Abstract
Several infrequent genetic polymorphisms in the SERPINA1 gene are known to substantially reduce concentration of alpha1-antitrypsin (AAT) in the blood. Since low AAT serum levels fail to protect pulmonary tissue from enzymatic degradation, these polymorphisms also increase the risk for early onset chronic obstructive pulmonary disease (COPD). The role of more common SERPINA1 single nucleotide polymorphisms (SNPs) in respiratory health remains poorly understood.
We present here an agnostic investigation of genetic determinants of circulating AAT levels in a general population sample by performing a genome-wide association study (GWAS) in 1392 individuals of the SAPALDIA cohort.
Five common SNPs, defined by showing minor allele frequencies (MAFs) >5%, reached genome-wide significance, all located in the SERPINA gene cluster at 14q32.13. The top-ranking genotyped SNP rs4905179 was associated with an estimated effect of β = −0.068 g/L per minor allele (P = 1.20*10−12). But denser SERPINA1 locus genotyping in 5569 participants with subsequent stepwise conditional analysis, as well as exon-sequencing in a subsample (N = 410), suggested that AAT serum level is causally determined at this locus by rare (MAF<1%) and low-frequent (MAF 1–5%) variants only, in particular by the well-documented protein inhibitor S and Z (PI S, PI Z) variants. Replication of the association of rs4905179 with AAT serum levels in the Copenhagen City Heart Study (N = 8273) was successful (P<0.0001), as was the replication of its synthetic nature (the effect disappeared after adjusting for PI S and Z, P = 0.57). Extending the analysis to lung function revealed a more complex situation. Only in individuals with severely compromised pulmonary health (N = 397), associations of common SNPs at this locus with lung function were driven by rarer PI S or Z variants. Overall, our meta-analysis of lung function in ever-smokers does not support a functional role of common SNPs in the SERPINA gene cluster in the general population.
Author Summary
Low levels of alpha1-antitrypsin (AAT) in the blood are a well-established risk factor for accelerated loss in lung function and chronic obstructive pulmonary disease. While a few infrequent genetic polymorphisms are known to influence the serum levels of this enzyme, the role of common genetic variants has not been examined so far. The present genome-wide scan for associated variants in approximately 1400 Swiss inhabitants revealed a chromosomal locus containing the functionally established variants of AAT deficiency and variants previously associated with lung function and emphysema. We used dense genotyping of this genetic region in more than 5500 individuals and subsequent conditional analyses to unravel which of these associated variants contribute independently to the phenotype's variability. All associations of common variants could be attributed to the rarer functionally established variants, a result which was then replicated in an independent population-based Danish cohort. Hence, this locus represents a textbook example of how a large part of a trait's heritability can be hidden in infrequent genetic polymorphisms. The attempt to transfer these results to lung function furthermore suggests that effects of common variants in this genetic region in ever-smokers may also be explained by rarer variants, but only in individuals with hampered pulmonary health.
Introduction
Alpha1-antitrypsin (AAT) is a serum marker for inflammation produced in the liver. Its main function is to inhibit neutrophil elastase and consequently protect pulmonary tissue. The SERPINA1 gene encoding the AAT protein is known to be polymorphic in the general population. The best studied single nucleotide polymorphisms (SNPs) causing a reduction in AAT serum levels are the protease inhibitor S (PI S, rs17580) and the protease inhibitor Z (PI Z, rs28929474) variants [1]. The loss of function mechanism is especially well investigated for the PI Z variant. The resulting amino acid change in AAT leads to the protein's intracellular polymerization in hepatocytes and therefore to a reduced level of secreted serum AAT [2]. Homozygosity for PI Z (PI ZZ genotype) with a frequency of about 0.01% in Caucasian populations [3] causes blood AAT levels below 30% of normal. This genotype is clearly associated with elevated chronic obstructive pulmonary disease (COPD) risk accounting for 1–2% of all cases [4], [5]. There is also strong evidence that accelerated lung function decline and increased obstructive disease risk can be caused by compound heterozygosity of PI Z and PI S (PI SZ genotype). The case is less clear for PI MZ, PI MS or PI SS genotypes (PI M standing for the normal allele), which cause a less pronounced reduction in AAT concentration, as previous studies produced inconsistent evidence [6]–[9].
Further of note, large-scale genome-wide association studies (GWAS) on COPD or on cross-sectional or longitudinal lung function have not identified the SERPINA1 gene to be a major genetic determinant [10]–[12]. But a recent GWAS on emphysema [13] and a comprehensive evaluation of candidate regions for lung function [14] reported rs4905179 and rs3748312, two common SNPs (minor allele frequencies (MAFs) >5%) located in the SERPINA gene cluster on 14q32.13, among their most strongly associated results. This locus encompasses SERPINA1 and ten other genes (SERPINA2 to SERPINA6 and SERPINA9 to SERPINA13) encoding extracellular ‘clade A’ serpins with very heterogeneous functions [15]. It is currently not known whether such association signals observed for this locus reflect a causal role of common variants or whether they are merely synthetic, reflecting effects of rarer causal variants [16]. Towards that aim, but also to detect further chromosomal loci of potential relevance to circulating levels of AAT, we first performed a GWAS on AAT serum level using a subset of the population-based Swiss Cohort Study of Air Pollution and Lung Disease in Adults (SAPALDIA) as discovery sample, and a second subset of SAPALDIA as well as an independent cohort, the Copenhagen City Heart Study (henceforth referred to as Copenhagen), as replication sample. We also conducted fine mapping analyses of the SERPINA1 gene in the SAPALDIA cohort. Finally, we meta-analyzed the lung function effect of common and low-frequent SERPINA1 SNPs previously observed to be associated with pulmonary health in ever-smokers, based on data provided by several population- and patient-based studies.
Results
The discovery population and the design used to determine AAT-associated genetic variants are depicted in Figure S1 and further described in the Materials and Methods section. A comparison between the characteristics of the genome-wide analyzed sample (SAPALDIA discovery arm, N = 1392) and the remainder of the SAPALDIA cohort (SAPALDIA replication arm, N = 4245) did not reveal substantial differences in AAT serum levels or covariate distribution (Table S1), although asthmatics were overrepresented (39.4%) in the SAPALDIA discovery arm and absent in the replication arm, which is due to previous study design [17]. The participants of the independent replication cohort Copenhagen (N = 8273) were on average five years older and had twice as many current smokers (Table S1). This was in line with substantially lower lung function levels (more than 800 mL lower forced expiratory volume in 1 second, FEV1, compared to both SAPALDIA subsets) and slightly elevated AAT blood levels (1.339 g/L vs. 1.257 and 1.255 g/L, respectively). The characteristics of the study populations contributing to the genetic association analyses with lung function are given in Table S2.
GWAS on AAT Serum Level
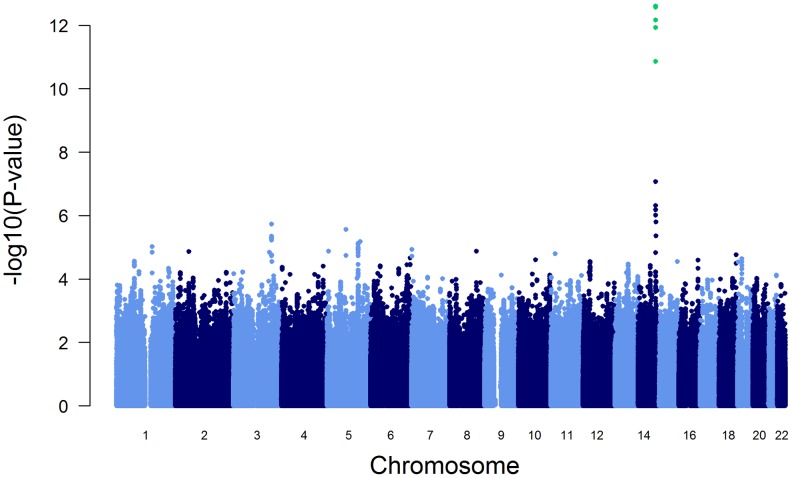
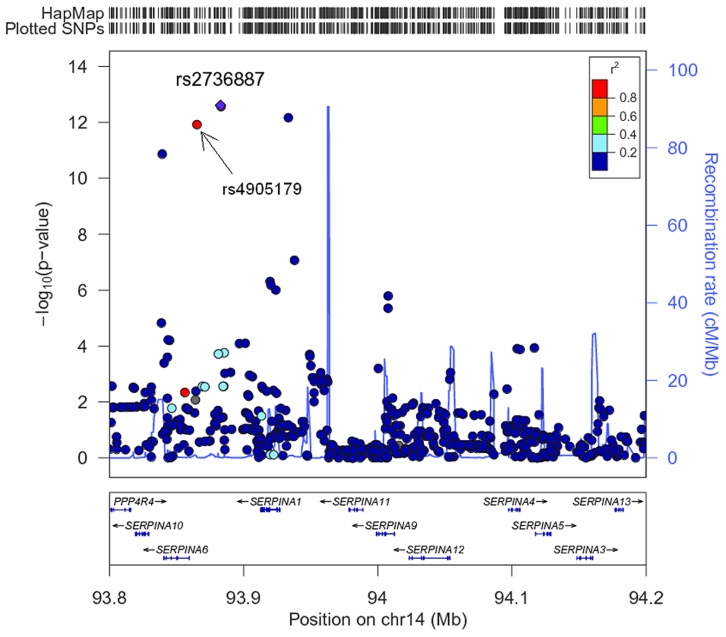
The association of more than 2.1 million genome-wide SNPs with AAT serum levels is shown in Figure 1. The ten most strongly associated SNPs were all located in the SERPINA gene cluster, half of them reached genome-wide significance (P<5*10−8, Table 1). The top 100 ranking SNPs are provided in Table S3. A regional association plot for the SERPINA gene cluster is shown in Figure 2. Both the top-ranking imputed SNP, rs2736887, and the top-ranking genotyped SNP, rs4905179, were located in close proximity to the SERPINA6 gene and approximately 33 kb and 50 kb downstream of SERPINA1 (effect estimates β = −0.071 and −0.068 g/L per minor allele; P = 2.48*10−13 and 1.20*10−12, respectively). Linkage disequilibrium (LD) between these two variants based on HapMap2 CEU (Utah residents with Northern and Western European ancestry) derived haplotype data [18] was strong (r2 = 0.88, D′ = 1), but Figure 2 suggests that the LD, expressed in r2 values, between the top-ranking SNP and the other SNPs in the region is generally modest. The genomic inflation factor lambda was low (λ = 1.02), suggesting minimal population stratification. The quantile-quantile plot (Q-Q plot) showed good adherence to null expectation and substantial positive deviation between observed and expected p-values for the top-ranking SNPs (Figure S2). In a sensitivity analysis adjusting for additional covariates, including high sensitivity C-reactive protein (hs-CRP), body mass index (BMI), passive smoking and alcohol intake, the genome-wide association results did not show an increase in the strength of the top-ranking loci, nor did they point to additional loci (data not shown). Even though this GWAS was enriched with asthma patients, GWAS stratification according to asthma status did not show heterogeneity for the top-ranking signals between participants with and without asthma (data not shown).
