeLa cells stably expressing α-tubulin-GFP and H2B-RFP was performed on a an xcellence IX81 microscope system (Olympus) using an UPLSAPO 40x/0.95 numerical aperture (NA) air objective lens (Olympus) and appropriate fluorescence filter and differential interference contrast (DIC) settings, respectively. Cells were kept at 37°C, 95% air, 5% CO2 with a microscope stage incubator (okolab). For fluorescence imaging, Z-stacks were acquired over time with step widths guaranteeing partially overlapping image sections. The images were spectrally unmixed and deconvolved using Wiener filtering. All fluorescence images are displayed as maximum intensity projections. Image processing was performed using xcellence imaging software (Olympus) and ImageJ (NIH, http://rsbweb.nih.gov/ij/).
Live cell microscopy of MEFs was performed using an Olympus IX-81 microscope (objective Olympus UPlanApo 40x NA 0,70) equipped with a motorized stand and a cell incubator for live microscopy. The microscope was connected to a Photometrics Cascade 1K camera (pixel dimension 8 μm).
Supplementary Material
Acknowledgements
We kindly acknowledge J. Ellenberg (EMBL, Heidelberg), B. Baum (MRC-LMCB, England) and D. Gerlich (IMBA, Austria) for their kind gifts of stable Hela cell lines. P. Di Fiore, O. Gruss, B. Mardin and A. Maccario are ackknowledged for helpful discussions, F. Milanesi for Eps8 purification, R. Geiß-Friedlander for Fbxw5 construct generation, U. Gern for excellent technical assistance, and B. Mardin, S. Barysch, A. Flotho and U. Winter for critical manuscript reading. This work was supported by the Deutsche Forschungsgemeinschaft (SPP136, ME2279/3-1; F.M and A.W.) and NoE RUBICON, by ALSAC and NIH (2R01GM069530) to B.A.S., by the Associazione Italiana per la Ricerca sul Cancro (AIRC) to G.S and A.D.; by the Italian Ministries of Education-University-Research (MIUR-PRIN), the International Association For Cancer Resear, and the European Research Council to G.S.; B.A.S. is an Investigator of the Howard Hughes Medical Institute. M.F.C. is an HHMI fellow of the Damon Runyon Cancer Research Foundation.
Footnotes
Author contribution
A.W. designed and carried out most experiments and wrote the manuscript. A.D. and G.S. carried out analysis of Eps8 knock-out MEFs and contributed to experimental design and manuscript writing. N.R. and G.H. carried out some of the experiments. J.B. contributed to the generation of recombinant SCFFbxw5 complexes. M.C. and B.S. provided expert help in the reconstitution of SCF E3 ligases and provided neddylated Cul1/Rbx1 complexes. H.L. provided expert help in time-lapse microscopy and generated the videos. H.U. carried out mass spectrometry analysis. F.M. guided the project, designed experiments, and wrote the manuscript.
References
- 1.Fazioli F, et al. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993;12:3799–3808. doi: 10.1002/j.1460-2075.1993.tb06058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Fiore PP, Scita G. Eps8 in the midst of GTPases. Int J Biochem Cell Biol. 2002;34:1178–1183. doi: 10.1016/s1357-2725(02)00064-x. [DOI] [PubMed] [Google Scholar]
- 3.Disanza A, et al. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol. 2004;6:1180–1188. doi: 10.1038/ncb1199. [DOI] [PubMed] [Google Scholar]
- 4.Disanza A, et al. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol. 2006;8:1337–1347. doi: 10.1038/ncb1502. [DOI] [PubMed] [Google Scholar]
- 5.Scita G, et al. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 1999;401:290–293. doi: 10.1038/45822. [DOI] [PubMed] [Google Scholar]
- 6.Scita G, et al. An effector region in Eps8 is responsible for the activation of the Rac-specific GEF activity of Sos-1 and for the proper localization of the Rac-based actin-polymerizing machine. J Cell Biol. 2001;154:1031–1044. doi: 10.1083/jcb.200103146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertzog M, et al. Molecular basis for the dual function of Eps8 on actin dynamics: bundling and capping. PLoS Biol. 2010;8:e1000387. doi: 10.1371/journal.pbio.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menna E, et al. Eps8 regulates axonal filopodia in hippocampal neurons in response to brain-derived neurotrophic factor (BDNF). PLoS Biol. 2009;7:e1000138. doi: 10.1371/journal.pbio.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yap LF, et al. Upregulation of Eps8 in oral squamous cell carcinoma promotes cell migration and invasion through integrin-dependent Rac1 activation. Oncogene. 2009;28:2524–2534. doi: 10.1038/onc.2009.105. [DOI] [PubMed] [Google Scholar]
- 10.Frittoli E, et al. The Signaling Adaptor Eps8 Is an Essential Actin Capping Protein for Dendritic Cell Migration. Immunity. 2011;35:388–399. doi: 10.1016/j.immuni.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manor U, et al. Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Curr Biol. 2011;21:167–172. doi: 10.1016/j.cub.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maa MC, Hsieh CY, Leu TH. Overexpression of p97Eps8 leads to cellular transformation: implication of pleckstrin homology domain in p97Eps8-mediated ERK activation. Oncogene. 2001;20:106–112. doi: 10.1038/sj.onc.1204069. [DOI] [PubMed] [Google Scholar]
- 13.Chen YJ, Shen MR, Maa MC, Leu TH. Eps8 decreases chemosensitivity and affects survival of cervical cancer patients. Mol Cancer Ther. 2008;7:1376–1385. doi: 10.1158/1535-7163.MCT-07-2388. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Patel V, Miyazaki H, Gutkind JS, Yeudall WA. Role for EPS8 in squamous carcinogenesis. Carcinogenesis. 2009;30:165–174. doi: 10.1093/carcin/bgn252. [DOI] [PubMed] [Google Scholar]
