Background: 3-Hydroxylation of proline residues in type I collagen is rare but important.
Results: 3-Hyp sites have been identified in both chains of mouse type I collagen in wild type and P3H1 null mice.
Conclusion: The absence of 3-Hyp does not alter the D-period of collagen fibrils, but alters the lateral growth of the fibrils.
Significance: Type I collagen prolyl 3-hydroxylation is tissue-specific.
Keywords: Collagen, Hydroxylysine, Hydroxyproline, Post translational modification, Protein synthesis, Tendon, Prolyl 3-hydroxylase 1, glycosyl hydroxylysine
Abstract
Type I collagen extracted from tendon, skin, and bone of wild type and prolyl 3-hydroxylase 1 (P3H1) null mice shows distinct patterns of 3-hydroxylation and glycosylation of hydroxylysine residues. The A1 site (Pro-986) in the α1-chain of type I collagen is almost completely 3-hydroxylated in every tissue of the wild type mice. In contrast, no 3-hydroxylation of this proline residue was found in P3H1 null mice. Partial 3-hydroxylation of the A3 site (Pro-707) was present in tendon and bone, but absent in skin in both α-chains of the wild type animals. Type I collagen extracted from bone of P3H1 null mice shows a large reduction in 3-hydroxylation of the A3 site in both α-chains, whereas type I collagen extracted from tendon of P3H1 null mice shows little difference as compared with wild type. These results demonstrate that the A1 site in type I collagen is exclusively 3-hydroxylated by P3H1, and presumably, this enzyme is required for the 3-hydroxylation of the A3 site of both α-chains in bone but not in tendon. The increase in glycosylation of hydroxylysine in P3H1 null mice in bone was found to be due to an increased occupancy of normally glycosylated sites. Despite the severe disorganization of collagen fibrils in adult tissues, the D-period of the fibrils is unchanged. Tendon fibrils of newborn P3H1 null mice are well organized with only a slight increase in diameter. The absence of 3-hydroxyproline and/or the increased glycosylation of hydroxylysine in type I collagen disturbs the lateral growth of the fibrils.
Introduction
Collagens are the most abundant proteins in vertebrate animals. Procollagen biosynthesis occurs in the rough endoplasmic reticulum of cells and involves a large number of posttranslational modifications (1). Two modifications of proline residues are known: 4(R)-hydroxyproline and 3(S)-hydroxyproline. The role of 4(R)-hydroxyproline is well established; it stabilizes the collagen triple helix, and the enzyme prolyl 4-hydroxylase is well characterized (2, 3). Although it has been known for a long time that type I collagen also contains 3-hydroxyproline residues (4), the enzyme activity was only partially characterized (5–7), and the enzymes were only recently identified (8). The prolyl 3-hydroxylases (P3Hs)2 belong to a family of three proteins, P3H1, P3H2, and P3H3, which share a common C-terminal dioxygenase domain, which is similar to lysyl hydroxylases and the α-subunit of prolyl 4-hydroxylase, and a unique N-terminal domain with four cysteine repeats (CXXXC) (8). This domain is homologous to cartilage-associated protein (CRTAP) and SC65. The function of this domain is not established, but it also contains tetratricopeptide repeat domains that are known to be important in protein-protein interactions (9, 10).
P3H1 forms a complex with CRTAP and cyclophilin B (CypB), and this complex was shown to 3-hydroxylate proline residue 986 of the α1-chain of type I collagen (8, 11). The P3H1·CRTAP·CypB complex is also a potent molecular chaperone for procollagen biosynthesis (12). The absence of P3H1 in mice leads to abnormalities in fibrillar collagen-rich tissues such as tendon, skin, and bone (13). Type I collagen from these mice lack the 3-hydroxyproline at position 986 and show increased glycosylation of hydroxylysine residues (13). Recessive osteogenesis imperfecta cases have been reported to be caused by mutations in CRTAP, P3H1, or CypB (14–18). The CRTAP null mice and the CypB null mice also show severe osteogenesis imperfecta, and both show the absence of 3-hydroxylation at position 986 (11, 19).
A comprehensive analysis of the location of 3-hydroxyproline sites in fibrillar collagens from human and bovine tissues showed the existence of additional sites (20). Four sites were identified in fibrillar collagens and termed the A1 site (position 986), the A2 site (position 944), the A3 site (position 707), and the A4 site (position 470). The α1-chain of type I collagen was shown to only contain a 3-hydroxyproline at the A1 site, which was 96–99% occupied. The α2-chain of type I collagen was 80% 3-hydroxylated at the A3 site. The α1-chain of type II collagen was 95–99% 3-hydroxylated at the A1 site and 10–87% 3-hydroxylated at the A2 site (20). The spatial arrangement of these sites, which are staggered by a D-period, suggested a fundamental role for these modifications in the supramolecular assembly by forming hydrogen bonds between adjacent collagen triple helices (20, 21). In rat tail tendon, a 3-Hyp-rich motif was found at the C-terminal end of the triple helix (22).
Glycosylation of hydroxylysine residues is a normal posttranslational modification observed in many collagens (23). The degree of glycosylation varies among different collagens (24, 25). For type I collagen, the degree of glycosylation also varies between different tissues.
Here we report that type I collagen extracted from wild type and P3H1 null mice tissues shows differences from the published data on human and bovine collagens, and we show that another prolyl 3-hydroxylase besides P3H1 also acts on type I collagen. We demonstrate that the D-period of collagen fibrils is not affected by the lack of 3-hydroxyproline or increasing amounts of hydroxylysine glycosylation. In addition, we see only small differences in the fibril diameter of newborn mice tail tendons as compared with the severe disorganization found in adult P3H1 null mice, implicating the lateral growth of fibrils in the phenotype.
EXPERIMENTAL PROCEDURES
Collagen Extraction from Tissues
Tendons and skin were taken from adult mice and pepsinized in 0.5 m acetic acid. Bone was first decalcified in 0.2 m EDTA in 50 mm Tris/HCl, pH 7.8, in a Pelco (Ted Pella, Inc., Redding, CA) 3450 microwave oven equipped with a 3430 variable power module. The high temperature limit set to 40 °C, and power was set to 99 watts. The EDTA solution was changed prior to each of eight 99-min cycles. Briefly, tissues were incubated at 4 °C in excess volumes of 0.5 m acetic acid with shaking for several hours. Pepsin was added to a final concentration of ∼1 mg/ml, and tissues were digested at 4 °C overnight. The solutions were centrifuged to remove insoluble material, and then NaCl was added to a final concentration of 0.7 m to precipitate collagens and the solution was incubated at 4 °C for 2–4 h. Precipitates were collected by centrifugation at 20,000 × g for 40 min and resuspended in 0.1 n acetic acid. Saturated Tris was added to bring collagen solutions to neutral pH, and then NaCl was added to a final concentration of 2.5 m to preferentially precipitate type I and III collagen. The solution was incubated at 4 °C for several hours and then centrifuged at 20,000 × g for 40 min to collect the collagen precipitates. The pellets were resolubilized in 0.1 m acetic acid, analyzed on SDS-PAGE gels, and lyophilized for further digestion and analysis. For the skin extracts, the type III collagen was precipitated with 1.6 m NaCl to separate it from type I collagen.
Collagen Digestion and MS Analysis
SDS-PAGE bands were subjected to in-gel digestion with trypsin using a protocol similar to that described by Ref. 26. Digest conditions were 13 ng/μl Promega trypsin in 100 mm ammonium bicarbonate at 37 °C for 18 h. Identification of tryptic peptides was performed on a Q-TOF Micro mass spectrometer (Waters, Billerica, MA) equipped with an electrospray ionization source. Data were collected with the MassLynx (version 4.1) data acquisition software (Waters) and processed using Mascot Distiller (Matrix Software, London, UK). High performance liquid chromatography was performed with a nanoACQUITY (Waters) system using a 75 μm × 100-mm 3-μm Atlantis dC18 column as the analytical column and a 180 μm × 20-mm 5-μm Symmetry C18 column as the trapping column. Chromatography mobile phases consisted of solvents A (0.1% formic acid and 99.9% water (v/v)) and B (0.1% formic acid and 99.9% acetonitrile (v/v)). Peptide samples were loaded onto the trapping column and equilibrated for 2 min in 99% solvent A followed by a 120-min gradient to 60% solvent A at a constant flow rate of 1 μl/min. Analysis was performed in survey scan mode. Tryptic peptides were identified from MS/MS spectra by a Mascot search against the National Center for Biotechnology Information (NCBI) nr database (peptide tolerance 1.0 Da, MS/MS tolerance 1.0 Da).