Figure 1. Manhattan plot of genome-wide -log(10) p-values for association with AAT serum level.
SNPs reaching genome-wide significance are shown in green. They all belong to the SERPINA gene cluster.
Table 1. The ten most strongly associated SNPs in the unconditional GWAS on AAT serum level in SAPALDIA (N = 1392).
| SNP | Chromosome | Position | Gene | Location | Determination | MAF | Imp-r2 | Allele Effect | P |
| rs2736887 | 14 | 93882733 | intergenic | imputed | 0.185 | 0.950 | 0.071 | 2.48E-13 | |
| rs926144 | 14 | 93883155 | intergenic | imputed | 0.186 | 0.950 | 0.071 | 2.72E-13 | |
| rs7151526 | 14 | 93933389 | SERPINA1 | 5′UTR | imputed | 0.065 | 0.769 | 0.116 | 6.78E-13 |
| rs4905179 | 14 | 93865245 | SERPINA6 | 5′UTR | genotyped | 0.180 | 1.000 | 0.068 | 1.20E-12 |
| rs11621961 | 14 | 93839229 | SERPINA6 | 3′UTR | genotyped | 0.355 | 0.945 | 0.052 | 1.37E-11 |
| rs17751837 | 14 | 93937997 | SERPINA1 | 5′UTR | genotyped | 0.097 | 0.995 | 0.063 | 8.56E-08 |
| rs1028580 | 14 | 93919635 | SERPINA1 | intron | imputed | 0.154 | 0.979 | 0.051 | 4.87E-07 |
| rs8010121 | 14 | 93920367 | SERPINA1 | intron | genotyped | 0.155 | 0.999 | 0.049 | 6.64E-07 |
| rs3748312 | 14 | 93924017 | SERPINA1 | intron | imputed | 0.148 | 0.846 | 0.053 | 9.84E-07 |
| rs17752593 | 14 | 94007781 | SERPINA9 | intron | genotyped | 0.129 | 0.997 | 0.053 | 1.59E-06 |
Abbreviations: AAT, alpha1-antitrypsin; GWAS, genome-wide association study; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Imp-r2 is an indicator for imputation quality. SNPs with MAF<0.05 or imp-r2<0.5 were excluded.
Chromosomal position is based on reference panel, NCBI build 36.3. Allele effects are shown in absolute numbers.
Figure 2. Regional plot for the SERPINA gene cluster (93.8–94.2 Mb on chromosome 14q32.13, reference panel: NCBI build 36.3).
Presented are -log(10) p-values and LD (r2) with top-ranking SNP rs2736887 (purple diamond) for all SNPs in this region. The blue line shows recombination rate.
Association of 1000 Genomes Imputed Data for the SERPINA Gene Cluster with AAT Serum Level
In order to further refine association signals in this region, we imputed additional SNPs on chromosome 14 using haplotype data from the 1000 Genomes Project (1000G) [19]. The 1000G imputation yielded a three times higher number of imputed variants with reasonable quality scores (imputation-r2>0.5) compared to HapMap-derived imputed variants. A region defined by 1 Mb up- and downstream of the SERPINA1 gene revealed 24 additional variants that were associated below a local significance level of P<3*10−5, adjusting for approximately 1800 SNPs covering a region of 2 Mb (Table S4). Among them, four low-frequent variants and one rare variant showed p-values reaching genome-wide significance level, and interestingly, none of them was in high LD (r2>0.8) with any other regional variant tested. The most strongly associated signal came from the PI Z variant, which is well known to be associated with reduced AAT serum levels (β = −0.620 g/L per minor allele, P = 4.61*10−43, MAF = 0.84%). The other well established causal polymorphism, the PI S variant, was less prominently ranked (β = −0.110 g/L per minor allele, P = 1.95*10−6, MAF = 5.70%) and exhibited an insufficient imputation quality (imputation-r2 = 0.45).
GWAS on AAT Serum Level, Conditional on PI S and PI Z Variants
Accuracy of the imputed PI S and PI Z results was confirmed by direct genotyping of the samples [20]. The discovery arm revealed 33 PI Z carriers, 111 PI S carriers and two compound heterozygous carriers of PI S and PI Z (MAF = 1.26% for PI Z and 4.06% for PI S, respectively). No homozygous PI S or PI Z genotypes were detected.
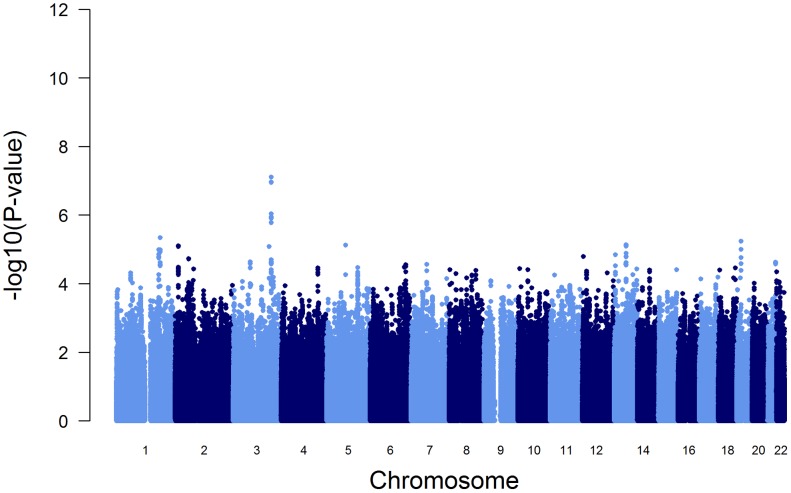
To test the influence of these variants on the initially reported GWAS results (Figure 1), we performed a conditional GWAS by additionally adjusting the regression models for the presence of PI S and PI Z alleles. We observed a drastic change in the association of the SERPINA gene cluster SNPs with AAT serum level (Figure 3 and Table 2). The strong signal on chromosome 14 observed in the original GWAS disappeared completely and the top-ranking imputed and genotyped SNPs (rs2736887 and rs4905179) were no longer significant (P = 0.44 and 0.31, respectively). In fact, no SNP was found near the SERPINA gene cluster among the 100 most strongly associated common variants (Table S5). In addition, the 1000G imputed data, comprising sequences 1 Mb up- and downstream of the SERPINA1 gene, did not show evidence of other independent AAT-associated SNPs. An alternative approach that excluded all PI S and PI Z carriers from the GWAS sample (N = 146), instead of adjusting for them, confirmed the results. Both analyses revealed an intergenic region on chromosome 3 with borderline genome-wide significance (top-ranking SNP rs2566347, β = −0.043 g/L per minor allele, P = 7.88*10−8, in the adjusted GWAS). The top SNPs in this region were located in proximity to MFSD1 and RARRES1, which are two genes with sparsely annotated function. The 1000G imputation of this region did not reveal further variants. In addition, we were unable to replicate this association signal in the SAPALDIA replication arm (N = 4245, β = −0.004 g/L per minor allele, P = 0.46).
Figure 3. Manhattan plot of genome-wide -log(10) p-values for association with AAT serum level, conditional on PI S and PI Z alleles.
Table 2. The ten most strongly associated SNPs in the GWAS on AAT serum level, conditional on PI S and PI Z alleles in SAPALDIA (N = 1392).
| SNP | Chromosome | Position | Gene | Location | Determination | MAF | Imp-r2 | Allele Effect | P |
| rs2566347 | 3 | 159974071 | intergenic | imputed | 0.192 | 0.998 | 0.043 | 7.88E-08 | |
| rs1560417 | 3 | 159972476 | intergenic | imputed | 0.200 | 0.998 | 0.042 | 1.11E-07 | |
| rs1560418 | 3 | 159972335 | intergenic | genotyped | 0.200 | 1.000 | 0.042 | 1.11E-07 | |
| rs1430414 | 3 | 159987697 | MFSD1 | 5′UTR | imputed | 0.137 | 0.984 | 0.045 | 9.26E-07 |
| rs6761989 | 3 | 159983253 | intergenic | imputed | 0.137 | 0.993 | 0.044 | 1.14E-06 | |
| rs17643917 | 3 | 159968433 | intergenic | imputed | 0.137 | 1.000 | 0.044 | 1.23E-06 | |
| rs17643860 | 3 | 159967954 | intergenic | imputed | 0.137 | 1.000 | 0.044 | 1.24E-06 | |
| rs17700475 | 3 | 159967627 | intergenic | genotyped | 0.137 | 1.000 | 0.044 | 1.25E-06 | |
| rs3863076 | 3 | 159969394 | intergenic | genotyped | 0.145 | 1.000 | 0.042 | 1.69E-06 | |
| rs2206593 | 1 | 184909052 | PTGS2 | 3′UTR | genotyped | 0.065 | 0.956 | 0.060 | 4.60E-06 |
Abbreviations: AAT, alpha1-antitrypsin; GWAS, genome-wide association study; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Imp-r2 is an indicator for imputation quality. SNPs with MAF<0.05 or imp-r2<0.5 were excluded.
Chromosomal position is based on reference panel, NCBI build 36.3. Allele effects are shown in absolute numbers.
