Abstract
Hypertension is a common hereditary syndrome with unclear pathogenesis. Chromogranin A (Chga), which catalyzes formation and cargo storage of regulated secretory granules in neuroendocrine cells, contributes to blood pressure homeostasis centrally and peripherally. Elevated Chga occurs in spontaneously hypertensive rat (SHR) adrenal glands and plasma, but central expression is unexplored. In this report, we measured SHR and Wistar–Kyoto rat (control) Chga expression in central and peripheral nervous systems, and found Chga protein to be decreased in the SHR brainstem, yet increased in the adrenal and the plasma. By re-sequencing, we systematically identified five promoter, two coding and one 3′-untranslated region (3′-UTR) polymorphism at the SHR (versus WKY or BN) Chga locus. Using HXB/BXH recombinant inbred (RI) strain linkage and correlations, we demonstrated genetic determination of Chga expression in SHR, including a cis-quantitative trait loci (QTLs) (i.e. at the Chga locus), and such expression influenced biochemical determinants of blood pressure, including a cascade of catecholamine biosynthetic enzymes, catecholamines themselves and steroids. Luciferase reporter assays demonstrated that the 3′-UTR polymorphism (which disrupts a microRNA miR-22 motif) and promoter polymorphisms altered gene expression consistent with the decline in SHR central Chga expression. Coding region polymorphisms did not account for changes in Chga expression or function. Thus, we hypothesized that the 3′-UTR and promoter mutations lead to dysregulation (diminution) of Chga in brainstem cardiovascular control nuclei, ultimately contributing to the pathogenesis of hypertension in SHR. Accordingly, we demonstrated that in vivo administration of miR-22 antagomir to SHR causes substantial (∼18 mmHg) reductions in blood pressure, opening a novel therapeutic avenue for hypertension.
INTRODUCTION
Hypertension is a common hereditary disorder whose pathogenesis remains largely obscure. Chromogranin A (CHGA = human and Chga = rodent) is a 48 kDa acidic protein crucial for regulated secretory granule biogenesis as well as cargo trafficking and storage in neurons, chromaffin cells and neuroendocrine tissues (1). CHGA is expressed in the central nervous system as well as the periphery, in sites such as the adrenal medulla, pancreatic islet cells and enteroendocrine cells (1). CHGA also functions as a pro-hormone that yields bioactive peptides capable of influencing a set of diverse traits, including blood pressure (BP) and catecholamine secretion (catestatin peptide) (2), insulin sensitivity and glucose homeostasis (pancreastatin peptide) (3) and vasodilation and inflammation (vasostatin peptide) (4).
Chromogranin A was first implicated in the pathogenesis of human essential (genetic) hypertension when heritable increases in plasma CHGA were observed in hypertensive subjects (5–7). Functional genetic variation at the human CHGA locus, in both the proximal promoter (8,9) and the 3′-untranslated region (3′-UTR) (9,10), is associated with essential hypertension (8,10,11) as well as hypertensive end-organ damage (12), and naturally occurring non-synonymous amino acid variation within the catestatin (11,13) and pancreastatin (14,15) regions change the potencies of these peptide hormones. Mahapatra et al. (16) used targeted ablation of the Chga locus in the mouse to demonstrate a direct relationship between Chga genetic variation and hypertension in vivo. More recently, Vaingankar et al. (17) used this same Chga knockout model of hypertension to create mouse lines with 0 (complete knockout), 1, 2, 3 or 4 copies of the Chga (or CHGA) gene per diploid organism. The relationship between Chga copy number and BP was U-shaped, wherein mice with 0 and 4 copies of Chga had the highest BPs, whereas mice with 2 copies of Chga had the lowest BP. Mice with one and three copies of Chga showed intermediate BP values. It thus appeared that an optimal level of Chga expression was necessary for normal BP homeostasis, with either too much or too little expression leading to BP elevation.
The spontaneously hypertensive rat (SHR) and its control strain, the Wistar–Kyoto rat (WKY), form the most widely studied inbred animal model of genetic hypertension. Previous studies reported overexpression of Chga in the plasma and the adrenal gland of SHR versus WKY (18) and in the adrenal gland of SHRSP versus WKY (19). Chga expression is widely distributed in the brain (20,21), including the brainstem with its cardiovascular control nuclei, such as the nucleus tractus solitarii (NTS). Since Chga plays an important role in BP homeostasis in both central and peripheral components of the nervous system, we investigated Chga in the brainstem (central nervous component) and adrenal gland (peripheral nervous component) of the SHR. Since genetic variation at the Chga locus on rat chromosome 6q32 (22) remains unexplored, we sought to identify cis-acting polymorphisms within the SHR Chga locus and determine their contributions to disease-mediating changes in Chga expression or function. Finally, we tested the hypothesis that a mutation within the Chga 3′-UTR which alters a microRNA-binding motif, or the proximal promoter, can lead to dysregulation of central Chga expression, ultimately contributing to the pathogenesis of hypertension in the SHR.
RESULTS
Chromogranin A expression in SHR: peripheral elevation versus central diminution
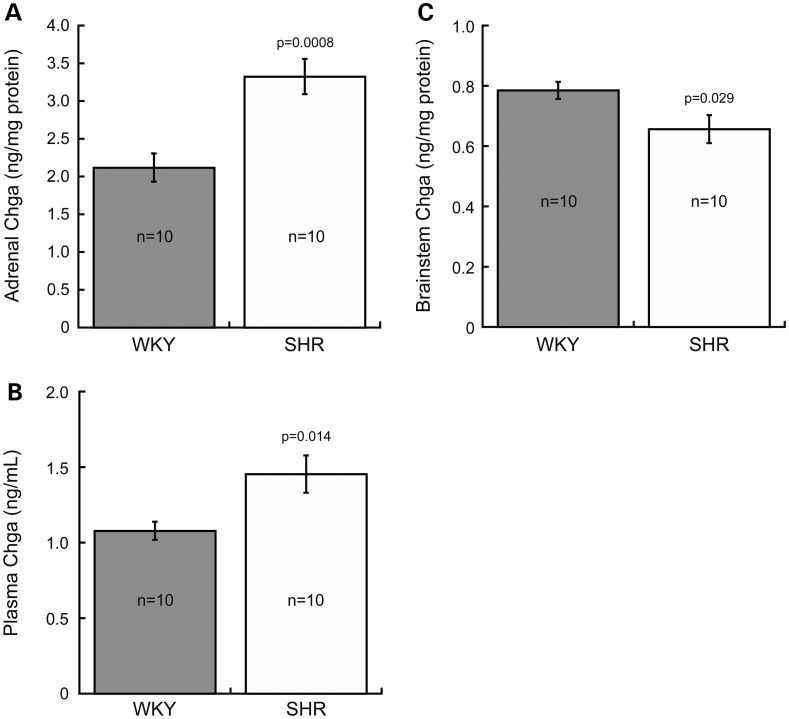
Chromogranin A (Chga) was measured in central (brainstem) and peripheral (adrenal gland and plasma) components of the nervous system in SHR and WKY rats (Fig. 1). Adrenal Chga protein was increased 1.57-fold in SHR (3.32 ± 0.23 ng/mg adrenal protein) compared with WKY (2.12 ± 0.19 ng/mg adrenal protein; P = 0.0008, Fig. 1A). Plasma Chga protein was also increased 1.35-fold in SHR (1.45 ± 0.12 ng/ml) compared with WKY (1.08 ± 0.06 ng/ml; P = 0.014, Fig. 1B). In contrast, brainstem Chga protein was decreased 0.83-fold in SHR (0.65 ± 0.05 ng/mg brainstem protein) compared with WKY (0.78 ± 0.03 ng/mg brainstem protein; P = 0.029, Fig. 1C).
Figure 1.
Chromogranin A (Chga) protein is elevated in peripheral (adrenal gland and plasma) but decreased central (brainstem) components of the nervous system of SHR. An enzyme immunoassay determined that Chga protein was significantly increased in SHR adrenal gland [by 1.57-fold, (A)] and plasma [by 1.35-fold, (B)], but decreased in SHR brainstem [by 0.83-fold, (C)]. Data were analyzed by an unpaired Student's t-test, and presented as mean ± standard error.
Genomics: polymorphism discovery across the rat Chga locus: SHR, WKY and BN strains
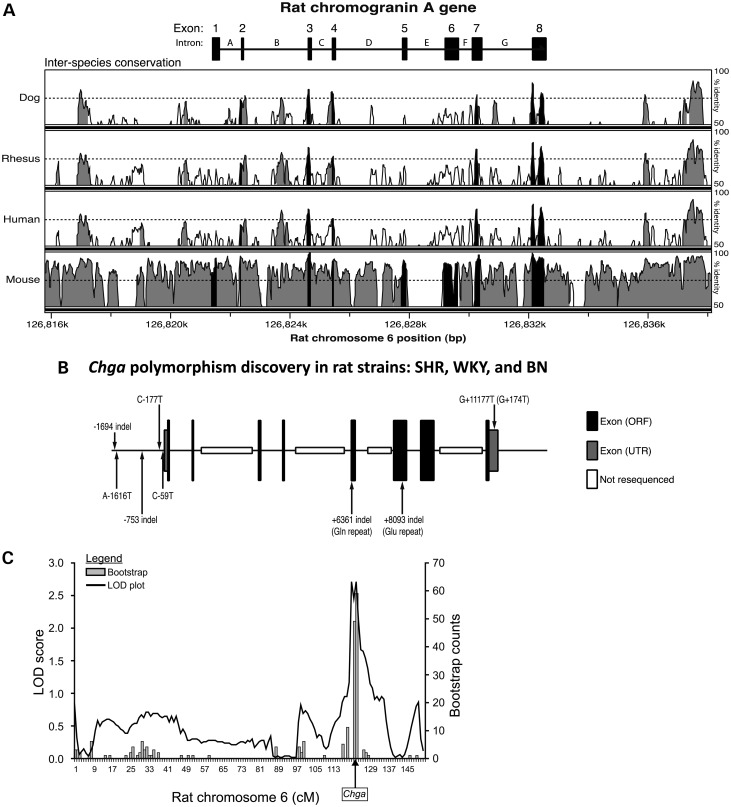
Targeted re-sequencing of the proximal promoter (∼2000 bp), 8 exons (including 5′- and 3′-UTRs) and intron/exon borders (splice junctions) of the Chga locus in SHR versus WKY and BN identified eight polymorphisms (Table 1, Fig. 2B). Five SHR polymorphisms were identified within the proximal promoter (−1694 Ins/Del; A-1616T; −753 Ins/Del; C-177T; C-59T), one within Exon 5 [ + 6361 Ins/Del (Gln repeat: SHR = 12, WKY = 20, BN = 19)], one within Exon 6 [+8093 Ins/Del (Glu repeat: SHR = 15, WKY = 16, BN = 16)] and one within the 3′-UTR of Exon 8 (G + 11177T or G + 174T with reference to the start of the 3′-UTR). The 3′-UTR variant G + 174T is found in NCBI dbSNP as rs13449558. No polymorphisms were detected within regions encoding bioactive peptides proteolytically derived from full-length Chga protein, including vasostatin, catestatin or pancreastatin. Fourteen polymorphisms were identified within the introns. These regions were not specific targets of re-sequencing and were thus not systematically covered (gaps are present in introns B, D, E and G; Fig. 2B). The single nucleotide polymorphism (SNP) within the 3′-UTR (G + 174T) occurred within a region of the Chga locus that displayed high inter-species conservation (Fig. 2A).
Table 1.
Polymorphism discovery across the rat Chga locus
| Polymorphism(s) |
Strain allele(s) |
||||
|---|---|---|---|---|---|
| Location | Domain | Type | BN (normotensive) | WKY (normotensive) | SHR (hypertensive) |
| −1694 Ins/Del | Promoter | Ins/Del | – | – | G |
| A-1616T | Promoter | SNP | A | A | T |
| −753 Ins/Del | Promoter | Ins/Del | 15 ‘A’ repeat | 15 ‘A’ repeat | 11 ‘A’ repeat |
| C-177T | Promoter | SNP | C | C | T |
| C-59T | Promoter | SNP | C | C | T |
| +6361 Ins/Del (Gln repeat) | Exon 5 | Ins/Del | 15 tri-nucleotide ‘CAG’ repeats (19 Gln repeat) | 16 tri-nucleotide ‘CAG’ repeats (20 Gln repeat) | 8 tri-nucleotide ‘CAG’ repeats (12 Gln repeat) |
| +8093 Ins/Del (Glu repeat) | Exon 6 | Ins/Del | GAG (16 Glu repeat) | GAG (16 Glu repeat) | – (15 Glu repeat) |
| G + 11177T (G + 174T) | Exon 8 (3′-UTR) | SNP | G | G | T |
A list of polymorphisms discovered through resequencing of the Chga locus in the SHR, WKY and BN rat strains is presented. Polymorphisms are numbered according to their position in relation to the mRNA cap site (transcriptional start site), which was designated as position ‘0’. Base pair position upstream of the cap site (i.e. in the promoter) was numbered negatively in the descending order. Base pair position downstream of the cap site was numbered in the ascending order. Coding region polymorphisms within exons 5 and 6 resulted in in-frame deletions of codons for glutamine and glutamic acid. Polymorphism G + 11177T in Exon 8 is also named G + 174T, which refers to its position relative to the beginning of the 3′-UTR.
Ins/Del, insertion/deletion; SNP, single nucleotide polymorphism; 3′-UTR, 3′-untranlsated region; Gln, glutamine (amino acid); Glu, glutamic acid (amino acid).