Quantification of glycosylated hydroxylysine residues in type I collagen was accomplished by initially identifying glycosylated peptides in the hydrazide extracted samples. These peptides were then located in raw MS data from nonextracted trypsin digests of type I collagen from P3H1 null and wild type mouse samples from skin, bone, and tail tendon. Extracted ion chromatograms were prepared for the peptides containing the hydroxylated lysine and unhydroxylated lysine of interest and the corresponding glycosylated peptides. Extracted ion chromatograms were integrated using the MassLynx software package. The average area ratios of the modified peptides for each lysine site are shown in Table 1. Verification of the identity of the peptides was achieved by examination of the MS/MS spectra for each peak of interest.
TABLE 1.
Relative abundance of lysine residues and posttranslationally modified lysine residues at position 174 of the α1-chain and α2-chain and position 219 of the α2-chain of type I collagen extracted from wild type and P3H1 null mice
The numbers represent the relative abundance of each species at each position and tissue when wild type and P3H1 null extracted collagens are compared. R indicates preceding amino-terminal sequence, and R′ indicates continuing C-terminal sequence.
| Mouse | Type I | Site | R-Lys | R-Lys(OH) | R-Lys(OH)-R′ | R-Lys(Gal)-R′ | R-Lys(GluGal)-R′ | |
|---|---|---|---|---|---|---|---|---|
| Tendon | WT | α1 | Lys-174 | 0.39 | 0.10 | 0.42 | 0.09 | 0.01 |
| Tendon | Null | α1 | Lys-174 | 0.20 | 0.09 | 0.46 | 0.19 | 0.06 |
| Tendon | WT | α2 | Lys-174 | 0.18 | 0.16 | 0.58 | 0.07 | 0.01 |
| Tendon | Null | α2 | Lys-174 | 0.13 | 0.16 | 0.44 | 0.20 | 0.07 |
| Tendon | WT | α2 | Lys-219 | 0.47 | 0.23 | 0.00 | 0.27 | 0.03 |
| Tendon | Null | α2 | Lys-219 | 0.31 | 0.36 | 0.00 | 0.23 | 0.10 |
| Skin | WT | α1 | Lys-174 | 0.19 | 0.25 | 0.20 | 0.31 | 0.04 |
| Skin | Null | α1 | Lys-174 | 0.17 | 0.24 | 0.17 | 0.22 | 0.23 |
| Skin | WT | α2 | Lys-174 | 0.33 | 0.16 | 0.13 | 0.26 | 0.12 |
| Skin | Null | α2 | Lys-174 | 0.19 | 0.05 | 0.11 | 0.43 | 0.23 |
| Skin | WT | α2 | Lys-219 | 0.23 | 0.27 | 0.00 | 0.07 | 0.43 |
| Skin | Null | α2 | Lys-219 | 0.30 | 0.32 | 0.00 | 0.07 | 0.30 |
| Bone | WT | α1 | Lys-174 | 0.30 | 0.25 | 0.25 | 0.16 | 0.04 |
| Bone | Null | α1 | Lys-174 | 0.17 | 0.17 | 0.25 | 0.29 | 0.13 |
| Bone | WT | α2 | Lys-174 | 0.10 | 0.13 | 0.39 | 0.26 | 0.08 |
| Bone | Null | α2 | Lys-174 | 0.18 | 0.14 | 0.17 | 0.34 | 0.17 |
| Bone | WT | α2 | Lys-219 | 0.35 | 0.43 | 0.00 | 0.11 | 0.11 |
| Bone | Null | α2 | Lys-219 | 0.18 | 0.46 | 0.00 | 0.16 | 0.23 |
N-terminal Sequencing
1 m Tris was added to solubilized collagen extracts to adjust the pH to 8, and the solutions were incubated at 50 °C for 1 h. The temperature was reduced to 37 °C, and sequencing grade trypsin was added. Collagen samples were incubated at 37 °C for 18 h. Trypsin digests of collagen were loaded onto Oasis HLB solid phase extraction cartridges, washed with 0.1 n acetic acid, and eluted with 40%/60% acetonitrile, 0.1% trifluoroacetic acid. After vacuum evaporation, the tryptic digests of collagen were reconstituted in 0.1% trifluoroacetic acid. Peptides were isolated by reverse phase HPLC on a 1090 liquid chromatograph (Agilent, Santa Clara, CA) with a 2.1 mm × 50-mm 5-μm ZORBAX 300 SBC-18 analytical column. Fractions were collected in 0.5-min intervals, dried by rotary evaporation, and reconstituted in 0.1% formic acid in water before analysis by LC-MS/MS using the method described above. Peptides in each fraction were identified by Mascot search using the conditions described above. HPLC conditions on the 1090 liquid chromatograph were adjusted so that a single chain contained peptides identified as containing a 3-Hyp modification from a single isoform of type I collagen. Two fractions, one containing the α1 A3 3-Hyp site and one containing the α2 A3 3-Hyp site, were analyzed by N-terminal sequencing. Peptide fractions containing the 3-Hyp site were applied to Applied Biosystems ProSorb cartridges (Applied Biosystems. Foster City, CA) and coated with 3 μl of diluted Biobrene solution (20 μg/μl) (Applied Biosystems). Edman sequencing was performed on an Applied Biosystems Procise 492 protein sequencer (Applied Biosystems). Sequencing data for these peptide fractions were processed using the SequencePro 2.1 software (Applied Biosystems). 3-Hyp and 4-Hyp phenylthiohydantoin standards were prepared by manual Edman chemistry using 3-Hyp and 4-Hyp dried standards purchased from Fluka Chemie (Buchs, Switzerland) and Aldrich, respectively. These were then added to phenylthiohydantoin-amino acid standard solution (Applied Biosystems) in appropriate concentrations for an injection standard.
Amino Acid Analysis
Acid hydrolysis was performed in 6 × 50-mm Pyrex culture tubes placed in Pico Tag reaction vessels fitted with a sealable cap (Eldex Laboratories, Inc., Napa, CA). Samples were placed in culture tubes, dried in a SpeedVac (GMI, Inc. Albertsville, MN), and then placed into a reaction vessel that contained 250 ml of 6 n HCl (Pierce) containing 2% phenol (Sigma-Aldrich). The vessel was then purged with argon gas and vacuumed using the automated evacuation work station Eldex hydrolysis/derivatization work station (Eldex Laboratories, Inc.). Closing the valve on the Pico Tag cap maintained the vacuum during hydrolysis at 110 °C for 24 h. The hydrolyzed samples were then dried in a Savant SpeedVac. The dried samples were dissolved in 100 ml of 0.02 n HCl containing an internal standard (100 μm norvaline; Sigma). Appropriate further dilutions were made using the same dilution solvent for concentrated samples. Analysis was performed by ion exchange chromatography with postcolumn ninhydrin derivatization and visible detection (440 nm/570 nm) with a Hitachi l-8800A amino acid analyzer (Hitachi High Technologies America, Inc., San Jose, CA) running the EZChrom Elite software (Scientific Software, Inc., Pleasanton, CA).
Mouse Tissue Preparation for RNA Extraction
Bone, skin, and tail tendon were isolated from wild type and P3H1 null 1-month-old mice and stored at −20 °C in RNAlater stabilization solution (Qiagen). Mouse hind limb bones, femur and tibia, were cut from both ends and extensively washed with PBS to remove bone marrow.
