Abstract
We have developed a yeast model system to address transcriptional repression by the retinoblastoma protein (pRB). When fused to the DNA-binding domain of Gal4p (DB-pRB), pRB can repress transcription of reporter genes containing Gal4p binding sites; the histone deacetylase activity encoded by yeast RPD3 is required for DB-pRB repression. Mutation of the LXCXE binding cleft in pRB, a region reported to be required for histone deacetylase recruitment, does not interfere with pRB-mediated repression. From these findings based on yeast experiments, we surmise that the small pocket region of pRB must contain an additional domain that confers histone deacetylase-dependent transcriptional repression. This hypothesis was verified by experiments examining pRB-dependent histone deacetylase association in mammalian cells. In addition to RPD3, repression by pRB in yeast requires MSI1, an ortholog of RbAp48, but not SIN3 or SAP30. By comparing the genetic requirements of DB-pRB repression in yeast to those of other DB-repressor fusions, we can suggest a mechanism by which pRB recruits histone deacetylase activity.
Since the discovery that the retinoblastoma protein could act as a transcriptional repressor (1–5), it has been of considerable interest to determine the mechanism by which this repression occurs. Several reports have indicated that retinoblastoma protein (pRB) can repress transcription by recruiting class I histone deacetylases (HDAC1, HDAC2, and HDAC3; refs. 6–9). However, other mechanisms of repression have been suggested and the relative contribution of different corepressors to pRB function remains unresolved (10–12).
The manner by which pRB recruits class I HDACs remains a topic of controversy. In one model, deacetylase recruitment occurs by direct interaction between pRB and HDAC1 or HDAC2 by means of the IXCXE motif present in the deacetylase (8). Many pRB-binding partners contain LXCXE or IXCXE motifs. This model does not account for recruitment of HDAC3, which does not have such a motif. Other experiments indicate that RBP1, an LXCXE-containing protein identified by virtue of its ability to interact with pRB (13, 14), is an intermediary in the interaction between pRB and HDAC (9). More recently, RBP1 has been identified as a component of the mSIN3A complex, indicating that pRB recruits the entire complex by virtue of its interaction with RBP1 (15). Both models would predict that mutation of the LXCXE binding cleft in pRB would abrogate HDAC interaction and possibly transcriptional repression. This prediction has been tested in three recent studies (16–18). pRB alleles deficient in LXCXE binding retain at least some ability to repress transcription in all three reports. However, different conclusions were reached concerning HDAC interaction. In one report, the pRB cleft mutants retained HDAC binding (18), whereas that interaction was shown to be reduced or lost in the two other reports by using similar pRB alleles (16, 17). The reasons for these discrepancies are not known. A fundamental difficulty with this analysis has been that it is not possible to assess cleanly the role of individual corepressor molecules because mammalian cells lacking these proteins do not exist.
The use of yeast as a model system for understanding the mechanisms by which heterologous transcriptional activators function has been both widespread and productive. The success of these experiments undoubtedly relies in large part on the high degree of conservation of basal transcription machinery between yeast and higher organisms. A large percentage of heterologous activators function in yeast, and experimental evidence suggests that they promote transcription in a mechanistically similar manner in both yeast and their endogenous organism (19–28). Transcriptional repressors have been tested to a much lesser extent.
We reasoned that if pRB could repress transcription in yeast, this genetically tractable system could be used to define the mechanism of pRB action. Because many mSIN3A components are conserved from yeast to humans, we can determine their relative contributions by testing pRB-mediated repression in strains bearing single gene deletions of each component. From this analysis, we show that pRB represses transcription in yeast when targeted to promoters by fusion to the DNA-binding domain (DB) of Gal4. This repression requires intact RPD3, the yeast ortholog of class I HDACs, and also MSI1, a potential ortholog of RbAp48. However, repression is not mediated through the LXCXE-binding cleft in pRB, indicating that the protein contains at least a second contact site within the pRB small pocket for recruiting histone deacetylase.
Materials and Methods
Yeast Strains and Plasmids.
The yeast strain MaV103 has been described (29); all yeast deletions were created and analyzed in this strain background. pPK104 was kindly supplied by P. Kaufman (Univ. of California, Berkeley, CA) and used to create Δmsi1∷hisG (30). Δsin3∷TRP1 and Δrpd3∷URA3 were created by γ-deletion with described plasmids (31, 32). Δrpd3∷TRP1 was derived from Δrpd3∷URA3 with selectable marker replacement (33). The Δume6∷LEU2 construct was generously provided by K. Struhl (Harvard Medical School, Boston) (34). Δcac2∷TRP1, ΔSAP18/APG16∷TRP1, Δsap30∷TRP1, Δhda1∷TRP1, and Δume1∷TRP1 strains were created with PCR-based disruption techniques. All PCR-based disruptions remove the entire ORF of the gene of interest. All gene deletions were confirmed by PCR.