Figure 2.
Chga locus on rat chromosome 6q32: Genomics. Inter-species sequence conservation, systematic polymorphism discovery across strains and cis-QTL. (A) Inter-species conservation at the Chga locus. Conservation of the rat Chga locus across dog, rhesus, human and mouse genomes is presented. Regions of the locus exceeding 70% conservation in a 100 bp sliding window analysis are colored either black (exons) or grey (upstream, intronic or downstream regions). The 70% conservation threshold is indicated with a horizontal dashed line. (B) Polymorphisms discovered through re-sequencing of the rat Chga locus. Results in the SHR, WKY and BN rat strains are also presented. Intronic SNPs and SNPs downstream of the final exon are not displayed. The Chga mRNA cap site (transcriptional start site) was designated as position ‘0’. Base pair position upstream of the cap site (i.e. in the promoter) was numbered negatively in the descending order. Base pair position downstream of the cap site in the exonic and intronic regions was numbered in ascending order. ORF, open reading frame; UTR, untranslated region. (C) The Chga locus as a cis-QTL. Mapping of adrenal Chga protein (by immunoassay) as a quantitative trait in the HXB/BXH RI strains resulted in a significant meiotic recombination (linkage) peak (LOD = 2.58, P = 0.00028) on rat chromosome-6q32 over the Chga locus (shown with an arrow). 2000 bootstraps refined the position of the QTL. The QTL accounted for ∼35% of trait variance. The SHR genotype was directionally associated with increased adrenal Chga protein. The x-axis is displayed in genetic (recombination) units of centimorgans (cM). Genome wide-significance was approached by 2000 permutations, establishing thresholds for significant (LRS = 16.12, LOD = 3.49) and suggestive (LRS = 9.64, LOD = 2.09) linkage.
SHR, WKY and BN sequences were directly aligned with the BN reference sequence at NCBI; the WKY and BN sequences were identical in the promoter and 3′-UTR regions, but differed at only one coding site (+6361 Ins/Del, Gln repeat) over the entire re-sequenced range.
Recombinant inbred strains: evidence for a genetic role of Chga in BP control in the SHR
Linkage analysis of adrenal Chga protein in HXB/BXH recombinant inbred (RI) strains revealed a peak (LOD = 2.58, P = 0.00028) on rat chromosome 6q32 (22) directly above the Chga locus (Fig. 2C). QTL peak location was confirmed by 2000 bootstraps. The LOD peak thus displayed suggestive linkage (by 2000 permutation tests, 2.09<LOD<3.49; see Fig. 2C legend), i.e. statistically expected to occur less than once at random in the genome (23). SHR genotype was associated with increased adrenal Chga protein. By variance components, we estimated that this cis-QTL accounted for 35% of the variability in the adrenal Chga protein trait (i.e. 35% heritability).
In the RI strains, adrenal Chga protein was directly correlated with multiple adrenal biochemical determinants of BP, including adrenal epinephrine (r = 0.492, P = 0.0051), norepinephrine (r = 0.481, P = 0.0064), dopamine (r = 0.427, P = 0.0179), phenylethanolamine N-methyltransferase enzymatic activity (Pnmt; r = 0.628, P = 0.00013), dopamine beta-hydroxylase enzymatic activity (Dbh; r = 0.510, P = 0.0034), chromogranin B protein (Chgb; r = 0.546, P = 0.0015) and corticosterone (r = 0.636, P = 0.00009) (Table 2). Paradoxically, in the RI adrenal glands Chga mRNA abundance was inversely correlated with Chga protein (r = −0.417, P = 0.0167), suggesting derangement in the expected coupling between transcription and translation for adrenal Chga (vide infra). Thus, a genetic control of Chga biosynthesis in cis seems to originate a cascade of tightly coupled catecholaminergic events in the RI strains that impinge on an autonomic control of the circulation (Supplementary Material, Fig. S1), with these elements: Chga gene → Chga protein → Dbh → Norepinephrine → Pnmt → Epinephrine.
Table 2.
Correlations of adrenal Chga protein with hypertensive disease biochemical phenotypes in the RI strains
| Hypertensive disease trait correlated with adrenal Chga protein in the HXB/BXH RI strains | Correlation coefficient (r) | Correlation P-value | Number of RI strains |
|---|---|---|---|
| Catecholamines | |||
| Adrenal epinephrine (μg/mg protein) | 0.492 | 0.00512* | 30 |
| Adrenal norepinephrine (μg/mg protein) | 0.481 | 0.00645* | 30 |
| Adrenal dopamine (μg/mg protein) | 0.427 | 0.01786* | 30 |
| Catecholamine biosynthetic enzymes | |||
| Adrenal Pnmt (nmol/h/mg protein) | 0.628 | 0.00013* | 30 |
| Adrenal Dbh (nmol/h/mg protein) | 0.510 | 0.00344* | 30 |
| Chromogranins | |||
| Adrenal Chgb (fragments/ml) | 0.546 | 0.00146* | 30 |
| Steroid hormones | |||
| Adrenal corticosterone (pg/mg protein) | 0.636 | 0.00009* | 30 |
Analysis of the HXB/BXH RI strains identified significant parametric (Pearson) correlations (*P < 0.05) of adrenal Chga protein with multiple adrenal biochemical regulators of blood pressure.
Pnmt (phenylethanolamine N-methyltransferase), Dbh (dopamine beta-hydroxylase), Chgb (chromogranin B).
Computational prediction of transcription factor binding in the Chga promoter
The ConSite algorithm predicted differential binding of transcription factors to the SHR and WKY Chga promoters in the regions harboring polymorphisms (Table 3). Each of the five polymorphisms was predicted to alter binding (as indicated by difference in the ConSite score between SHR and WKY) of at least one transcription factor: −1694 Ins/Del (Hand1, Pbx1), A-1616T (Irf1, Sox17), −753 Ins/Del (Foxd3), C-177T (E2f1, Brachyury) and C-59T (Foxi1, Mycn, Rreb1).
Table 3.
Computational prediction of transcription factor and microRNA binding to polymorphic regions within the Chga locus
| Polymorphism | Location in gene | Predicted TF | WKY ConSite score | SHR ConSite score |
| −1694 Ins/Del | Promoter | Hand1 (MA0092, +) | 5.828 | 6.090 |
| Pbx1 (MA0070, −) | 8.101 | 7.716 | ||
| A-1616T | Promoter | Irf1 (MA0050, −) | 7.822 | n/a |
| Sox17 (MA0078, +) | 7.255 | n/a | ||
| −753 Ins/Del | Promoter | Foxd3 (MA0041, −) | 9.407 | 8.520 |
| C-177T | Promoter | E2f1 (MA0024, −) | 6.381 | n/a |
| Brachyury (MA0009, −) | 7.749 | 12.466 | ||
| C-59T | Promoter | Foxi1 (MA0042, +) | n/a | 8.300 |
| Mycn (MA0104, −) | 4.675 | n/a | ||
| Rreb1 (MA0073, −) | 8.543 | 9.205 | ||
| Polymorphism | Location in gene | Predicted microRNA | WKY free energy | SHR free energy |
| G + 174T | 3′UTR | rno-miR-22 | −22.9 kcal/mol | −23.71 kcal/mol |
| rno-miR-320 | n/a | −22.6 kcal/mol |
The ConSite algorithm predicted differential binding (indicated by difference in score) of 10 transcription factors to regions within the Chga promoter containing the five polymorphisms discovered through resequencing. Transcription factor JASPAR accession number and DNA strand binding orientation (+ or −) are listed in parentheses following the transcription factor symbol. The MicroInspector tool predicted differential stability of hybridization (indicated by difference in free energy) between two microRNAs and the region of the Chga mRNA 3′-UTR containing the G + 174T polymorphism. A lower free energy indicates a more stable interaction between microRNA and mRNA.
TF, transcription factor; n/a, not applicable (no binding or hybridization was predicted).
Promoter-luciferase reporter assays: effect of Chga promoter polymorphisms on transcription
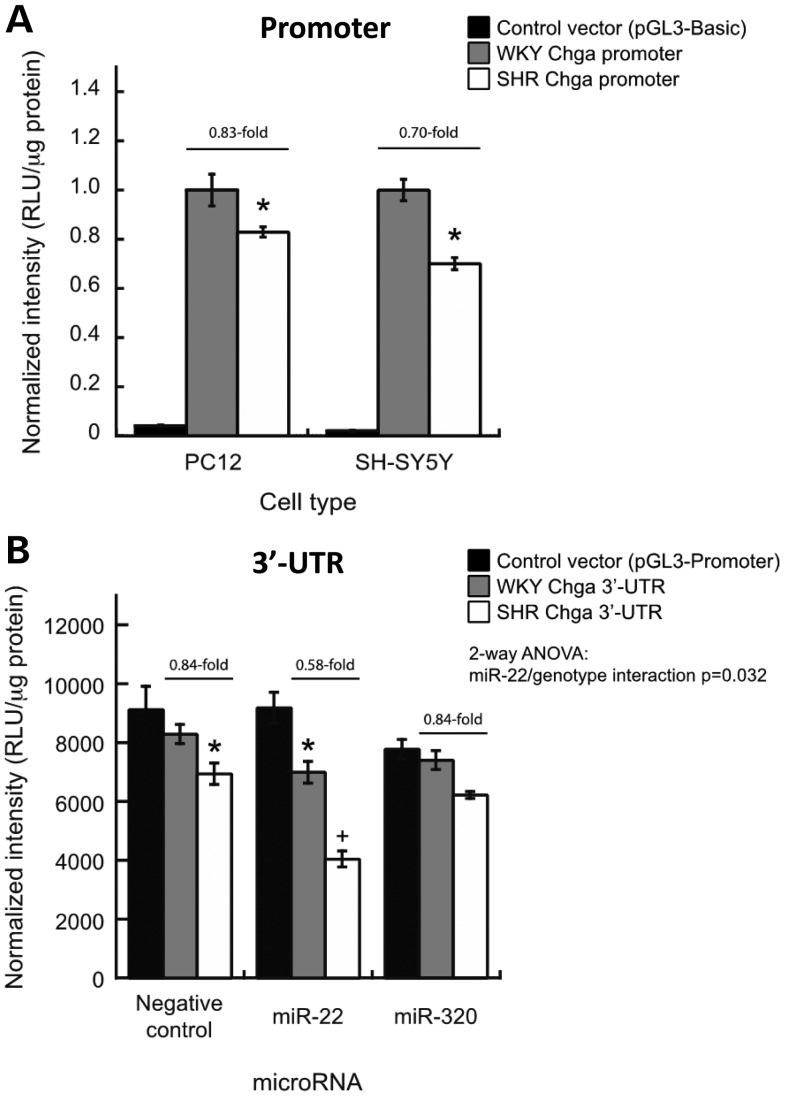
Promoter-luciferase reporter assays demonstrated that the basal transcriptional activity of the SHR Chga promoter was 0.83-fold less than that of the WKY Chga promoter in rat PC12 (chromaffin) cells (P < 0.05, Fig. 3A).
Figure 3.
Functional polymorphism (luciferase reporter) assays: SHR promoter and 3′-UTR polymorphisms decrease Chga gene expression in cultured cells. Luciferase reporter assays were used to determine whether polymorphisms within the SHR Chga promoter and 3′-UTR affect gene expression. Note that WKY and BN promoter and 3′-UTR sequences are identical. (A) Chga promoter. The basal transcriptional activity of the SHR Chga promoter was less than that of the WKY Chga promoter in both rat pheochromocytoma PC12 cells and human neuroblastoma SH-SY5Y cells (*P < 0.05 versus WKY basal expression). (B) Chga 3′-UTR. In PC12 cells, co-transfection of synthetic precursors for microRNAs predicted to hybridize differentially to the polymorphic region in the Chga 3′-UTR revealed a significant (two-way ANOVA) genotype-by-microRNA interaction for miR-22. Data are presented as mean ± standard error. *P < 0.05 versus WKY-negative control. +P < 0.05 versus SHR-negative control.
Since the basal promoter activity differed between strains, we performed luciferase assays with co-transfection of expression plasmids for the transcription factors predicted to differentially bind to the polymorphic regions. In PC12 cells, two transcription factors enhanced the expression of the WKY Chga promoter significantly more than the SHR Chga promoter: Foxi1 at C-59T (two-way ANOVA, P = 0.024) and Brachyury at C-177T (two-way ANOVA, P = 0.010). The remaining transcription factors (E2f1, Foxd3, Hand1, Irf1, Mycn, Pbx1, Rreb1 and Sox17) did not interact significantly with the polymorphism genotype (as determined by two-way ANOVA) in luciferase experiments in PC12 cells (data not shown). None of the transcription factors affected the expression of the pGL3-Basic control vector.
Quantitative RT–PCR determined that Foxi1 and Brachyury were not expressed in vivo in the SHR or WKY brainstem or the adrenal gland (data not shown). We, therefore, confirmed reduced basal expression of the SHR Chga promoter in a different neuroendocrine cell type: human neuroblastoma SH-SY5Y cells (Fig. 3A; 0.70-fold reduction, P < 0.05).
Computational prediction of microRNA binding in the Chga 3′-UTR
The MicroInspector algorithm was used to predict differences in hybridization free energy (and therefore the stability of the microRNA–mRNA interaction) between microRNAs and the SHR and WKY Chga 3′-UTRs in the region harboring the G + 174T polymorphism. Two microRNAs, rno-miR-22 and rno-miR-320, were predicted to form more stable duplexes (and therefore have lower free energy when hybridized) with the SHR Chga mRNA 3′-UTR than with the WKY Chga mRNA 3′-UTR (Table 3).