Real-time Quantitative PCR
Total mouse RNA was extracted from bone, skin, and tail tendon of wild type and P3H1 null mice using RNeasy fibrous tissue mini kit (Qiagen). RNA quality was validated by 260/280 absorption ratio, and concentration was estimated. Each representative RNA was reverse-transcribed using Superscript reverse transcriptase III (Invitrogen) according to the manufacturer's instructions with random hexamers (Invitrogen). Resulting cDNA was used as the template in real-time quantitative PCR experiments with the P3H1, P3H2, or P3H3 or GAPDH gene-specific oligonucleotides and the iQ SYBR Green supermix according to the manufacturer's instructions (Bio-Rad). The following primer pairs were used for each gene amplification: P3H1, CCCCCAGCCTACACGTTCCG and TGCCCCCTGGTGACAGCCTTC; P3H2, ATGCTAAAACCGTGACTGCC and CGCTCCAGTTCTCGGTAAAG; P3H3, AGAGGGCCTACTACCAGTTG and GTGTTGGGTTTGCCACAAAG; GAPDH, CCACCCAGAAGACTGTGGAT and TTCAGCTCTGGGATGACCTT.
Quantitative PCR data were measured and analyzed using the iQ5 multicolor real-time PCR detection system and software version 2.0 (Bio-Rad). Each PCR reaction was 25 μl. Because SYBR Green binding is not sequence-specific, careful validation of the primer pairs was undertaken to ensure that only the target gene sequence was amplified. To verify the specificity of each amplicon, a single gel band of the expected size was amplified for each primer pair, and melting curve analysis confirmed the presence of a single PCR product. -Fold differences in target genes were normalized to GAPDH expression in each tissue where indicated (ΔΔCT method). Data analysis was performed using the iQ5 optical system software version 2.0.
Analysis of Glycosylation
To verify the presence of sugar modifications, the glycosylated peptides were isolated using hydrazide chemistry (27). Galactose oxidase was bound to Sepharose 4B under basic conditions. Type I collagen from wild type and P3H1 null mouse tendon and bone was digested with trypsin at 37 °C for 18 h. The lyophilized digests were reconstituted in 0.1 m sodium phosphate buffer, pH 7.2, containing 0.15 m NaCl. 115 units of catalase (from bovine liver) and peroxidase (from horseradish) were added in a Bio-Rad Poly-Prep column and allowed to incubate overnight at 37 °C. The flow-through of the columns containing oxidized and unoxidized tryptic peptides of collagen was collected and adjusted to pH 5.5 with 1 m HCl. The hydrazide resin was washed with 0.1 m sodium acetate, pH 5.5, containing 0.15 m NaCl, and the flow-through was added to the resin in a Poly-Prep column. The solution was incubated overnight at 37 °C. The columns were then washed with 0.1 m sodium acetate, pH 5.5, containing 0.15 m NaCl followed by 0.15 m NaCl in water, then methanol, and finally water. The washed hydrazide gel bound to the oxidized galactose containing peptides was incubated with 0.1% formic acid for 1 h at 37 °C. The flow-through containing the oxidized peptides was collected and reduced with 1 mm sodium borohydride for 1 h at room temperature under basic conditions. The solution was then lyophilized, and the residue was analyzed by LC-MS using the method described above. Peptides containing glycosylation sites observed in the tryptic digests were also present in the hydrazide extracted samples.
P3H1 Null Mice
P3H1 null mice were purchased from Deltagen (San Mateo, CA). Directed knockouts were created in which exons 1–3 (nucleotides 15–817) of the mouse P3H1 or lepre1 (leprecan 1) gene (NCBI Reference sequence number: NM_019782.2) were deleted. Then a LacZ-Neo cassette was inserted into the area of the target gene that was deleted. P3H1 or lepre1 (leprecan 1) gene inactivation was verified in mice by RNA preparation from tissues of null mice followed by reverse transcription and PCR with primer sets spanning the length of the target gene. No P3H1 transcripts or gene expression were detected, as compared with normal levels of the target gene expressed in wild type animals. Mice were bred multiple generations into a C57B6 background prior to analysis to verify phenotype effects, and data reported are from 8th to 10th generation (13).
Electron Microscopy Analysis of P5 Tendon
Freshly obtained tendons were fixed in cacodylate-buffered 1.5% glutaraldehyde, 1.5% paraformaldehyde containing 0.05% tannic acid (w/v) and then rinsed, exposed to 1% osmium tetroxide, and dehydrated in a grade series of ethanol to 100%. Fixed tissues were rinsed in propylene oxide and infiltrated and embedded in Spurr's epoxy. 80-nm ultrathin sections were mounted on Formvar-coated single hole slot grids and stained in ethanolic uranyl acetate followed by Reynold's lead citrate. Stained sections were examined using an FEI Tecnai G2 electron microscope operated at 120 kV and photographed using either an FEI Eagle 2K camera or an AMT 2K camera. Magnifications were calibrated using a grating replica (Ted Pella catalogue number 603).
Small Angle X-ray Diffraction
The small angle x-ray diffraction measurements were carried out in the BL40B2 beamline of SPring-8 (Hyogo, Japan). Collagen fibers were dissected from mouse tail and immersed in phosphate-buffered saline. A picked collagen thread (diameter 0.3–0.8 mm) was clamped on a small metal holder and quickly set to the goniometer on the small angle x-ray diffraction equipment with the collagen fiber axis vertical. The two-dimensional small angle x-ray diffraction patterns were recorded with RAXIS IV++ (Rigaku Co., Japan). The camera length and x-ray wavelength λ were set to 1151 mm and 0.1 nm, respectively. The scattering intensity was accumulated for 30–120 s and all collagen samples were wet well during measurement. One-dimensional diffraction profiles were calculated by circular integrating the two-dimensional data over 20 degree fan-shaped sectors including the low angle meridional diffraction pattern as the function of scattering vector s, where s = 2sinθ/λ (θ = half of the scattering angle). Each higher order diffraction peak (from 2nd to 36th) of the D-stagger of collagen samples was fitted with Pseudo Voigt peak function to estimate full width of half-maximum (FWHM).