The rpd3 mutant alleles, H150A/H151A and H188A, were graciously provided by D. Kadosh (Harvard Medical School, Boston) and K. Struhl (35). DB-pRB (amino acids 302–928), DB-pRBΔ22 (amino acids 289–928, missing exon 22), DB-pRBΔ300 (amino acids 289–301 and 395–928), DB-p107, and DB-p130 have been described (36). The RB9 allele has been described (18) and contains the following amino acid changes: I753A, N757A, and M761A. A cDNA for PML was provided by K. S. Chang (Univ. of Texas, M. D. Anderson Cancer Center, Houston). DB-Mad and DB-PML were created by fusing the entire ORF of the respective genes to the sequence encoding Gal4p DB (amino acids 1–147) in plasmid pPC97 (29).
Western Blot Analysis.
Yeast cells were grown to mid-log phase in selective media, harvested, washed, and resuspended in 200 μl of breaking buffer (100 mM Tris⋅HCl, pH 8.0/20% glycerol/1mM EDTA/0.1% Triton X-100/5 mM MgCl2/10 mM β-mercaptoethanol/1 mM phenylmethylsulfonyl fluoride plus protease inhibitors). Glass beads (50–75 μl) were added and cells were vortexed at 4° for 10 min. Debris was removed by centrifugation, and 100 μg of remaining cell extract was separated by electrophoresis in SDS/gels. Blots were probed with the α-pRB antibody XZ105 (29).
β-Galactosidase Assays.
All β-galactosidase assays were performed according to standard techniques (37). All experiments were repeated multiple times. The units of activity are presented as [A400/(A600 × time in min)] × 1,000. All assays were conducted on cells freshly transformed with the reporter plasmid pRS314–17d80lacZdNeo, which was generated and kindly provided by R. Morse (Wadsworth Center, Albany, NY) (38).
Immunoprecipitation.
pRB and HDAC expression constructs were transfected into C33A cells as described (18). Immunoprecipitations were also carried out as previously reported.
Results and Discussion
The yeast two-hybrid strain MaV103 was used in this study (29) and contains a deletion of GAL4 and an integrated HIS3 reporter gene under the control of Gal4p binding sites (GAL1∷HIS3) (Fig. 1A). The transcriptional levels of GAL1∷HIS3 correlate closely with the levels of resistance to the His3p inhibitor 3-aminotriazole (3-AT) in MaV103 (29). For example, MaV103 cells that express wild-type Gal4p form colonies in 2–3 days on plates containing 3-AT concentrations up to 150 mM. In contrast, cells that express the DB of Gal4p in the absence of any activation domain take 5 days to form colonies in the presence of 10 mM 3-AT. This background growth is caused by the basal level of expression of GAL1∷HIS3 and provides an assay for potential repressors fused to DB.
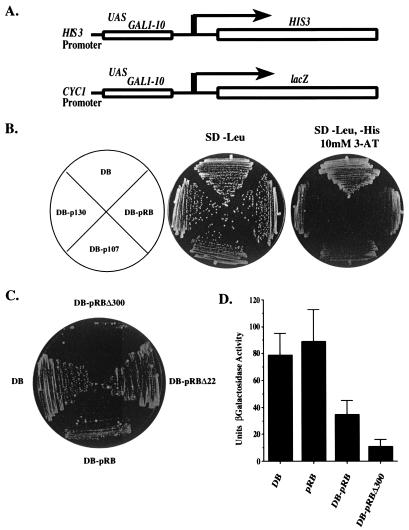
Figure 1.
pRB family members repress transcription in yeast. (A) Two reporters were used in the majority of repression assays. In the reporter used for growth assays, the UAS region from the HIS3 promoter is replaced with a region from the GAL1 promoter containing multiple Gal4p-binding sites (14). The reporter plasmid pRS314–17d80lacZdNeo (Lower) was used for all quantitation assays (19). (B) pRB family members repress transcription of the GAL1∷HIS3 reporter. Strain MaV103 was transformed with ARS-CEN plasmids expressing DB, DB-pRB, DB-p107, and DB-p130 and assayed for growth on either nonselective media (SD, −Leu) after 3 days or selective media (SD, −Leu, −His, 10 mM 3-AT) after 5 days. All growth assays were repeated several times with identical results. (C) Mutant alleles of pRB confer different levels of repression. Repression was assayed on SD, −Leu, −His, 10 mM 3-AT after 5 days. (D) pRB represses transcription in quantitative assays. All β-galactosidase assays were performed according to standard techniques. Activity units are presented as [A400/(A600 × time in minutes)] × 1,000. Data generated are the mean result and standard deviation of five independent cultures from one experiment including each DB construct. These results are representative of several independent experiments.