3′-UTR-luciferase reporter assays: effect of the Chga 3′-UTR polymorphism on gene expression
Initial 3′-UTR-luciferase reporter assays showed that the basal level of mRNA stability and translational efficiency conferred by the SHR Chga 3′-UTR was 0.84-fold (−16%) less than that conferred by the WKY Chga 3′-UTR in PC12 cells (P < 0.05). We then performed the luciferase assays with co-transfection of synthetic precursors for the two microRNAs predicted to hybridize the region harboring G + 174T. MicroRNA miR-22 displayed a significant interaction with the polymorphism genotype (two-way ANOVA; P = 0.032), and reduced the luciferase signal of the SHR 3′-UTR construct significantly more than the WKY 3′-UTR construct (Fig. 3B). MicroRNA miR-320 had no effect on SHR or WKY 3′-UTR construct expression (Fig. 3B). Neither of the microRNA precursors affected the pGL3-Promoter control vector (Fig. 3B). Polymorphism G + 174T was predicted to alter the conformation and free energy of binding between SHR Chga 3′-UTR and miR-22 (Supplementary Material, Fig. S2). RT–PCR analysis determined that miR-22 was expressed in vivo in SHR and WKY brainstems and adrenal glands (vide infra, under mRNAs).
Computational genome-wide prediction of miR-22 targets using the TargetScan algorithm identified 343 potential transcripts, with binding scores (total context + score) ranging from −0.70 to −0.02 (Supplementary Material, Table S2).
Computational analysis of secondary structural changes in Chga protein
Inter-species conservation (across mammals: Homo sapiens, Macaca mulatta, Bos taurus, Canis familiaris, Equus caballus, Mus musculus, Rattus norvegicus) of the regions harboring the glutamine (+6361 Ins/Del, Exon 5) and glutamic acid (+8093 Ins/Del, Exon 6) Ins/Del polymorphisms was determined: the glutamine Ins/Del was present only in rodents (rat and mouse), whereas the glutamic acid Ins/Del was present only in rat.
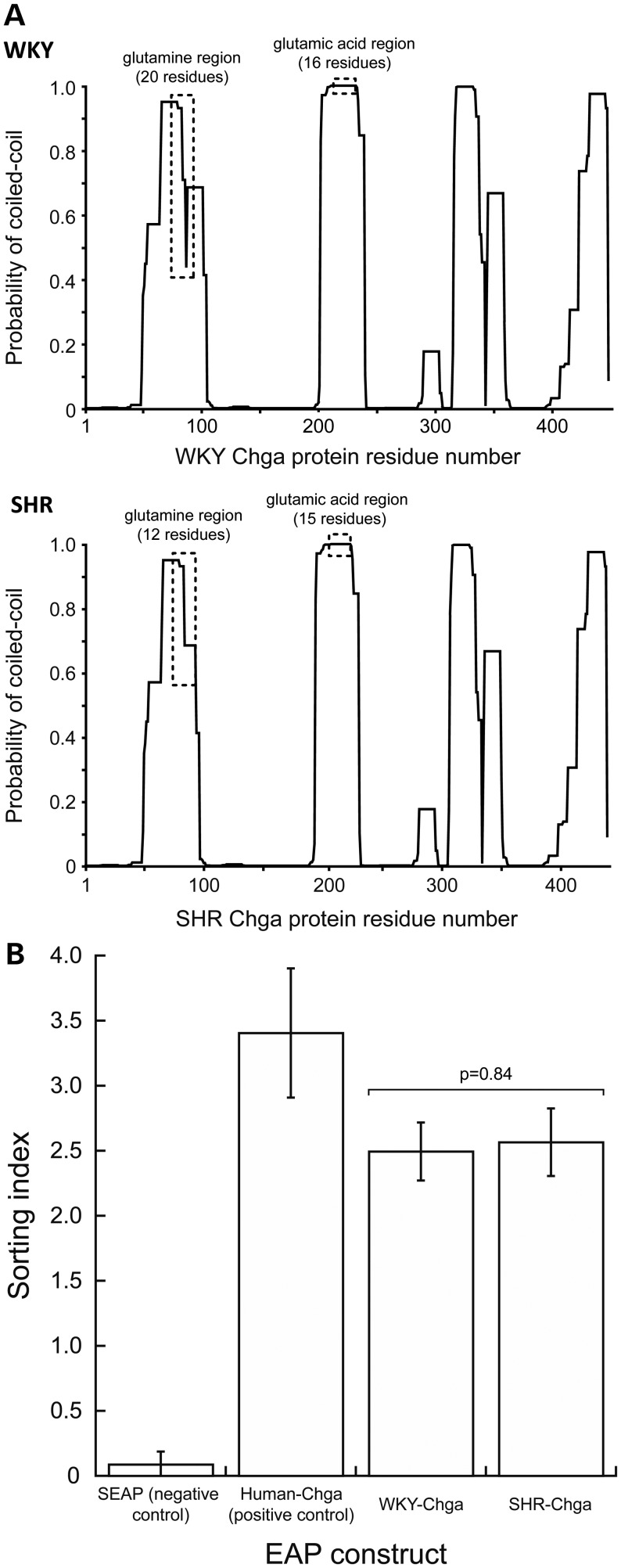
Since human Chga protein may assume a coiled-coil structure (24), and coiled coils usually contain a repeated pattern of hydrophobic and polar residues, we computed the effect of the glutamine (polar residue) and glutamic acid Ins/Dels on the ability of SHR and WKY Chga proteins to assume coiled-coil structures (Fig. 4A). The glutamine Ins/Del was predicted to increase the probability of SHR protein to form a coiled coil by ∼25%, yet decrease the number of residues involved in a coiled-coil segment by eight. The glutamic acid Ins/Del was predicted to have no effect on the coiled-coil structure of SHR Chga protein.
Figure 4.
Effect of coding region polymorphisms. (A) Chga protein coiled-coil structure. Since the human CHGA protein exhibits a coiled-coil structure, the probability of the WKY (top) and SHR (bottom) Chga proteins to also assume a coiled-coil structure was computed with the COILS algorithm. The probability of coiled-coil structure is presented as a function of amino acid residue position in the Chga protein. The glutamine repeat polymorphism was predicted to increase the probability of SHR protein to assume a coiled-coil structure by ∼25%, yet decrease the number of residues involved in a coiled-coil segment by eight. The glutamic acid polymorphism was predicted to have no effect on the coiled-coil structure of SHR Chga protein. WKY and BN sequences at Chga, though indentical in the promoter, 3′-UTR, and oligo-Glu repeat, differed at one coding site, the oligo-Gln repeat (+6361 Ins/Del, Gln repeat, Table 1), where SHR displayed (Gln)12, with WKY at (Gln)16 and BN at (Gln)15. (B) Chimeric (in-frame) reporter assays: SHR coding polymorphisms do not affect trafficking and secretion of Chga protein into the regulated secretory pathway. EAP secretion and activity assays were performed with SHR and WKY Chga in-frame cDNA/EAP reporter constructs to determine whether SHR coding polymorphisms affected trafficking and secretion of Chga protein in the regulated secretory pathway. The secreted embryonic alkaline phosphatase construct, which enters the constitutive secretory pathway, was a negative control for the SHR and WKY Chga constructs. The human construct, which has previously been tested and validated (56), served as a positive control for correct trafficking and regulated secretion of Chga. No difference was detected in the Rs of SHR or WKY constructs to either barium (a potent stimulus of regulated secretion) or calcium (negative control for barium) stimulation (data not shown). Rs = (EAP activity in supernatant)/(EAP activity in supernatant + EAP activity in cell lysate). No difference was detected in the sorting index of SHR (2.57 ± 0.26) and WKY (2.49 ± 0.22) Chga protein (P = 0.84). Sorting index (SI) = (Rsbarium – Rscalcium)/Rscalcium. Data were analyzed with an unpaired Student's t-test, and presented as mean ± standard error.
EAP chimeric reporter assays: effect of Chga coding polymorphisms on regulated secretion
Embryonic alkaline phosphatase (EAP) secretion and activity assays were performed with SHR and WKY Chga cDNA-EAP reporter constructs to determine whether the glutamine Ins/Del (predicted to increase coiled-coil probability by ∼25% and decrease the number of residues within a coiled coil by 8) affected trafficking and secretion of Chga protein by the regulated secretory pathway. Relative secretion (Rs) is defined as the fraction of total EAP activity that is found in the supernatant after stimulation: Rs = (EAP activity in supernatant)/(EAP activity in supernatant + EAP activity in lysate). Sorting index (SI) is defined as the difference between Rs of barium (strong exocytosis stimulation) and calcium (control)-treated cells, normalized by the Rs of calcium-treated cells: SI = (Rsbarium–Rscalcium)/Rscalcium. No difference was detected in Rs or SI of SHR (2.57 ± 0.26) and WKY (2.49 ± 0.22) Chga proteins (P = 0.84, Fig. 4B).
RNA transcript abundance: quantitative RT–PCR
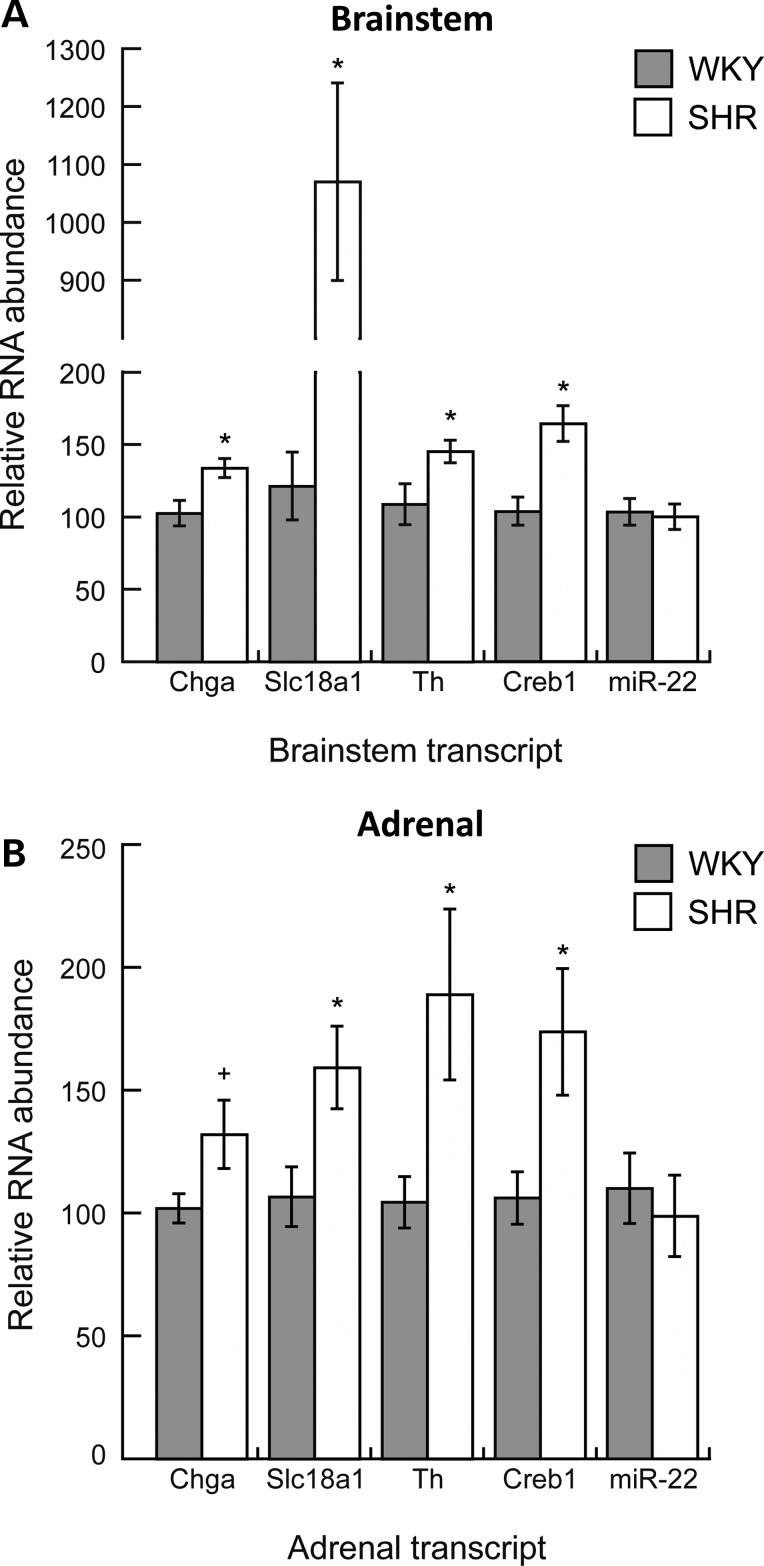
qRT–PCR probed in vivo mRNA and microRNA expression profiles in central (brainstem) and peripheral (adrenal gland) components of the nervous system in the SHR. We probed the following transcripts: chromogranin A (Chga); solute carrier family 18, member 1 (Slc18a1 or Vmat1; an amine transporter located in the membrane of catecholamine storage vesicles); tyrosine hydroxylase (Th; the rate-limiting enzyme in catecholamine biosynthesis); cAMP-responsive element-binding protein 1 [Creb1; transcription factor that regulates the expression of Chga (1), Slc18a1/Vmat1 (25) and Th (26,27)] and miR-22. In the brainstem, Chga (+1.3-fold, P = 0.01), Slc18a1/Vmat1 (+8.8-fold, P < 0.0001), Th (+1.3-fold, P = 0.037) and Creb1 (+1.6-fold, P < 0.001) mRNAs were elevated in the SHR, whereas miR-22 transcript was unchanged (Fig. 5A). In the adrenal gland, Chga [+1.3-fold, P = 0.055 (marginally significant)], Slc18a1/Vmat1 (+1.5-fold, P = 0.021), Th (+1.8-fold, P = 0.032) and Creb1 (+1.6-fold, P = 0.026) mRNAs were elevated in the SHR, whereas miR-22 transcript showed no change (Fig. 5B). The marginally significant elevation of Chga mRNA in the adrenal gland we observed (P = 0.055) is consistent with previous reports of a significant elevation of Chga mRNA in the SHR adrenal gland (18).