- 15.Xu M, et al. Epidermal Growth Factor Receptor Pathway Substrate 8 (Eps8) Is Overexpressed in Human Pituitary Tumors: Role in Proliferation and Survival. Endocrinology. 2009;150:2064–2071. doi: 10.1210/en.2008-1265. [DOI] [PubMed] [Google Scholar]
- 16.Welsch T, Endlich K, Giese T, Buchler MW, Schmidt J. Eps8 is increased in pancreatic cancer and required for dynamic actin-based cell protrusions and intercellular cytoskeletal organization. Cancer Lett. 2007;255:205–218. doi: 10.1016/j.canlet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Welsch T, et al. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp Cell Res. 2010;316:1914–1924. doi: 10.1016/j.yexcr.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 20.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin- RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 23.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 24.Puklowski A, et al. The SCF-Fbxw5 E3-ubiquitin ligase is regulated by Plk4 and targets HsSAS-6 to control centrosome duplication. Nat Cell Biol. 2011 doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 25.Pagan J, Pagano M. FBXW5 controls centrosome number. Nat Cell Biol. 2011;13:888–890. doi: 10.1038/ncb2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Won KA, Reed SI. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 27.Takizawa CG, Morgan DO. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr Opin Cell Biol. 2000;12:658–665. doi: 10.1016/s0955-0674(00)00149-6. [DOI] [PubMed] [Google Scholar]
- 28.Swaminathan S, et al. RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly. J Cell Biol. 2004;164:965–971. doi: 10.1083/jcb.200309126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J, et al. WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 2008;22:866–871. doi: 10.1101/gad.1624008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Zhou P. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Li T, Pavletich NP, Schulman BA, Zheng N. High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 2005;398:125–142. doi: 10.1016/S0076-6879(05)98012-9. [DOI] [PubMed] [Google Scholar]
- 32.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Provenzano C, et al. Eps8, a tyrosine kinase substrate, is recruited to the cell cortex and dynamic F-actin upon cytoskeleton remodeling. Exp Cell Res. 1998;242:186–200. doi: 10.1006/excr.1998.4095. [DOI] [PubMed] [Google Scholar]
- 36.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 37.Matthews HK, et al. Changes in ect2 localization couple actomyosin- dependent cell shape changes to mitotic progression. Dev Cell. 2012;23:371–383. doi: 10.1016/j.devcel.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 39.Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009;19:174–179. doi: 10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Kunda P, Pelling AE, Liu T, Baum B. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol. 2008;18:91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- 41.Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci U S A. 2007;104:7939–7944. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedzinski J, et al. Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature. 2011;476:462–466. doi: 10.1038/nature10286. [DOI] [PubMed] [Google Scholar]
- 43.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21:816–824. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D'Angiolella V, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skaar JR, Pagan JK, Pagano M. SnapShot: F box proteins I. Cell. 2009;137:1160–1160. e1161. doi: 10.1016/j.cell.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 46.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 47.Wu G, et al. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 48.Hao B, et al. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 50.Yang CS, et al. FBW2 targets GCMa to the ubiquitin-proteasome degradation system. J Biol Chem. 2005;280:10083–10090. doi: 10.1074/jbc.M413986200. [DOI] [PubMed] [Google Scholar]
- 51.Deshaies RJ, Ferrell JE., Jr. Multisite phosphorylation and the countdown to S phase. Cell. 2001;107:819–822. doi: 10.1016/s0092-8674(01)00620-1. [DOI] [PubMed] [Google Scholar]
- 52.Song L, Rape M. Substrate-specific regulation of ubiquitination by the anaphase-promoting complex. Cell Cycle. 2011;10:52–56. doi: 10.4161/cc.10.1.14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croce A, et al. A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol. 2004;6:1173–1179. doi: 10.1038/ncb1198. [DOI] [PubMed] [Google Scholar]
- 54.Paluch E, Sykes C, Prost J, Bornens M. Dynamic modes of the cortical actomyosin gel during cell locomotion and division. Trends Cell Biol. 2006;16:5–10. doi: 10.1016/j.tcb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 55.Stewart MP, et al. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 2011;469:226–230. doi: 10.1038/nature09642. [DOI] [PubMed] [Google Scholar]
- 56.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133:841–851. doi: 10.1016/j.cell.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bear JE. Follow the monomer. Cell. 2008;133:765–767. doi: 10.1016/j.cell.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 58.Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin- specific protease 25. Mol Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 59.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petroski MD, Deshaies RJ. In vitro reconstitution of SCF substrate ubiquitination with purified proteins. Methods Enzymol. 2005;398:143–158. doi: 10.1016/S0076-6879(05)98013-0. [DOI] [PubMed] [Google Scholar]
- 61.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 62.Shevchenko A, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci U S A. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.