There are no yeast ORFs that bear significant homology to pRB family members, and heterologous expression of the human pRB protein does not alter the yeast growth rate (39). However, pRB expressed in yeast is subject to cell cycle dependent phosphorylation by yeast CDKs (39) and is capable of interacting with mammalian binding partners such as E2F subunits (29), suggesting that it is not functionally impaired. By targeting pRB to a yeast reporter, we reasoned that it might be possible to address the mechanism by which pRB confers repression in a genetically tractable system. The large pocket domain of pRB (amino acids 302–928) was fused to DB (DB-pRB) and expressed in MaV103 cells from the monocopy vector pPC97 to maintain low expression levels. This large pocket domain of pRB is known to contain the sequences necessary for pRB-mediated repression in mammalian cells (5, 40). Compared with cells that express only DB, MaV103 cells expressing DB-pRB display little or no growth at 10 mM 3-AT levels after a 5-day incubation (Fig. 1B), indicating that DB-pRB can repress transcription of the GAL1∷HIS3 reporter gene. Similar results were seen when the entire ORF of pRB was fused to Gal4DB (data not shown). Likewise, two other pRB family members, p107 and p130, inhibit colony growth when fused to DB, indicating that they also can repress transcription in yeast (Fig. 1B).
To test the relevance of this assay, we used a tumor-derived pRB mutant missing exon 22 (pRBΔ22), which is defective in transcriptional repression in mammalian assays (41). Unlike DB-pRB, DB-pRBΔ22 is unable to repress transcription of GAL1∷HIS3 (Fig. 1C), although DB-pRB and DB-pRBΔ22 are expressed to the same extent in MaV103 cells (29). In the course of these experiments, we also observed that an N-terminally truncated allele of pRB large pocket lacking residues 302–394 (DB-pRBΔ300) is more potent than DB-pRB in its ability to repress transcription. The domain deleted in DB-pRBΔ300 contains several potentially inhibitory cyclin-dependent kinase (CDK) phosphorylation sites. It is possible that these sites can be phosphorylated by yeast CDKs, thus limiting DB-pRB's transcriptional repression activity.
To quantify DB-pRB repression, we used an episomal reporter with Gal4p-binding sites controlling the expression of lacZ (38) (Fig. 1A). Expression of DB-pRB reduces the β-galactosidase activity generated from this reporter approximately 3-fold, whereas that of DB-pRBΔ300 confers an 8-fold reduction (Fig. 1D). This experiment shows that pRB can repress transcription from multiple yeast reporter genes. Furthermore, expression of pRB alone does not confer repression, indicating that fusion to the DB of Gal4p and subsequent targeting to the reporter is required for pRB activity.
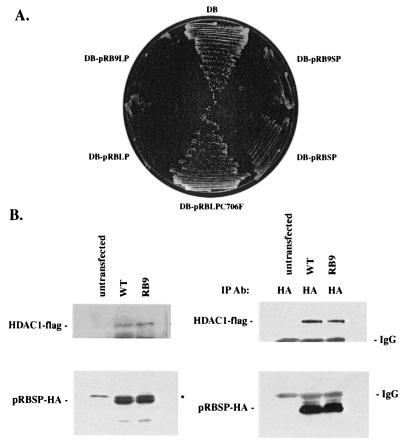
Several reports have concluded that pRB can repress transcription at least in part by recruiting mammalian class I histone deacetylases (6–8). Because human HDAC1 is 60% identical to yeast Rpd3p, we reasoned that pRB-dependent repression in yeast might be mediated through recruitment of RPD3. Binding to HDAC is reported to be mediated through the LXCXE-like sequences present in HDAC1 and HDAC2 or indirectly with the LXCXE-containing protein RBP1 serving as an intermediary. Therefore, we tested whether an allele of pRB with a dysfunctional LXCXE-binding cleft retains the ability to repress transcription in yeast. Cells expressing DB-pRBLP (DB-pRB Large Pocket, amino acids 379–928) or DB-pRB9LP, which fails to interact with viral LXCXE-containing proteins (Fig. 2A) were tested for growth on SD, −His media containing 10 mM 3-AT (Fig. 2A). We find that both constructs reduce growth to an equal extent, suggesting that pRB-mediated repression in yeast does not require a functional LXCXE binding cleft. Further, DB-pRBSP (amino acids 379–792), which contains the minimal sequences necessary for repression in mammalian cells, represses transcription in yeast. Mutation of the LXCXE-binding cleft does not block repression. Similar results were obtained by using β-galactosidase assays to measure expression from the lacZ reporter (data not shown). Therefore, we conclude that in yeast, pRBSP must contain another motif sufficient to confer transcriptional repression. Consistent with this finding, yeast RPD3 does not have an LXCXE sequence, and there are no yeast proteins that bear resemblance to RBP1.