Figure 5.
Endogenous gene expression by quantitative RT–PCR: The measurement of transcript abundance in central and peripheral tissues of the SHR nervous system. We performed quantitative RT–PCR to determine in vivo mRNA and microRNA expression profiles in central (brainstem) and peripheral (adrenal gland) components of the SHR nervous system for the following transcripts: Chga, Slc18a1/Vmat1, Th, Creb1 and miR-22. Data were normalized to beta-actin in the same sample, and analyzed with an unpaired Student's t-test, and presented as mean ± standard error. *P < 0.05 versus WKY (significant); +P = 0.055 versus WKY (marginally significant). Normalization of miR-22 abundance in the brainstem by a snoRNA (SNORD61) instead of beta-actin resulted in 0.76-fold (P < 0.05) decreased miR-22 in SHR compared with WKY; beta-actin normalization resulted in no difference. Normalization of miR-22 abundance in the adrenal gland by SNORD61 was consistent with beta-actin normalization, resulting in no difference between SHR and WKY.
Inhibition of microRNA-22 in vivo: BP measurement and biochemical assays
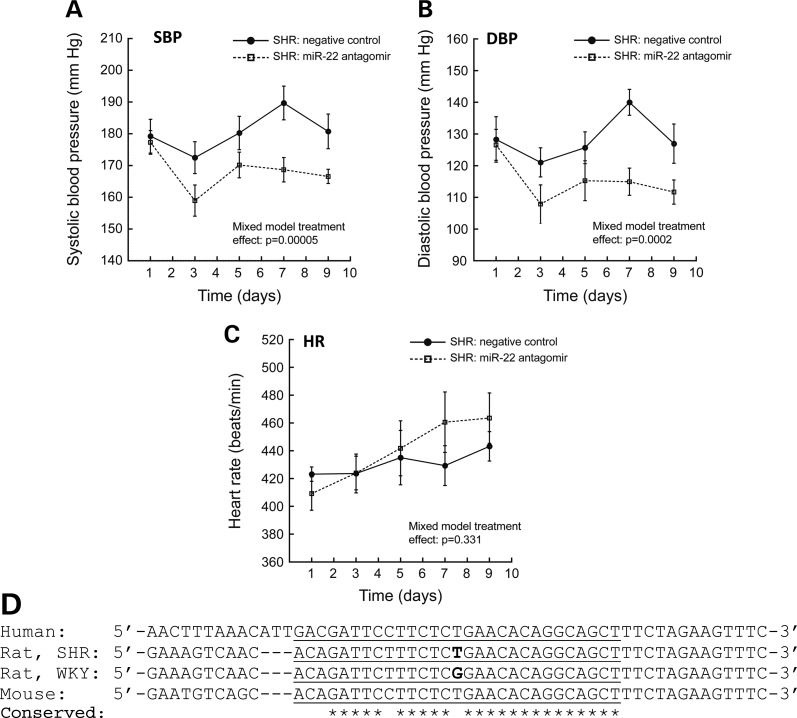
Administration of miR-22 antagomir (via IP injection; dose of 25 mg/kg every other day) resulted in an 11 mmHg reduction of systolic blood pressure (SBP) (177 mmHg on Day 1 to 166 mmHg on Day 9) (Fig. 6A) and a 15 mmHg reduction of diastolic blood pressure (DBP) (126 mmHg on Day 1, to 111 mmHg on Day 9) (Fig. 6B) in the SHR after 9 days (four doses). The initial dose of antagomir resulted in rapid (within 48 h) and substantial ∼18 mmHg decreases in both SBP (177 mmHg on Day 1, to 159 mmHg on Day 3) and DBP (126 mmHg on Day 1 to 108 mmHg on Day 3). Treatment with miR-22 antagomir significantly reduced both SBP (F = 74.5, P = 0.00005) and DBP (F = 15.3, P = 0.0002) compared with negative control oligonucleotide. MicroRNA-22 antagomir administration had no differential effect on heart rate (HR) (F = 0.956, P = 0.331) (Fig. 6C).
Figure 6.
MicroRNA-22 inhibition in vivo and conservation across species. (A–C) MicroRNA-22 inhibition in vivo: miR-22 antagomir decreases blood pressure in the SHR. SHR were administered either miR-22 antagomir (n = 10) or scrambled sequence negative control (n = 9) via IP injection. Animals received IP doses (at 25 mg/kg) on Days 1, 3, 5, and 7. MicroRNA-22 antagomir treatment significantly reduced both systolic (A) and diastolic (B) blood pressure in the SHR (versus negative control group). Antagomir administration had substantial acute (∼18 mmHg reduction within 48 h) and sustained (∼13 mmHg reduction after 9 days) effects on blood pressure. MicroRNA-22 antagomir administration had no effect on heart rate (C). Data were analyzed with a linear mixed effects model in SPSS. (D) Inter-species conservation of the miR-22 binding site in the 3′-UTR region of the rat Chga locus. The predicted site of miR-22 hybridization (as it would occur in transcribed Chga mRNA) is underlined in the DNA sequence of the Chga locus. Location of the SHR/WKY G + 174T SNP (SHR allele = ‘T’; WKY allele = ‘G’) is shown in bold text. Individual bases conserved across human, rat and mouse are indicated with *. The miR-22 binding site is well conserved, indicating its likelihood of miR-22 regulation of CHGA in humans. Note that WKY and BN 3′-UTR sequences are identical.
We measured Chga protein and mRNA abundance in the brainstem, adrenal gland and plasma in SHR after 9 days of the administration of miR-22 antagomir or negative control (Table 4). Brainstem Chga protein increased 1.33-fold (+33%, P = 0.027), whereas adrenal Chga protein decreased 0.52-fold (−48%, P = 0.01), and plasma Chga protein did not change. Neither brainstem nor adrenal Chga mRNA abundance changed after miR-22 antagomir administration.
Table 4.
Effect of miR-22 antagomir treatment on Chga mRNA and protein abundance in the SHR
| Analyte | Brainstem fold change (antagomir/control) | Adrenal gland fold change (antagomir/control) | Plasma fold change (antagomir/control) |
|---|---|---|---|
| Chga protein | 1.33-fold (P = 0.027) | 0.52-fold (P = 0.01) | No change |
| Chga mRNA | No change | No change | Not applicable |
We measured Chga protein and Chga mRNA transcript abundance fold change in SHR after 9 days of treatment with miRNA-22 antagomir or negative control. Statistically significant P-values (<0.05) are shown in parentheses following fold-change values. Fold change >1 indicates increased abundance in the miR-22 antagomir treatment group; fold change <1 indicates decreased abundance in the miR-22 antagomir treatment group. Data were analyzed with Student's t-test.
A blood plasma chemistry panel revealed no adverse effects of in vivo miR-22 antagomir treatment on carbohydrate metabolism (glucose), kidney function [blood urea nitrogen (BUN), creatinine], electrolytes (sodium, phosphorus) or liver function (alkaline phosphatase, albumin, total protein, globulin, total bilirubin) in the SHR (Table 5). Treatment with miR-22 antagomir actually resulted in modest declines in hepatocyte glutamic pyruvic transaminase (control: 68 ± 5 U/l, antagomir: 55 ± 2 U/l; P = 0.02) and pancreatic amylase (control: 846 ± 17 U/l, antagomir: 729 ± 23 U/l; P = 0.0008).
Table 5.
Chemistry panel results indicate no adverse effects of in vivo miR-22 antagomir treatment
| Chemistry panel marker | SHR treated with control (mean ± SEM) | SHR treated with antagomir (mean ± SEM) | P-value |
|---|---|---|---|
| General metabolism | |||
| Glucose (mg/dl) | 189 ± 5 | 196 ± 3 | 0.23 |
| Kidney function | |||
| BUN (mg/dl) | 20 ± 1 | 20 ± 1 | 0.70 |
| Creatinine (mg/dl) | 0.2 ± 0.01 | 0.2 ± 0.02 | 0.34 |
| Electrolytes | |||
| Sodium (mEq/l) | 162 ± 1 | 164 ± 1 | 0.33 |
| Phosphorus (mg/dl) | 9.9 ± 0.2 | 9.5 ± 0.2 | 0.13 |
| Liver function | |||
| Alkaline phosphatase (U/l) | 29 ± 10 | 21 ± 6 | 0.49 |
| Albumin (g/dl) | 4.7 ± 0.1 | 4.7 ± 0.1 | 0.89 |
| SGPT (ALT) (U/l) | 68 ± 5 | 55 ± 2 | 0.02* |
| Total protein (g/dl) | 5.8 ± 0.1 | 5.8 ± 0.1 | 0.46 |
| Globulin (g/dl) | 1.1 ± 0.1 | 1.1 ± 0.1 | 0.75 |
| Total bilirubin (mg/dl) | 0.2 ± 0.01 | 0.2 ± 0.02 | 0.34 |
| Pancreas function | |||
| Amylase (U/l) | 846 ± 17 | 729 ± 23 | 0.0008* |
A standard chemistry panel was performed on plasma from SHR treated with miR-22 antagomir or negative control. Data were analyzed with Student's t-test. Statistically significant differences (P < 0.05) are indicated with *.
BUN, blood urea nitrogen; SGPT, serum glutamic pyruvic transaminase.
DISCUSSION
Central versus peripheral expression of chromogranin A
Chromogranin A plays a necessary role in the formation and cargo storage of regulated secretory granules in neuroendocrine cells (16). In the central nervous system (brainstem), neuronal Chga-granules store catecholamines and may release these neurotransmitters to regulate sympathetic outflow to the periphery (11). We observed a 0.83-fold decrease in Chga protein in the brainstem of SHR (Fig. 1C), likely leading to impaired or reduced ability of SHR to store catecholamines in this brain region vital to BP regulation. In fact, it was previously shown that the norepinephrine content was decreased in the NTS (in the medulla oblongata of the brainstem) of SHR versus WKY (28), and in the medulla oblongata (brainstem) of SHRSP versus WKY (29).
In adrenal medullary chromaffin cells of the peripheral nervous system, Chga-granules store catecholamines and release these endocrine hormones to stimulate and support sympathetic activity throughout the periphery. We observed 1.57-fold elevation of Chga protein in the adrenal gland of SHR (Fig. 1A), which is consistent with previous reports of elevated adrenal Chga protein in SHR versus WKY (18,19). Increased abundance of Chga suggests that the SHR adrenal gland has acquired additional capacity to store catecholamines in regulated secretory granules. Indeed, elevated adrenal norepinephrine in SHR versus WKY (18,30) and in SHRSP versus WKY (19,30) has been reported in the literature. We also observed a 1.35-fold elevation of Chga protein in the plasma of SHR (Fig. 1B), consistent with previous investigations of SHR versus WKY (18), which indicates that peripheral sympathetic activity is enhanced in the SHR.
Catecholamines, such as norepinephrine, have quite different, indeed opposite effects on BP (31) when administered peripherally (e.g. pressor/hypertensive effects when given intravenously) versus centrally (e.g. depressor/hypotensive effects when given within/onto the NTS in the brainstem, likely mediated by α2-adrenergic receptors) (32). Since Chga triggers the formation of catecholamine storage vesicles in the regulated pathway (16) and binds catecholamines within the soluble core of such vesicles (33), changes in catecholamine storage/availability mediated by variation in Chga abundance in the SHR would be predicted to have opposite consequences in central and peripheral compartments, as follows: ↓ Central Chga → ↓ Central catecholamines → ↑ BP; ↑ Peripheral Chga → ↑ Peripheral catecholamines → ↑ BP. Indeed, the catecholamine release-inhibitory catestatin fragment of Chga exhibits opposite peripheral versus central effects on BP: decreasing BP when given intravenously (16), yet increasing BP when given centrally (34).
Thus, the changes we observed in SHR Chga expression (↓ Central → ↑ Peripheral) would each be expected to yield increases in BP. The question now turned to whether and how Chga genetic variation in cis might achieve such effects.
BP is influenced by genetic cis-regulation of the Chga locus in the SHR
Using RI strain meiotic linkage, we first established that adrenal Chga protein in the SHR was under substantial genetic regulation in cis at the Chga locus (Fig. 2C), and then demonstrated that these genetically determined levels of Chga significantly predict biochemical regulators of BP, including catecholamines, catecholamine synthetic enzymes, catecholamine storage proteins and steroid hormones (Table 2), suggesting that cis-regulation of Chga expression (Fig. 2C) controls a sequential pathway of catecholaminergic events (Supplementary Material, Fig. S1). While the RI linkage and cis-QTL data are based on adrenal expression, additional brainstem-specific linkage analysis would further support genetic regulation of the Chga locus in cis, and elucidate the role of tissue-specific regulation.
