Abstract
Although oxidative stress is a key aspect of innate immunity, little is known about how host-restricted pathogens successfully repair DNA damage. Base excision repair (BER) is responsible for correcting nucleobases damaged by oxidative stress, and is essential for bloodstream infection caused by the human pathogen, Neisseria meningitidis. We have characterised meningococcal BER enzymes involved in the recognition and removal of damaged nucleobases, and incision of the DNA backbone. We demonstrate that the bi-functional glycosylase/lyases Nth and MutM share several overlapping activities and functional redundancy. However MutM and other members of the GO system, which deal with 8-oxoG, a common lesion of oxidative damage, are not required for survival of N. meningitidis under oxidative stress. Instead, the mismatch repair pathway provides back-up for the GO system, while the lyase activity of Nth can substitute for the meningococcal AP endonuclease, NApe. Our genetic and biochemical evidence show that DNA repair is achieved through a robust network of enzymes that provides a flexible system of DNA repair. This network is likely to reflect successful adaptation to the human nasopharynx, and might provide a paradigm for DNA repair in other prokaryotes.
INTRODUCTION
A challenge faced by all organisms is to preserve integrity of their DNA in the face of assault by environmental agents. This is particularly critical for pathogens that exist for the majority of their lifecycle in intimate association with their mammalian hosts and are subject to oxidative stress as a key component of the immune system. Failure to repair oxidative nucleobase damage results in aberrant cellular functioning, genome instability during DNA replication, and the accumulation of heritable deleterious mutations (Friedberg et al., 2006). Base excision repair (BER) is the primary mechanism by which living cells repair DNA lesions following oxidative damage (Bjelland & Seeberg, 2003). However, there are remarkably few studies detailing the pathways and mechanisms of repair of oxidative DNA damage in pathogenic microbes.
Neisseria meningitidis is a leading cause of meningitis and sepsis (Tzeng & Stephens, 2000), and has a host-restricted lifestyle with no reservoir outside the human nasopharynx (Cartwright et al., 1987). In this niche, the meningococcus has evolved exquisite mechanisms to avoid the immune system by expressing surface antigens that perfectly mimic host molecules (Finne et al., 1983), undergo rapid antigenic variation (Plant & Jonsson, 2003, Merz & So, 2000, Deitsch et al., 2009) and imitate host carbohydrates to recruit human complement regulators (Schneider et al., 2009). This highly evolved adaptation to the human host will also occur at the level of metabolism (Exley et al., 2005) and during repair of oxidative DNA damage. Indeed, in comparison to the widely studied Escherichia coli, the meningococcus has a limited repertoire of BER enzymes and lacks an SOS response to damaged DNA (Black et al., 1998, Davidsen & Tonjum, 2006). These features, along with its genetic tractability, make the meningococcus an excellent organism for understanding the function and integration of enzymes involved in DNA repair in a host adapted pathogen.
BER is conventionally thought to be organised into three distinct pathways that are initiated by recognition of different damaged nucleobases, and which then proceed independently and are characterised by co-operative activity between enzymes and hand-off of DNA substrates from enzyme to enzyme in an ordered fashion (Wiederhold et al., 2004, Friedberg et al., 2006). In the first pathway, mono-functional glycosylases cleave the glycosidic bond linking the damaged nucleobase to the DNA backbone, leaving an apurinic/apyrimidinic (AP) site. AP sites are substrates for AP endonucleases, which incise the DNA backbone, leaving a 3′-hydroxyl (3′-OH, a substrate for DNA ligase) and 5′-deoxyribose phosphate (5′-dRP), which is converted into a 5′-phosphate (Friedberg et al., 2006). The two remaining pathways involve bi-functional glycosylases, enzymes which eliminate damaged nucleobases, incise the DNA strand through their lyase activity, and generate different products from AP endonucleases. Bi-functional glycosylases of the Nth family use β-elimination to yield a 3′-unsaturated aldehyde (3′-Ald), while β-δ-elimination by members of the MutM/Fpg family generate a 3′-phosphate (3′-PO4) (Friedberg et al., 2006); both these 3′ blocking lesions must be processed before successful DNA repair and replication can be completed.
Current understanding of BER in the meningococcus is limited, even though this process is required for disseminated bloodstream infection (Carpenter et al., 2007), a pre-requisite in pathogenesis. N. meningitidis has two AP endonuclease paralogues, but only one of these, NApe, is a functional AP endonuclease (Carpenter et al., 2007). The other enzyme, NExo, shares 3′-deoxyribose phosphodiesterase (3′-RPase, which processes 3′-Ald) activity with NApe (Carpenter et al., 2007), but has evolved to be a specialised 3′-phosphatase (Silhan et al., 2011). Additionally, the bacterium is known to possess the bi-functional glycosylase, MutM (also called Fpg), which excises 8-oxoG (Tibballs et al., 2009), and MutY which is an anti-mutator responsible for excising adenine (A) mismatched with 8-oxoG (Davidsen et al., 2005).
Here we sought to further understand how the meningococcus withstands oxidative stress and nucleobase damage. Our work reveals overlap of enzyme activity and functional redundancy at the level of elimination of damaged nucleobases by the bi-functional glycosylases Nth and MutM, as well as during strand incision at AP sites. This contrasts with the known specialization of enzyme function during the processing of 3′ blocking lesions generated following strand incision (Silhan et al., 2011). There was no evidence of co-operativity between the bi-functional glycosylases with enzymes (NApe and NExo) that process 3′ blocking lesions that would indicate the enzymes act in discrete pathways. We also examined whether other DNA repair pathways, including nucleotide excision and mismatch repair pathways, might contribute to protection against reactive oxygen species, and found that they provide further back-up for BER. Therefore meningococcal BER is organised as an integrated, robust network of enzymes that share overlapping functions, and the route of repair is determined by the relative abundance and specific activity of enzymes rather than along ordered pathways. The network model is likely to provide a paradigm for repair in other prokaryotic pathogens.
RESULTS
Nth and MutM share redundant functions during survival against oxidative stress
N. meningitidis possesses two putative bi-functional glycosylases, MutM and Nth (Davidsen & Tonjum, 2006). E. coli MutM was initially named Fpg because formamidopyrimidine (FAPY) was its first characterised substrate (Chetsanga & Lindahl, 1979, Boiteux et al., 1989). However, as the E. coli and meningococcal enzymes both recognise other DNA lesions including 8-oxoG (Tibballs et al., 2009, Morland et al., 2002), we refer to this enzyme as MutM (Nghiem et al., 1988). Nothing is known about the activity of the putative meningococcal Nth (encoded by NMB0533), which shares 72% amino acid identity with the E. coli enzyme that recognises a broad range of oxidised pyrimidines (Dizdaroglu et al., 1993).
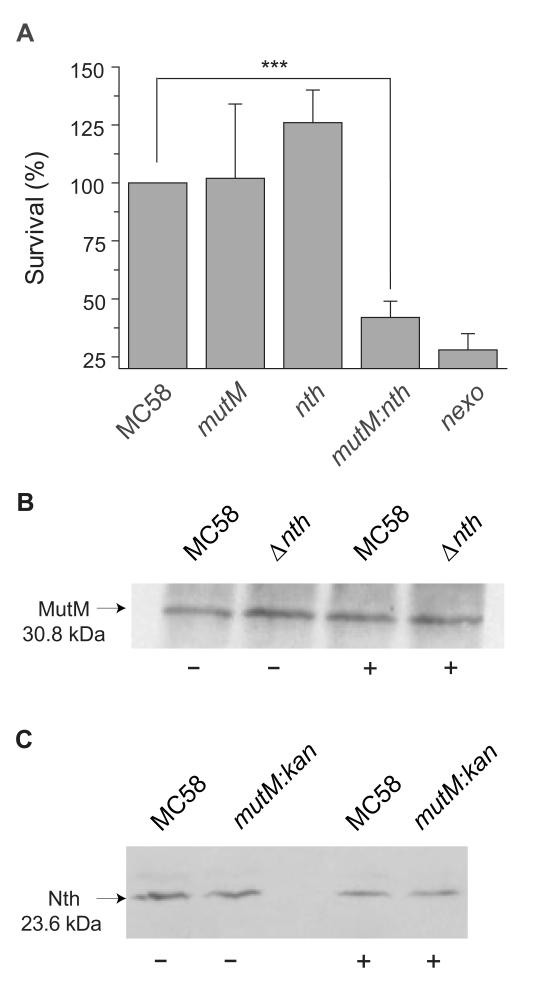
To define the contribution of Nth and MutM to the ability of N. meningitidis to withstand oxidative stress, we constructed strains lacking one or both enzymes, and examined their survival following exposure to 0.4 mM H2O2 over 30 min (Fig. 1A). Deletion of either MutM or Nth alone did not affect the survival of N. meningitidis in the face of oxidative stress. However, deletion of both enzymes led to a significant decrease in the ability of bacteria to withstand oxidative killing; MC58ΔmutMΔnth was recovered at 42% of the level of the wild-type strain (S.D. 6.8%, p<0.0001) following exposure to H2O2. This indicates that Nth and MutM are functionally redundant under these conditions, with the enzymes able to compensate for each other.