Replication in the Copenhagen City Heart Study
The effect of rs4905179, the top genotyped SNP in our GWAS, on AAT serum levels was tested for replication in Copenhagen (Table 3). The minor allele was associated with β = −0.097 g/L (P<0.0001, N = 8332). As observed in the GWAS, adjustment for PI S and Z polymorphisms resulted in a complete loss of this signal (β = 0.003 g/L, P = 0.57, N = 8273).
Table 3. Minor allele effects of PI S, PI Z and rs4905179 on AAT serum levels in the Copenhagen City Heart Study.
| SNP | N | MAF (genotyped) | Allele Effect (g/L) | 95% Confidence Intervals | P |
| rs17580 (PI S) | 8338 | 0.029 | −0.188 | −0.211 to −0.165 | <0.0001 |
| rs28929474 (PI Z) | 8338 | 0.027 | −0.492 | −0.514 to −0.470 | <0.0001 |
| rs4905179 | 8332 | 0.186 | −0.097 | −0.107 to −0.087 | <0.0001 |
| rs4905179, adjusted for PI S and Z | 8273 | 0.186 | 0.003 | −0.007 to 0.013 | 0.57 |
Abbreviations: MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Impact of Common and Low-Frequent SERPINA1 Genetic Variants on AAT Serum Level
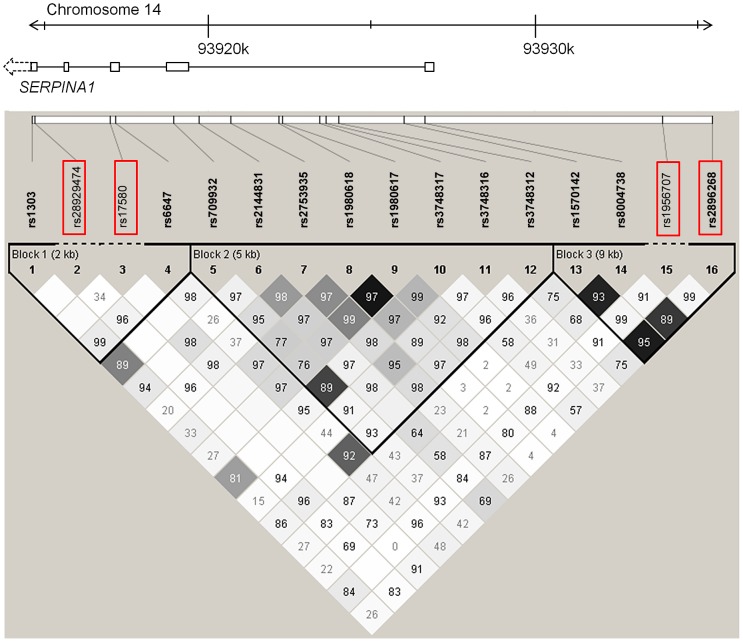
In a first fine mapping step, 16 SERPINA1 SNPs (see Materials and Methods section for a description of the SNP selection) were successfully genotyped in 5569 SAPALDIA subjects (discovery and replication arm combined). The genotype results in the discovery arm allowed us to compare allele frequencies with imputed results derived from the 1000G data. Table S6 shows that the agreement was very high. Stepwise conditional regression analyses were then applied to evaluate the independent effects of each of these SNPs on AAT serum levels (Table 4). The PI Z variant was most strongly associated with circulating levels of AAT. The PI S variant remained strongly associated after conditioning on PI Z. Two variants located in the 5′ non-coding gene region (rs2896268 and rs1956707) were marginally associated with the phenotype in two further steps after conditioning on PI S and PI Z. The total variance of AAT explained by statistical models increased from 8.8% (model with only non-genetic factors) to 32.6% (adding PI S and PI Z alleles), and to 32.8% adding rs2896268 and rs1956707. Based on genotype data from the SAPALDIA cohort, the SERPINA1 gene contains three haplotype blocks using D′-based block definition (Figure 4). The AAT deficiency variants PI S and PI Z are located in block 1, while rs2896268 and rs1956707 are located in block 3, roughly 8 kb upstream of exon 1.
Table 4. Common and low-frequent SERPINA1 SNPs and their association with AAT serum level, univariate and conditional on significantly associated SNPs (N = 5569a), in SAPALDIA.
| SNP | Location | Position | Selectionb | MAF | Univariatec | Conditionald | ||
| Allele Effect | P | Allele Effect | P | |||||
| rs2896268 | 5′UTR | 93935461 | C | 0.495 | 0.006 | 0.10 | 0.013 | 4.1E-05 |
| rs1956707 | 5′UTR | 93933946 | C | 0.038 | 0.016 | 0.10 | 0.029 | 5.0E-04 |
| rs8004738 | exon 1 | 93926667 | D | 0.490 | 0.005 | 0.15 | 0.001 | 0.81 |
| rs1570142 | intron 1 | 93926015 | A,B,C | 0.488 | 0.005 | 0.19 | 0.001 | 0.88 |
| rs3748312 | intron 1 | 93924017 | A,B | 0.153 | 0.035 | 2.4E-12 | 0.002 | 0.70 |
| rs3748316 | intron 1 | 93923617 | A,C | 0.181 | 0.011 | 0.02 | 0.002 | 0.63 |
| rs3748317 | intron 1 | 93923432 | A | 0.158 | 0.020 | 5.5E-05 | 0.004 | 0.36 |
| rs1980617 | intron 1 | 93922287 | A | 0.389 | 0.030 | 5.0E-16 | 0.003 | 0.31 |
| rs1980618 | intron 1 | 93922176 | A,C | 0.383 | 0.030 | 1.3E-16 | 0.005 | 0.15 |
| rs2753935 | intron 1 | 93920690 | A | 0.435 | 0.012 | 9.7E-04 | 0.004 | 0.24 |
| rs2144831 | intron 1 | 93919723 | A,C | 0.240 | 0.021 | 9.8E-07 | 0.002 | 0.71 |
| rs709932 | exon 2 | 93918954 | A,C | 0.169 | 0.021 | 1.2E-05 | 0.003 | 0.50 |
| rs6647 | exon 3 | 93917168 | A,C | 0.200 | 0.026 | 7.2E-09 | 0.003 | 0.51 |
| rs17580 (PI S) | exon 3 | 93917015 | D | 0.041 | 0.210 | 1.9E-127 | 0.218 | 1.9E-163 |
| rs28929474 (PI Z) | exon 5 | 93914700 | D | 0.013 | 0.483 | 1.3E-240 | 0.482 | 5.2E-269 |
| rs1303 | exon 5 | 93914596 | A,C | 0.247 | 0.015 | 4.7E-04 | 0.001 | 0.89 |
Abbreviations: AAT, alpha1-antitrypsin; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Chromosomal position is based on reference panel, NCBI build 36.3.
Includes subjects for whom all the 16 SNPs have been successfully genotyped.
SNP selection was based on extreme trait sequence data (A), tagging SNPs according to HapMap (B), TAMAL software (C) and publication about functionality (D); see Materials and Methods for a more detailed description.
Univariate analyses were adjusted for non-genetic factors only (sex, age, recruiting area and current smoking status). Allele effects are shown in absolute numbers and P<0.005 was considered statistically significant.
In a forward selection approach of stepwise regression, the four SNPs in bold contributed statistically significantly to the variability in AAT serum levels and were included in the final statistical model. Allele effects and p-values refer to this final model.
Figure 4. LD plot among common and low-frequent SNPs in the SERPINA1 gene within the SAPALDIA study.
Shading represent r2 values, whereas numbers represent D′ values (no number equals D′ = 1). Red framed SNPs are independently associated with AAT serum levels after forward selection stepwise regression modeling. Rs17580 is the PI S variant and rs28929474 is the PI Z variant.
Impact of Rare SERPINA1 Genetic Variants on AAT Serum Level
In a second fine mapping step, exon sequencing was performed in 410 subjects with low AAT levels that were independent of the presence of PI S or PI Z alleles [21]. 16 additional SERPINA1 variants (two deletions and 14 SNPs) were detected, of which all but one had already been described [21]–[29] (Table S7). Three of the SNPs were synonymous, and five had no accession numbers in public databases (as of April 1st, 2013). Most of the non-synonymous SNPs have already been described as potentially lowering AAT serum level, and computational tools only classified one of them as no damaging to the protein's tertiary structure. In order to estimate the phenotypic influence of these rare variants, we compared mean AAT blood levels, adjusted for sex, age, study center, current smoking, as well as for the presence of PI S and Z alleles, between samples without rare variants (N = 346) and those with a single rare variant (N = 63) or more than one (N = 1). The subjects with rare variants had a lower adjusted mean AAT level (0.904 g/L, 95% CI 0.884 to 0.924 g/L) compared to those without rare variants (0.992 g/L, 95% CI 0.984 to 1.000 g/L, P<0.001). Although this difference is small, the range covers the recently proposed upper limit of intermediate AAT deficiency (0.92 g/L), a value with some clinical relevance [20]. AAT levels of carriers of synonymous mutations or non-synonymous mutations without predicted damaging consequences to protein structure (N = 20) were not different from those carrying no rare variants (0.985 vs. 0.990 g/L, P = 0.77). Assuming that unsequenced samples were negative for mutations with predicted deleterious functional effects, the total variance of explained AAT further increased from 32.8% to 35.4% (based on a statistical model adding all rare mutations with predicted damaging consequences to the protein structure).