Pertinent to our results, in a previous rat genetic F2 intercross of a salt-sensitive hypertension model (BN/SsNHsdxSS/JrHsdMcwi; as described in (35), and archived at the Rat Genome Database <http://rgd.mcw.edu>), the Chga locus [on rat chromosome 6q22; (22)] lay within the linkage confidence interval for mean arterial pressure, as BP-QTL-211 (LOD = 3.66).
Proximal promoter polymorphisms decrease SHR Chga promoter activity
Next, we sought to identify cis-acting Chga polymorphisms that might contribute to differential expression of Chga protein and, therefore, regulation of BP. We re-sequenced the Chga locus and demonstrated that the C-59T and C-177T polymorphisms disrupted binding sites for Foxi1 and Brachyury, respectively, consistent with the decreased basal transcriptional activity of the SHR Chga promoter. However, since neither Foxi1 nor Brachyury mRNA transcripts were expressed in SHR or WKY brainstem or adrenal gland in vivo, we confirmed the reduced basal activity of the SHR promoter in a different cell type, SH-SY5Y (Fig. 3A). Foxi1 was expressed endogenously in PC12 cells (25) but not in SH-SY5Y cells (NCBI Gene Expression Omnibus Dataset, GDS1548; http://www.ncbi.nlm.nih.gov/sites/geo). The endogenous expression of Brachyury in PC12 cells is unknown; however, Brachyury is not expressed endogenously in SH-SY5Y cells (NCBI Gene Expression Omnibus Dataset, GDS1548). Perhaps Foxi1 and Brachyury do not contribute to the reduced basal activity of the SHR promoter. Additional experiments may elucidate the mechanism whereby functional SNPs within the SHR promoter reduce basal transcriptional activity.
3′-UTR polymorphism decreases SHR Chga gene expression
The observation that RI adrenal gland Chga mRNA abundance was inversely correlated with Chga protein (r = −0.366, P = 0.046), suggested derangement in the expected coupling between transcription and translation for Chga; since microRNA binding to the 3′-UTR can control mRNA translation, we explored the 3′-UTR sequences of Chga. The G + 174T mutation was demonstrated to lie within a miR-22 binding site and provide a plausible mechanism to explain the decreased mRNA translational efficiency (and, therefore, decreased gene expression) conferred by the SHR 3′-UTR (Fig. 3B). We probed the expression of miR-22 in vivo and determined that it was indeed expressed in both SHR and WKY brainstems as well as adrenal glands (Fig. 5).
Chga coding region polymorphisms lack detectable effects on secretory trafficking
Even though the glutamine Ins/Del was computationally predicted to change the coiled-coil structure of SHR Chga protein (Fig. 4A), the fact that the glutamine repeat region existed only in rodents and was not conserved across species (rat, mouse, dog, rhesus and human) suggested that this region is not crucial for Chga protein function. Indeed, EAP secretion and activity assays confirmed that the glutamine deletions in SHR Chga did not affect the trafficking or regulated release of the protein (Fig. 4B).
Potential contribution of unknown or uncharacterized polymorphisms
It was conceivable that functional transcriptional elements were present upstream of the ∼2-kb proximal promoter region that was re-sequenced in SHR and WKY. For example, the highly conserved region ∼4 kb upstream of Chga exon-1 (Fig. 2A) might function in transcriptional regulation. The sequence of this region in the SHR and WKY strains remains unknown, and our promoter-luciferase reporter constructs did not contain or test the function of this upstream region. Nor was our study designed to detect enhancer elements located long distances from the Chga locus.
The effects of the 14 intronic polymorphisms were not examined. Of interest were polymorphisms A + 1113G within Intron-B and T + 3961C within Intron-C, which occur within regions of high inter-species conservation (Fig. 2A). We did not target the entire introns for re-sequencing, and additional or unknown functional polymorphisms might exist within un-sequenced intronic regions.
Chga as a contributor to the pathogenesis of hypertension in the SHR
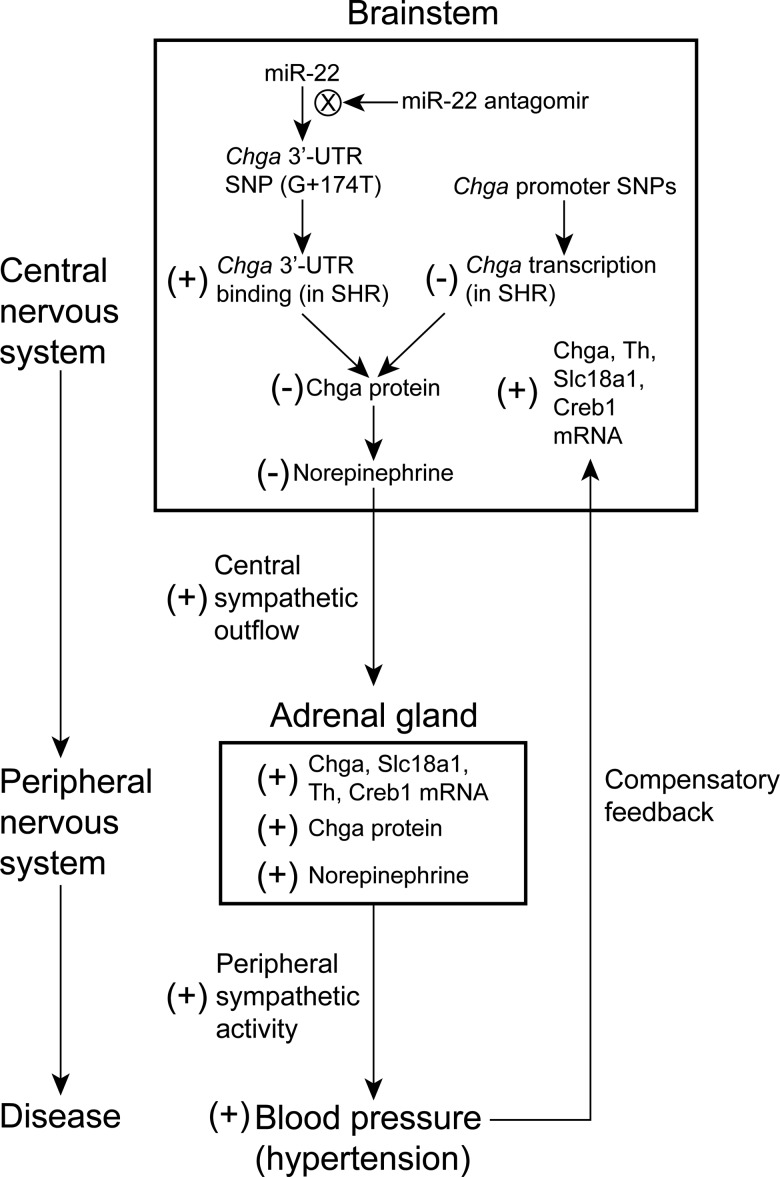
We present the hypothesis that the 3′-UTR and promoter Chga polymorphisms contribute to reduced Chga protein expression in the brainstem of SHR, ultimately eventuating in development of systemic hypertension (Fig. 7). We are not the first to hypothesize involvement of the brainstem in the pathogenesis of hypertension in the SHR. Takami et al. (29) hypothesized that since norepinephrine was decreased in the NTS in the medulla oblongata of SHR, and since norepinephrine has an inhibitory effect in the NTS on systemic BP elevation, ‘the suppression of negative feedback due to a decrease in the activity of inhibitory neurons in the medulla oblongata appears to be involved in the development and progression’ of hypertension in the SHR (29); we build upon the conclusions of Takami et al. (29) to include a genetic basis at the Chga locus.
Figure 7.
Hypothetical schema illustrating the contribution of the Chga 3′-UTR G + 174T polymorphism to the pathogenesis of hypertension in the SHR. The Chga 3′-UTR SNP (G + 174T) increases binding of miR-22 to the SHR Chga mRNA transcript, which decreases Chga protein abundance in the brainstem (with no change in the Chga mRNA level) and initiates a cascade of events that increase blood pressure. The effect of treatment with miR-22 antagomir is to reduce binding of miR-22 to the Chga 3-UTR, which increases the abundance of Chga protein in the brainstem (without change in Chga mRNA) and, ultimately, leads to reduction in blood pressure.
We therefore propose that the 3′-UTR and promoter Chga polymorphisms lead to reduced Chga protein expression in the SHR brainstem (Fig. 1C) via reduced mRNA transcription (Fig. 3A) and altered binding of miR-22 (Fig. 3B), and consequently reduced norepinephrine levels (28,29). It is likely that reduced mRNA translation also contributes to reduced Chga protein level. Since norepinephrine acts to reduce BP in the brainstem (especially in the NTS of the medulla oblongata), reduction of norepinephrine in this region should then lead to enhanced central sympathetic outflow to the periphery. Aberrant storage and secretion of norepinephrine in the brainstem might also affect baroreceptor function. Elevation of adrenal Chga protein (Fig. 1A), adrenal Chga (18), Slc18a1, Th, and Creb1 mRNAs (Fig. 5), adrenal norepinephrine (18,19,30) and plasma Chga protein (Fig. 1B) support the notion of increased peripheral sympathetic stimulation and activity. Indeed, Chga expression is up-regulated by ganglionic sympathetic transmitters, such as nicotinic–cholinergic agonists and neuropeptides (1), and previous studies have reported enhanced peripheral sympathetic activity in the SHR (18,36–38). The ultimate effect of the 3′-UTR and promoter Chga polymorphisms would then be elevated BP (hypertension) stemming from increased central sympathetic outflow from the brainstem.
Although the Chga 3′-UTR and promoter SNPs are expected to be operative in the adrenal gland as well, enhanced central sympathetic outflow arising from the brainstem is likely to stimulate adrenal Chga gene expression, resulting in our observations of increased Chga adrenal mRNA and protein. Lack of cis-regulation for adrenal Chga mRNA in the RI strains supports a primary role for central control of adrenal Chga mRNA and protein expression. In addition, elevated BP is proposed to provide a negative feedback to the central nervous system, leading to compensatory increases in Chga, Slc18a1, Creb1 and Th mRNAs in the brainstem (Fig. 5A). Apparently, the compensatory signals are not strong enough to overcome the combined pro-hypertensive effect of both the promoter and 3′-UTR Chga SNPs, resulting in dominance of the primary over compensatory effects. The main effect of SNPs in the SHR promoter is to reduce transcriptional activity (Fig. 3A). Elucidation of the temporal characteristics of Chga, Slc18a1, Creb1 and Th transcript alterations in the brainstem would prompt additional investigation. Normalization of miR-22 abundance in the brainstem by a snoRNA (SNORD61) instead of beta-actin resulted in 0.76-fold (P < 0.05) decreased miR-22 in SHR compared with WKY; beta-actin normalization resulted in no difference. In either case, the decreased level of Chga protein in the SHR brainstem suggests that the effect of the G + 174T 3′-UTR SNP is dominant over the effect of miR-22 expression. Normalization of miR-22 abundance in the adrenal gland by SNORD61 was consistent with beta-actin normalization, resulting in no difference between SHR and WKY.
Seemingly paradoxical differences in effects of SNPs on gene expression in cell culture and in vivo are frequently observed in complex trait genetics. Here, we observe an effect of the Chga 3′-UTR and promoter SNPs to reduce gene expression in cell culture, yet Chga protein is overexpressed in the SHR adrenal gland and the plasma. Only through consideration of central (brainstem) versus peripheral (adrenal gland and plasma) Chga expression were we able to reconcile these seemingly opposing observations. This concept of central versus peripheral actions and the expression of physiological pathways and their candidate genes loom large in the complex trait genetics literature. We have already noted the opposing effects of catecholamines on BP in the central versus peripheral pathways (31). Other transmitter systems have opposing central and peripheral effects on BP. In humans, genetic variation in the neuropeptide Y receptor type 1 (NPY1R) promoter and 3′-UTR that decreased gene expression in cell culture predicted increased BP in vivo (39), and genetic variation in the human dopamine beta-hydroxylase (DBH) promoter had directionally opposite effects on gene expression in cell culture and on plasma DBH activity, urine epinephrine and BP in vivo (40). Similar effects in the central and peripheral nervous systems have also been reported for polymorphisms in human peptide YY (PYY) and obesity (41). In each case (NPY1R, DBH and PYY), the deleterious effects of specific genetic determinants were explained through consideration of their reciprocal effects in the central and peripheral nervous systems.
Administration of miR-22 antagomir in vivo leads to a rapid yet prolonged decrease in SBP and DBP in the SHR (∼18 mmHg reduction with 48 h; sustained reduction after 9 days), without change in the HR or the reflex tachycardia response (Fig. 6). Such striking and effective reductions in BP further implicate the Chga locus in the pathogenesis of hypertension in the SHR, and suggest therapeutic applications for miR-22 antagomir. Computational, genome-wide analysis identified additional targets potentially regulated by miR-22 (Supplementary Material, Table S2), though the exact number of genes regulated in vivo is unknown. Undoubtedly, miR-22 regulates the expression of at least some other genes, and the contribution of these additional effects to BP homeostasis remains unexplored.