Figure 1.
Sensitivity of strains to oxidative stress and enzyme levels.
(A) Bacteria were incubated in 0.4 mM H2O2 or PBS at RT for 30 min, and the number of surviving bacteria determined by plating to solid media. The percentage survival was calculated by comparing the number of CFU recovered in the presence and absence of H2O2, and shown as the percentage survival relative to the wild-type strain, MC58. Error bars show the S.D. of assays performed in triplicate at least (*** indicates p < 0.0001, 1-column Student’s t test); a mutant lacking NExo is included for comparison. (B and C) Western blot analysis with anti-MutM and Nth antibodies showing protein levels in N. meningitidis incubated with (+) or without (−) H2O2. Whole-cell lysates of N. meningitidis MC58 or strains lacking mutM or nth were separated by 12% SDS–PAGE and transferred to membranes, which were incubated with anti-MutM (Panel B, 1:5000 dilution) or anti-Nth (Panel C, 1:3000 dilution) sera. Strains are shown above each lane, and the predicted molecular mass of each protein is indicated.
Next we examined whether the expression of Nth and MutM is affected by conditions of oxidative stress or the presence/absence of the other enzyme. Cellular levels of Nth and MutM in the wild-type strain MC58, grown with or without 0.4 mM H2O2, were analysed by Western blot analysis using polyclonal immune sera raised against the recombinant proteins; the levels of these enzymes per bacterium determined by quantitative Western blot analysis. MutM was detected at approximately 120 molecules per cell (S.E., 56) and Nth at 182 molecules per cell (S.E., 32) with no significant change in the amount of either enzyme after bacteria were exposed to oxidative stress or following removal of the other bi-functional glycosylase (Fig. 1B and C). These data are consistent with the constitutive presence of the 3′ processing enzymes NApe and NExo in the meningococcus (Carpenter et al., 2007), and the absence of an SOS response (Black et al., 1998, Davidsen & Tonjum, 2006). Further analysis of NApe and NExo levels (not shown) demonstrate that NApe is the most abundant of these enzymes reaching levels of approximately 840 molecules per cell (S.E., 120), while NExo (287 per cell, S.E. 46) is present at similar albeit lower levels.
Nth and MutM are bi-functional glycosylases that remove a broad range of oxidised nucleobases
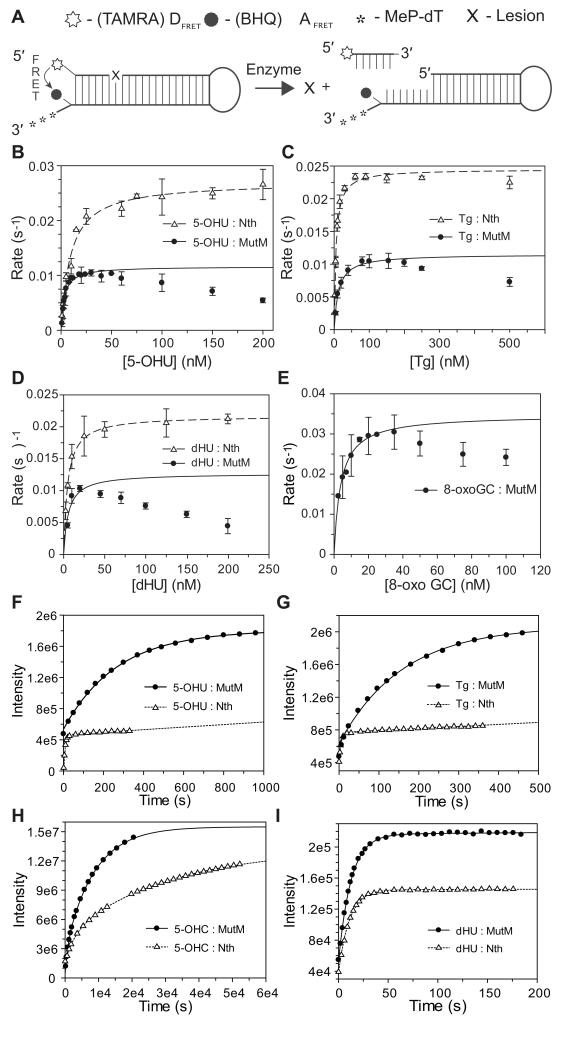
To further understand the contribution of bi-functional glycosylases to resistance against oxidative stress, the meningococcal genes encoding MutM and Nth were cloned and expressed in E. coli, and the recombinant proteins purified by anion exchange chromatography. The activity of the enzymes was assessed in an assay employing DNA hairpins containing molecular beacons (Fig. 2A). In their unmodified form, a TAMRA acceptor at the 5′ end of the DNA substrates is in immediate proximity to a BHQII quencher at the 3′ end, which prevents fluorescence. Cleavage of the base lesion and incision of the DNA backbone liberates the 5′ end with the TAMRA group, which then emits fluorescence.
Figure 2.
Catalytic properties of Nth and MutM.
(A) Substrates used for assays of bi-functional glycosylases. A damaged base (X) is removed by glycosylase activity, while lyase activity cleaves the DNA strand. After cleavage, the six base strand of DNA with a 5′ fluorescent TAMRA FRET donor (DFRET) is no longer in close proximity to the BHQ2 FRET acceptor (AFRET), allowing fluorescence and indicating product release. (B, C, D, and E) Kinetics of MutM (closed circles) and Nth (open triangles) with various DNA lesions. Data where plotted as initial rates against the concentration of substrate and fitted to the Michaelis-Menten’s equation. Data points that are lowered by substrate-product inhibition were omitted from the fit as these provide low and inaccurate estimates of rates. Error bars show the S.D. of at least three measurements. Determination of the reaction rates under steady state conditions (i.e. at least 10-fold excess of substrate to enzyme) for MutM was problematic due to product inhibition; 8-oxoG·C is a substrate for MutM but not Nth (not shown). (F, G, H and I) Single-turnover kinetics of Nth and MutM. Fluorescent hairpin substrates containing the lesions indicated were mixed with 1 μM of enzyme, and reactions followed using a FluoroMax®-3 fluorimeter. Data were fitted using Grafit® 6. Key rate constants are displayed in Table 1. Different saturation levels of Nth and MutM are caused by quenching of fluorescence by binding of Nth. Panels F and G show that reaction rates are faster with Nth (open triangles) than MutM (closed circles) with 5-OHU and Tg under-single turnover conditions (performed with an at least 10-fold excess of enzyme to substrate). (H and I) Nth and MutM display similar rate constants with dHU, and limited activity against 5-OHU but only over several hours.
The activities of Nth and MutM were characterised with a variety of DNA lesions that are characteristic of oxidative damage (Fig. 2, Table 1); since the assay with hairpins requires cleavage of the DNA backbone to generate a signal, it is evident that both enzymes possess both glycosylase and lyase activities. Comparison of the kcat/KM values reveals up to a ten-fold difference between the substrates tested, but all are physiologically relevant rates. Nth and MutM have similar activities against 5′-OH substituted uracil (5′-OHU, Fig. 2B) and the ring saturated pyrimidines, thymine glycol (Tg, Fig. 2C) and di-hydro-uracil (dHU, Fig. 2D). The activity of MutM (but not Nth) displays substantial product inhibition (Fig. 2B-E), with lowered reaction rates in the presence of higher concentrations of substrate (when product accumulates); this is likely to result from binding of MutM to damaged and undamaged DNA, whereas Nth only recognises damaged DNA (Silhan et al., 2011). Both enzymes fail to remove 5′-OH cytosine (5′-OHC, not shown), which may reflect their inability to discriminate 5′-OHC from native T, which has a 5′-methyl group (Wibley et al., 2003).
Table 1.