RESULTS
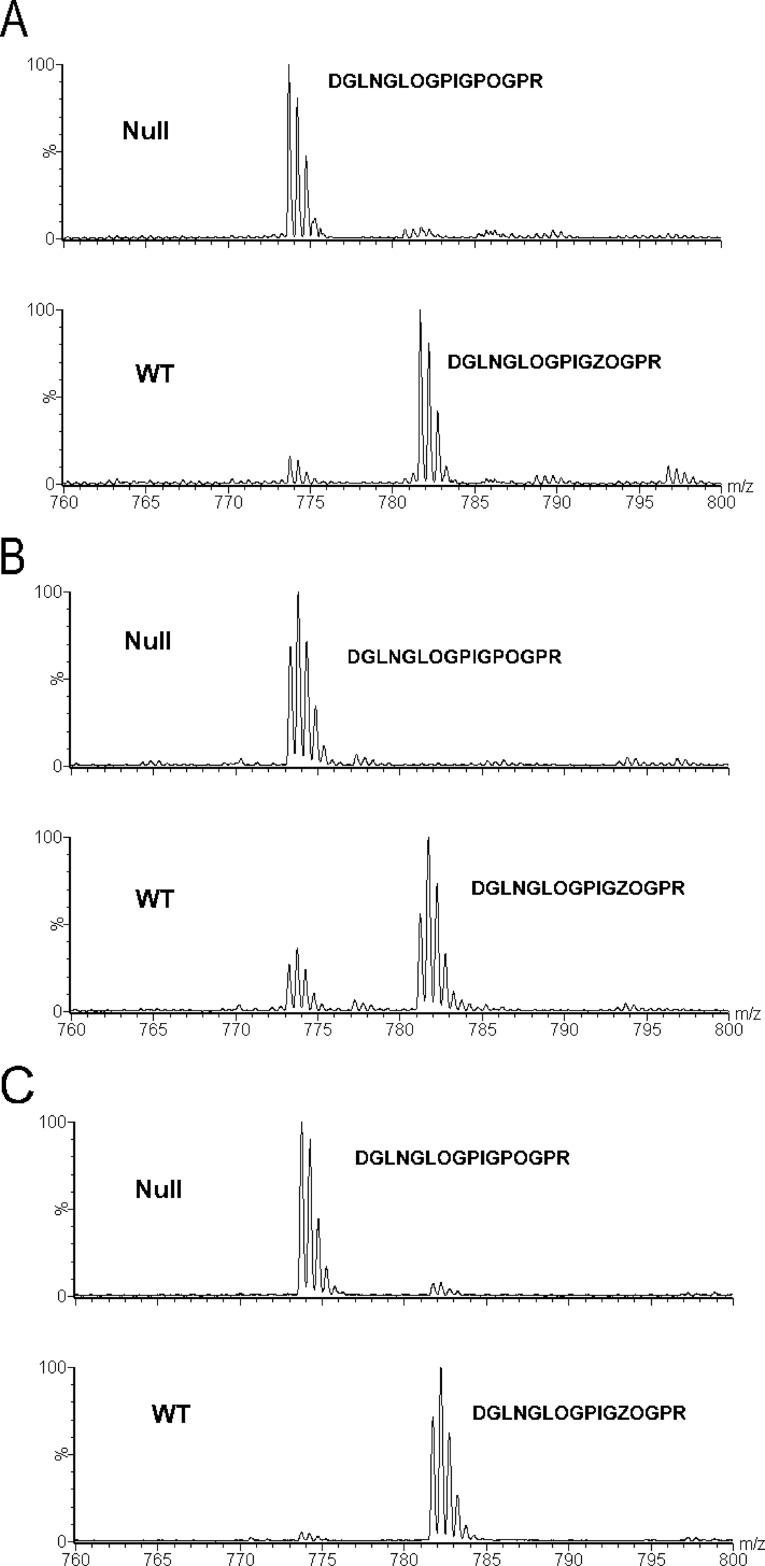
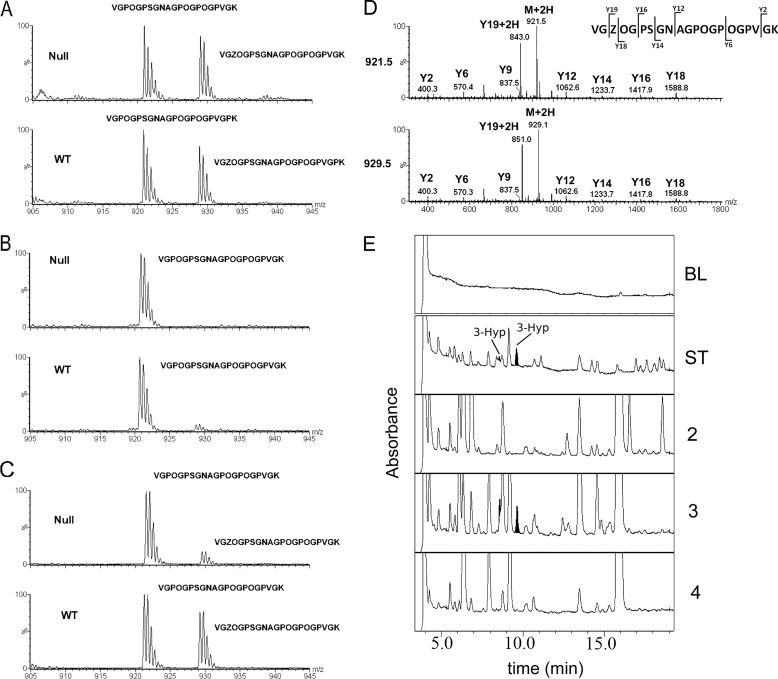
The A1 site (Pro-986) of the α1-chain of type I collagen is 3-hydroxylated almost completely in tendon and bone and to a high degree in skin of wild type mice. Type I collagen extracted from the same tissues of the P3H1 null mice show a complete absence of 3-hydroxylation at this site (Fig. 1). A tissue-specific 3-hydroxylation was found for the A3 site (Pro-707) in the α1-chain. In tendon and bone of wild type mice, this site was about 50% 3-hydroxylated, whereas in skin, the 3-hydroxylated peptide was not detectable. The only difference found in tissues extracted from P3H1 null mice at this site was the lesser extent of 3-hydroxylation in bone (Fig. 2).
FIGURE 1.
Analysis of the A1 site (Pro-986) of the α1-chain of type I collagen. A–C, the α1-chain of type I collagen extracted from tendon (A), skin (B), and bone (C) was analyzed by mass spectrometry. The occurrences of the tryptic peptides DGLNGLOGPIGPOGPR (mass = 1548 Da) and DGLNGLOGPIGZOGPR (mass = 1564 Da) are shown from wild type and P3H1 null mice. O is 4(R)Hyp, and Z is 3(S)Hyp. Panel A was previously published in Ref. 13.
FIGURE 2.
Analysis of the A3 site (Pro-707) of the α1-chain of type I collagen. A–C, the α1-chain of type I collagen extracted from tendon (A), skin (B), and bone (C) was analyzed by mass spectrometry. The occurrences of the tryptic peptides VGPOGPSGNAGPOGPOGPVGK and VGZOGPSGNAGPOGPOGPVGK are shown from wild type and P3H1 null mice. D, MS/MS spectra of these peptides of the α1-chain of type I collagen are shown. The Y19 + 2H ion shows a mass difference of 8 daltons, corresponding to the hydroxylation of Pro-707 (A3 site). Sequencing identifies this hydroxylation as 3-hydroxyproline. E, N-terminal sequencing cycles for the A3 site are shown. 3-Hyp residues are highlighted to confirm the presence in the cycle. BL, base line; ST, standards; the cycle numbers are indicated (2–4). O is 4(R)Hyp, and Z is 3(S)Hyp.
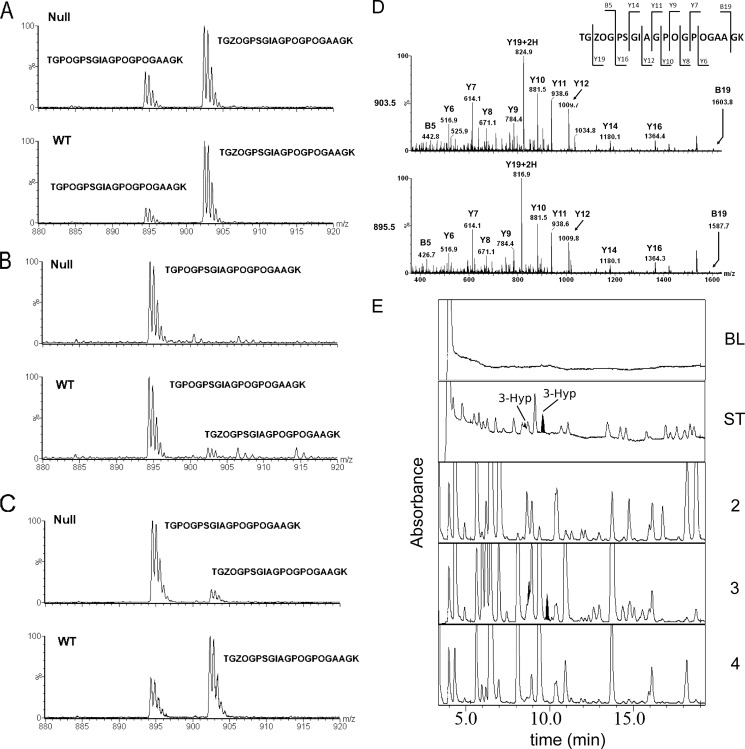
The α2-chain of type I collagen lacked 3-hydroxylation of the A1 site in all tissues because the corresponding sequence is Gly-Pro-Ala instead of the required Gly-Pro-Hyp. In tendon and bone of wild type mice, a substantial amount of 3-hydroxylation was observed at the A3 site (Pro-707). Skin had very little 3-hydroxylation at this site. In tendon extracted from P3H1 null mice, no significant reduction in 3-hydroxylation was observed. However, in bone, a significant decrease in 3-hydroxylation at this site was found (Fig. 3) in knockouts. Schematic representation of 3-hydroxylation sites in both chains of type I collagen is shown in Fig. 4.