Figure 2.
A pRB LXCXE-binding cleft mutant represses transcription in yeast. (A) For each strain, the pRB allele listed was fused to Gal4DB on an ARS-CEN vector and transformed into wild-type yeast. The ability of each pRB allele to repress transcription was determined by growth on SD, −His, 10 mM 3-AT plates after 5 days. (B) pRB9SP (pRB9 Small Pocket) interacts with HDAC1 in mammalian cells. HDAC1-flag was cotransfected with either pRBSP or pRB9SP in C33A cells. Western blots reflect protein expression levels (Left). Immunoprecipitations with antibodies to pRB are shown (Right). * marks a nonspecific band.
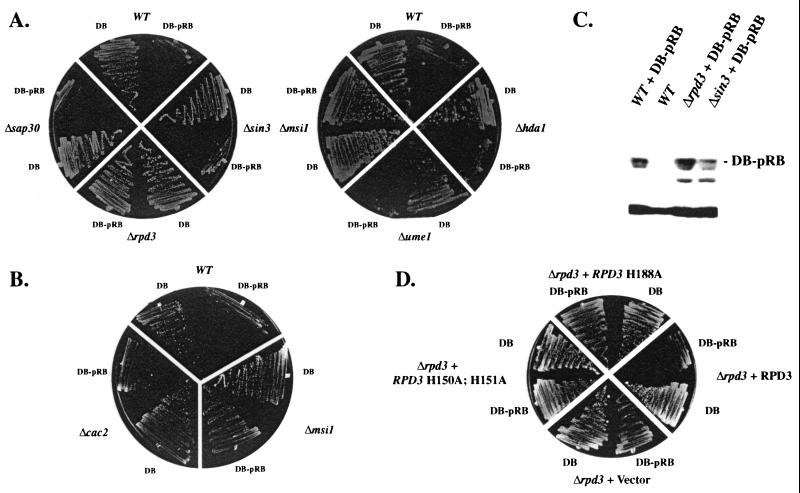
We then tested whether repression by pRB in yeast occurs through histone deacetylase recruitment. The yeast RPD3 gene was deleted in MaV103 (Δrpd3), and growth on 10 mM 3-AT was determined. DB-pRB is unable to repress transcription in the absence of Rpd3p showing a genetic requirement for this histone deacetylase (Fig. 3A). In contrast, another histone deacetylase, HDA1, that is less related to HDAC1–3 is not required for repression by DB-pRB. Furthermore, yeast cells expressing mutant alleles of RPD3 that specifically lack deacetylase activity are unable to support pRB-mediated repression (Fig. 3D). One trivial explanation for the lack of repression by DB-pRB in the absence of RPD3 is that the fusion protein is destabilized. To rule out this possibility, we performed Western blot analysis to determine DB-pRB levels in different strain backgrounds. We found that DB-pRB steady-state levels remained constant in wild-type, Δrpd3, and Δsin3 strains (Fig. 3C). In addition, we found that DB fusions to other HDAC-independent transcriptional repressors retained the ability to repress transcription in the absence of Rpd3p, indicating that DB access to the GAL1∷HIS3 reporter is not significantly affected in the Δrpd3 background (data not shown). Finally, we tested repression of the GAL1∷lacZ reporter by DB-pRB in the Δrpd3 background. In accord with results generated on the GAL1∷HIS3 reporter, DB-pRBΔ300 does not reduce lacZ expression in the absence of Rpd3p (see Fig. 4B). This result establishes that deacetylase activity is required for transcriptional repression by pRB in yeast.
Figure 3.
Histone deacetylase components required for transcriptional repression by pRB. (A) pRB-mediated repression in strains lacking deacetylase components. Either Gal4DB- or DB-pRB-expressing ARS-CEN vectors were transformed into strains deleted for components important for RPD3-dependent histone deacetylase; repression was determined by growth on SD, −His, 10 mM 3-AT plates after 5 days. White lines through plates are drawn for convenience. For each image, eight strains were from the same plate and were photographed at the same time. (B) MSI1/CAC3, but not CAC2, is required for repression by DB-pRB. Plates were treated exactly as described in A. (C) DB-pRB protein levels are unaltered in the Δrpd3 and Δsin3 strain backgrounds. Extracts loaded in each lane were equilibrated for total protein concentration. (D) A Δrpd3 strain was transformed with either DB or DB-pRB. Representative transformants were then transformed with vector, wild-type RPD3, or mutant alleles of RPD3 that specifically lack deacetylase activity. The SD, −His, 10 mM 3-AT plate was photographed after 5 days growth.