Glycosylase/lyase activity of Nth and MutM
| STO | Steady-state |
Relative | ||||
|---|---|---|---|---|---|---|
| Enzyme | Substrate | kcl (s−1) | KM (nM) | kcat (s−1) | kcat/KM | kcat/KM |
| Nth | dHU | 0.12 | 4.9 | 0.022 | 0.0045 | 1.0 |
| Tg | 0.18 | 5.3 | 0.025 | 0.0047 | 1.0 | |
| 5′-OHU | 0.12 | 9.4 | 0.027 | 0.0029 | 0.6 | |
| 5′-OHC | 3.4 × 10−4 | ND* | ND* | |||
| 8-oxoG·C | ND* | |||||
| 8-oxoG·A | ND* | |||||
|
| ||||||
| MutM | dHU | 0.08 | 5.7 | 0.013 | 0.0023 | 0.5 |
| Tg | 0.007 | 12.4 | 0.012 | 0.0010 | 0.2 | |
| 5′-OHU | 0.04 | 3 | 0.012 | 0.0040 | 0.9 | |
| 5′-OHC | 1 × 10−4 | ND* | ND* | |||
| 8-oxoG·C | 0.1 | 4 | 0.035 | 0.0088 | 1.9 | |
| 8-oxoG·A | 9 × 10−4 | ND* | ND* | |||
ND, no activity detected: STO, Single Turn Over conditions: kcl, rate constant for base removal and substrate cleavage:
fitted to the double exponential rate
We also assayed the kinetic activity of the enzymes under single-turnover condition of their complete reaction cycles (i.e. glycosylase and lyase activities, and product release, Fig. 2F-I and Table 1). The reaction cycles for Nth with 5-OHU, Tg, and dHU were very rapid, with rate constants for removal of the damaged base and cleavage of the substrate (kcl) of 0.12 (± 0.01) s−1, 0.12 (± 0.02) s−1, and 0.18 (± 0.02) s−1, respectively. In contrast, the activity of MutM was highly dependent on the lesion; dHU removal was marginally slower than Nth, while excision of 5-OHU and Tg were three- and ten-fold slower than Nth, respectively.
One clear distinction in the lesions recognised by MutM and Nth is 8-oxoG·C, a primary product following oxidative damage. While we found that MutM has glycosylase/lyase activity against 8-oxoG·C (Fig. 2E), Nth failed to remove this lesion (Table 1), which is consistent with previous results in E. coli and N. meningitidis (Morland et al., 2002, Tibballs et al., 2009). MutM excised 8-oxoG·C with the highest rate constant observed (kcl, 0.1 ± 0.006 s−1) for this enzyme with any substrate tested. On the other hand, 8-oxoG·A was excised by MutM but at a much lower rate (kcl, < 0.0009 s−1) than 8-oxoG·C (Table 1).
Lack of co-operation between glycosylases and 3′ processing enzymes
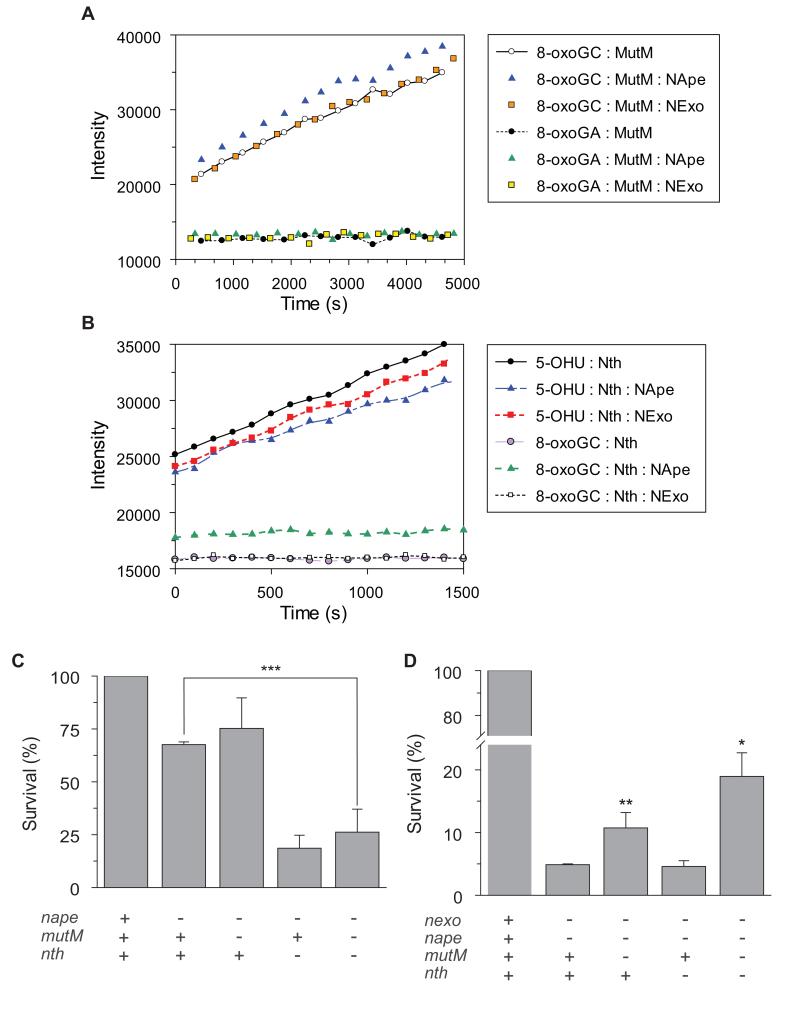
There has been considerable interest regarding communication between enzymes during BER. In an ‘ordered pathway model’, each enzyme interacts with a specific downstream enzyme so that substrates are handed-off and repair proceeds in a sequential manner. This is particularly pertinent where enzyme activity is limited by binding to its product (for example with MutM), and many studies have suggested that the activity of glycosylases can increase in the presence of the downstream AP endonucleases (Farez-Vidal et al., 2001b, Hill et al., 2001, Parikh et al., 1998, Pope et al., 2002a, Cortazar et al., 2007a, Zharkov et al., 2000, Sung & Mosbaugh, 2000, Vidal et al., 2001a, Waters et al., 1999, Fitzgerald & Drohat, 2008). N. meningitidis has two members of the Exonuclease III/Xth family, NApe and NExo, which specialise in the processing of apurinic/apyrimidinic (AP) sites, and 3′-PO4 blocking lesions, respectively (Silhan et al., 2011). We therefore examined the activity of MutM and Nth against representative substrates in the presence of NExo or NApe. When MutM was reacted with either 8-oxoG·C or 8-oxoG·A, the addition of a ten-fold greater concentration of NExo or NApe did not increase the turnover of the substrate (Fig. 3A). The same result was also observed with Nth, where the addition of an excess of NExo or NApe did not increase the turnover of 5′-OHU or 8-oxoG·C (Fig. 3B). Furthermore addition of NApe or NExo to reactions containing MutM did not relieve the product inhibition observed with this glycosylase (not shown). Therefore there was no biochemical evidence of co-operation between the bi-functional glycosylases and the 3′ processing enzymes.
Figure 3.
Lack of co-operation between glycosylase and 3′ processing enzymes.
(A and B) Steady-state glycosylase assays were performed in the presence or absence of NExo or NApe. Hairpin substrates (100 nM) were mixed with 10 nM NExo, NApe or reaction buffer alone. After 5 mins, MutM (A) or Nth (B) was added to reactions at a final concentration of 1 nM. Time points are shown after the addition of the glycosylase. Genetic analysis of interactions between bi-functional glycosylases and 3′ processing enzymes. (C) NApe and Nth share redundant functions in the meningococcus; removal of Nth but not MutM impairs the survival of a nape mutant in the presence of oxidative stress while loss of MutM or both bi-functional glycosylases improves the survival of cells lacking both 3′ processing enzymes (D). Bacteria were incubated with H2O2 (0.4 mM for C and 0.1 mM for D) or PBS for 30 min and then recovered by plating to solid media. The percentage killing was calculated by comparing the number of CFU in the presence and the absence of the oxidizing agent. + and − indicate the presence and absence of enzymes, respectively. Results show the percentage survival of strains relative to the nexo and nape mutants. Error bars show the SD of assays performed in triplicate on three occasions (***, **, and *indicate p < 0.001, 0.003, and 0.04, respectively, 1-column Student’s t test).
Nth is an efficient DNA lyase that can substitute for NApe
To further understand how Nth and MutM are integrated in meningococcal BER, we performed genetic analysis to determine the contribution of combinations of enzymes to bacterial survival under conditions of oxidative stress. We examined the effect of removing Nth and/or MutM in NApe-deficient bacteria. Removal of MutM from bacteria lacking NApe did not have any impact on survival, whereas the ΔnapeΔnth double mutant demonstrated only 25% survival compared to the mutant lacking only NApe (Fig. 3C). This synergistic effect suggests that Nth, but not MutM, can compensate for NApe in the meningococcus. This probably occurs through the efficient lyase activity of Nth (Fig. 2), which can act on AP sites (Silhan et. al., 2011), participating in the strand incision of AP sites during BER, while the product inhibition observed with MutM fails to perform this function in vivo.