Common and Low-Frequent SERPINA1 SNPs Previously Associated with Lung Function
Results from a previous GWAS on emphysema [13] and a large-scale evaluation of candidate loci on lung function [14] pointed to a role of common variants in the SERPINA gene cluster. The SNPs rs4905179 (associated with emphysema in smokers [13]) and rs3748312 (associated with cross-sectional lung function among ever smokers [14]) were strongly associated with AAT in our study (Tables 1 and 4), but both signals disappeared upon adjustment for the low-frequent variants PI S and Z. In order to clarify whether the association of the two common SNPs with pulmonary health could also be explained by effects of the rarer SNPs, we conducted a meta-analysis for cross-sectional lung function in ever-smokers across 17 studies with a total sample size of N = 24,446 (Table S2). We included nine studies which had contributed to the original finding on lung function [14] and had available genotypes or 1000G imputed genotype data on PI S and Z. The meta-analysis in cohorts of general population study design showed that rs4905179 was not associated with lung function in ever-smokers (P = 0.90 in the fixed-effect meta-analysis, N = 20,153, Figure 5). Yet smaller studies recruited within population isolates showed a trend for the rare allele to be associated with low lung function (random-effect P = 0.02, N = 1623, Figure 5), and in contrast to the association with AAT serum levels, adjusting for PI S and Z alleles did not modify the association of rs4905179 with lung function (Figure 6). For the second common SNP, rs3748312, we could nominally replicate the statistically significant allele effect on FEV1 in the general population of ever-smokers (P = 0.02, N = 15,450), and the stronger effect that was published [14] seems to be driven by population isolates (Figure 7). Again, as for rs4905179, the associations were not dependent on S and Z alleles (Figure 8). Meta-analyses of the associations of PI S and Z alleles with lung function revealed no consistent associations between these functional AAT level determining variants and reduced FEV1 (Figures S3 and S4). Remarkably, the significant associations of rs4905179 and rs3748312 with lung function assessed in two additional studies with patients featuring compromised pulmonary health and undergoing lung resection, showed evidence for synthetic associations of the common SNPs with lung function that are consistent with our results for circulating AAT (Table 5). The minor alleles were associated with lower lung function and the association completely disappeared when conditioned on the presence of PI S and Z alleles.
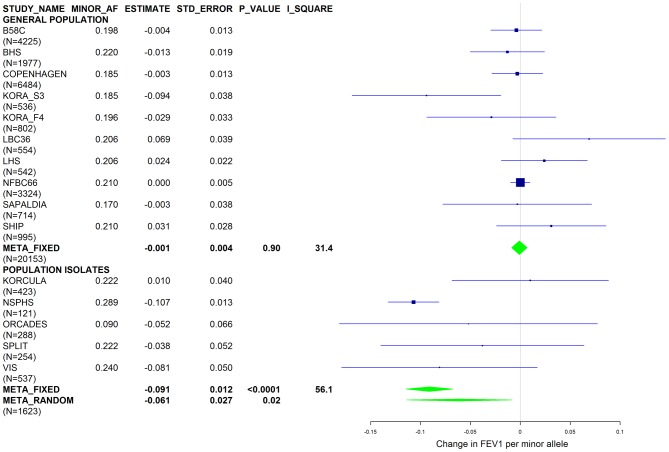
Figure 5. Forest plot of meta-analyzed results for the effect per minor allele of rs4905179 on FEV1 in ever-smokers, adjusted for sex, age, height and population stratification factors.
Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to fixed effect meta-analyses.
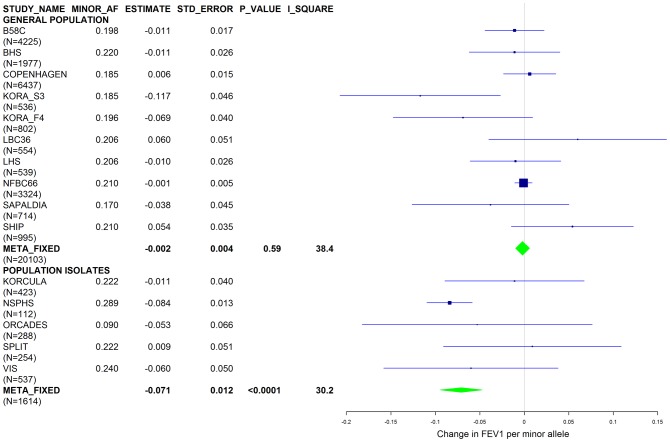
Figure 6. Forest plot of meta-analyzed results for the effect per minor allele of rs4905179 on FEV1 in ever-smokers, adjusted for sex, age, height, population stratification factors and the presence of PI S and Z alleles.
Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to fixed effect meta-analyses.
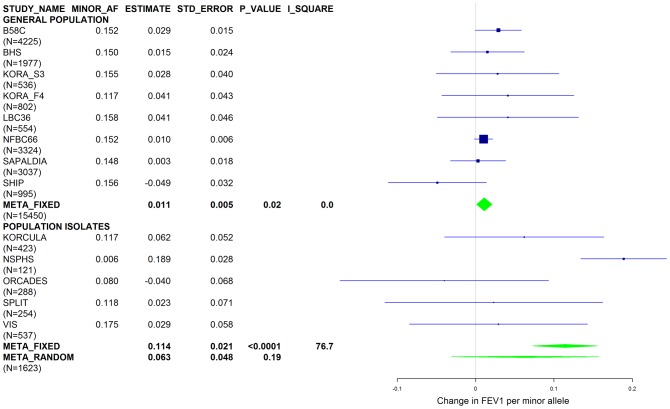
Figure 7. Forest plot of meta-analyzed results for the effect per minor allele of rs3748312 on FEV1 in ever-smokers, adjusted for sex, age, height and population stratification factors.
Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to fixed effect meta-analyses.
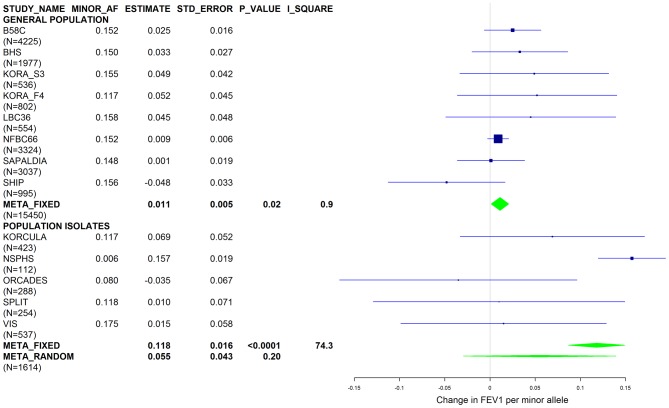
Figure 8. Forest plot of meta-analyzed results for the effect per minor allele of rs3748312 on FEV1 in ever-smokers, adjusted for sex, age, height, population stratification factors and the presence of PI S and Z alleles.
Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to fixed effect meta-analyses.
Table 5. Minor allele effects on FEV1 of low-frequent and common SNPs in the SERPINA gene cluster in ever-smokers undergoing lung resection.
| SNP | MAF | Imp-r2 | Allele Effect (L) | P | MAF | Imp-r2 | Allele Effect (L) | P |
| Groningen (N = 133) | University of British Columbia, UBC (N = 264) | |||||||
| rs17580 (PI S) | 0.055 | 0.72 | 0.42 | 0.10 | 0.079 | 0.76 | −0.15 | 0.29 |
| rs28929474 (PI Z) | 0.123 | 0.98 | −0.68 | <0.001 | 0.032 | 0.98 | −0.81 | <0.0001 |
| rs4905179 | 0.299 | 1.00 | −0.22 | 0.04 | 0.249 | naa | −0.23 | 0.002 |
| rs4905179, adjusted for PI S and Z | −0.07 | 0.65 | −0.08 | 0.36 | ||||
| rs3748312 | 0.233 | 0.95 | −0.34 | 0.008 | 0.148 | 0.94 | −0.16 | 0.07 |
| rs3748312, adjusted for PI S and Z | 0.16 | 0.37 | 0.09 | 0.35 | ||||
Abbreviations: FEV1, forced expiratory volume in one second; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Imp-r2 is an indicator for imputation quality. The analyses were adjusted for age, sex and height.
Rs4905179 was genotyped in UBC.
Discussion
We present here the first GWAS on circulating AAT blood levels. Our results confirm that genetic variation in the SERPINA1 gene is a strong determinant of serum AAT levels. Fine mapping of SERPINA1 and subsequent stepwise regression analyses further revealed that the associations with common variants in the SERPINA locus could be attributed to rarer variants previously identified to be causally linked with AAT deficiency.
There is an ongoing debate about whether rare variants are responsible for the missing heritability observed in GWAS on many complex outcomes [30]. We show here an example in which the polymorphisms PI S and PI Z seem to account for basically all observable effects of common variants in the SERPINA gene cluster on AAT serum level. The top-ranking genotyped SNP in our GWAS, rs4905179, was in low r2-based LD with PI S (r2 = 0.18) and PI Z (r2 = 0.06), reflecting in part the unequal allele frequencies of these SNPs. However, PI S and PI Z showed very high LD in terms of D′ with the GWAS top signals (e.g. D′ = 0.95 and 0.96, respectively, with rs4905179) and generally with many common variants in this locus (Figure 4), suggesting little genetic recombination. This proof-of-principle approach, revealing that signals of common variants in fact merely reflect rarer variants, has recently also been shown for some of the loci regulating low-density lipoprotein (LDL) cholesterol [31], [32]. Yet for other loci linked to LDL cholesterol, as well as for loci influencing other traits, both common and low-frequent variants contributed independently of the original GWAS signal to the phenotypic trait [31], [33], [34].
Using regional 1000G imputation within the top-ranking loci can allow the identification of additional association signals of stronger size to support the initial GWAS top result, as observed here for the SERPINA cluster, but not for the locus near MFSD1, an association which was not confirmed in the SAPALDIA replication arm. The resequencing strategy of the GWAS-identified locus in a sample with low AAT concentrations yielded in the identification of rare variants being strongly associated with reduced AAT blood levels. Such an accumulation of rare variants in the extreme range of the respective phenotype has also been reported by others [35], [36]. As for the relative contribution of genetic variants on the phenotype, we confirmed that effect sizes of PI S and Z on AAT serum levels were comparably strong, explaining alone a high proportion of the total variability (24.2%). We estimated that rare variants explained at least another 2% in our population-based sample, but since we did not sequence the entire SAPALDIA sample for rare variants, we cannot reliably quantify this contribution. In terms of blood markers, similar examples exist in which one genetic variant could explain well above 5% of the phenotype's variability (e.g. lipoprotein(a) [37], bilirubin [38] or adiponectin [39]), but for many other markers like serum lipid levels, only variants with small effects have been detected so far [40].