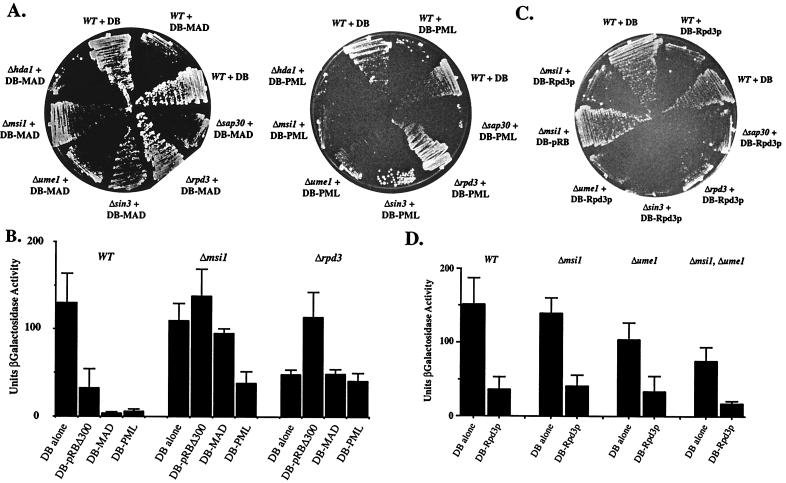
Figure 4.
The role of RbAp48 orthologs in RPD3-dependent transcriptional repression. (A) Repression by Mad or PML in strains lacking deacetylase components. Strains were transformed constructs expressing the GAL4 DB fused to either full-length human Mad or PML, and repression was assayed by determining growth after 5 days on SD, −His, 10 mM 3-AT media. (B) Quantitation of repression by pRB, Mad, and PML in Δmsi1 and Δrpd3 strains. This experiment was performed exactly as described in Fig. 1D. (C) Strains bearing deletions in deacetylase components were transformed with either vector or a construct expressing GAL4DB fused to full-length yeast RPD3; repression was assayed after 5 days on SD, −His, 10 mM 3-AT media. A representative plate is imaged. This experiment was performed several times. (D) Quantitation of repression by DB-Rpd3p in strains lacking RbAp48 orthologs. Experiment was performed as in Fig. 1D.
These findings suggest that the SP of pRB contains another domain independent of the LXCXE-binding cleft that can serve as an interaction surface for histone deacetylases. To test this possibility directly, we expressed RB9SP, which contains the SP of pRB and mutations that abolish LXCXE binding identical to those used in RB9LP (18), as well as HDAC1 in SAOS-2 human osteosarcoma cells, and assayed binding by coimmunoprecipitation. We found that this mutant version of pRBSP retains the ability to interact with HDAC1 in mammalian cells at levels comparable to that of the pRB wild-type small pocket (Fig. 2B).
To test whether other protein components of the deacetylase complex are required for transcriptional repression by DB-pRB in yeast, strains were generated with deletions of the genes encoding Rpd3p (HDAC)-associated proteins including Sin3p, Sap30p, and the RbAp48-related proteins Msi1p and Ume1p/Wtm3p (42). We found that only MSI1 (RbAp48) is required for repression by DB-pRB (Fig. 3A). DB-pRB protein levels are similar in both wild-type and Δmsi1 (RbAp48) strain backgrounds (data not shown). Mammalian RbAp48 has been identified as a component of the HDAC complex (43), although the link between MSI1 and RPD3 in yeast has not been confirmed. Moreover, Msi1p has been shown to be a stoichiometric component of the yeast chromatin assembly factor complex (CAC; ref. 30). Therefore, it was possible that loss of CAC function might abrogate pRB-mediated repression, suggesting a nondeacetylase-mediated mechanism to explain the observation that Δmsi1 (RbAp48) strains are refractory to pRB-mediated repression. To test this possibility, we deleted CAC2, a gene encoding another component of the chromatin assembly complex. DB-pRB represses transcription in the Δcac2 strain background (Fig. 3B), linking Msi1p (RbAp48) more closely to Rpd3p-mediated histone deacetylation. In addition to RPD3, MSI1 is required for repression by other pRB family members (data not shown). Other components thought to be important for HDAC-dependent repression such as Sin3p, Ume1p/Wtm3p, and Sap30p are dispensable in the DB-pRB repression assay (Fig. 3A). However, as shown below, many components are involved in the activity of other HDAC-dependent repressors.