Additionally, the effect of removing Nth and MutM from the ΔnapeΔnexo double mutant was examined. The ΔnapeΔnexo strain is more impaired for survival in the presence of oxidative stress than the single Δnexo deletion (Fig. 1), consistent with these enzymes having specialised functions within the cell. In this background, deletion of MutM (ΔnapeΔnexoΔmutM) actually lead to a two-fold increase in survival of the bacterium (Fig. 3D). This is consistent with MutM generating an increased burden of 3′-PO4 lesions, which reduces survival in the ΔnapeΔnexo strain. There was also significant increase in bacterial survival following exposure to oxidative stress of the ΔnapeΔnexo mutant following removal of both bi-functional glycosylases MutM and Nth with an increased recovery of 330% (p <0.0096), consistent with there being less toxic blocking lesions in the absence of these glycosylases in a cell deficient in 3′ processing enzymes. Whereas the absence of MutM or Nth did not lead to increase in survival in the Δnexo strain (not shown) suggesting that NApe shares a role(s) with NExo, possibly in dealing with direct oxidative strand incision.
The role of MutM and the GO system
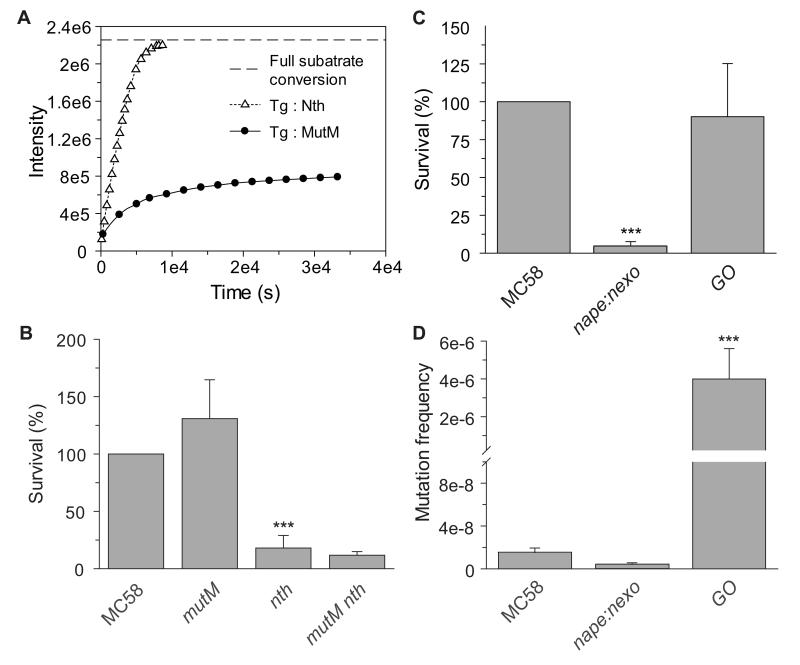
A notable difference between the two bi-functional glycosylases is that MutM is inhibited by its product (Fig. 4A shows an example of Nth and MutM with Tg); reactions with this enzyme typically occur at reduced rates with higher substrate concentrations, due to the accumulation of the product which inhibits turnover of the enzyme. MutM is thus product inhibited, with affinity for the product limiting the dissociation rate, consistent with other observations with abasic DNA substrates. This suggests that when the bacterium is in situations of high levels of oxidative stress, Nth will continue to function efficiently and mediate DNA repair, while MutM activity would be impeded by the increased abasic site concentrations in the cell. To evaluate this possibility, nth and mutM mutants were tested for their survival under conditions of high oxidative stress from a combination of oxidising agents, H2O2 and paraquat (Fig. 4B). Loss of Nth leads to sensitivity to oxidative stress unlike MutM, which is dispensable for survival, consistent with the higher turnover of Nth being able to process higher amounts of oxidative DNA lesions (Fig. 2 and Fig. 4B), although this enzyme might be able to recognise lethal lesions that are not dealt with by MutM.
Figure 4.
Distinct functions of Nth and MutM, and the contribution of the GO system to survival and mutation frequencies
(A) 100 nM of Tg DNA hairpin was mixed with 1 nM Nth or MutM. Increase in fluorescence demonstrates product formation and its release from the enzyme-product complex. MutM (black-dots) has an apparent burst phase, then a short steady-state phase, and after first few hundred seconds, the reaction is further slowed by product inhibition. Nth (open triangles) has standard steady-state profile allowing simple estimation of initial rates, with no evidence of product inhibition. (B) Strains (indicated below each bar) were incubated in the presence of 0.2 mM H2O2 and 15 mM paraquat, or PBS at room temperature for 30 min and then plated to solid media. Percentage killing was calculated by comparing the number of CFU recovered in the presence and the absence of oxidizing agents, and are given as the percentage survival relative to MC58. Error bars show the S.D. of assays performed on at least three occasions; *** indicates p < 0.0001. The GO system is dispensable for the survival under oxidative stress, but is necessary for maintenance of the rate of spontaneous mutation. (C) Strains (indicated: strain GO lacks MutM, MutT, and MutY) were incubated in the presence of 0.4 mM H2O2 or PBS at room temperature for 30 min and then plated to solid media. Percentage killing was calculated by comparing the number of CFU recovered in the presence and the absence of oxidizing agents, and are given as the percentage survival relative to MC58. (D) Single colonies of bacteria were grown on solid media for 24 hrs then suspensions were prepared and plated to medium with or without rifampicin. The ratio of rifampicin-resistant cells to the total number of cells was calculated; strains are indicated under each lane. Error bars show the S.D. of assays performed on at least three occasions; *** indicates p < 0.0005.
Surprisingly, MutM which is the only BER enzyme shown to excise the most common oxidative lesion 8-oxoG·C (a substrate not recognised by Nth), yet is not necessary for survival following exposure to even high levels of oxidative stress. Therefore, we examined whether other BER enzymes are responsible for this functional redundancy. In eukaryotes and prokaryotes, 8-oxoG is primarily dealt with by the GO system (Michaels & Miller, 1992), which includes MutM (OGG1 in eukaryotes) along with MutT and MutY. MutT decreases the pools of the damaged unincorporated nucleotide 8-oxodGTP, while MutY removes adenine from 8-oxoG:A mismatches following incorporation of 8-oxoG into DNA (Lu et al., 1996, Bulychev et al., 1996). We examined the survival of strain lacking all components of the GO system on exposure to H2O2. Removal of MutT and MutY from MC58ΔmutM did not significantly increase sensitivity to oxidative stress, demonstrating that neither of these enzymes contributes to the survival of the mutM mutant (Fig. 4C). Surprisingly we found that the neisserial GO system is entirely dispensable for bacterial survival following exposure to extensive oxidative stress (not shown), unlike in other bacteria (Sanders et al., 2010). This is in marked contrast to a strain lacking 3′ processing enzymes (Carpenter et al., 2007), MC58ΔnapeΔnexo, which is highly sensitive to the effects of reactive oxygen species (Fig. 4C).
We also assessed the mutation rates of strains lacking enzymes of the GO system by measuring the frequency of emergence of rifampicin resistance. While removal of MutM or MutT alone did not affect the mutation rate compared with MC58, there was a 40- and 110-fold increase in the mutation frequency of MC58ΔmutY and MC58ΔmutYΔmutM, respectively compared with the wild-type strain (not shown), consistent with previous findings (Davidsen et al, 2005a). In the absence of all compenents of the GO system (MC58ΔmutYΔmutMΔmutT), the mutation frequency to 340-fold more than MC58 (Fig. 4D), demonstrating that all members of the GO system function together to maintain the mutation frequency in the meningococcus.
Cross-talk between DNA repair systems in N. meningitidis
As the GO system is dispensable for the survival of the meningococcus under oxidative stress (Figure 4C), we reasoned that other DNA repair systems, including mismatch repair (MMR) and nucleotide excision repair (NER), might contribute to the repair of ROS-mediated DNA damage. The primary function of the MMR is to remove base-base mismatches or small insertions-deletions loops arising during replication (Li, 2008, Yamada et al., 2002), whereas, NER is thought to be the most widespread repair pathway and deals mainly with UV-mediated damage (van Houten et al., 2002, Seeberg, 1978).
To determine whether NER and/or MMR can serve a function in excising a ROS-generated DNA lesions we constructed a series of mutants deficient in BER enzymes as well as key MMR or NER enzymes. NER typically consists of UvrA, UvrB, UvrC, and the UvrD helicase; dimeric UvrA associates with UvrB forming a recognition complex that searches for structural abnormalities in the DNA helix (Goosen and Moolenaar, 2008, Thiagalingham and Grossman, 1993). In the case of the MMR, MutS is responsible for recognizing abnormal base-base complexes on the daughter strand(Yang et al., 2000). Therefore we constructed strains lacking UvrA or MutS to examine the roles of NER and MMR, respectively, during the repair of oxidative DNA damage.