Association patterns between SERPINA1 variants and circulating AAT did not translate to according associations with lung function level in a straightforward manner. Lung function is a complex phenotype associated with numerous genetic variants [11], [12]. Studies on the associations of SERPINA1 polymorphisms with lung function and COPD have produced mixed results. It is well accepted that severe AAT deficiency caused by PI null mutations or by the presence of two PI Z alleles puts a subgroup of carriers at higher risk of emphysema and COPD, especially when smoking [5]. Studies on COPD found suggestive evidence for an association with heterozygous status for the PI Z allele [6], [8], but we observed no associations in our meta-analysis between PI Z and lung function level in ever-smokers. In the SAPALDIA general population sample, we had previously reported that an effect of the PI Z allele on lung function decline is restricted to persistent smokers and primarily observed for forced expiratory flow 25–75% [9]. Other variation in or close to the SERPINA1 gene has been proposed to play a role for pulmonary health. First, a haplotype pattern of five common SNPs was reported to be more frequent in COPD cases than in controls in a study with limited statistical power [41]. The only SNP which was also separately associated with COPD in that analysis was not associated with reduced serum levels in our study (rs8004738, Table 4). Second, the minor allele of rs4905179, which was the top signal in the current AAT GWAS, was positively associated with emphysema assessed by chest tomography in three independent cohorts consisting of smoking COPD patients without severe AAT deficiency (PI ZZ) [13]. Finally, the minor allele of the intronic SNP rs3748312 was positively associated with lung function in ever-smokers from different population-based studies of the SpiroMeta Consortium [14]. The association of these two SNPs with lung function in ever-smokers was heterogeneous across studies in our meta-analysis. Dependency on PI S and Z was limited to studies in patients with lung resection (Groningen, UBC), consistent with the notion that SERPINA1 may only confer risk in selected population subgroups.
There are several possible explanations for the poor translation of genetic association patterns with serum AAT to lung function and for the heterogeneity of associations between SERPINA1 variants and lung function. First, lung function is influenced by mechanisms in addition to protease-antiprotease disequilibrium. Second, the contribution of the SERPINA1 gene variants as a determinant of lung function likely depends on both the evolutionary pressure in isolated populations and the prevalence of effect modifiers in the respective study populations. These include smoking and smoking intensity, and likely other markers of inflammation. AAT itself plays a dual role in its relationship with lung function. While chronic AAT deficiency is etiologically associated with adverse pulmonary health, individuals with lung function impairment in fact exhibit higher AAT levels for a given genetic background due to AAT's role as an acute-phase inflammation marker [42], [43]. Third, tissue-specific regulation of the SERPINA1 locus may play an important role. Serum AAT levels are driven by SERPINA1 expression, protein formation and secretion in hepatocytes, so that regulatory SNPs associated with serum AAT likely reflect processes in the liver. One way to infer causality of potentially regulatory SNPs is by testing if they are simultaneously associated with health outcome and gene expression in the relevant tissue [44]. We therefore conducted a look-up in an expression quantitative loci (eQTL) database of lung tissue [45], but could not find any common variant which was significantly associated in cis with the transcripts deriving from the SERPINA1 locus. In a recent study on networks of blood metabolites, the SNPs rs11628917 and rs1884549 were the most strongly associated blood and liver eQTLs with respect to SERPINA1 expression [46]. They both lie in the 3′ untranslated region of SERPINA1, but were not associated with blood AAT in our GWAS (P = 0.80 and P = 0.21, respectively). Moreover, we could not detect epistasis between those variants and the deleterious coding variants PI S and Z in terms of AAT serum levels. The absence of such an interaction does not point to regulatory function of the common SNPs [47] and argues in favor of tissue-specific heterogeneity. Forth, the role of SERPINA1 in selected subgroups of persons exhibiting accelerated lung function decline or COPD needs to be considered from a perspective beyond genetic variation, as a recent study investigating epigenetic mechanisms of disease revealed methylation status of the SERPINA1 gene to be most strongly associated with cross-sectional lung function and COPD [48].
The strength of this study is that it combines the report of a GWAS on AAT serum levels with meta-analyses of the associations of some of the GWAS top variants with lung function. The effects of the underlying functional variants are thoroughly investigated resulting in the hitherto largest meta-analysis of PI S and Z on FEV1 in ever-smokers. Ascertainment and study design of the many participating studies were sufficiently diverse to informatively address heterogeneity in association of common and rarer variants in the SERPINA gene cluster with lung function. The strength of the discovery sample is the population-based study design and the detailed characterization of the participants. Sex, age and smoking are important modifiers of AAT blood levels in the general population [43] and were included in all regression models. More refined smoking variables covering smoking intensity were not included as this information is less complete than smoking status in SAPALDIA and would lower the sample size. By excluding samples with elevated hs-CRP values we avoided the masking of AAT deficiencies due to a chronic or acute inflammation. On the methodological side, conditional analysis is a well-established tool for identifying independent signals within a certain locus [38], [49], [50]. Furthermore, 1000G imputation was able to point to the causal variant demonstrating its reliability to correctly assign alleles close to the 1% MAF threshold.
The limitations of this investigation include firstly the small sample size of the GWAS discovery arm, resulting in a high susceptibility to false negative findings. We calculated 63% power to detect SNPs with an allele effect of 0.1 g/L AAT serum level ( = 2.4% of the phenotypic variance) to a genome-wide significance level of 5*10−8. However, if we define the clinically important threshold of AAT as the upper limit of intermediate AAT deficiency, which has been recently suggested as 0.92 g/L [20], we have more than 99.9% power to detect such a large-impact variant. Nevertheless, genes that contribute to AAT serum levels with smaller effects than SERPINA1 were likely to be missed. This could be a reason why neither SNPs in interleukin 6 (IL-6) nor in hepatocyte nuclear factor 1α (HNF-1α)/HNF-4, both important regulators of AAT expression [51], were associated with circulating AAT concentrations. Furthermore, by sequencing only the coding region of SERPINA1, rare variants in introns and outside the gene could not be determined. Another potential limitation of our GWAS on AAT serum level is the overrepresentation of asthmatics in the discovery sample. Asthma patients usually show higher levels of inflammatory markers in their lungs. However, we did not find heterogeneity in the effects of the most strongly associated SNPs when comparing asthmatics with non-asthmatics. Moreover, AAT mean values between the discovery and the replication arm were not significantly different, as participants with elevated hs-CRP had been excluded.
In conclusion, our study confirms the SERPINA1 locus as the major genetic determinant of AAT blood levels. Methodologically, it represents a powerful example how low-frequent variants, separated by several kilobases from the top-ranking GWAS signals, can create purely synthetic associations which do not add to the variance of the respective outcome. In terms of lung function, our data do not support a functional role of any common SNP in the SERPINA cluster in the general population.
Materials and Methods
Ethics Statement
SAPALDIA was approved by the Swiss Academy of Medical Sciences, the national ethics committee for clinical research (UREK, Project Approval Number 123/00) and the Cantonal Ethics Committees for each of the eight examination areas (Ethics commissions of the cantons Aargau, Basel, Geneva, Grisons, Ticino, Valais, Vaud and Zurich). Participants were required to give written consent before any part of the health examination was conducted either globally (for all health examinations) or separately for each investigation. For ethics statements of the additional studies contributing to this work, see Table S8.
Study Population
SAPALDIA
In 1991, a random sample of 9651 adults, aged 18–60 years, from eight areas in Switzerland responded to a questionnaire about respiratory health, occupational and lifestyle exposures. 99.0% of them also underwent spirometry testing [52]. Eleven years later, 8047 persons were reassessed and 6058 subjects provided blood samples and consented to DNA analysis [53]. In the present study, we used a subgroup of the second survey (N = 1640) that underwent genotyping in the context of the GWAS on asthma by the GABRIEL consortium [17]. This sample included all asthmatics (positive answer to the question “Have you ever had asthma?” at either survey) as well as a random sample of non-asthmatic controls. 248 participants were removed due to several reasons, including elevated levels of the inflammatory marker hs-CRP (>10 mg/L, N = 54), leading to a discovery arm of 1392 individuals (Figure S1). The discovery arm contained 548 (39.4%) self-declared asthmatics, whereas there were no self-declared asthmatics in the replication arm. Both the discovery and the replication sample were submitted to a first step of fine mapping of SERPINA1 resulting in 5569 individuals from whom all the selected SNPs could be successfully determined. In a second step, a subsample with abnormally low AAT measurements additionally underwent SERPINA1 exon sequencing.
Additional Studies
The populations are briefly described in Table S8.
Phenotype Measurements
AAT serum levels in SAPALDIA were determined by latex-enhanced immunoturbidimetric assays (Roche Diagnostics, on a Roche Cobas Integra analyzer) with interassay coefficients of variation below 5% and lower detection rate of 0.21 g/L. Serum concentrations in Copenhagen were measured by immunoturbidimetric assays (Thermo Scientific, on a Thermo Scientific Konelab analyzer) with coefficients of variation below 5% and lower detection rate of 0.10 g/L.
Lung function was measured in all participating studies by spirometry without bronchodilation ([52] and Table S8). In the patient-based studies, in which a lung resection was carried out (Groningen, UBC), lung function measurements were carried out prior to the intervention.
Genotyping
SAPALDIA
Genomic DNA was extracted from blood samples using the Puregene DNA Isolation Kit (Gentra Systems). Genotyping of the GWA-bound subset was performed on the Illumina Human 610quad array. Asthmatic and non-asthmatic samples were tested in random blinded order to avoid systematic array-related artifacts. 567,589 autosomal SNPs were satisfactorily genotyped (mean call rate: 99.7%). 69,892 were excluded from analysis due to violation of Hardy Weinberg Equilibrium (HWE, P<10−4), low call rate (<97%) or MAF<5%.
Genotyping of GWAS finding rs2566347 on chromosome 3 in the SAPALDIA replication arm was carried out using the MassARRAY iPLEX Gold (Sequenom).
The SNPs selected in the first fine mapping step were genotyped by polymerase chain reaction (PCR) with fluorescently labeled Taq-Man probes (Vic or Fam labels) on a Light Cycler 480 (Roche Diagnostics). All SNPs were in HWE (P>0.01) [20].
Additional Studies
PI S and Z genotypes were determined by PCR in Copenhagen and LHS as previously described [54], [55]. The SNP rs4905179 was genotyped in the course of GWAS projects in B58C, BHS, Copenhagen, Korcula, KORA S3, KORA F4, LBC36, LHS, NSPHS, ORCADES, Split, UBC, and Vis.