The fact that MSI1 but not SIN3 or another RbAp48-related gene, UME1, is required for DB-pRB transcriptional repression suggests that those potential recruitment factors might display specificity for different HDAC-repressed genes in vivo. To test this notion, we analyzed the requirement of Msi1p for the activity of Ume6p, a yeast transcriptional repressor of meiotic genes (44) that physically interacts with and functionally requires Sin3p (34). We analyzed Ume6p repression by using another reporter gene, SPAL10∷URA3, which contains Ume6p-binding sites (29) (see Fig. 5A, which is published as supplemental data on the PNAS web site, www.pnas.org). Levels of expression of SPAL10∷URA3 inversely correlate with the levels of resistance to the drug 5-fluoroorotic acid (5-FOA) (45). Thus, the UME6 wild-type MaV103 strain is resistant to 0.2% 5-FOA, whereas the Δume6 derivative is sensitive. Strains bearing deletions in genes encoding HDAC components were plated on 5-FOA-containing media and assayed for growth (Fig. 5B, which is published as supplemental data). In agreement with previous reports, we find that RPD3 and SIN3 are required for Ume6-mediated repression (34). However, unlike for DB-pRB, Ume6p-mediated repression does not depend on MSI1 (RbAp48), but instead requires the family member UME1, as reported (42). These findings suggest that components of the histone deacetylase complex are specifically used in a repressor-dependent manner.
To make a more direct comparison of the requirements of pRB repression, we created DB fusions to Mad (DB-Mad) and PML (DB-PML), two mammalian proteins shown to recruit histone deacetylase (46–48), and tested their ability to repress the GAL1∷HIS3 reporter gene. For DB-Mad, we find that RPD3, SIN3, and MSI1 (RbAp48) are required for transcriptional repression (Fig. 4A). These observations are in agreement with those reported for mammalian cells in which Mad physically interacts with SIN3A (46). In the case of PML, RPD3, but not SIN3, is required for repression. Neither of these two repressors is functionally impaired by deletion of another yeast, deacetylase HDA1. In addition, we tested repression of the lacZ reporter gene by DB-Mad, DB-PML, and DB-pRB. All three repressors reduce the levels of β-galactosidase in the wild-type background and fail to function in the absence of RPD3 (Fig. 4B). Deletion of MSI1 (RbAp48) abrogates repression by DB-pRB and DB-Mad, but affects DB-PML to a lesser extent. Together, this set of experiments demonstrates that histone deacetylase components such as Sin3p and Msi1p (RbAp48) are differentially required for repressor-mediated recruitment of histone deacetylase. This finding substantiates the notion that, in mammalian cells, SIN3 functions in the recruitment process specifically for a subset of repressors. In addition, these findings are consistent with RbAp48 acting as a bridging factor between pRB and HDAC.
We imagine two possibilities to explain the importance of MSI1 (RbAp48) for pRB-dependent repression. Either Msi1p acts as a bridging or specificity factor important for linking pRB to Rpd3p, or MSI1 is important for the enzymatic activity of Rpd3p on histones, a function proposed for RbAp48 (49, 50). Given that MSI1 is not required for repression by Ume6p, the latter possibility cannot be strictly true. However, because Ume6p-dependent repression does require another RbAp48-related protein Ume1p, it may be that Rpd3p requires a RbAp48-like factor to access histones. To examine these possibilities, we fused Rpd3p to DB (DB-Rpd3p) and tested the transcriptional repression of GAL1∷HIS3 mediated by this fusion protein. A similar fusion has been shown to confer repression in yeast in a manner independent of SIN3, but other proposed deacetylase subunits have not been examined (34). In this situation, all proteins that function solely to recruit Rpd3p should be dispensable, but proteins important for histone deacetylation should be required. We transformed the DB-Rpd3p-encoding construct into the previously tested deletion backgrounds and determined growth capacity in the presence of 10 mM 3-AT (Fig. 4C). Deletions strains expressing on Gal4DB have similar growth rates under these conditions (Fig. 2A; data not shown). We find that DB-Rpd3p represses transcription of GAL1∷HIS3, and that this repression is not impaired in strains lacking SIN3, MSI1, UME1, or any of the other components reported to be involved in histone deacetylation. We also examined repression by DB-Rpd3p in quantitative assays and found similar results (Fig. 4D). Moreover, in this assay, we examined repression by DB-Rpd3p in a Δmsi1, Δume1 double-mutant strain. Even in this background, DB-Rpd3p was active in repression to a level similar to that seen in the wild-type setting. This finding calls into question the hypothesis that RbAp48 function is generally important for HDAC-mediated deacetylation of histones. However, we cannot rule out this possibility, because there are two other gene products, Wtm1p and Wtm2p, that are structurally related to Ume1p and may participate in Ume6-dependent repression, albeit to a lesser extent than Ume1p (42). It may be that these proteins can provide the RbAp48 function for DB-Rpd3p in the absence of MSI1 and UME1. Nevertheless, the finding that MSI1 is specifically required for repression by DB-pRB and not DB-Rpd3p indicates that RbAp48 plays a more direct role in the recruitment of HDAC by the pRB family of proteins.