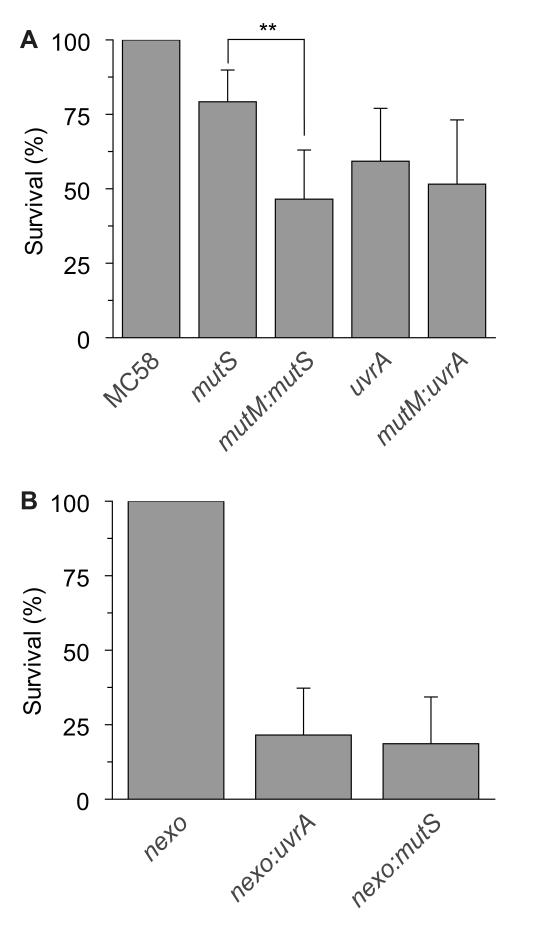
The mutS single mutant showed no difference in survival compared with MC58, whereas removing MutS from strain lacking mutM resulted in a significant reduction in survival in the presence of H2O2 and paraquat (Fig. 5A), indicating that the MMR provides a back-up system for the recognition and removal of 8-oxo G, which is not recognised by Nth (Table 1); deletion of MutT in a ΔmutM:mutS background did not further impair survival in the face of oxidative stress (not shown). In contrast, the uvrA mutant displayed reduced survival under oxidative stress compared with the parental strain (Fig. 5A), although there was no further decrease in the survival when mutM was removed from the uvrA mutant (ΔmutM:uvrA).
Figure 5.
Contribution of the MMR and NER to the repair of oxidative DNA damage.
(A) Strains (indicated below each bar) were incubated in the presence of 0.2 mM H2O2 and 15 mM paraquat or PBS at room temperature for 30 min and then plated to solid media. Percentage killing was calculated by comparing the number of CFU recovered in the presence or absence of oxidizing agents, and are given as the percentage survival relative to MC58. Error bars show the SD of assays performed in triplicate (** indicates p < 0.01, Students t test). (B) Strains (indicated below each bar) were incubated in the presence of 0.4 mM H2O2 or PBS at room temperature for 30 min and then plated to solid media. Percentage survival was calculated by comparing the number of CFU recovered in the presence and the absence of oxidizing agents, and are given as the percentage survival relative to nexo mutant.
To reveal whether NER and /or MMR serve as back-up systems during BER beyond the stage of base recognition and excision, we tested the sensitivity against H2O2 of mutants lacking UvrA or MutS together with NExo, which processes the 3′-PO4 and aldehyde groups producing by MutM and Nth, respectively. Of note, both Δnexo:uvrA and Δnexo:mutS were dramatically attenuated for the survival, recovered at 14 % of the level of the nexo mutant (p<0.0025 and p<0.0018, respectively) following exposure to H2O2, indicating that both MMR and NER are involved in dealing with oxidized DNA lesions (Fig. 5B).
DISCUSSION
N. meningitidis inhabits the human nasopharynx, and is in virtually constant contact with its host; person-to-person transmission occurs via droplet spread, and the bacterium does not have a significant environmental reservoir. Therefore the meningococcus is unlikely to be exposed to high level UV radiation for sustained periods, and the major threat to genome integrity will be oxidative nucleobase damage, generated by the human immune system and primarily dealt with by BER. The organisation of BER enzymes into an integrated network, and their constitutive expression in the absence of an SOS response is likely to reflect the successful adaption of this important pathogen to continued exposure to oxidative stress, and its evolution to suit the specific environmental challenges it faces in its habitat within the human host.
N. meningitidis has only two bi-functional DNA glycosylases, Nth and MutM, which can remove oxidised nucleobases from the genome. These enzymes have considerable overlap in function, recognising many of the same lesions, similar characteristics of binding to AP sites (Silhan et al., 2011), and both possess lyase activity. However there are also significant differences, as only MutM can remove 8-oxoG·C from the DNA backbone, while the product of Nth is a 3′-Ald, whereas MutM leaves a 3′-PO4. Furthermore, their kinetic behaviours are distinct. Nth exhibits efficient enzymatic turnover, whereas MutM is inhibited by its product. These are key when considering mechanisms to repair oxidative DNA damage.
Our biochemical data suggest that when the bacterium is in situations of high levels of oxidative stress, for instance within activated phagocytic cells, Nth will continue to function efficiently and mediate DNA repair, while MutM would be product inhibited by the increased concentration of abasic sites in the cell. Indeed, MutM homologues in other bacteria are extensively inhibited by multiple, adjacent DNA lesions (Cunniffe et al., 2007, Weinfeld et al., 2001). This is supported by the finding that an nth mutant but not the strain lacking MutM is more sensitive to a combination of oxidising agents, H2O2 and paraquat, when compared to the wild-type strain. In E. coli, the accumulation of AP sites also initiates the SOS response, which leads to the processing of AP sites by either RuvA-dependent recombination repair or trans-lesion synthesis, in addition to BER (Otterlei et al., 2000). This makes it difficult to interpret phenotypic responses of BER mutants in E. coli.
In contrast, N. meningitidis lacks an SOS response, and the analysis of mutants provides clear phenotypic changes for the loss of enzymes that can be explained by their function and integration with other activities in the cell. Thus the meningococcus is an attractive organism for a comprehensive study of BER in a host-restricted pathogen. For instance, removal of MutM, or MutM and Nth from a strain lacking NApe and NExo (enzymes which deal with 3′ blocking lesions) improved the capacity of the bacterium to survive oxidative stress. This indicates that cells lacking enzymes that process 3′ lesions survive better in the absence of enzymes (i.e. MutM and Nth) that generate these cytotoxic lesions, and is consistent with observations in Salmonella typhimurium (Richardson et al., 2009). It is also notable that there was no increase in the survival of the mutM/nexo mutant compared to a nexo single mutant in the face of oxidative stress (not shown). This indicates that even in the absence of MutM (which generates 3′-PO4), NExo is crucial for cell survival. The likely reason is the continued need for NExo to repair lesions occurring spontaneously or through direct oxidative strand cleavage, which give rise to single strand breaks with 3′-PO4 or other 3′ lesions such as 3′ glycolate (Takemoto et al., 1998).
In addition to MutM, N. meningitidis possesses two other enzymes, MutT and MutY, that prevent or correct 8-oxoG mismatches. MutT decreases the pools of the damaged unincorporated nucleotide 8-oxodGTP, while MutY removes adenine from 8-oxoG·A mismatches following incorporation of 8-oxoG into DNA (Lu et al., 1996, Bulychev et al., 1996). These enzymes can act in concert to deal with 8-oxoG, an abundant product of oxidative DNA damage (Michaels and Miller, 1992). To our knowledge, we have constructed the first prokaryote devoid of all components of the GO system, and found that the strain does not exhibit impaired survival in the presence of oxidative stress. This is in contrast to other bacteria such as Pseudomonas aeruginosa, in which removal of even a single member of the GO system and exposure to high level of ROS in human lungs significantly affects bacterial survival (Morero and Argarana, 2009). Similarly, the absence of the bi-functional glycosylase MutM impairs the ability of the photosynthetic bacterium Synechococcus elongatus and Mycobacterium smegmatis to deal with DNA lesions arising from oxidative stress (Jain et al., 2007, Mühlenhoff, 2000). We examined the GO system in N. meningitidis in relation to its effect on mutation; the results are very clear. When the GO system is missing, N. meningitidis is still viable and its survival is not impaired after exposure to oxidative stress (Fig. 4C). However, the mutation rate of N. meningitidis lacking the GO system increases dramatically, demonstrating that GO system acts as an anti-mutator, and that its main role is to exclude 8-oxoG mismatches from the DNA backbone, which do not block DNA replication and are thus not cytotoxic (Davidsen et al., 2005).
In N. meningitidis, analysis of strains lacking MutS indicates that the functional redundancy of MutM in the meningococcus is likely to be due to the contribution of the MMR system, but not NER. Instead, we found that NER has a MutM-independent role in protection against oxidative stress, similar to the gonococcus in which this pathway contributes to survival in the presence of low amounts of oxidizing agents (LeCuyer et al., 2010). However, we found that the phenotype of the meningococcal uvrA mutant was more pronounced when exposed extensive oxidative stress (Fig. 5A), which might reflect the different niches inhabited by these related pathogens where the gonococcus is likely to be exposed to less UV light than the meningococcus.