FIGURE 3.
Analysis of the A3 site (Pro-707) of the α2-chain of type I collagen. A–C, the α2-chain of type I collagen extracted from tendon (A), skin (B), and bone (C) was analyzed by mass spectrometry. The occurrences of the tryptic peptides TGPOGPSGIAGPOGPOGAAGK and TGZOGPSGIAGPOGPOGAAGK are shown from wild type and P3H1 null mice. D, MS/MS spectra of the TGZOGPSGIAGPOGPOGAAGK and TGPOGPSGIAGPOGPOGAAGK peptides of the α2-chain of type I collagen are shown. The Y19 + 2H ion shows a mass difference of 8 daltons, corresponding to the hydroxylation of Pro-707 (A3 site). E, N-terminal sequencing identifies this hydroxylation as 3-hydroxyproline. 3-Hyp residues are highlighted to confirm the presence in the cycle. BL, base line; ST, standards; the cycle numbers are indicated (2–4). O is 4(R)Hyp, and Z is 3(S)Hyp.
FIGURE 4.
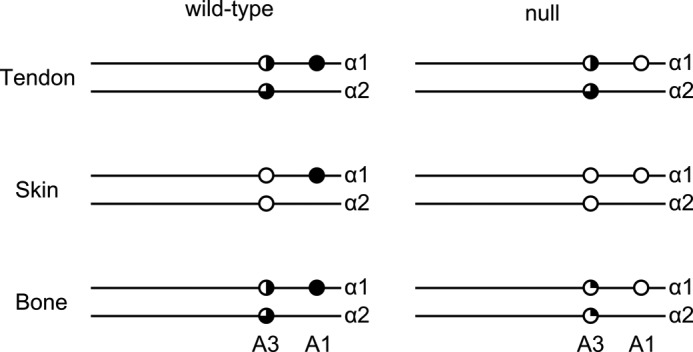
Schematic representation of the A1 and A3 3-hydroxylation ratios of type I collagen from skin, tendon and bone of wild type and P3H1 null mice. White circles indicate no 3-hydroxylation, and black circle indicates full 3-hydroxylation. Partial 3-hydroxylation is indicated by blackened quarters.
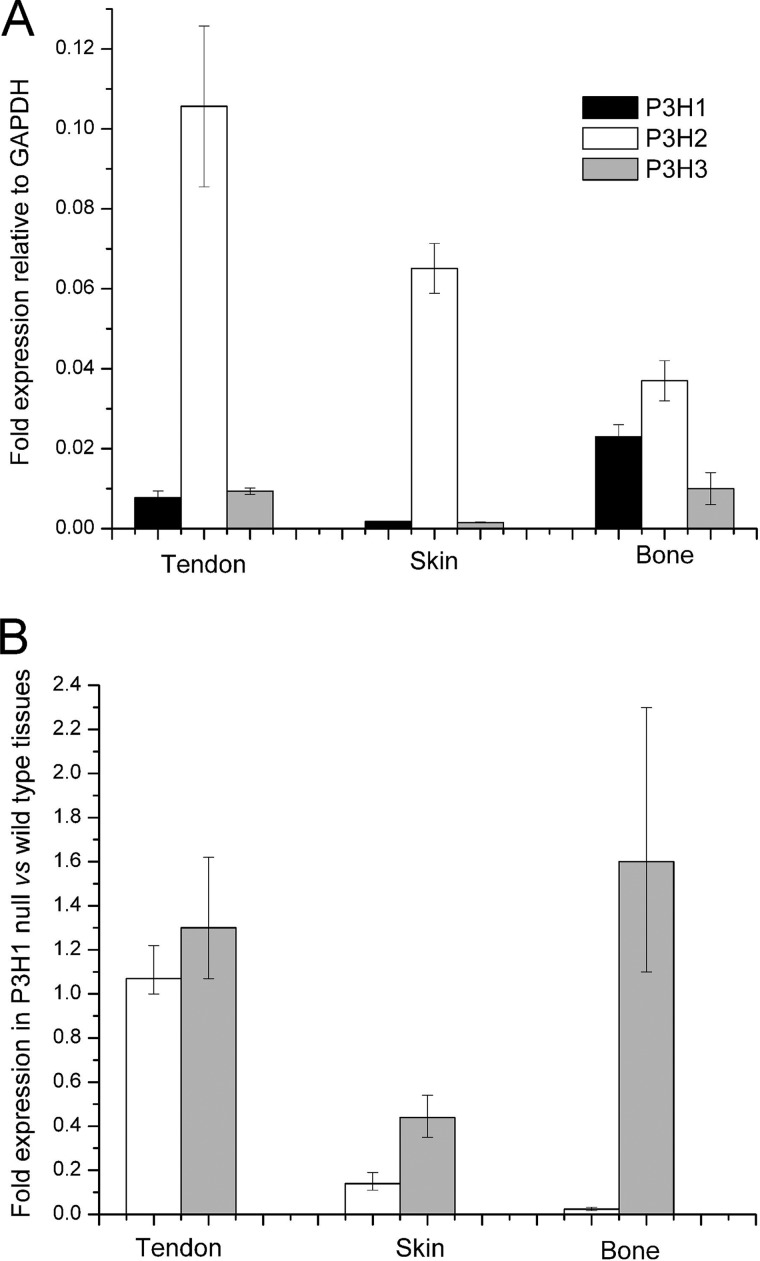
To investigate the tissue-specific expression levels of the P3H1, P3H2, and P3H3 enzymes in bone, skin, and tendon of mutant and wild type mice, quantitative real-time PCRs were performed. -Fold expression relative to GAPDH of the three enzymes in wild type tissues is shown in Fig. 5A. The most highly expressed enzyme type was P3H2. This finding is consistent with the previously published data for bone and skin (28). P3H1 was up-regulated in bone as compared with skin and tendon.
FIGURE 5.
Graphical representation of real-time quantitative PCR analysis of adult mouse bone, skin, and tendon. Data are represented as the -fold change differences. A shows relative target gene expression to GAPDH signal. B shows -fold change expression of P3H2 and P3H3 genes in P3H1 null relative to wild type control tissues. All experiments were performed in triplicate, and mean values are represented on each graph. P3H1 gene expression is represented by the black boxes, P3H2 is in white, and P3H3 is in gray. Error bars indicate S.D.
-Fold expression of the P3H2 and P3H3 in the P3H1 null tissues relative to the wild type controls is represented in Fig. 5B. A dramatic 40-fold reduction in the P3H2 expression level was observed in bone of the P3H1 null. In mutant skin, the P3H2 expression level was also reduced to about one-fifth of that of the control. However, in tendon of the P3H1 null mice, expression of the P3H2 was 100% preserved as compared with wild types. Expression of the P3H3 was slightly up-regulated in P3H1 null bone and tendon but down-regulated in skin.