There have been several models proposed to explain the mechanism by which pRB recruits class I histone deacetylases to target promoters. Originally, it was reported that pRB could bind directly to HDAC1 (8). In vitro experiments demonstrated that HDAC1 bound to pRB in a manner dependent on its IXCXE motif, which closely resembles the LXCXE motif present in many pRB-binding proteins. Moreover, it was reported recently that RbAp48 is important for pRB-dependent repression, and that its interaction with pRB depended on HDAC1 or another LXCXE-containing protein as a linking factor (51). This model cannot entirely explain the pRB/HDAC/RbAp48 interactions for two reasons. First, pRB interacts with HDAC1, HDAC2, and HDAC3, yet only HDAC1 and HDAC2 have the IXCXE motif (9). Therefore, the interaction with HDAC3 must occur by a different mechanism. Second, dramatic mutation of the LXCXE-binding cleft in pRB does not significantly reduce the level of HDAC1 association (18). In another report, RBP1, an LXCXE-containing protein known to interact with pRB, is proposed to be a linking factor between pRB and HDAC1 (9). Recent findings by Lai et al. (15) indicate that RBP1 is a bona fide member of the mSIN3A complex and recruits the complex to pRB-bound promoters. However, their results also indicate that there may be other mechanisms in addition to RBP1 through which HDAC is recruited.
In yeast, none of these proposed mechanisms are likely to explain recruitment of Rpd3p to reporters by pRB. Rpd3p does not have any sequences resembling an LXCXE motif, and there are no proteins that bear similarity to RBP1. We propose that Msi1p (RbAp48) can serve as a bridging protein that promotes the pRB/Rpd3p interaction. This proposal is based on the observation that apart from Rpd3p, Msi1p (RbAp48) is the only known HDAC-associated protein that is required for repression by pRB. Moreover, Msi1p is not required when Rpd3p is directly targeted to reporters by means of fusion to Gal4DB. Therefore, Msi1p mediates recruitment of Rpd3p rather than its enzymatic activity. Finally, our experiments demonstrate that the LXCXE-binding cleft on pRB is not essential for repression by pRB, indicating that there must be another contact site for histone deacetylase within the small pocket. From these findings, we suggest that in mammalian cells, pRB may contact the Sin3 complex through two independent mechanisms, one that is LXCXE-dependent and one that is not. Such a dual recruitment mechanism would be likely to confer stronger contacts with the corepressor complex.
Supplementary Material
Acknowledgments
The authors thank K. S. Chang, D. Kadosh, P. Kaufman, R. Morse, T. Nagase, and C. Sardet, R. Strich, and K. Struhl for reagents and strains. B.K.K. and F.A.D. have received support from the Leukemia Society of America. This work was funded by a National Institutes of Health grant (to E.H.). E.H. is an American Cancer Society Research Professor.