Base pair hybridization between miR-22 and the SHR or WKY Chga 3′-UTR is imperfect (i.e. it contains two mismatches in canonical Watson–Crick pairing; Supplementary Material, Figure S2), suggesting that the mechanism whereby miR-22 decreases Chga protein expression involves translational repression (perhaps by steric hindrance) rather than degradation of target mRNA (which would be expected only for perfect match microRNA:mRNA base pairing) (42). Our observation in the brainstem of increased Chga protein but unchanged Chga mRNA after miR-22 antagomir treatment is consistent with this mechanism of translational repression. Increased Chga protein abundance in the brainstem after miR-22 antagomir administration is consistent with inhibition of miR-22 in this tissue; however, additional studies could reveal more extensive patterns of tissue and cell type-specific uptake of miR-22 antagomir in the SHR after IP administration. It has previously been reported that a microRNA antagomir administered intravenously through the tail vein in the mouse penetrated the brain to exert its regulatory effects (43). It was also conceivable that increased Chga protein in the brainstem resulted, in part, from feedback of a primary effect of antagomir in the periphery (adrenal gland); however, reduced Chga adrenal protein after antagomir treatment seems incompatible with this idea.
A parsimonious working model (Fig. 7) of a mechanistic pathway for miR-22 antagomir-mediated BP reduction proposes a primary effect in the brainstem to increase Chga protein (without effect on brainstem Chga mRNA abundance), which would decrease sympathetic outflow to the periphery (and therefore lead to reduced adrenal Chga protein and catecholamine levels), and, ultimately, lower BP. Indeed, we observed significantly reduced adrenal mRNA abundance across the catecholamine biosynthetic pathway: Creb1, Th, Dbh and Pnmt expression decreased after the antagomir. The action of miR-22 is unlikely to be entirely specific for Chga, and the role of additional miR-22 targets [such as estrogen receptor alpha (44) or phosphatase and tensin homolog (45)] in reducing BP in vivo may prompt further investigation.
The miR-22 motif in the Chga mRNA 3′-UTR is broadly conserved across species: mouse, rat and human (Fig. 6D), suggesting that inhibition of miR-22 might constitute a viable therapeutic target even in human hypertension. Genetic variation at the human miR-22 locus has been associated with panic disorder and miR-22 regulates several anxiety candidate genes (46), suggesting broader applications for miR-22 therapy acting through reduction in sympathetic activity. Lack of adverse effects of miR-22 antagomir on general metabolism in the SHR (as indexed by the chemistry panel) is an initial indicator of the safety of miR-22 antagomir therapy (Table 5); however, additional investigations into potential oncogenic functions of miR-22 (47,48) may be indicated.
CONCLUSIONS AND PERSPECTIVES
Chromogranin A has been implicated in both the heritability (6) and pathogenesis of human essential hypertension (9), leading us to probe the Chga locus in the SHR, the most widely studied animal model of genetic hypertension. Human CHGA 3′-UTR and promoter polymorphisms that showed association with essential hypertension (8–10) were not directly homologous to promoter and 3′-UTR polymorphisms located in the SHR Chga locus. Nonetheless, existence of functional Chga polymorphisms in both human essential hypertension and the SHR model argues for the pertinence of Chga in the SHR for diagnostic or pathophysiological insights in the human disease. Additional experiments, such as generation of congenic strains wherein the hypertensive and normotensive Chga loci are interchanged across strains, might further buttress a genetic role for Chga in the pathogenesis of hypertension in the SHR. As novel genetic associations are established with hypertension, identification of precise functional variants and elucidation of their molecular mechanisms will become even more important in understanding disease pathophysiology. Substantial reduction in BP in the SHR after miR-22 antagomir highlights a novel potential avenue for treatment of hypertension.
MATERIALS AND METHODS
Rat strains
Animal studies were performed with age-matched, adult (12–17 weeks) male SHR and WKY rat strains from Charles River Laboratories (Wilmington, MA. USA). Rats were studied according to a protocol approved by the Animal Subjects Committee of the University of California at San Diego, and research was conducted in accordance with institutional guidelines.
Transcripts by quantitative real-time polymerase chain reaction (qRT–PCR)
Isoflurane was used for terminal anesthesia of SHR and WKY rats. Adrenal glands, brainstem and whole blood (centrifuged to collect plasma) were isolated from each rat, immediately frozen in liquid nitrogen, then stored at −80°C. Total RNA extraction and quantitative RT–PCR experiments were performed by the Animal Clinical Chemistry and Gene Expression Laboratory at the University of North Carolina (Chapel Hill, NC, USA). Total RNA was extracted using the Applied Biosystems (ABI) 6700 automated nucleic acid workstation and quantitative RT–PCR was performed on mRNA → cDNA with the ABI-7700, as previously described (49). Cycle threshold (Ct) values were used to calculate the amount of amplified PCR product. Messenger RNAs were normalized to beta-actin mRNA, while microRNAs were normalized to either beta-actin mRNA or to snoRNA-U61 (SNORD61) small nucleolar RNA. Data were analyzed with Student's t-test.
Chromogranin A protein assay
Frozen adrenal glands and brainstem were homogenized in 10 mm TRIS (pH = 7.0) buffer (at 4°C) and then centrifuged at 13 000 G (at 4°C) for 1 min to clear debris. The supernatant was retained for immunoassays. Measurement of chromogranin A protein in the adrenal gland, brainstem and plasma was performed with a rat enzyme immunoassay kit (epitope: Arg-Ser-Met-Arg-Leu-Ser-Phe-Arg-Ala-Arg-Gly-Tyr-Gly-Phe-Arg-Asp-Pro-Gly-Leu-Gln-Leu; Cat. # EK-053-29, Phoenix Pharmaceuticals, Inc., Burlingame, CA, USA). Tissue protein concentration was determined using the Bio-Rad Protein Assay (coomassie blue dye absorbance shift; based on the Bradford method) (Bio-Rad, Hercules, CA, USA). In some experiments, we measured adrenal Chga by radiommunoassay based on the epitope CHGA324–337 (or ‘WE-14’) (50). Data were analyzed with Student's t-test.
Systematic polymorphism discovery by re-sequencing rat chromogranin A
The nucleotide sequences of the chromogranin A locus (Chga) in the SHR, WKY or BN rat strains were determined by capillary (Sanger dideoxy) re-sequencing with an Applied Biosystems ABI-3100 Genetic Analyzer, as previously described (51), and directly aligned with the Brown Norway (BN) reference sequence at NCBI. Primers are listed in Supplementary Material, Table S1. Polymorphisms were identified and visually confirmed in the raw sequencing data using the EditView 1.0.1 software for Macintosh OS-9 (Applied Biosystems).
Computational prediction of transcription factor and microRNA binding
Predictions for transcription factors that bind the Chga promoter regions harboring polymorphisms were made with ConSite (http:/www.phylofoot.org/consite) (52), a tool for identification of cis-regulatory elements in genomic sequences. Predictions for microRNAs that hybridize the Chga 3′-UTR region containing polymorphisms were performed with MicroInspector (http://bioinfo.uni-plovdiv.bg/microinspector) (53), a tool for detection of microRNA binding sites within RNA sequences. TargetScan (http://www.targetscan.org) (54) was used to predict potential target transcripts for miR-22.
Construction of promoter and 3′-UTR luciferase reporter plasmids
The following PCR primers were used to amplify a ∼1.8 kb Chga proximal promoter fragment from SHR and WKY genomic DNA: 5′-TAAAGGTACCAGCACACCTAAACTGTGAC-3′ (forward primer; KpnI site underlined) and 5′-GAGCAAGCTTGTGCGGAAAGAGAG-3′ (reverse primer; HindIII site underlined). The promoter fragment was inserted between KpnI and HindIII sites in the pGL3-Basic vector (Promega), which lacks eukaryotic promoter and enhancer sequences, and contains the cDNA for firefly luciferase. The following PCR primers were used to amplify the 333 bp Chga 3′-UTR from SHR and WKY genomic DNA: 5′-ACGGGCTAGCGGCACTGGCTGGTGGGGTCCGGCCA-3′ (forward primer; NheI site underlined) and 5′-AAAGGCTAGCGAAGAGCCCAAAGCAGGTTTATTCT-3′ (reverse primer; NheI site underlined). The 3′-UTR was inserted into the XbaI site of the pGL3-Promoter vector (Promega), just downstream (3′) of the cDNA for firefly luciferase driven by the SV40 promoter. Correct insertion and orientation of Chga promoter and 3′-UTR was confirmed by DNA sequencing. Plasmid DNA for transfection was prepared and purified with the QIAfilter Plasmid Midi Kit (Qiagen).
Chga promoter and 3′-UTR luciferase reporter transfection and activity assays
The rat pheochromocytoma cell line PC12, and the human neuroblastoma cell line SH-SY5Y, were grown and transfected with TransFectin (Bio-Rad) (for SH-SY5Y experiments), Superfect (Qiagen) (for PC12 promoter–reporter experiments) or DharmaFECT Duo (Dharmacon) (for PC12 3′-UTR-reporter experiments) using established protocols.
Promoter-luciferase reporter experiments were performed with co-transfection of Chga promoter-luciferase reporter plasmid DNA and mammalian cDNA expression plasmid DNA (pCMV-promoter; into SH-SY5Y cells: 24-well plates, 250 ng reporter plasmid DNA; into PC12 cells: 12-well plates, 1 μg reporter DNA, 50 ng expression plasmid DNA) for the following transcription factors: pcDNA3.1(-) (empty vector, negative control; Invitrogen, Carlsbad, CA, USA), E2f1 (Mus musculus; MMM1013-9201426, Open Biosystems, Huntsville, AL, USA), Foxd3 (Xenopus tropicalis; MXT1765-98077348, Open Biosystems), Foxi1 (Mus musculus; MMM1013-64167, Open Biosystems), HAND1 (Homo sapiens; SC122690, OriGene, Rockville, MD, USA), Irf1 (Rattus norvegicus; MRN1768-9510505, Open Biosystems), MYCN (Homo sapiens; MHS1010-57504, Open Biosystems), PBX1 (Homo sapiens; MHS4768-99609490, Open Biosystems), Creb1 (Mus musculus; MMM1013-98477947, Open Biosystems), Sox17 (Danio rerio; MDR1734-97029554, Open Biosystems) and Brachyury (Mus musculus; MMM1013-99829478, Open Biosystems). Cells co-transfected with transcription factor plasmids were harvested and lysed 20–24 h after transfection for the reporter assay.
3′-UTR-luciferase reporter experiments were performed with co-transfected Chga 3′-UTR-luciferase reporter plasmid DNA (PC12 cells: 24-well plates; 250 ng reporter plasmid DNA), and synthetic microRNA precursor (PC12 cells: 30 nm) for either rno-miR-22 (PM10203, Ambion), rno-miR-320 (PM11621, Ambion) or negative control (CN-001000-01-05, Dharmacon). Cells co-transfected with microRNAs were incubated for 24–48 h before harvest for reporter activity.
After transfection and subsequent incubation, cells were treated with lysis buffer (300 μl per well in 12-well plates; 125 μl per well in 24-well plates) [0.1 m phosphate buffer (K2HPO4 + KH2PO4) (pH 7.8), 1 mm DTT and 0.1% Triton-X 100]. The bioluminescent activity of luciferase in 50 μl of cell lysate was determined using the AutoLumat LB 953 luminometer (EG&G Berthold, Nashua, NH, USA) to measure light emission (incubation time = 0 s, measure time = 10 s, temperature = 25°C) after the addition of assay buffer [100 μl per sample; 100 mm Tris–acetate (pH 7.8), 10 mm Mg-acetate, 1 mm EDTA (pH 8.0), 3 mm ATP and 100 μM luciferin (Sigma-Aldrich)]. As an internal control for varying cell number in each well, the total protein content was measured in cell lysate using the Bio-Rad Protein Assay (coomassie blue dye absorbance shift; based on the Bradford method) (Bio-Rad, Hercules, CA, USA). Luciferase activity was expressed as the normalized ratio of (relative light unit)/(total protein) or (RLU/μg protein). Data were analyzed with analysis-of-variance (ANOVA), followed by pair wise t-tests corrected for multiple comparisons (Bonferroni). Significance of genotype–transcription factor interaction and genotype–microRNA interaction was assessed with two-way ANOVA (dependent variable = luciferase activity; two independent variables = genotype and transcription factor/microRNA).
Chga cDNA/EAP chimera reporter plasmid construction and EAP secretion and activity assay
The following PCR primers were used to amplify the 1389 bp rat Chga cDNA from a Norway rat (Rattus norvegicus) cDNA clone (NCBI GenBank accession: NM_021655):
5′-ATGCCTCGAGGCCACCATGCGCTCCTCCGCGGCTTTGG-3′ (forward primer; XhoI site underlined; Kozak sequence in bold; translation start site underlined) and 5′-ATGCGGTACCCTCCCCGTCGCTAAGCCTGCAG-3′ (reverse primer; KpnI site underlined). The amplified Chga cDNA was inserted between XhoI and KpnI sites in an EAP chemiluminescent reporter vector, pEAP-N2, which contains the CMV promoter and a truncated domain (EAP) of full-length human secreted embryonic alkaline phosphatase (55). The BN rat Chga cDNA sequence was identical to the WKY Chga cDNA sequence. The SHR Chga cDNA-EAP reporter was constructed by PCR amplifying the WKY cDNA-EAP construct with mutagenesis primers (designed to delete eight glutamine residues or 24 bp) [5′-AAAGAGGAAGAGGAGGAGGAGAAAGAGGAGAAGGCGATCGCCAGAGAGAAGGCT-3′ (forward primer) and 5′-CTCCTCCTCTTCCTCTTTCTCCTCTTGCTTGGCTTTTCTGGCTTGCTGCTGGGCACTGGGACC-3′, reverse primer)], treating the PCR product with DpnI to digest WKY template DNA, and then ligating the linear SHR PCR product with the Rapid DNA Ligation Kit (Roche, Indianapolis, IN, USA). Sequence of the SHR and WKY Chga cDNA was confirmed by DNA sequencing (ABI 3100). Plasmid DNA was prepared and purified for transfection with the QIAfilter Plasmid Midi Kit (Qiagen).