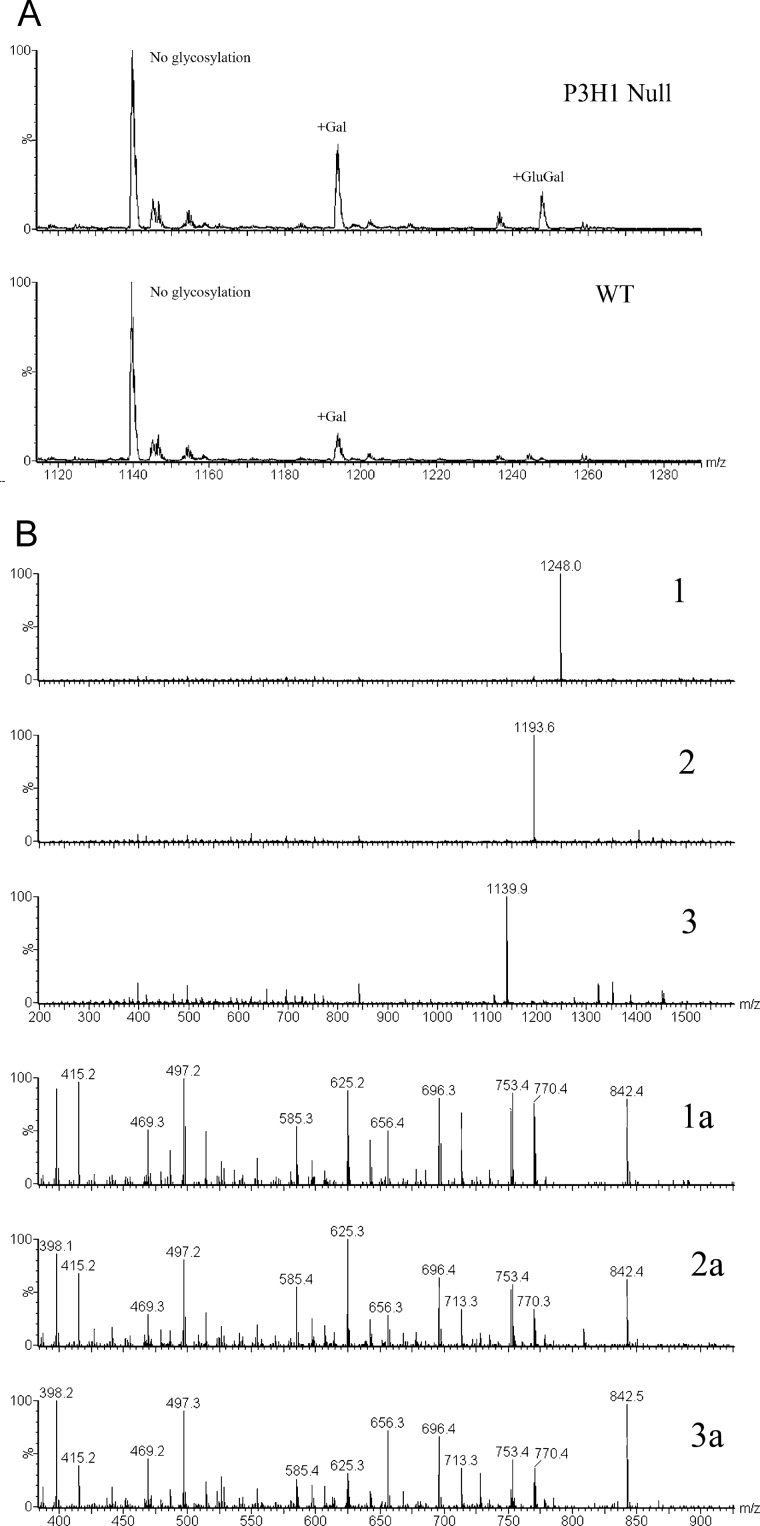
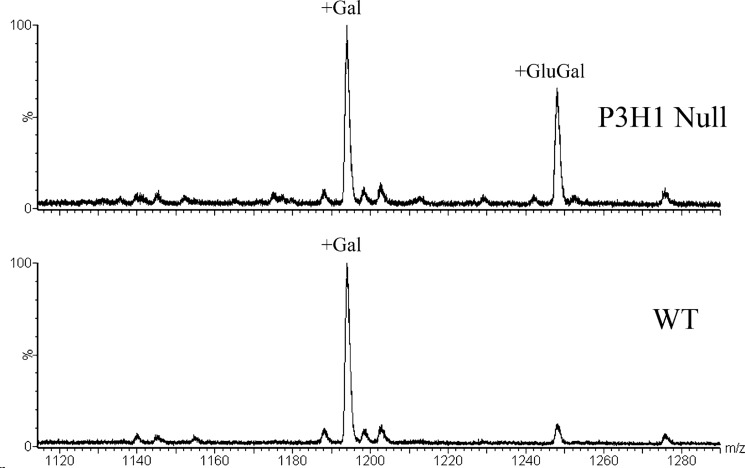
Several glycosylation sites were identified in mouse type I collagen. It was shown previously that type I collagen in P3H1 null mice has increased levels of hydroxylysine and glycosylated hydroxylysines (13). However, it was not clear whether this increase is due to additional hydroxylation and glycosylation sites or due to an increased occupancy of existing modifications. Fig. 6 shows a comparison of the posttranslational modification of Lys-174 of the α1-chain of type I collagen of wild type and P3H1 null mice. There is a clear increase in the amount of galactosyl hydroxylysine and the existence of glucosyl galactosyl hydroxylysine at this position in the P3H1 null mouse. The increase in these glycosylations is confirmed by the analysis of tryptic peptides of the α1-chain of tendon type I collagen using hydrazide extraction to enrich for glycopeptides. Fig. 7 shows the relative occurrence of the galactosyl hydroxylysine and the glucosyl galactosyl hydroxylysine at this position in wild types and P3H1 nulls. Table 1 lists the occupancies of hydroxylysine modifications for a number of locations and tissues.
FIGURE 6.
MS spectra of trypsin digested mouse tendon type I collagen. A, the peptide GNDGAVGAAGPPGPTGPTGPPGFPGAVGAK(174)GEAGPQGAR is shown for P3H1 null and wild type extracts from tendon. The relative abundance of glycosylated peptide in collagen extracted from P3H1 null mice is much higher. B, MS/MS data for the glycosylated peptide GNDGAVGAAGPPGPTGPTGPPGFPGAVGAK(174)GEAGPQGAR. Spectra labeled 1, 2, and 3 show the partially fragmented 3+ charge state precursor ions for the glucosyl galactosyl, galactosyl, and non-sugar-containing peptides of Lys-174 respectively. Spectra labeled 1a, 2a, and 2c show an expanded view of the previous peptides at the low mass region containing the same identifying fragments for all three peptides.
FIGURE 7.
MS spectra of trypsin digested mouse tendon type I collagen purified by hydrazide extraction. Only the glycosylated variants of the peptide GNDGAVGAAGPPGPTGPTGPPGFPGAVGAK(174)GEAGPQGAR are observed by this method. The relative abundance of the two forms of glycosylation from one extract can be compared, but not the ratio between two extracts.
Tendons from adult wild type and P3H1 null mice were analyzed by small angle x-ray scattering. X-ray diffraction patterns of both wild type and P3H1 null mice tendon show the set of meridional reflections parallel to the fiber axis. These reflections provide information about the axial order of the collagen fibrils. From this experiment, the D-period of the fibrils was determined to be 66.16 and 66.11 nm, for wild type and P3H1 null, respectively. Because there is no significant difference between them, it is suggested that the basic molecular packing of collagen molecules along the fibril axis is the same. Fig. 8 shows the s2 dependence of FWHMs for diffraction peaks of D-period of two mouse types. The line broadening of the diffraction peaks is due to not only the instrumental broadening but also to the small size of diffracting coherent domains and to the lattice distortions generated by imperfections. Because the instrumental broadening does not change between different samples measured under the same condition, the larger FWHMs indicate that the coherent domain size is smaller and/or the lattice distortions are larger. When the line width is plotted against s2, the value obtained by extrapolation to s = 0 can be interpreted in terms of the crystal size effect, whereas the slope of such a plot can be related to the extent of lattice distortion (29). We easily perceive that the two extrapolated values from line width are almost the same (slightly larger for P3H1 null), but the slope of P3H1 null mice data is larger than that of wild type. The larger slope of P3H1 null mouse is caused by the larger D-stagger distortions. In other words, collagen fibrils of null mouse tendon formed more disordered packing as compared with wild type tendon.
FIGURE 8.