The role of the MMR in the repair of oxidatively damaged bases is much more clearly established than NER and most likely due to the removal of oxidized guanine residues. In E. coli, MMR system acts on 8-oxoG (Wyrzykowski and Volkert, 2003), while mouse embryonic stem cells deficient in MMR and H. pylori mutS mutants accumulate increased levels of 8-oxoG (Dantzer et al., 2003, Wang et al., 2005). In previous work in the meningococcus using a disc diffusion assay, neither mutS nor mutM:mutS deficient strains exhibited enhanced sensitivity to H2O2 compared with the wild-type strains (Davidsen et al., 2007). As this method relies on measuring zone of growth inhibition (rather than changes in bacterial recovery), it might not have been sufficiently sensitive to detect the influence of MMR on survival.
Conceptually the ordered pathway model of DNA repair is attractive as it involves defined, sequential steps, with the protection of reaction intermediates and co-operative enzyme activity. This allows sequestration of the potentially toxic intermediates, passing them along without allowing these molecules to trigger cell cycle arrest, necrotic cell death, or apoptosis as shown in mammalian systems (Prasad et al., 2010). This model has received widespread support, with reports of enhanced DNA glycosylase activity in the presence of AP endonucleases (Farez-Vidal et al., 2001a, Hill et al., 2001, Parikh et al., 1998, Pope et al., 2002b, Cortazar et al., 2007b, Zharkov et al., 2000, Sung & Mosbaugh, 2000, Vidal et al., 2001b, Waters et al., 1999, Fitzgerald & Drohat, 2008). However, detailed examination of these effects is rare, with human TDG providing the exception (Fitzgerald & Drohat, 2008, Waters et al., 1999). Alternatively, in a network model several enzymes share the capacity of processing of specific lesions, and the likelihood that a certain lesion being processed by a certain enzyme is dependent on the relative abundance and efficiency of each enzyme. A network model would provide a more robust foundation for DNA repair, as it provides degeneracy within a flexible system; genetic networks are known to offer robustness against mutations (Wagner, 2000).
Our and others’ data (Hatahet et al., 1994, Blaisdell & Wallace, 2001, Matsumoto et al., 2001, Senturker et al., 1998) indicate significant overlap in the spectrum of DNA lesions processed by MutM and Nth, demonstrating that at the initiation of DNA repair there is degeneracy in enzyme function, as seen in other organisms (Slupphaug et al., 2003). The subsequent processing of AP sites is typically viewed as being defined by the initiating DNA glycosylase (Slupphaug et al., 2003, Fortini et al., 1999), as part of an ‘ordered pathway’. However, processing in N. meningitidis is not delineated by the initial DNA lesion, since we demonstrate that MutM and Nth are functionally redundant, and Nth can substitute for NApe in cleavage of AP sites. Furthermore, the biochemical data lend no support for a pathway model; addition of NApe or NExo to reactions catalysed by MutM or Nth did not enhance their activity at stoichiometric or even higher concentrations.
Therefore BER in the meningococcus operates as a network of enzymes with overlapping functions during the recognition and removal of damaged nucleobase, with both NER and MMR contributing to repair of oxidative DNA damage. The likelihood that any given lesion is processed by a given enzyme depends on the amount of enzymes in the cell, their defined substrates and rates of activity, and the extent of DNA damage. Knowledge of biochemical characteristics and cellular levels of enzymes together with genetic data on their interaction enables us to construct a model for repair of oxidative DNA damage in the meningococcus (Fig. 6), which highlights the efficiency and abundance of the specialised 3′ processing enzymes, NApe and Nexo, compared with the bi-functional glycosylases. This could reflect the role of NApe and NExo during repair of direct DNA damage, or functional redundancy of the glycosylases. The network of enzymes would provide a robust foundation for repair, as it provides degeneracy within flexible system, and protection against cytotoxic mutations (Wagner, 2000). Discrete sub-pathways of BER in N. meningitidis exist only at the level of processing of 3′ lesions, which must be removed before DNA repair can be completed. It is evident from inspection of the relative abundance and activities of enzymes that efficient repair of 3′ lesions is critical to the meningococcus in BER, and mediated by highly specialised and effective enzymes. These must deal with BER intermediates that are not only generated by glycosylases, but also arise through direct oxidative damage to DNA (Fig. 6). Given the degeneracy observed in BER and the role of other repair systems by the meningococcus and other prokaryotes (Hatahet et al., 1994, Blaisdell & Wallace, 2001, Matsumoto et al., 2001, Senturker et al., 1998), the network model of BER is likely to provide a paradigm for DNA repair in many prokaryotes.
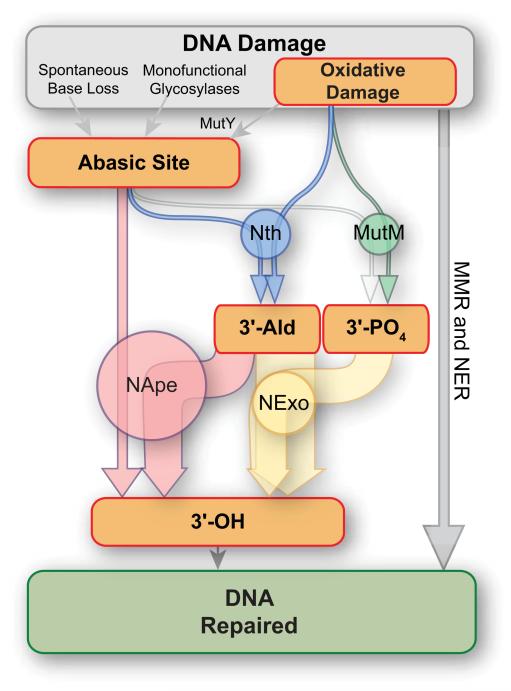
Figure 6.
Model of the network of BER in N. meningitidis.
Oxidative DNA damage is processed by MutY, Nth or MutM, to generate abasic sites, 3′-Ald or 3′- PO4, respectively. The 3′-Ald or 3′-PO4 groups block DNA polymerisation, and thence replication, and must be converted to a 3′-OH before repair can be completed. The width of arrows is proportional to the kcat/KM of the reactions (except for MutY) while the area of the circles is proportional to the abundance of each enzyme in the meningococcus.
EXPERIMENTAL PROCEDURES
Bacterial strains, growth and construction of mutants
Bacterial strains used in this study are listed in the Supplementary data. Genomic DNA from strain MC58 (serogroup B) was used as the template in PCRs to amplify genes from N. meningitidis. Reactions were performed with Pfu polymerase (Promega) and appropriate primers; details of all primers are given in the Supplemental data. The mutM mutant was generated by transposon mutagenesis with a kanamycin resistance cassette inserted at nt. 1607 of the NMB1295, which is predicted to encode a MutM homologue (www.jmrc.org). The number of bacteria was calculated by measuring the O.D. A260 of an aliquot of a suspension of cells in lysis buffer (1% SDS, 0.1M NaOH), and confirmed by plating dilutions to solid media and counting CFUs the following day.
To delete nth, mutM, and nexo, the following strategy was employed. DNA fragments of between 650 and 1200 bp flanking the target gene were amplified by PCR with oligonucleotide primers. The products were digested with appropriate restriction enzymes and ligated into pUC19 (Yanisch-Perron et al., 1985). Finally, antibiotic resistance genes were amplified from pDG1728 (for spc) (Guerout-Fleury et al., 1996) or pGCC4 (erm) (Mehr et al., 2000) and inserted between the upstream and downstream fragments to yield the constructs for transforming N. meningitidis. Constructs for other gene deletions were generated by PCR with primers given in Supplementary data. Double, triple and quadruple mutants used in this study were generated using genomic DNA from single mutants to transform other strains. Transformants were analyzed by PCR and Western blotting to confirm that allelic replacement had occurred and the protein is no longer expressed (not shown).
Determining the rate of spontaneous mutation
For each experiment, 10 colonies of each strain were streaked on BHI agar and grown overnight. A suspension of bacteria was made in PBS and adjusted to 2 × 1010 CFU/ml. Next, 100 μl of each suspension was spread onto BHI agar containing 3 μg/ml rifampicin (Sigma–Aldrich). Serial dilutions were plated on BHI-agar without antibiotic to determine the number of the viable bacteria. Rifampicin-resistant colonies were counted after 24 hr incubation at 37° C in 5% CO2.