Rat PC12 pheochromocytoma cells (from ATCC) [grown in F-12K (Invitrogen) with 2.5% heat-inactivated fetal bovine serum (Gemini Bioproducts, Woodland, CA, USA), 15% heat-inactivated horse serum (Gemini Bioproducts), penicillin (100 U/ml), streptomycin (100 μg/ml) and l-glutamine (0.292 mg/ml)] were transfected (at ∼50% confluence, 1 day after splitting 1:3) with Chga cDNA/EAP reporter plasmid DNA [SHR or WKY; 2.5 μg supercoiled DNA per well; 6-well polystyrene plates (coated with poly-l-lysine (Sigma) and rat collagen (Upstate)), 3.48-cm diameter wells, Corning Inc., Corning, NY] using the GenePorter2 Transfection Reagent (cationic lipid, Genlantis, San Diego, CA). After transfection, cells were incubated for 30 h to allow sufficient time for the Chga-EAP chimera to be transcribed, translated, and shuttled through the Golgi into regulated secretory granules.
Two hours before performing the EAP secretion activity assays, the cells were incubated with tritiated norepinephrine (3H-NE; 0.5 μCi per 1 ml cell media). The assay was performed by stimulating the cells with 700 μl of calcium secretion buffer (a negative control; 150 mm NaCl, 5 mm KCl, 2 mm CaCl2, 10 mm HEPES pH 7.4) or barium secretion buffer (a potent exocytotic secretory stimulus; 150 mm NaCl, 5 mm KCl, 2 mm BaCl2, 10 mm HEPES pH 7.4), and collecting cellular supernatant and lysate fractions after a 15-min secretory incubation. Tritiated norepinephrine (read for 2 min) and chemiluminescent EAP activity (read for 10 s) were measured in both cell supernatant and cell lysate fractions (Phospha-Light System, Applied Biosystems). Final EAP activity is expressed as the normalized ratio of (EAP activity)/(3H-NE counts per minute) or (RLU/cpm), and was used to compute ‘relative secretion’ and ‘sorting index.’ We previously described EAP activity assays in additional detail (56).
Rs is defined as the fraction of total EAP activity that is found in the supernatant: Rs = (EAP activity in supernatant)/(EAP activity in supernatant + EAP activity in lysate). SI is defined as the difference between Rs of barium- and calcium-treated cells normalized by the Rs of calcium treated cells: SI = (Rsbarium – Rscalcium)/Rscalcium.
Chga sequence conservation and coiled-coil prediction
Conservation of the Chga locus sequence across rat, mouse, dog, rhesus and human was determined with the VISTA browser (http://pipeline.lbl.gov/cgi-bin/gateway2) (57). Conservation of specific Chga protein domains across mammals (Homo sapiens, Macaca mulatta, Bos taurus, Canis familiaris, Equus caballus, Mus musculus and Rattus norvegicus) was determined by ClustalW alignment of the protein sequence downloaded from the University of California, Santa Cruz, genome browser (http://genome.ucsc.edu) (58).
The probability of SHR and WKY Chga protein to assume a 3D coiled-coil conformation was computed with the COILS algorithm (http://www.ch.embnet.org/software/COILS_form.html) (59). In brief, the algorithm searches for strongly amphipathic regions that display a pattern of alternating hydrophilic and hydrophobic residues repeated every seven residues (heptad repeat residues a →g, with a and d being hydrophobic). The minimum window was two such heptads, over 14 residues. The probability of coiled-coil structure is presented as a function of amino acid position in the mature Chga protein.
Inhibition of microRNA-22 by synthetic antagomir in vivo
Male SHR age 10–11 weeks, weight 210–240 g; from Charles River) were administered either a negative control (n = 9) [LNA (‘locked’ nucleic acid) oligonucleotide scrambled sequence with phosphorothioate backbone: 5′-ACGTCTATACGCCCA-3′; Exiqon, Woburn, MA, USA] or rno-miR-22 antagomir (n = 10) (LNA oligonucleotide with phosphorothioate backbone: 5′-CTTCAACTGGCAGCT-3′; Exiqon). Oligonucleotides were delivered via i.p. injection (every other day for a total of four injections; Days 1, 3, 5 and 7) at a dose of 25 mg/kg body weight. Blood pressure measurement preceded antagomir administration by 1–2 h each day to allow a recovery period. We used the CODA high-throughput rodent tail-cuff system (Kent Scientific; Torrington, CT, USA) to measure BP 1 time per day (average of 20 readings in 1 session) on Days 1, 3, 5, 7 and 9. Pentobarbital sodium (‘Fatal-Plus’) was used for terminal anesthesia of rats after four doses (9 days) of miR-22 antagomir or negative control. Adrenal glands, brainstem and whole blood (which was centrifuged to collect plasma) were isolated from each rat, immediately frozen in liquid nitrogen, then stored at −80°C. Quantitative real-time polymerase chain reaction (qRT–PCR) and chromogranin A protein immunoassay (Cat. # EK-053-29, Phoenix Pharmaceuticals, Inc.) were performed as described above. A linear mixed effects model for fixed effects (implemented in SPSS-17 for Mac OS-X) was used to test for the effect of treatment (negative control versus miR-22 antagomir) on SBP, DBP and HR. Data are presented as mean ± SEM.
Blood chemistry panel during miR antagomir treatment
We performed a chemistry panel on the plasma from SHR treated with negative control or miR-22 antagomir. We assayed glucose, BUN, creatinine, sodium, phosphorus, alkaline phosphatase, albumin, serum glutamic pyruvic transaminase (SGPT), total protein, globulin, total bilirubin and amylase using the VetScan VS2 comprehensive diagnostic profile (#500–1038; Abaxis, Union City, CA, USA). Data were analyzed with Student's t-test.
Linkage and correlation analysis of Chga in rat RI strains
Linkage analysis was performed in the HXB/BXH RI set of rat strains derived from SHR and BN progenitor strains (60), as previously described (61). Experiments were performed in 29 RI strains (HXB and BXH at >F60) in accordance with the Animal Protection Law of the Czech Republic (311/1997) and were approved by the Ethics Committee of the Institute of Physiology, Czech Academy of Sciences, Prague. Animals were male and age matched at 6 weeks.
Adrenal glands were harvested, immediately frozen and stored at −80°C. One adrenal gland per animal was used for Chga protein quantification, whereas the other was used for mRNA extraction, as previously described (61). Adrenal glands for the Chga protein assay were homogenized in 1.9 ml of 10 mm MES buffer (pH = 6.0) with a Tissuemizer (Tekmar, Cincinnati, OH, USA), and then spun at 13 000g for 1 min to clear debris. Supernatants were saved, divided into aliquots and placed immediately on dry-ice to freeze. Chga protein was quantified in RI strain adrenal glands with a radioimmunoassay that recognized rat Chga. Radioimmunoassay results were normalized to wet tissue weight or milligram protein.
We used a SNP genotype-based linkage map (62), and the QTL Cartographer (63) and Map Manager QTX (64) software packages to detect the presence of QTLs for adrenal Chga protein across the rat genome. Permutation analyses (65) were used to compute the probabilistic significance of linkage and to correct P-values for genome-wide multiple testing. The two-LOD support interval method (66) and the bootstrap test (67) were implemented to estimate the 95% confidence interval for the size of chromosomal segments containing QTLs, as previously described (61). The LOD score significance was estimated as described by Nyholt (23), with the calculator available at <http://genepi.qimr.edu.au/general/daleN/>.
We searched the online data repository at <www.genenetwork.org> (68) (where our RI strain adrenal biochemical phenotypes are deposited) for evidence of meiotic linkage for Chga protein (adrenal) in the HXB/BXH RI strains. We also computed Pearson correlations between adrenal Chga protein and biochemical hypertensive disease traits using the statistical tools available at <www.genenetwork.org>.
SUPPLEMENTARY MATERIAL
FUNDING
Funding was provided by the National Center for Advancing Translational Science (NCATS), UL1-TR000100; Comprehensive Research Center of Excellence in Minority Health and Health Disparities (CRCHD), MD00020; NIH/NIDDK DK007671 Nephrology Training Grant; Department of Veterans Affairs (S.K.M., D.T.O.); National Institutes of Health (DA011311 to S.K.M.; DK094894 and DK60702 to D.T.O.; HL10881 to G.W.S.-S.; HL58120 to S.K.M. & D.T.O.; HL108629 to S.V.) and grants P301/10/0290 and P301/12/0696 from the Grant Agency of the Czech Republic to M.P.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health, and the Department of Veterans Affairs. Plasmid DNA sequencing was performed by the DNA Sequencing Shared Resource, UCSD Cancer Center, which is funded in part by NCI Cancer Center Support Grant # 2 P30 CA023100-23.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Taupenot L., Harper K.L., O'Connor D.T. The chromogranin-secretogranin family. N. Engl. J. Med. 2003;348:1134–1149. doi: 10.1056/NEJMra021405. [DOI] [PubMed] [Google Scholar]
- 2.Mahapatra N.R. Catestatin is a novel endogenous peptide that regulates cardiac function and blood pressure. Cardiovasc. Res. 2008;80:330–338. doi: 10.1093/cvr/cvn155. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Margalet V., Gonzalez-Yanes C., Najib S., Santos-Alvarez J. Metabolic effects and mechanism of action of the chromogranin A-derived peptide pancreastatin. Regul. Pept. 2010;161:8–14. doi: 10.1016/j.regpep.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Helle K.B. Chromogranins A and B and secretogranin II as prohormones for regulatory peptides from the diffuse neuroendocrine system. Results Probl. Cell Differ. 2010;50:21–44. doi: 10.1007/400_2009_26. [DOI] [PubMed] [Google Scholar]
- 5.O'Connor D.T. Chromogranin A: implications for hypertension. J. Hypertens. Suppl. 1984;2:S147–S150. [PubMed] [Google Scholar]
- 6.Takiyyuddin M.A., Parmer R.J., Kailasam M.T., Cervenka J.H., Kennedy B., Ziegler M.G., Lin M.C., Li J., Grim C.E., Wright F.A., et al. Chromogranin A in human hypertension. Influence of heredity. Hypertension. 1995;26:213–220. doi: 10.1161/01.hyp.26.1.213. [DOI] [PubMed] [Google Scholar]
- 7.O'Connor D.T. Plasma chromogranin A. Initial studies in human hypertension. Hypertension. 1985;7:I76–I79. doi: 10.1161/01.hyp.7.3_pt_2.i76. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Rao F., Rodriguez-Flores J.L., Mahapatra N.R., Mahata M., Wen G., Salem R.M., Shih P.A., Das M., Schork N.J., et al. Common genetic variants in the chromogranin A promoter alter autonomic activity and blood pressure. Kidney Int. 2008;74:115–125. doi: 10.1038/ki.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahu B.S., Sonawane P.J., Mahapatra N.R. Chromogranin A: a novel susceptibility gene for essential hypertension. Cell Mol. Life Sci. 2010;67:861–874. doi: 10.1007/s00018-009-0208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Rao F., Rodriguez-Flores J.L., Mahata M., Fung M.M., Stridsberg M., Vaingankar S.M., Wen G., Salem R.M., Das M., et al. Naturally occurring human genetic variation in the 3′-untranslated region of the secretory protein chromogranin A is associated with autonomic blood pressure regulation and hypertension in a sex-dependent fashion. J. Am. Coll. Cardiol. 2008;52:1468–1481. doi: 10.1016/j.jacc.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao F., Wen G., Gayen J.R., Das M., Vaingankar S.M., Rana B.K., Mahata M., Kennedy B.P., Salem R.M., Stridsberg M., et al. Catecholamine release-inhibitory peptide catestatin (chromogranin A(352–372)): naturally occurring amino acid variant Gly364Ser causes profound changes in human autonomic activity and alters risk for hypertension. Circulation. 2007;115:2271–2281. doi: 10.1161/CIRCULATIONAHA.106.628859. [DOI] [PubMed] [Google Scholar]
- 12.Salem R.M., Cadman P.E., Chen Y., Rao F., Wen G., Hamilton B.A., Rana B.K., Smith D.W., Stridsberg M., Ward H.J., et al. Chromogranin A polymorphisms are associated with hypertensive renal disease. J. Am. Soc. Nephrol. 2008;19:600–614. doi: 10.1681/ASN.2007070754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahata S.K., Mahata M., Wen G., Wong W.B., Mahapatra N.R., Hamilton B.A., O'Connor D.T. The catecholamine release-inhibitory “catestatin” fragment of chromogranin a: naturally occurring human variants with different potencies for multiple chromaffin cell nicotinic cholinergic responses. Mol. Pharmacol. 2004;66:1180–1191. doi: 10.1124/mol.104.002139. [DOI] [PubMed] [Google Scholar]
- 14.O'Connor D.T., Cadman P.E., Smiley C., Salem R.M., Rao F., Smith J., Funk S.D., Mahata S.K., Mahata M., Wen G., et al. Pancreastatin: Multiple actions on human intermediary metabolism in vivo, variation in disease, and naturally occurring functional genetic polymorphism. J. Clin. Endocrinol. Metab. 2005;90:5414–5425. doi: 10.1210/jc.2005-0408. [DOI] [PubMed] [Google Scholar]