Imputation
SAPALDIA
We have carried out genome-wide imputation from 60 CEU HapMap2 (release 22, NCBI build 36) reference panels [18] using MACH 1.0.16 [56] resulting in 2,588,592 autosomal HapMap-based SNPs. 2,168,668 SNPs fulfilled the quality criteria, which are as mentioned above for genotyped SNPs and additionally consisted of an imputation-r2>0.5.
Further imputation was carried out in the most promising loci using 566 EUR reference haplotypes from the August 2010 release of 1000G on the MACH (pre-phasing) and Minimac-omp programs. SNPs with an imputation-r2>0.5 and MAF>0.1% passed the quality check.
Additional Studies
Rs4905197 was imputed based on 1000G reference panels in Groningen, NFBC66, and SHIP (imputation-r2≥0.99). Rs3748312, rs17580 (PI S) and rs28929474 (PI Z) were 1000G imputed in B58C, BHS, Groningen, Korcula, KORA S3, KORA F4, LBC36, NFBC66, NSPHS, ORCADES, Split, SHIP, UBC, and Vis (imputation-r2 0.86–0.98, 0.69–0.83, and 0.82–0.98, respectively).
SERPINA1 SNP Selection for Fine Mapping in SAPALDIA
In an attempt to find AAT modifying SERPINA1 gene variants acting independently of each other, a multiple strategy to optimally cover the gene was applied. Sequencing of the whole SERPINA1 gene in 25 unrelated samples from the Italian registry of AAT deficiency which demonstrated extreme phenotypes was used to identify common SNPs not present in HapMap. Extreme phenotypes consisted of 11 samples with AAT>1.60 g/L and hs-CRP <8 mg/L, 3 samples with PI ZZ or PI SZ genotype and AAT<0.20 g/L, 2 samples with PI MZ genotype and AAT<0.60 g/L, as well as 9 non-carriers of PI S or PI Z alleles with blood levels >0.65 and <1.10 g/L. In these 25 samples, a total of 129 mutations were identified in the SERPINA1 gene. After removing SNPs which were monomorphic in our data, SNPs deviating from HWE or lying in high LD with an adjacent marker (D′>0.8 and r2>0.4 according to JLIN [57]), we finally obtained a list of 22 common SNPs (Table 4, selection A). In a second strategy, HapMap CEU data was used to select tagging SNPs (Haploview 3.32) [58], resulting in 8 polymorphisms (selection B). Third, TAMAL [59] was used to identify promising SNPs in the region of the SERPINA1 gene (selection C). Pairwise LD and the feasibility of designing a corresponding TaqMan assay reduced the number of SNPs to 13. Two established (PI S and Z) and one suggestive (rs8004738 [60]) functional SNPs were added (selection D), resulting in 16 SNPs used in the conditional analysis. Three of them were already part of the SNP array genotyped for the GWAS. The 5 SNPs in coding regions (exons 2–5) were all non-synonymous.
Exon Sequencing in SAPALDIA
We sequenced 410 individuals with abnormally low AAT levels with the Sanger chain-termination method. Different thresholds according to the deficiency genotypes and hs-CRP values were applied to define an abnormally low AAT concentration (PI MM: 1.13 g/L if hs-CRP >8 mg/L and 1.00 g/L if hs-CRP ≤8 mg/L; PI MS: 0.85 g/L; PI MZ: 0.65 g/L) [21]. The cut-off of 1.13 g/L was earlier reported to be the best to differentiate AAT-deficient patients from healthy individuals [61]. Since exon 1 is non-coding, the sequencing procedure was only applied to exons 2 to 5.
Statistical Analysis
AAT serum levels were only marginally skewed to the right, and a log-transformation of these data was omitted since it led to a stronger deviation from normality. Student's t-test was used to compare adjusted mean AAT levels between different subgroups of the sequenced samples. The genome-wide association of 2.17 million quality-controlled SNPs with serum AAT levels was assessed using fixed effects linear regression with ProbABEL [62]. An additive genetic model was applied and the association was adjusted for sex, age, study center, dichotomous current smoking status, as well as population stratification factors. To account for population stratification, we relied on previously inferred ancestry-informative principal components using EIGENSTRAT 2.0 software [63] and HapMap data, as well as additional reference European samples [64]. Cryptic relatedness was detected based on identity-by-state (IBS) analysis. Influence of additional suggestive determinants of AAT, such as hs-CRP, BMI, alcohol intake and passive smoking was assessed in a sensitivity analysis. We also performed genome-wide analysis conditioned on the functionally established PI S and PI Z variants. Bonferroni correction for multiple testing was applied, resulting in P<5*10−8 to designate genome-wide significance, taking account of one million independent tests for common variants across the genome. For the SNPs imputed by using 1000G reference samples, we considered a three times lower p-value as adequate as roughly three times more SNPs on chromosome 14 passed an imputation-r2 threshold of 0.5 (219,471 1000G-derived variants vs. 82,296 HapMap2-derived variants). Applying this to a 2 Mb chromosomal stretch (with approximately 600 HapMap2-derived SNPs) resulted in a significance threshold of roughly 3*10−5.
For the replicated SNP in the SAPALDIA replication arm, as well as for the lung function analysis, a two-sided p-value of 0.05 was considered significant. We investigated heterogeneity between asthmatics and non-asthmatics in the discovery arm by testing for a difference between the two effects, using a chi-square test with one degree of freedom.
Replication analysis for AAT in Copenhagen, as well as association analyses of the 16 genotyped SERPINA1 SNPs in both the SAPALDIA discovery and replication arm, was carried out applying the same statistical model as in the GWAS apart from the adjustment for population stratification factors. Stepwise conditional analyses were conducted by testing each SNP for AAT association after including at each step the most significantly associated SNP in the model. As some of these SNPs turned out to be in unexpectedly high LD, we applied a threshold level for statistical significance of P = 0.005, accounting for approximately ten independent tests [65].
To be as close as possible to the calculations carried out in the original publication [14], multivariate linear regression models for lung function analyses were used adjusted for sex, age, height and population stratification factors (if available).
All the SAPALDIA regression analyses were performed with STATA 12.1 IC.
Further Software
Manhattan, Q-Q and forest plots were created with the help of R 2.15.1 (www.r-project.org). Regional association plots were drawn using LocusZoom [66]. Pairwise LD was calculated for HapMap2 and 1000G CEU data using SNAP [67]. The LD plot was produced with HaploView 4.2 [58]. The effect of non-synonymous SNPs on protein structure was predicted by SIFT [68]. Finally, Quanto 1.2.4 (hydra.usc.edu/gxe/) was used for power calculations for the GWAS.
Supporting Information
SAPALDIA study design for the determination of AAT associated genetic variants.a consisting of subjects with abnormally low AAT levels independent of PI S or Z alleles (see Materials and Methods).
(TIF)
Q-Q plot of genome-wide -log(10) p-values for association with AAT serum level.
(TIF)
Forest plot of meta-analyzed results for the effect per minor allele of rs17580 (PI S) on FEV1 in ever-smokers, adjusted for sex, age, height and population stratification factors. Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to the fixed effect meta-analyses.
(TIF)
Forest plot of meta-analyzed results for the effect per minor allele of rs28929474 (PI Z) on FEV1 in ever-smokers, adjusted for sex, age, height and population stratification factors. Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to the fixed effect meta-analyses.
(TIF)
Characteristics of SAPALDIA follow-up participants belonging to the discovery (N = 1392) and replication arm (N = 4245), and of participants of the Copenhagen City Heart Study (N = 8273).
(DOC)
Characteristics of study populations contributing to the SNP association analyses with FEV1.
(XLS)
The top 100 ranking SNPs associated with AAT serum level in SAPALDIA (N = 1392).
(DOC)
SERPINA regional variants based on 1000 Genomes imputation reaching statistical significance for the association with AAT serum level in SAPALDIA (N = 1392).
(DOC)
The top 100 ranking SNPs associated with AAT serum level, conditional on PI S and Z alleles in SAPALDIA (N = 1392).
(DOC)
Accuracy of 1000 Genomes based imputation in the SERPINA1 region in SAPALDIA (N = 1392).
(DOC)
Further variants in the SERPINA1 coding region, present in a SAPALDIA subsample with abnormally low AAT serum levels (N = 410).
(DOC)
Descriptions and acknowledgments of individual studies contributing to the SNP association analyses with FEV1.
(XLS)
Acknowledgments
The SAPALDIA Team:
Study directorate: T Rochat (p), NM Probst Hensch (e/g), JM Gaspoz (c), N Künzli (e/exp), C Schindler (s).
Scientific team: JC Barthélémy (c), W Berger (g), R Bettschart (p), A Bircher (a), G Bolognini (p), O Brändli (p), C Brombach (n), M Brutsche (p), L Burdet (p), M Frey (p), U Frey (pd), MW Gerbase (p), D Gold (e/c/p), E de Groot (c), W Karrer (p), R Keller (p), B Knöpfli (p), B Martin (pa), D Miedinger (o), U Neu (exp), L Nicod (p), M Pons (p), F Roche (c), T Rothe (p), E Russi (p), P Schmid-Grendelmeyer (a), A Schmidt-Trucksäss (pa), A Turk (p), J Schwartz (e), D. Stolz (p), P Straehl (exp), JM Tschopp (p), A von Eckardstein (cc), E Zemp Stutz (e).
Scientific team at coordinating centers: M Adam (e/g), PO Bridevaux (p), D Carballo (c), E Corradi (exp), I Curjuric (e), J Dratva (e), A Di Pasquale (s), L Grize (s), D Keidel (s), S Kriemler (pa), A Kumar (g), M Imboden (g), N Maire (s), A Mehta (e), F Meier (e), H Phuleria (exp), E Schaffner (s), GA Thun (g) A Ineichen (exp), M Ragettli (exp), M Ritter (exp), T Schikowski (e), G Stern (pd), M Tarantino (s), M Tsai (exp), M Wanner (pa)
(a) allergology, (c) cardiology, (cc) clinical chemistry, (e) epidemiology, (exp) exposure, (g) genetic and molecular biology, (m) meteorology, (n) nutrition, (o) occupational health, (p) pneumology, (pa) physical activity, (pd) pediatrics, (s) statistics.
Administrative staff: C Gabriel, R Gutknecht.
The study could not have been done without the help of the study participants, technical and administrative support and the medical teams and field workers at the local study sites.