Generation of immune sera and Western Blot Analysis
Adult female mice (BALB/c, Harlan) received 25 μg of recombinant protein on three occasions (on days 0, 21, and 35); the protein was mixed with an equal volume of incomplete Freunds adjuvant and given via the subcutaneous route. Immune sera was obtained on day 42. Proteins were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions and transferred onto polyvinylidene fluoride membranes (Millipore, Bedford, MA). Membranes were blocked for 1.5 hr at room temperature in Tris-buffered saline (TBS) containing 5 % (wt./vol.) fat-free dry milk, and incubated first with primary antibodies overnight at 4°C, then with a horseradish-peroxidase-conjugated anti-mouse secondary antibody (1:1000) for 90 min. Specific proteins were visualized using chemiluminescence according to the manufacturer’s instructions (GE Healthcare UK). The intensity of bands was determined with a Lynx video densitometer (Biological Vision). The amount of protein in N. meningitidis was determined by comparing signals obtained from serial dilutions of whole cell lysates of bacteria with those from known amounts of recombinant protein determined by the method of Bradford.
Survival under oxidative stress
Strains were grown overnight, harvested into PBS and the bacterial suspensions adjusted to 109 colony forming units (CFU)/ml. Subsequently, 100 μl of the bacterial suspension was incubated with either 0.4 mM (Sigma) for 30 min, or 0.2 mM H2O2 and 15 mM paraquat (Sigma) for 30 min at room temperature. The same number of bacteria was incubated in PBS alone under the same conditions. After half an hour, serial dilutions were plated onto BHI medium. The number of CFU were counted the next day and the survival of each strain was expressed as the proportion of surviving bacteria after incubation with the oxidative agent(s) to the number recovered after incubation in PBS only. Assays were performed in triplicate on at least three independent occasions. Results were analysed for significance by 1-column Student’s t test as indicated in the Figure legends.
Expression and purification of Nth and MutM
For expression of recombinant proteins, mutM (NMB1295) was amplified from MC58 using oligonucleotides MutMpET1 and MutMpET2 and the product inserted into pET-21a(+) using the NdeI and HinDIII restriction sites in the primers. nth (NMB0533) was amplified using oligonucleotides Nth1 and Nth2 then ligated into pProEX-HTb (Invitrogen) using BamHI and HinDIII sites in the primers. A termination codon was added at the end of the nth gene using site-directed mutagenesis and oligonucleotides NthStop1 and NthStop2. Final fusion protein then contained N-terminal polyhistidine tag with a cleavage site for TEV protease. E. coli Rosetta™(DE3) pLysS strain was used for protein expression.
Cultures were typically grown in 3 × 500 ml of LB medium to an O.D. A600 of 0.6 at 37°C, cooled to room temperature and protein expression induced by the addition of 0.5 mM IPTG. Cells were harvested by centrifugation. Pellets containing Nth were resuspended in PBS, 50 mM imidazole pH 8.0, 0.5 M NaCl and 2 mM β-mercaptoethanol (β-ME) and sonicated, while MutM pellets were resuspended in PBS, 2 mM β-ME, 1 mM EDTA; sonicates were clarified by centrifugation. His-tagged Nth was purified on a metal affinity chromatography column loaded with Chelating Sepharose® Fast Flow (GE Healthcare) equilibrated with Ni2+ and equilibrated with PBS, 50 mM imidazole pH 8.0, 0.5 M NaCl and 2 mM β-ME. Nth was eluted with same buffer with 0.5 M imidazole. Fractions containing protein were pooled and purified on a HiLoad 26/60 GF Superdex75 column (GE Healthcare) equilibrated with buffer (25 mM Tris pH 7.5, 0.5 M NaCl and 2 mM β-ME). The polyhistidine tag was removed by overnight digestion with TEV protease (Invitrogen) at 4°C. Undigested Nth was removed on a Chelating Sepharose column while the digested protein was dialyzed against 25 mM Tris 7.5, 0.5 M NaCl, 1 mM EDTA, 2 mM DTT and 10% (v/v) glycerol. Clarified MutM lysates were subjected to tandem anion/cation exchange chromatography on columns filled with DE52 resin (Whatman Ltd) or SP Sepharose (GE Healthcare) on an ÄKTA purifier (GE Healthcare). The system was equilibrated with 20 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, and 2 mM β-ME. MutM was eluted with the same buffer containing a gradient of NaCl, and the protein eluted at approximately 375 mM NaCl. Fractions containing MutM were further purified using a HiLoad 26/60 GF Superdex75 column equilibrated 25 mM Tris 7.5, 0.5 M NaCl, 1 mM EDTA, 10% (v/v) glycerol, and 2 mM β-ME.
Glycosylase and lyase activity
All DNA hairpin substrates had a 5′ TAMRA fluorescent label with the sequence 5′-GAC TAA XAA TGA CTG CGG TTA CGC AGT CAT T1T TAG TC2 *T*T*TT-3′, where X denotes the damaged base and 1 denotes the base opposite (either an A for dHU, Tg, and 5-OHU, a C for 8-oxoG, or a G for 5-OHC), 2 denotes the BHQ2dT quencher, while * is methyl phosphonate (Eurogentec Ltd). For instance, the substrate 8-oxodG·A had an A in position X and 8-oxodG at 1. Continual fluorescence-based assays were carried out with a POLARstar Omega microplate reader (BMG Labtech) or a FluoroMax-3 fluorimeter (Horiba Scientific). Assays were performed in 50 mM Tris pH 7.5, 125 mM NaCl, 1 mM EDTA, 2 mM β-ME, 0.1 mg/ml BSA at 25°C. Steady state reactions were initiated by mixing equal volumes of substrate and enzyme for the Omega reader, or 450 μl of substrate with 50 μl of enzyme for FluoroMax3. Final concentrations of enzymes were at least one hundred times lower than substrate. Initial rates were estimated from the linear phase of four to eight reactions performed on separate occasions, then plotted against the substrate concentration. Initial rates were utilised due to the substantial product inhibition observed particularly with MutM. Data were fitted to the Michaelis–Menten equation using Grafit 6 (Erithacus software). Single-turnover reactions were carried out in the same buffer at 25°C. Typically at least a 10-fold excess of an enzyme over substrate was used to determine single turnover rates. Reactions were initiated by mixing 100 nM of substrate with 1 μM enzyme. Data were fitted to either a single exponential rate; a single exponential with a slope (Nth), or a double exponential for reactions with 5′-OHC. All data fitting was performed using Grafit 6 (Erithacus software).
Supplementary Material
Acknowledgments
This work was supported by Programme Grants from the Wellcome Trust (to CMT, GB and PF) and the Medical Research Council (to CMT).