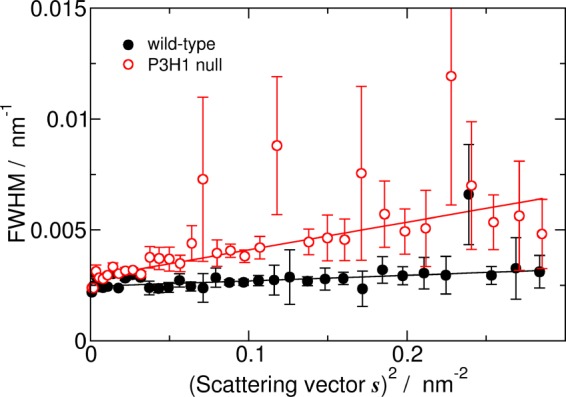
s2 dependence of FWHMs for diffraction peaks of D-stagger. The FWHMs of low angle meridional x-ray diffraction peaks were estimated by fitting with pseudo Voigt peak function. The black closed circles represent the FWHM from diffraction of wild type mouse tendon, whereas the red open circles represent that of P3H1 null mouse tendon. Error bars indicate S.D.
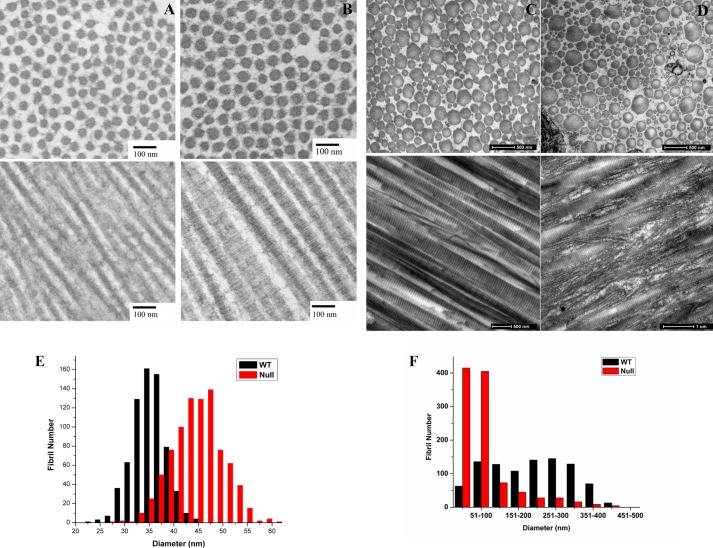
Tail tendons from newborn wild type and P3H1 null mice (P5) were analyzed by electron microscopy. The diameter of these fibrils is rather uniform, and the P3H1 null mouse tendons have a slightly larger diameter (Fig. 9). There is no difference in the repeat period of the fibrils when measured from electron microscopy images (data not shown). The tendon fibrils of adult P3H1 null mice show a complete disorganization with large fibrils splitting into smaller structures and the merging of smaller fibrils into larger ones.
FIGURE 9.
Electron micrographs of tendon fibrils of wild type and P3H1 null mice. A and B, tendon fibrils of wild type (A) and P3H1 null (B) mice at P5 in cross sections and longitudinal sections and the distribution of measured diameters from the cross sections underneath. The length of the bars in the micrographs corresponds to 100 nm. C and D, tendon fibrils of adult wild type (C) and P3H1 null mice (D) in cross sections and longitudinal sections. The length of the bars corresponds to 500 nm. E and F, the diameter distribution from cross sections is given for P5 (E) and for adult mice (F). Panels C and D are similar to fields published previously, and panel F is taken from Fig. 2E in Ref. 13.
DISCUSSION
The proline 3-hydroxylation patterns of the mouse α1- and α2-chains of type I collagen extracted from tendon, skin, and bone were analyzed and are summarized in Fig. 4. The A1 site (Pro-986) of the α1-chain is fully 3-hydroxylated. The lack of 3-hydroxylation of proline residue 986 in type I collagen extracted from P3H1 null tissues clearly demonstrates that P3H1 is solely responsible for the modification of this site. The A3 site exhibited a tissue-specific degree of 3-hydroxylation and variable disappearance in the absence of the P3H1. This places one of the other two enzymes, P3H2 or P3H3, in the position to fulfill the function of this site 3-hydroxylation. In a recent study (30) based on cell culture experiments, the suppression of P3H2 by siRNA was associated with the sharp decrease in 3-hydroxylation of the A3 site in the α-2 chain of type I collagen. Our quantitative real-time PCR results, which summarize tissue-specific expression patters of the prolyl 3-hydroxylase family members, lead to the same conclusion. As shown in Fig. 5B, the expression level of P3H2 drops dramatically in the P3H1 null bone where 3-Hyp at the A3 site is drastically reduced. At the same time, P3H2 is fully expressed in the P3H1 null tendon where the 3-hydroxylation level of the A3 site remains very similar to that of the wild type. In the P3H1 null bone, the expression of P3H3 is slightly up-regulated, perhaps in attempt to compensate for the loss of the P3H2. In P3H1 null skin, both P3H2 and P3H3 expression levels are somewhat reduced. This, however, does not have an effect for modification of the A3 site because it already lacks 3-hydroxylation in the normal skin (Fig. 4). Thus, P3H2 plays an important role in modification of the A3 site in type I collagen.
The increase in glycosylation of hydroxylysine, observed quantitatively by base hydrolysis and amino acid analysis in P3H1 null mice (13), was analyzed at Lys-174 of the α1-chain and Lys-174 and Lys-219 of the α2-chain in various tissues. In almost all cases, a significant increase of glucosyl galactosyl hydroxylysine was found. In many cases, the level of galactosyl hydroxylysine is also increased. This analysis is consistent with the quantitation previously published (32). The major increase in glycosylation occurs in bone as compared with other tissues, and it seems that this increase occurs at normally modified hydroxylysines.
Full occupancy of the A1 site in the α-1 chain of type I collagen along with the osteogenesis imperfecta phenotypes in the absence of such modification is suggestive of its importance for collagen fibrillogenesis. The concept of fibril molecular packing in which the subunits are molecular dimers in register and staggered axially by D-periods was proposed (31). Recently, this concept was proposed to be strengthened by interactions involving 3-Hyp (20) because of the D-periodic spacing of the 3-Hyp sites in collagen. Additionally, limited evidence was found for the role of 3-Hyp at 986 position in dimerization of collagen triple helices (21). According to this concept, one would expect at least a somewhat disturbed D-period in the collagen fibrils where the A1 site is not 3-hydroxylated. However, our data clearly demonstrate no difference in the D-spacing between wild type and P3H1 null collagen fibrils, both by small angle x-ray diffraction and by electron microscopy. Moreover, EM images of P5 tendon demonstrate that collagen fibrils of the mutant mice lack any disorganization. In fact, these fibrils are perfectly normal despite a slight increase in the diameter. Both findings disagree with the previously described concept (20, 21).
Tendon fibrils apparently form normally during embryonic development even in the absence of the 3-Hyp at the A1 site. Their diameter is likely increased due to bulkier collagen chains carrying more sugars. It is during postnatal growth when the disorganization is observed. Adult tendon appears to be composed of extremely disorganized fibrils that are branching and fusing as seen by EM. During the postnatal maturation process, the lateral fusion of smaller fibrils (40–50 nm) into larger ones (<200 nm) occurs. The conclusion is that the P3H1 null immature tendon fibrils fail to fuse properly.
The bone and tendon embryonic development shows distinct differences in P3H1 null mice. Although tendon appears almost normal at birth, the degree of bone mineralization is already severely affected at this stage. Simultaneously, 3-hydroxylation of the A3 site of type I collagen in bone is strongly affected in the P3H1 null mice. Thus, it is possible that the 3-Hyp located at the A3 site could play a role during initial stages of fibrillogenesis. On the other hand, type I collagen is least posttranslationally modified in bone as compared with other tissues. The low abundance of sugars might be a prerequisite for the tight packing and mineralization of type I collagen in bone. It is possible that the excess glycosylation in bone results in more severe disturbances than the lack of 3-hydroxyproline.
Acknowledgments
We thank Sara Tufa and Richard Sinclair for technical assistance.
This work was supported by grants from Shriners Hospital for Children (to H. P. B. and D. R. K.).