- 15.Zhang K., Rao F., Wen G., Salem R.M., Vaingankar S., Mahata M., Mahapatra N.R., Lillie E.O., Cadman P.E., Friese R.S., et al. Catecholamine storage vesicles and the metabolic syndrome: The role of the chromogranin A fragment pancreastatin. Diabetes Obes. Metab. 2006;8:621–633. doi: 10.1111/j.1463-1326.2006.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahapatra N.R., O'Connor D.T., Vaingankar S.M., Hikim A.P., Mahata M., Ray S., Staite E., Wu H., Gu Y., Dalton N., et al. Hypertension from targeted ablation of chromogranin A can be rescued by the human ortholog. J. Clin. Invest. 2005;115:1942–1952. doi: 10.1172/JCI24354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaingankar S.M., Li Y., Biswas N., Gayen J., Choksi S., Rao F., Ziegler M.G., Mahata S.K., O'Connor D.T. Effects of chromogranin A deficiency and excess in vivo: biphasic blood pressure and catecholamine responses. J. Hypertens. 2010;28:817–825. doi: 10.1097/HJH.0b013e328336ed3e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connor D.T., Takiyyuddin M.A., Printz M.P., Dinh T.Q., Barbosa J.A., Rozansky D.J., Mahata S.K., Wu H., Kennedy B.P., Ziegler M.G., et al. Catecholamine storage vesicle protein expression in genetic hypertension. Blood Press. 1999;8:285–295. doi: 10.1080/080370599439508. [DOI] [PubMed] [Google Scholar]
- 19.Schober M., Howe P.R., Sperk G., Fischer-Colbrie R., Winkler H. An increased pool of secretory hormones and peptides in adrenal medulla of stroke-prone spontaneously hypertensive rats. Hypertension. 1989;13:469–474. doi: 10.1161/01.hyp.13.5.469. [DOI] [PubMed] [Google Scholar]
- 20.Somogyi P., Hodgson A.J., DePotter R.W., Fischer-Colbrie R., Schober M., Winkler H., Chubb I.W. Chromogranin immunoreactivity in the central nervous system. Immunochemical characterisation, distribution and relationship to catecholamine and enkephalin pathways. Brain Res. 1984;320:193–230. doi: 10.1016/0165-0173(84)90007-9. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor D.T., Frigon R.P. Chromogranin A, the major catecholamine storage vesicle soluble protein. Multiple size forms, subcellular storage, and regional distribution in chromaffin and nervous tissue elucidated by radioimmunoassay. J. Biol. Chem. 1984;259:3237–3247. [PubMed] [Google Scholar]
- 22.Mahata S.K., Kozak C.A., Szpirer J., Szpirer C., Modi W.S., Gerdes H.H., Huttner W.B., O'Connor D.T. Dispersion of chromogranin/secretogranin secretory protein family loci in mammalian genomes. Genomics. 1996;33:135–139. doi: 10.1006/geno.1996.0171. [DOI] [PubMed] [Google Scholar]
- 23.Nyholt D.R. All LODs are not created equal. Am. J. Hum. Genet. 2000;67:282–288. doi: 10.1086/303029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mosley C.A., Taupenot L., Biswas N., Taulane J.P., Olson N.H., Vaingankar S.M., Wen G., Schork N.J., Ziegler M.G., Mahata S.K., et al. Biogenesis of the secretory granule: chromogranin A coiled-coil structure results in unusual physical properties and suggests a mechanism for granule core condensation. Biochemistry. 2007;46:10999–11012. doi: 10.1021/bi700704r. [DOI] [PubMed] [Google Scholar]
- 25.Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Lim J., Yang C., Hong S.J., Kim K.S. Regulation of tyrosine hydroxylase gene transcription by the cAMP-signaling pathway: involvement of multiple transcription factors. Mol. Cell Biochem. 2000;212:51–60. [PubMed] [Google Scholar]
- 27.Lewis-Tuffin L.J., Quinn P.G., Chikaraishi D.M. Tyrosine hydroxylase transcription depends primarily on cAMP response element activity, regardless of the type of inducing stimulus. Mol. Cell Neurosci. 2004;25:536–547. doi: 10.1016/j.mcn.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Fujino K. Brain catecholamines in spontaneously hypertensive and DOCA-salt hypertensive rats. Acta. Med. Okayama. 1984;38:325–340. doi: 10.18926/AMO/30309. [DOI] [PubMed] [Google Scholar]
- 29.Takami T., Ito H., Suzuki T. Decreased norepinephrine content in the medulla oblongata in severely hypertensive rats. Clin. Exp. Pharmacol. Physiol. 1993;20:161–167. doi: 10.1111/j.1440-1681.1993.tb01664.x. [DOI] [PubMed] [Google Scholar]
- 30.Nakada T., Koike H., Katayama T., Watanabe H., Yamori Y. Increased adrenal epinephrine and norepinephrine in spontaneously hypertensive rats treated with hyperbaric oxygen. Hinyokika Kiyo. 1984;30:1357–1366. [PubMed] [Google Scholar]
- 31.Davies D.S., Reid J.L. British Pharmacological Society. Central action of drugs in blood pressure regulation : proceedings of an International Symposium on Central Actions of Drugs in the Regulation of Blood Pressure. Pitman Medical, Tunbridge Wells [Eng.]; 1975. [Google Scholar]
- 32.Reid J.L., Rubin P.C., Howden C.W. Central α2-adrenoceptors and blood pressure regulation in man: Studies with guanfacine (BS 100–141) and azepexole (BHT 933) Br. J. Clin. Pharmacol. 1983;15(Suppl. 4):463S–469S. [Google Scholar]
- 33.Videen J.S., Mezger M.S., Chang Y.M., O'Connor D.T. Calcium and catecholamine interactions with adrenal chromogranins. Comparison of driving forces in binding and aggregation. J. Biol. Chem. 1992;267:3066–3073. [PubMed] [Google Scholar]
- 34.Gaede A.H., Pilowsky P.M. Catestatin in rat RVLM is sympathoexcitatory, increases barosensitivity, and attenuates chemosensitivity and the somatosympathetic reflex. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;299:R1538–R1545. doi: 10.1152/ajpregu.00335.2010. [DOI] [PubMed] [Google Scholar]
- 35.Stoll M., Cowley A.W., Jr, Tonellato P.J., Greene A.S., Kaldunski M.L., Roman R.J., Dumas P., Schork N.J., Wang Z., Jacob H.J. A genomic-systems biology map for cardiovascular function. Science. 2001;294:1723–1726. doi: 10.1126/science.1062117. [DOI] [PubMed] [Google Scholar]
- 36.Nagatsu T., Kato T., Numata Y., Keiko I., Umezawa H. Serum dopamine beta-hydroxylase activity in developing hypertensive rats. Nature. 1974;251:630–631. doi: 10.1038/251630a0. [DOI] [PubMed] [Google Scholar]
- 37.Grobecker G., Roizen M.F., Weise V., Saavedra J.M., Kopin I.J. Letter: Sympathoadrenal medullary activity in young, spontaneously hypertensive rats. Nature. 1975;258:267–268. doi: 10.1038/258267a0. [DOI] [PubMed] [Google Scholar]
- 38.Nagaoka A., Lovenberg W. Plasma norepinephrine and dopamine-beta-hydroxylase in genetic hypertensive rats. Life Sci. 1976;19:29–34. doi: 10.1016/0024-3205(76)90370-2. [DOI] [PubMed] [Google Scholar]
- 39.Wang L., Rao F., Zhang K., Mahata M., Rodriguez-Flores J.L., Fung M.M., Waalen J., Cockburn M.G., Hamilton B.A., Mahata S.K., et al. Neuropeptide Y(1) Receptor NPY1R discovery of naturally occurring human genetic variants governing gene expression in cella as well as pleiotropic effects on autonomic activity and blood pressure in vivo. J. Am. Coll. Cardiol. 2009;54:944–954. doi: 10.1016/j.jacc.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y., Wen G., Rao F., Zhang K., Wang L., Rodriguez-Flores J.L., Sanchez A.P., Mahata M., Taupenot L., Sun P., et al. Human dopamine beta-hydroxylase (DBH) regulatory polymorphism that influences enzymatic activity, autonomic function, and blood pressure. J. Hypertens. 2010;28:76–86. doi: 10.1097/HJH.0b013e328332bc87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shih P.A., Wang L., Chiron S., Wen G., Nievergelt C., Mahata M., Khandrika S., Rao F., Fung M.M., Mahata S.K., et al. Peptide YY (PYY) gene polymorphisms in the 3′-untranslated and proximal promoter regions regulate cellular gene expression and PYY secretion and metabolic syndrome traits in vivo. J. Clin. Endocrinol. Metab. 2009;94:4557–4566. doi: 10.1210/jc.2009-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 43.Wang H., Chiu M., Xie Z., Liu Z., Chen P., Liu S., Byrd J.C., Muthusamy N., Garzon R., Croce C.M., et al. Synthetic microRNA cassette dosing: pharmacokinetics, tissue distribution and bioactivity. Mol. Pharm. 2012;9:1638–1644. doi: 10.1021/mp2006483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandey D.P., Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol. Cell Biol. 2009;29:3783–3790. doi: 10.1128/MCB.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bar N., Dikstein R. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS ONE. 2010;5:e10859. doi: 10.1371/journal.pone.0010859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muinos-Gimeno M., Espinosa-Parrilla Y., Guidi M., Kagerbauer B., Sipila T., Maron E., Pettai K., Kananen L., Navines R., Martin-Santos R., et al. Human microRNAs miR-22, miR-138–2, miR-148a, and miR-488 Are Associated with Panic Disorder and Regulate Several Anxiety Candidate Genes and Related Pathways. Biol. Psychiatry. 2011;69:526–533. doi: 10.1016/j.biopsych.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Zhang J., Yang Y., Yang T., Liu Y., Li A., Fu S., Wu M., Pan Z., Zhou W. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br. J. Cancer. 2010;103:1215–1220. doi: 10.1038/sj.bjc.6605895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong J., Yu D., Wei N., Fu H., Cai T., Huang Y., Wu C., Zheng X., Du Q., Lin D., et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277:1684–1694. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- 49.Kim H.S., Lee G., John S.W., Maeda N., Smithies O. Molecular phenotyping for analyzing subtle genetic effects in mice: application to an angiotensinogen gene titration. Proc. Natl. Acad. Sci. U S A. 2002;99:4602–4607. doi: 10.1073/pnas.072083799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stridsberg M., Eriksson B., Oberg K., Janson E.T. A panel of 11 region-specific radioimmunoassays for measurements of human chromogranin A. Regul. Pept. 2004;117:219–227. doi: 10.1016/j.regpep.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 51.Friese R.S., Schmid-Schonbein G.W., O'Connor D.T. Systematic polymorphism discovery after genome-wide identification of potential susceptibility loci in a hereditary rodent model of human hypertension. Blood Press. 2011;20:222–231. doi: 10.3109/08037051.2011.566012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandelin A., Wasserman W.W., Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rusinov V., Baev V., Minkov I.N., Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taupenot L., Harper K.L., O'Connor D.T. Role of H+-ATPase-mediated acidification in sorting and release of the regulated secretory protein chromogranin A: evidence for a vesiculogenic function. J. Biol. Chem. 2005;280:3885–3897. doi: 10.1074/jbc.M408197200. [DOI] [PubMed] [Google Scholar]
- 56.Courel M., Rodemer C., Nguyen S.T., Pance A., Jackson A.P., O'Connor D.T., Taupenot L. Secretory granule biogenesis in sympathoadrenal cells: identification of a granulogenic determinant in the secretory prohormone chromogranin A. J. Biol. Chem. 2006;281:38038–38051. doi: 10.1074/jbc.M604037200. [DOI] [PubMed] [Google Scholar]
- 57.Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 60.Pravenec M., Klir P., Kren V., Zicha J., Kunes J. An analysis of spontaneous hypertension in spontaneously hypertensive rats by means of new recombinant inbred strains. J. Hypertens. 1989;7:217–221. [PubMed] [Google Scholar]
- 61.Jirout M.L., Friese R.S., Mahapatra N.R., Mahata M., Taupenot L., Mahata S.K., Kren V., Zidek V., Fischer J., Maatz H., et al. Genetic regulation of catecholamine synthesis, storage and secretion in the spontaneously hypertensive rat. Hum. Mol. Genet. 2010;19:2567–2580. doi: 10.1093/hmg/ddq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saar K., Beck A., Bihoreau M.T., Birney E., Brocklebank D., Chen Y., Cuppen E., Demonchy S., Dopazo J., Flicek P., et al. SNP and haplotype mapping for genetic analysis in the rat. Nat. Genet. 2008;40:560–566. doi: 10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Basten C.J., Weir B.S., Zeng Z.B. QTL Cartographer: A Reference Manual and Tutorial for QTL Mapping. Raleigh, NC: Department of Statistics, North Carolina State University; 2002. [Google Scholar]
- 64.Manly K.F., Cudmore R.H., Jr, Meer J.M. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- 65.Doerge R.W., Churchill G.A. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Ooijen J.W. Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- 67.Visscher P.M., Thompson R., Haley C.S. Confidence intervals in QTL mapping by bootstrapping. Genetics. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J., Williams R.W., Manly K.F. WebQTL: web-based complex trait analysis. Neuroinformatics. 2003;1:299–308. doi: 10.1385/NI:1:4:299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.