References
- Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Black CG, Fyfe JA, Davies JK. Absence of an SOS-like system in Neisseria gonorrhoeae. Gene. 1998;208:61–66. doi: 10.1016/s0378-1119(97)00653-7. [DOI] [PubMed] [Google Scholar]
- Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, Bichara M, Fuchs RP, Laval J. Excision of the imidazole ring-opened form of N-2-aminofluorene-C(8)-guanine adduct in poly(dG-dC) by Escherichia coli formamidopyrimidine-DNA glycosylase. Carcinogenesis. 1989;10:1905–1909. doi: 10.1093/carcin/10.10.1905. [DOI] [PubMed] [Google Scholar]
- Bulychev NV, Varaprasad CV, Dorman G, Miller JH, Eisenberg M, Grollman AP, Johnson F. Substrate specificity of Escherichia coli MutY protein. Biochemistry. 1996;35:13147–13156. doi: 10.1021/bi960694h. [DOI] [PubMed] [Google Scholar]
- Carpenter EP, Corbett A, Thomson H, Adacha J, Jensen K, Bergeron J, Kasampalidis I, Exley R, Winterbotham M, Tang C, Baldwin GS, Freemont P. AP endonuclease paralogues with distinct activities in DNA repair and bacterial pathogenesis. EMBO J. 2007;26:1363–1372. doi: 10.1038/sj.emboj.7601593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright KA, Stuart JM, Jones DM, Noah ND. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol Infect. 1987;99:591–601. doi: 10.1017/s0950268800066449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007a;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair. 2007b;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Cunniffe SM, Lomax ME, O’Neill P. An AP site can protect against the mutagenic potential of 8-oxoG when present within a tandem clustered site in E. coli. DNA Repair (Amst) 2007;6:1839–1849. doi: 10.1016/j.dnarep.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Bjørås M, Luna L, Klungland A, Seeberg E. Comparative analysis of 8-oxoG:C, 8-oxoG:A, A:C and C:C DNA repair in extracts from wild type or oxoG DNA glycosylase deficient mammalian and bacterial cells. DNA Repair (Amst) 2003;2:707–18. doi: 10.1016/s1568-7864(03)00041-7. [DOI] [PubMed] [Google Scholar]
- Davidsen T, Bjoras M, Seeberg EC, Tonjum T. Antimutator role of DNA glycosylase MutY in pathogenic Neisseria species. J. Bacteriol. 2005;187:2801–2809. doi: 10.1128/JB.187.8.2801-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen T, Tonjum T. Meningococcal genome dynamics. Nat Rev Microbiol. 2006;4:11–22. doi: 10.1038/nrmicro1324. [DOI] [PubMed] [Google Scholar]
- Davidsen T, Tuven HK, Bjoras M, Rodland EA, Tonjum T. Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J Bacteriol. 2007;189:5728–5737. doi: 10.1128/JB.00161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M, Laval J, Boiteux S. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry. 1993;32:12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- Exley RM, Shaw J, Mowe E, Sun YH, West NP, Williamson M, Botto M, Smith H, Tang CM. Available carbon source influences the resistance of Neisseria meningitidis against complement. J Exp Med. 2005;201:1637–1645. doi: 10.1084/jem.20041548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farez-Vidal ME, Gallego C, Ruiz-Perez LM, Gonzalez-Pacanowska D. Characterization of uracil-DNA glycosylase activity from Trypanosoma cruzi and its stimulation by AP endonuclease. Nucleic Acids Res. 2001a;29:1549–1555. doi: 10.1093/nar/29.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farez-Vidal ME, Gallego C, Ruiz-Perez LM, Gonzalez-Pacanowska D. Characterization of uracil-DNA glycosylase activity from Trypanosoma cruzi and its stimulation by AP endonuclease. Nucleic Acids Research. 2001b;29:1549–1555. doi: 10.1093/nar/29.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2:355–357. doi: 10.1016/s0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Fitzgerald ME, Drohat AC. Coordinating the initial steps of base excision repair apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. J Biol Chem. 2008;283:32680–32690. doi: 10.1074/jbc.M805504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J Biol Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, D. C.: 2006. [Google Scholar]
- Goosen N, Moolenaar GF. Repair of UV damage in bacteria. DNA Repair (Amst) 2008;7:353–379. doi: 10.1016/j.dnarep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Kumar P, Varshney U. A distinct role of formamidopyrimidine DNA glycosylase (MutM) in down-regulation of accumulation of G, C mutations and protection against oxidative stress in mycobacteria. 2007;6:1774–1785. doi: 10.1016/j.dnarep.2007.06.009. [DOI] [PubMed] [Google Scholar]
- LeCuyer BE, Criss AK, Seifert HS. Genetic characterization of the nucleotide excision repair system of Neisseria gonorrhoeae. J Bacteriol. 2010;192:665–673. doi: 10.1128/JB.01018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- Lu AL, Yuen DS, Cillo J. Catalytic mechanism and DNA substrate recognition of Escherichia coli MutY protein. J Biol Chem. 1996;271:24138–24143. doi: 10.1074/jbc.271.39.24138. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Zhang QM, Takao M, Yasui A, Yonei S. Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA. Nucleic Acids Res. 2001;29:1975–1981. doi: 10.1093/nar/29.9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehr IJ, Long CD, Serkin CD, Seifert HS. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics. 2000;154:523–532. doi: 10.1093/genetics/154.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz AJ, So M. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu Rev Cell Dev Biol. 2000;16:423–457. doi: 10.1146/annurev.cellbio.16.1.423. [DOI] [PubMed] [Google Scholar]
- Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J. Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morero NR, Argaraña CE. Pseudomonas aeruginosa deficient in 8-oxodeoxyguanine repair system shows a high frequency of resistance to ciprofloxacin. FEMS Microbiol Lett. 2009;290:217–226. doi: 10.1111/j.1574-6968.2008.01411.x. [DOI] [PubMed] [Google Scholar]
- Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U. The FAPY-DNA glycosylase (Fpg) is required for survival of the cyanobacterium Synechococcus elongatus under high light irradiance. FEMS Microbiol Lett. 2000;187:127–132. doi: 10.1111/j.1574-6968.2000.tb09148.x. 2000. [DOI] [PubMed] [Google Scholar]
- Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: a mutator locus in Escherichia coli that generates G.C----T.A transversions. Proc Natl Acad Sci U S A. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M, Kavli B, Standal R, Skjelbred C, Bharati S, Krokan HE. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 2000;19:5542–5551. doi: 10.1093/emboj/19.20.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998a;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant L, Jonsson AB. Contacting the host: insights and implications of pathogenic Neisseria cell interactions. Scand J Infect Dis. 2003;35:608–613. doi: 10.1080/00365540310016349. [DOI] [PubMed] [Google Scholar]
- Pope MA, Porello SL, David SS. Escherichia coli apurinic-apyrimidinic endonucleases enhance the turnover of the adenine glycosylase MutY with G:A substrates. J Biol Chem. 2002;277:22605–22615. doi: 10.1074/jbc.M203037200. [DOI] [PubMed] [Google Scholar]
- Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channelling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR, Soliven KC, Castor ME, Barnes PD, Libby SJ, Fang FC. The Base Excision Repair system of Salmonella enterica serovar typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog. 2009;5:e1000451. doi: 10.1371/journal.ppat.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders LH, Sudhakaran J, Sutton MD. The GO system prevents ROS-induced mutagenesis and killing in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2009;294:89–96. [Google Scholar]
- Schneider MC, Prosser BE, Caesar JJ, Kugelberg E, Li S, Zhang Q, Quoraishi S, Lovett JE, Deane JE, Sim RB, Roversi P, Johnson S, Tang CM, Lea SM. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458:890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberg E. Reconstitution of an Escherichia coli repair endonuclease activity from the separated uvrA+ and uvrB+/uvrC+ gene products. Proc Natl Acad Sci U S A. 1978;75:2569–2573. doi: 10.1073/pnas.75.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senturker S, Auffret van der Kemp P, You HJ, Doetsch PW, Dizdaroglu M, Boiteux S. Substrate specificities of the ntg1 and ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhan J, Nagorska K, Zhao G, Jensen K, Freemont PS, Tang CM, Baldwin GS. Specialization of an Exonuclease III family enzyme in the repair of 3′ DNA lesions during base excision repair in the human pathogen Neisseria meningitidis. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Sung JS, Mosbaugh DW. Escherichia coli double-strand uracil-DNA glycosylase: involvement in uracil-mediated DNA base excision repair and stimulation of activity by endonuclease IV. Biochemistry. 2000;39:10224–10235. doi: 10.1021/bi0007066. [DOI] [PubMed] [Google Scholar]
- Takemoto T, Zhang QM, Matsumoto Y, Mito S, Izumi T, Ikehata H, Yonei S. 3′-blocking damage of DNA as a mutagenic lesion caused by hydrogen peroxide in Escherichia coli. J Radiat Res (Tokyo) 1998;39:137–144. doi: 10.1269/jrr.39.137. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Grossman L. The multiple roles for ATP in the Escherichia coli UvrABC endonuclease-catalyzed incision reaction. J Biol Chem. 1993;268:18382–18389. [PubMed] [Google Scholar]
- Tibballs KL, Ambur OH, Alfsnes K, Homberset H, Frye SA, Davidsen T, Tonjum T. Characterization of the meningococcal DNA glycosylase Fpg involved in base excision repair. BMC microbiology. 2009;9:7. doi: 10.1186/1471-2180-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Stephens DS. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2000;2:687–700. doi: 10.1016/s1286-4579(00)00356-7. [DOI] [PubMed] [Google Scholar]
- Van Houten B, Eisen JA, Hanawalt PC. A cut above: Discovery of an alternative excision repair pathway in bacteria. Proc Natl Acad Sci USA. 2002;99:2581–2583. doi: 10.1073/pnas.062062599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Research. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Robustness against mutations in genetic networks of yeast. Nat Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- Wang G, Alamuri P, Humayun MZ, Taylor DE, Maier RJ. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol Microbiol. 2005;58:166–76. doi: 10.1111/j.1365-2958.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- Waters TR, Gallinari P, Jiricny J, Swann PF. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem. 1999;274:67–74. doi: 10.1074/jbc.274.1.67. [DOI] [PubMed] [Google Scholar]
- Weinfeld M, Rasouli-Nia A, Chaudhry MA, Britten RA. Response of base excision repair enzymes to complex DNA lesions. Radiat Res. 2001;156:584–589. doi: 10.1667/0033-7587(2001)156[0584:robere]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wyrzykowski J, Volkert MR. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J Bacteriol. 2003;185:1701–1704. doi: 10.1128/JB.185.5.1701-1704.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada NA, Smith GA, Castro A, Roques CN, Boyer JC, Farber RA. Relative rates of insertion and deletion mutations in dinucleotide repeats of various lengths in mismatch repair proficient mouse and mismatch repair deficient human cells. Mutat Res. 2002;499:213–225. doi: 10.1016/s0027-5107(01)00282-2. [DOI] [PubMed] [Google Scholar]
- Yang W, Junop MS, Ban C, Obmolova G, Hsieh P. DNA mismatch repair: from structure to mechanism. Cold Spring Harb Symp Quant Biol. 2000;65:225–232. doi: 10.1101/sqb.2000.65.225. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J Biol Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.