- P3H
- prolyl 3-hydroxylase
- CRTAP
- cartilage-associated protein
- CypB
- cyclophilin B
- 3-Hyp
- 3-hydroxyproline
- 4-Hyp
- 4-hydroxyproline
- FWHM
- full width of half-maximum
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1. Myllyharju J., Kivirikko K. I. (2004) Collagens, modifying enzymes and their mutations in humans, flies, and worms. Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 2. Myllyharju J. (2003) Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 22, 15–24 [DOI] [PubMed] [Google Scholar]
- 3. Pihlajaniemi T., Myllylä R., Kivirikko K. I. (1991) Prolyl 4-hydroxylase and its role in collagen synthesis. J. Hepatol. 13, Suppl, 3, S2–S7 [DOI] [PubMed] [Google Scholar]
- 4. Ogle J. D., Arlinghaus R. B., Lgan M. A. (1962) 3-Hydroxyproline, a new amino acid of collagen. J. Biol. Chem. 237, 3667–3673 [PubMed] [Google Scholar]
- 5. Risteli J., Tryggvason K., Kivirikko K. I. (1977) Prolyl 3-hydroxylase: partial characterization of the enzyme from rat kidney cortex. Eur. J. Biochem. 73, 485–492 [DOI] [PubMed] [Google Scholar]
- 6. Risteli J., Tryggvason K., Kivirikko K. I. (1978) A rapid assay for prolyl 3-hydroxylase activity. Anal. Biochem. 84, 423–431 [DOI] [PubMed] [Google Scholar]
- 7. Tryggvason K., Majamaa K., Risteli J., Kivirikko K. I. (1979) Partial purification and characterization of chick-embryo prolyl 3-hydroxylase. Biochem. J. 183, 303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vranka J. A., Sakai L. Y., Bächinger H. P. (2004) Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J. Biol. Chem. 279, 23615–23621 [DOI] [PubMed] [Google Scholar]
- 9. Blatch G. L., Lässle M. (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939 [DOI] [PubMed] [Google Scholar]
- 10. D'Andrea L. D., Regan L. (2003) TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 11. Morello R., Bertin T. K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F. H., Vranka J., Bächinger H. P., Pace J. M., Schwarze U., Byers P. H., Weis M., Fernandes R. J., Eyre D. R., Yao Z., Boyce B. F., Lee B. (2006) CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127, 291–304 [DOI] [PubMed] [Google Scholar]
- 12. Ishikawa Y., Wirz J., Vranka J. A., Nagata K., Bächinger H. P. (2009) Biochemical characterization of the prolyl 3-hydroxylase 1·cartilage-associated protein·cyclophilin B complex. J. Biol. Chem. 284, 17641–17647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vranka J. A., Pokidysheva E., Hayashi L., Zientek K., Mizuno K., Ishikawa Y., Maddox K., Tufa S., Keene D. R., Klein R., Bächinger H. P. (2010) Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin, and bones. J. Biol. Chem. 285, 17253–17262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baldridge D., Schwarze U., Morello R., Lennington J., Bertin T. K., Pace J. M., Pepin M. G., Weis M., Eyre D. R., Walsh J., Lambert D., Green A., Robinson H., Michelson M., Houge G., Lindman C., Martin J., Ward J., Lemyre E., Mitchell J. J., Krakow D., Rimoin D. L., Cohn D. H., Byers P. H., Lee B. (2008) CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum. Mutat. 29, 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barnes A. M., Chang W., Morello R., Cabral W. A., Weis M., Eyre D. R., Leikin S., Makareeva E., Kuznetsova N., Uveges T. E., Ashok A., Flor A. W., Mulvihill J. J., Wilson P. L., Sundaram U. T., Lee B., Marini J. C. (2006) Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N. Engl. J. Med. 355, 2757–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cabral W. A., Chang W., Barnes A. M., Weis M., Scott M. A., Leikin S., Makareeva E., Kuznetsova N. V., Rosenbaum K. N., Tifft C. J., Bulas D. I., Kozma C., Smith P. A., Eyre D. R., Marini J. C. (2007) Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat. Genet. 39, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Dijk F. S., Nesbitt I. M., Zwikstra E. H., Nikkels P. G., Piersma S. R., Fratantoni S. A., Jimenez C. R., Huizer M., Morsman A. C., Cobben J. M., van Roij M. H., Elting M. W., Verbeke J. I., Wijnaendts L. C., Shaw N. J., Högler W., McKeown C., Sistermans E. A., Dalton A., Meijers-Heijboer H., Pals G. (2009) PPIB mutations cause severe osteogenesis imperfecta. Am. J. Hum. Genet. 85, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Willaert A., Malfait F., Symoens S., Gevaert K., Kayserili H., Megarbane A., Mortier G., Leroy J. G., Coucke P. J., De Paepe A. (2009) Recessive osteogenesis imperfecta caused by LEPRE1 mutations: clinical documentation and identification of the splice form responsible for prolyl 3-hydroxylation. J. Med. Genet. 46, 233–241 [DOI] [PubMed] [Google Scholar]
- 19. Choi J. W., Sutor S. L., Lindquist L., Evans G. L., Madden B. J., Bergen H. R., 3rd, Hefferan T. E., Yaszemski M. J., Bram R. J. (2009) Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet. 5, e1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weis M. A., Hudson D. M., Kim L., Scott M., Wu J. J., Eyre D. R. (2010) Location of 3-hydroxyproline residues in collagen types I, II, III and V/XI implies a role in fibril supramolecular assembly. J. Biol. Chem. 285, 2580–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hudson D. M., Kim L. S., Weis M., Cohn D. H., Eyre D. R. (2012) Peptidyl 3-hydroxyproline binding properties of type I collagen suggest a function in fibril supramolecular assembly. Biochemistry 51, 2417–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eyre D. R., Weis M., Hudson D. M., Wu J. J., Kim L. (2011) A novel 3-hydroxyproline (3Hyp)-rich motif marks the triple-helical C terminus of tendon type I collagen. J. Biol. Chem. 286, 7732–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spiro R. G. (1969) Characterization and quantitative determination of the hydroxylysine-linked carbohydrate units of several collagens. J. Biol. Chem. 244, 602–612 [PubMed] [Google Scholar]
- 24. Dreisewerd K., Rohlfing A., Spottke B., Urbanke C., Henkel W. (2004) Characterization of whole fibril-forming collagen proteins of types I, III, and V from fetal calf skin by infrared matrix-assisted laser desorption ionization mass spectrometry. Anal. Chem. 76, 3482–3491 [DOI] [PubMed] [Google Scholar]
- 25. Henkel W., Dreisewerd K. (2007) Cyanogen bromide peptides of the fibrillar collagens I, III, and V and their mass spectrometric characterization: detection of linear peptides, peptide glycosylation, and cross-linking peptides involved in formation of homo- and heterotypic fibrils. J. Proteome Res. 6, 4269–4289 [DOI] [PubMed] [Google Scholar]
- 26. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 27. Taga Y., Kusubata M., Ogawa-Goto K., Hattori S. (2012) Development of a novel method for analyzing collagen O-glycosylations by hydrazide chemistry. Mol. Cell. Proteomics 11, M111.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vranka J., Stadler H. S., Bächinger H. P. (2009) Expression of prolyl 3-hydroxylase genes in embryonic and adult mouse tissues. Cell Struct. Funct. 34, 97–104 [DOI] [PubMed] [Google Scholar]
- 29. Hosemann R., Bagchi S. N. (eds). (1962) Direct Analysis of Diffraction by Matter, North-Holland, Amsterdam [Google Scholar]
- 30. Fernandes R. J., Farnand A. W., Traeger G. R., Weis M. A., Eyre D. R. (2011) A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens. J. Biol. Chem. 286, 30662–30669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eyre D. R., Paz M. A., Gallop P. M. (1984) Cross-linking in collagen and elastin. Annu. Rev. Biochem. 53, 717–748 [DOI] [PubMed] [Google Scholar]
- 32. Taga Y., Kusubata M., Ogawa-Goto K., Hattori S. (2013) Site-specific quantitative analysis of overglycosylation of collagen in osteogenesis imperfecta using hydrazide chemistry and SILAC. J. Proteome Res. 12, 2225–2232 [DOI] [PubMed] [Google Scholar]