Local fieldworkers: Aarau: S Brun, G Giger, M Sperisen, M Stahel. Basel: C Bürli, C Dahler, N Oertli, I Harreh, F Karrer, G Novicic, N Wyttenbacher. Davos: A Saner, P Senn, R Winzeler, Geneva: F Bonfils, B Blicharz, C Landolt, J Rochat. Lugano: S Boccia, E Gehrig, MT Mandia, G Solari, B Viscardi. Montana: AP Bieri, C Darioly, M Maire. Payerne: F Ding, P Danieli A Vonnez. Wald: D Bodmer, E Hochstrasser, R Kunz, C Meier, J Rakic, U Schafroth, A Walder.
Acknowledgments for additional studies are presented in Table S8.
Funding Statement
This SAPALDIA project was supported by the Swiss National Science Foundation (grants: 33CS30_134276/1, 33CSCO-108796, 3247BO-104283, 3247BO-104288, 3247BO-104284, 3247-065896, 3100-059302, 3200-052720, 3200-042532, 4026-028099, 3233-054996, PDFMP3-123171); the Federal Office for Forest, Environment and Landscape; the Federal Office of Public Health; the Federal Office of Roads and Transport; the canton's government of Aargau, Basel-Stadt, Basel-Land, Geneva, Luzern, Ticino, Valais and Zurich; the Swiss Lung League; the canton's Lung League of Basel Stadt, Basel Landschaft, Geneva, Ticino, Valais and Zurich; Schweizerische Unfallversicherungsanstalt (SUVA); Freiwillige Akademische Gesellschaft; UBS Wealth Foundation; Talecris Biotherapeutics GmbH; Grifols; Abbott Diagnostics; Kedrion S.p.A.; IRCCS (Istituto di ricovero e cura a carattere scientifico) Foundation San Matteo Hospital; and Cariplo Foundation 2006 projects. Genotyping in the GABRIEL framework was supported by European Commission (018996) and Wellcome Trust (WT 084703MA). Individual studies: The British 1958 Birth Cohort DNA collection was funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. Genotyping for the B58C-WTCCC subset was funded by the Wellcome Trust grant 076113/B/04/Z. The B58C-T1DGC genotyping utilized resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418. B58C-T1DGC GWAS data were deposited by the Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research (CIMR), University of Cambridge, which is funded by Juvenile Diabetes Research Foundation International, the Wellcome Trust and the National Institute for Health Research Cambridge Biomedical Research Centre; the CIMR is in receipt of a Wellcome Trust Strategic Award (079895). The B58C-GABRIEL genotyping was supported by a contract from the European Commission Framework Programme 6 (018996) and grants from the French Ministry of Research. The 1994–95 Busselton Health Study is funded by Healthway. The Copenhagen City Heart Study is funded by the Danish Heart Foundation and the Danish Lung Foundation. The CROATIA-Korcula, CROATIA-Vis and CROATIA-Split studies in the Croatian islands of Korcula and Vis and mainland city of Split were supported by grants from the Medical Research Council (UK); the Ministry of Science, Education, and Sport of the Republic of Croatia (grant number 108-1080315-0302); and the European Union framework program 6 European Special Populations Research Network project (contract LSHG-CT-2006-018947). The eQTL-Study, including for this project the University of British Columbia and the University of Groningen, was funded by Merck Research Laboratories. The KORA research platform (KORA, Cooperative Health Research in the Region of Augsburg) was initiated and financed by the Helmholtz Zentrum München - German Research Center for Environmental Health which is funded by the German Federal Ministry of Education and Research and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. Further support was provided by the Competence Network ASCONET, subnetwork COSYCONET (FKZ 01GI0882). The Lothian Birth Cohort 1936 data collection was funded by Research Into Ageing (Ref. 251). The work was undertaken by The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (Ref. G0700704/84698). The whole genome association study was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) (Ref. BB/F019394/1). Funding from the BBSRC, Engineering and Physical Sciences Research Council (EPSRC), Economic and Social Research Council (ESRC) and Medical Research Council (MRC) is gratefully acknowledged. The Lung Health Study was supported by contract N01-HR-46002 from the Division of Lung Diseases of the National Heart, Lung, and Blood Institute. The rs4905179 data were from Gene-Environment Association Studies (GENEVA). The Northern Finland Birth Cohort 1966 received financial support from the Academy of Finland (project grants 104781, 120315, 129269, 1114194, 24300796, Center of Excellence in Complex Disease Genetics and SALVE), University Hospital Oulu, Biocenter, University of Oulu, Finland (75617), NHLBI grant 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), NIH/NIMH (5R01MH63706:02), ENGAGE project and grant agreement HEALTH-F4-2007-201413, and the Medical Research Council, UK (G0500539, G0600705, G1002319, PrevMetSyn/SALVE). The DNA extractions, sample quality controls, biobank up-keeping and aliquotting was performed in the National Public Health Institute, Biomedicum Helsinki, Finland and supported financially by the Academy of Finland and Biocentrum Helsinki. The Northern Swedish Population Health Study was funded by the Swedish Medical Research Council (Project K2007-66X-20270-01-3), and the Foundation for Strategic Research. NSPHS, as part of European Special Populations Research Network, was also supported by European Commission Sixth Framework Programme Specific Targeted Research Projects Grant 01947 (LSHG-CT-2006-01947). The Orkney Complex Disease Study was supported by the Chief Scientist Office of the Scottish Government, the Royal Society, and the European Union Framework Programme 6 EUROSPAN project (contract LSHG-CT-2006-018947). The Study of Health in Pomerania is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grants no. 01ZZ9603, 01ZZ0103, and 01ZZ0403), the Ministry of Cultural Affairs, as well as the Social Ministry of the Federal State of Mecklenburg-West Pomerania, and the networks ‘Greifswald Approach to Individualized Medicine’ (GANI_MED, grant no. 03IS2061A) and COSYCONET (grant no. 01GI0883), both funded by the Federal Ministry of Education and Research. Genome-wide data have been supported by the Federal Ministry of Education and Research (grant no. 03ZIK012) and a joint grant from Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania. The University of Greifswald is a member of the ‘Center of Knowledge Interchange’ program of the Siemens AG and the Caché Campus program of the InterSystems GmbH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brantly ML, Wittes JT, Vogelmeier CF, Hubbard RC, Fells GA, et al. (1991) Use of a highly purified alpha 1-antitrypsin standard to establish ranges for the common normal and deficient alpha 1-antitrypsin phenotypes. Chest 100: 703–708. [DOI] [PubMed] [Google Scholar]
- 2. Lomas DA, Evans DL, Finch JT, Carrell RW (1992) The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature 357: 605–607. [DOI] [PubMed] [Google Scholar]
- 3. Luisetti M, Seersholm N (2004) Alpha1-antitrypsin deficiency. 1: epidemiology of alpha1-antitrypsin deficiency. Thorax 59: 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lieberman J, Winter B, Sastre A (1986) Alpha 1-antitrypsin Pi-types in 965 COPD patients. Chest 89: 370–373. [DOI] [PubMed] [Google Scholar]
- 5. American Thoracic Society/European Respiratory Society statement: standards for the diagnosis and management of individuals with alpha-1 antitrypsin deficiency. Am J Respir Crit Care Med 168: 818–900. [DOI] [PubMed] [Google Scholar]
- 6. Hersh CP, Dahl M, Ly NP, Berkey CS, Nordestgaard BG, et al. (2004) Chronic obstructive pulmonary disease in alpha1-antitrypsin PI MZ heterozygotes: a meta-analysis. Thorax 59: 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dahl M, Hersh CP, Ly NP, Berkey CS, Silverman EK, et al. (2005) The protease inhibitor PI*S allele and COPD: a meta-analysis. Eur Respir J 26: 67–76. [DOI] [PubMed] [Google Scholar]
- 8. Sorheim IC, Bakke P, Gulsvik A, Pillai SG, Johannessen A, et al. (2010) alpha-Antitrypsin protease inhibitor MZ heterozygosity is associated with airflow obstruction in two large cohorts. Chest 138: 1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thun GA, Ferrarotti I, Imboden M, Rochat T, Gerbase M, et al. (2012) SERPINA1 PiZ and PiS Heterozygotes and Lung Function Decline in the SAPALDIA Cohort. PLoS One 7: e42728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilk JB, Shrine NR, Loehr LR, Zhao JH, Manichaikul A, et al. (2012) Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med 186: 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Artigas MS, Loth DW, Wain LV, Gharib SA, Obeidat M, et al. (2011) Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet 43: 1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Imboden M, Bouzigon E, Curjuric I, Ramasamy A, Kumar A, et al. (2012) Genome-wide association study of lung function decline in adults with and without asthma. J Allergy Clin Immunol 129: 1218–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kong X, Cho MH, Anderson W, Coxson HO, Muller N, et al. (2011) Genome-wide Association Study Identifies BICD1 as a Susceptibility Gene for Emphysema. Am J Respir Crit Care Med 183: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Obeidat M, Wain LV, Shrine N, Kalsheker N, Artigas MS, et al. (2011) A comprehensive evaluation of potential lung function associated genes in the SpiroMeta general population sample. PLoS One 6: e19382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, et al. (2006) An overview of the serpin superfamily. Genome Biol 7: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB (2010) Rare variants create synthetic genome-wide associations. PLoS Biol 8: e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, et al. (2010) A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 363: 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferrarotti I, Thun GA, Zorzetto M, Ottaviani S, Imboden M, et al. (2012) Serum levels and genotype distribution of alpha1-antitrypsin in the general population. Thorax 67: 669–674. [DOI] [PubMed] [Google Scholar]
- 21. Zorzetto M, Russi E, Senn O, Imboden M, Ferrarotti I, et al. (2008) SERPINA1 gene variants in individuals from the general population with reduced alpha1-antitrypsin concentrations. Clin Chem 54: 1331–1338. [DOI] [PubMed] [Google Scholar]
- 22. Miyake K, Suzuki H, Oka H, Oda T, Harada S (1979) Distribution of alpha 1-antitrypsin phenotypes in Japanese: description of Pi M subtypes by isoelectric focusing. Jinrui Idengaku Zasshi 24: 55–62. [DOI] [PubMed] [Google Scholar]