Abbreviations
- pRB
retinoblastoma protein
- HDACn
histone deacetylases
- 3-AT
3-aminotriazole
- DB
DNA-binding domain
- CAC
chromatin assembly factor complex
- SD
yeast synthetic media
- PML
promyelocytic leukemia protein
References
- 1.Weintraub S J, Prater C A, Dean D C. Nature (London) 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 2.Adnane J, Shao Z, Robbins P D. J Biol Chem. 1995;270:8837–8843. doi: 10.1074/jbc.270.15.8837. [DOI] [PubMed] [Google Scholar]
- 3.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sellers W R, Rodgers J W, Kaelin W G., Jr Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weintraub S J, Chow K N B, Luo R X, Zhang S H, He S, Dean D C. Nature (London) 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Luo R X, Postigo A A, Dean D C. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 8.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Nature (London) 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 9.Lai A, Lee J M, Yang W M, DeCaprio J A, Kaelin W G, Jr, Seto E, Branton P E. Mol Cell Biol. 1999;19:6632–6641. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunaief J L, Strober B E, Guha P A, Khavari A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 11.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meloni A R, Smith E J, Nevins J R. Proc Natl Acad Sci USA. 1999;96:9574–9579. doi: 10.1073/pnas.96.17.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Defeo-Jones D, Huang P S, Jones R E, Haskell K M, Vuocolo G A, Hanobik M G, Huber H E, Oliff A. Nature (London) 1991;352:251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- 14.Fattaey A R, Helin K, Dembski M S, Dyson N, Harlow E, Vuocolo G A, Hanobik M G, Haskell K M, Oliff A, Defeo-Jones D, et al. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- 15.Lai A, Kennedy B K, Barbie D A, Bertos N R, Yang X J, Theberge M C, Tsai S C, Seto E, Zhang Y, Kuzmichev A, et al. Mol Cell Biol. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T T, Wang J Y. Mol Cell Biol. 2000;20:5571–5580. doi: 10.1128/mcb.20.15.5571-5580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahiya A, Gavin M R, Luo R X, Dean D C. Mol Cell Biol. 2000;20:6799–6805. doi: 10.1128/mcb.20.18.6799-6805.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dick F A, Sailhamer E, Dyson N. Mol Cell Biol. 2000;20:3715–3727. doi: 10.1128/mcb.20.10.3715-3727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Struhl K. Nature (London) 1988;332:649–650. doi: 10.1038/332649a0. [DOI] [PubMed] [Google Scholar]
- 20.Fields S, Jang S K. Science. 1990;249:1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- 21.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 22.Seneca S, Punyammalee B, Sureau A, Perbal B, Dvorak M, Crabeel M. Oncogene. 1993;8:2335–2342. [PubMed] [Google Scholar]
- 23.Mak P, Fuemkranz H A, Ge R, Karathanasis S K. Gene. 1994;145:129–133. doi: 10.1016/0378-1119(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian T, D'Sa-Eipper C, Elangovan B, Chinnadurai G. Nucleic Acids Res. 1994;22:1496–1499. doi: 10.1093/nar/22.8.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shim Y H, Bonner J J, Blumenthal T. J Mol Biol. 1995;253:665–676. doi: 10.1006/jmbi.1995.0581. [DOI] [PubMed] [Google Scholar]
- 26.Brachmann R K, Yu K, Eby Y, Pavletich N P, Boeke J D. EMBO J. 1998;17:1847–1859. doi: 10.1093/emboj/17.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Como C J, Prives C. Oncogene. 1998;16:2527–2539. doi: 10.1038/sj.onc.1202041. [DOI] [PubMed] [Google Scholar]
- 28.Epstein C B, Attiyeh E F, Hobson D A, Silver A L, Broach J R, Levine A J. Oncogene. 1998;16:2115–2122. doi: 10.1038/sj.onc.1201734. [DOI] [PubMed] [Google Scholar]
- 29.Vidal M, Brachmann R, Fattaey A, Harlow E, Boeke J D. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufman P D, Kobayashi R, Stillman B. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 31.Vidal M, Gaber R F. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidal M, Strich R, Esposito R E, Gaber R F. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidal M, Gaber R F. Yeast. 1994;10:141–149. doi: 10.1002/yea.320100202. [DOI] [PubMed] [Google Scholar]
- 34.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 35.Kadosh D, Struhl K. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal M, Braun P, Chen E, Boeke J D, Harlow E. Proc Natl Acad Sci USA. 1996;93:10321–10326. doi: 10.1073/pnas.93.19.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose M D, Winston F, Hieter P. In: Methods in Yeast Genetics: A Laboratory Course Manual. Rose M D, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. p. 93. [Google Scholar]
- 38.Ryan M P, Jones R, Morse R H. Mol Cell Biol. 1998;18:1774–1782. doi: 10.1128/mcb.18.4.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatakeyama M, Brill J A, Fink G R, Weinberg R A. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 40.Chow K N, Dean D C. Mol Cell Biol. 1996;16:4862–4868. doi: 10.1128/mcb.16.9.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaelin W G, Jr, Pallas D C, DeCaprio J A, Kaye F J, Livingston D M. Cell. 1991;64:521–532. doi: 10.1016/0092-8674(91)90236-r. [DOI] [PubMed] [Google Scholar]
- 42.Pemberton L F, Blobel G. Mol Cell Biol. 1997;17:4830–4841. doi: 10.1128/mcb.17.8.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 44.Strich R, Slater M R, Esposito R E. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 46.Ayer D E, Lawrence Q A, Eisenmann R N. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 47.Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, et al. Nature (London) 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 48.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 49.Parthun M R, Widom J, Gottschling D E. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 50.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Curr Biol. 1998;15:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 51.Nicolas E, Morales V, Magnanhi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.