- 23. Nukiwa T, Takahashi H, Brantly M, Courtney M, Crystal RG (1987) alpha 1-Antitrypsin nullGranite Falls, a nonexpressing alpha 1-antitrypsin gene associated with a frameshift to stop mutation in a coding exon. J Biol Chem 262: 11999–12004. [PubMed] [Google Scholar]
- 24. Graham A, Kalsheker NA, Newton CR, Bamforth FJ, Powell SJ, et al. (1989) Molecular characterisation of three alpha-1-antitrypsin deficiency variants: proteinase inhibitor (Pi) nullcardiff (Asp256----Val); PiMmalton (Phe51----deletion) and PiI (Arg39----Cys). Hum Genet 84: 55–58. [DOI] [PubMed] [Google Scholar]
- 25. Holmes MD, Brantly ML, Crystal RG (1990) Molecular analysis of the heterogeneity among the P-family of alpha-1-antitrypsin alleles. Am Rev Respir Dis 142: 1185–1192. [DOI] [PubMed] [Google Scholar]
- 26. Okayama H, Brantly M, Holmes M, Crystal RG (1991) Characterization of the molecular basis of the alpha 1-antitrypsin F allele. Am J Hum Genet 48: 1154–1158. [PMC free article] [PubMed] [Google Scholar]
- 27. Faber JP, Poller W, Weidinger S, Kirchgesser M, Schwaab R, et al. (1994) Identification and DNA sequence analysis of 15 new alpha 1-antitrypsin variants, including two PI*Q0 alleles and one deficient PI*M allele. Am J Hum Genet 55: 1113–1121. [PMC free article] [PubMed] [Google Scholar]
- 28. Poller W, Merklein F, Schneider-Rasp S, Haack A, Fechner H, et al. (1999) Molecular characterisation of the defective alpha 1-antitrypsin alleles PI Mwurzburg (Pro369Ser), Mheerlen (Pro369Leu), and Q0lisbon (Thr68Ile). Eur J Hum Genet 7: 321–331. [DOI] [PubMed] [Google Scholar]
- 29. Fra AM, Gooptu B, Ferrarotti I, Miranda E, Scabini R, et al. (2012) Three new alpha1-antitrypsin deficiency variants help to define a C-terminal region regulating conformational change and polymerization. PLoS One 7: e38405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gibson G (2011) Rare and common variants: twenty arguments. Nat Rev Genet 13: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanna S, Li B, Mulas A, Sidore C, Kang HM, et al. (2011) Fine mapping of five Loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet 7: e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oosterveer DM, Versmissen J, Defesche JC, Sivapalaratnam S, Yazdanpanah M, et al. (2013) Low-density lipoprotein receptor mutations generate synthetic genome-wide associations. Eur J Hum Genet 21: 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, et al. (2010) Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet 42: 1049–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, et al. (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43: 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, et al. (2010) Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet 42: 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coassin S, Schweiger M, Kloss-Brandstatter A, Lamina C, Haun M, et al. (2010) Investigation and functional characterization of rare genetic variants in the adipose triglyceride lipase in a large healthy working population. PLoS Genet 6: e1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kronenberg F, Utermann G (2013) Lipoprotein(a): resurrected by genetics. J Intern Med 273: 6–30. [DOI] [PubMed] [Google Scholar]
- 38. Lin JP, Schwaiger JP, Cupples LA, O'Donnell CJ, Zheng G, et al. (2009) Conditional linkage and genome-wide association studies identify UGT1A1 as a major gene for anti-atherogenic serum bilirubin levels–the Framingham Heart Study. Atherosclerosis 206: 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heid IM, Wagner SA, Gohlke H, Iglseder B, Mueller JC, et al. (2006) Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes 55: 375–384. [DOI] [PubMed] [Google Scholar]
- 40. Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, et al. (2009) Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet 41: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chappell S, Daly L, Morgan K, Guetta Baranes T, Roca J, et al. (2006) Cryptic haplotypes of SERPINA1 confer susceptibility to chronic obstructive pulmonary disease. Hum Mutat 27: 103–109. [DOI] [PubMed] [Google Scholar]
- 42. Silverman EK, Province MA, Rao DC, Pierce JA, Campbell EJ (1990) A family study of the variability of pulmonary function in alpha 1-antitrypsin deficiency. Quantitative phenotypes. Am Rev Respir Dis 142: 1015–1021. [DOI] [PubMed] [Google Scholar]
- 43. Senn O, Russi EW, Schindler C, Imboden M, von Eckardstein A, et al. (2008) Circulating alpha1-antitrypsin in the general population: determinants and association with lung function. Respir Res 9: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, et al. (2008) Genetics of gene expression and its effect on disease. Nature 452: 423–428. [DOI] [PubMed] [Google Scholar]
- 45. Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, et al. (2012) Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet 8: e1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Inouye M, Ripatti S, Kettunen J, Lyytikainen LP, Oksala N, et al. (2012) Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet 8: e1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lappalainen T, Montgomery SB, Nica AC, Dermitzakis ET (2011) Epistatic selection between coding and regulatory variation in human evolution and disease. Am J Hum Genet 89: 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Qiu W, Baccarelli A, Carey VJ, Boutaoui N, Bacherman H, et al. (2012) Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am J Respir Crit Care Med 185: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, et al. (2010) Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467: 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, et al. (2011) Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet 43: 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rollini P, Fournier RE (1999) The HNF-4/HNF-1alpha transactivation cascade regulates gene activity and chromatin structure of the human serine protease inhibitor gene cluster at 14q32.1. Proc Natl Acad Sci USA 96: 10308–10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Martin BW, Ackermann-Liebrich U, Leuenberger P, Kunzli N, Stutz EZ, et al. (1997) SAPALDIA: methods and participation in the cross-sectional part of the Swiss Study on Air Pollution and Lung Diseases in Adults. Soz Praventivmed 42: 67–84. [DOI] [PubMed] [Google Scholar]
- 53. Ackermann-Liebrich U, Kuna-Dibbert B, Probst-Hensch NM, Schindler C, Felber Dietrich D, et al. (2005) Follow-up of the Swiss Cohort Study on Air Pollution and Lung Diseases in Adults (SAPALDIA 2) 1991–2003: methods and characterization of participants. Soz Praventivmed 50: 245–263. [DOI] [PubMed] [Google Scholar]
- 54. Dahl M, Tybjaerg-Hansen A, Lange P, Vestbo J, Nordestgaard BG (2002) Change in lung function and morbidity from chronic obstructive pulmonary disease in alpha1-antitrypsin MZ heterozygotes: A longitudinal study of the general population. Ann Intern Med 136: 270–279. [DOI] [PubMed] [Google Scholar]
- 55. Sandford AJ, Chagani T, Weir TD, Connett JE, Anthonisen NR, et al. (2001) Susceptibility genes for rapid decline of lung function in the Lung Health Study. American Journal of Respiratory and Critical Care Medicine 163: 469–473. [DOI] [PubMed] [Google Scholar]
- 56. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Carter KW, McCaskie PA, Palmer LJ (2006) JLIN: a java based linkage disequilibrium plotter. BMC Bioinformatics 7: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 59. Hemminger BM, Saelim B, Sullivan PF (2006) TAMAL: an integrated approach to choosing SNPs for genetic studies of human complex traits. Bioinformatics 22: 626–627. [DOI] [PubMed] [Google Scholar]
- 60. Chappell S, Hadzic N, Stockley R, Guetta-Baranes T, Morgan K, et al. (2008) A polymorphism of the alpha1-antitrypsin gene represents a risk factor for liver disease. Hepatology 47: 127–132. [DOI] [PubMed] [Google Scholar]
- 61. Gorrini M, Ferrarotti I, Lupi A, Bosoni T, Mazzola P, et al. (2006) Validation of a rapid, simple method to measure alpha1-antitrypsin in human dried blood spots. Clin Chem 52: 899–901. [DOI] [PubMed] [Google Scholar]
- 62. Aulchenko YS, Struchalin MV, van Duijn CM (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 64. Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, et al. (2008) Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet 16: 1413–1429. [DOI] [PubMed] [Google Scholar]
- 65. Li J, Ji L (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95: 221–227. [DOI] [PubMed] [Google Scholar]
- 66. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, et al. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SAPALDIA study design for the determination of AAT associated genetic variants.a consisting of subjects with abnormally low AAT levels independent of PI S or Z alleles (see Materials and Methods).
(TIF)
Q-Q plot of genome-wide -log(10) p-values for association with AAT serum level.
(TIF)
Forest plot of meta-analyzed results for the effect per minor allele of rs17580 (PI S) on FEV1 in ever-smokers, adjusted for sex, age, height and population stratification factors. Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to the fixed effect meta-analyses.
(TIF)
Forest plot of meta-analyzed results for the effect per minor allele of rs28929474 (PI Z) on FEV1 in ever-smokers, adjusted for sex, age, height and population stratification factors. Studies based on population isolates with a high degree of cryptic relatedness are presented separately. Effect estimates of meta-analyses are shown with green diamonds. I2 is a measure of the heterogeneity between studies. Random effect meta-analyses are included if I2>0.5. Study weights (blue squares) correspond to the fixed effect meta-analyses.
(TIF)
Characteristics of SAPALDIA follow-up participants belonging to the discovery (N = 1392) and replication arm (N = 4245), and of participants of the Copenhagen City Heart Study (N = 8273).
(DOC)
Characteristics of study populations contributing to the SNP association analyses with FEV1.
(XLS)
The top 100 ranking SNPs associated with AAT serum level in SAPALDIA (N = 1392).
(DOC)
SERPINA regional variants based on 1000 Genomes imputation reaching statistical significance for the association with AAT serum level in SAPALDIA (N = 1392).
(DOC)
The top 100 ranking SNPs associated with AAT serum level, conditional on PI S and Z alleles in SAPALDIA (N = 1392).
(DOC)
Accuracy of 1000 Genomes based imputation in the SERPINA1 region in SAPALDIA (N = 1392).
(DOC)
Further variants in the SERPINA1 coding region, present in a SAPALDIA subsample with abnormally low AAT serum levels (N = 410).
(DOC)
Descriptions and acknowledgments of individual studies contributing to the SNP association analyses with FEV1.
(XLS)