Key Points
B cells appear in zebrafish by 3 weeks of development, supporting previous data that this is the transition point to adult hematopoiesis.
Shifting sites of B-cell development likely occur in all jawed vertebrates.
Abstract
Teleost fish are among the most ancient vertebrates possessing an adaptive immune system with B and T lymphocytes that produce memory responses to pathogens. Most bony fish, however, have only 2 types of B lymphocytes, in contrast to the 4 types available to mammals. To better understand the evolution of adaptive immunity, we generated transgenic zebrafish in which the major immunoglobulin M (IgM+) B-cell subset expresses green fluorescence protein (GFP) (IgM1:eGFP). We discovered that the earliest IgM+ B cells appear between the dorsal aorta and posterior cardinal vein and also in the kidney around 20 days postfertilization. We also examined B-cell ontogeny in adult IgM1:eGFP;rag2:DsRed animals, where we defined pro-B, pre-B, and immature/mature B cells in the adult kidney. Sites of B-cell development that shift between the embryo and adult have previously been described in birds and mammals. Our results suggest that this developmental shift occurs in all jawed vertebrates. Finally, we used IgM1:eGFP and cd45DsRed;blimp1:eGFP zebrafish to characterize plasma B cells and investigate B-cell function. The IgM1:eGFP reporter fish are the first nonmammalian B-cell reporter animals to be described. They will be important for further investigation of immune cell evolution and development and host-pathogen interactions in zebrafish.
Introduction
All vertebrate animals possess an adaptive immune system with lymphocytes that can proliferate in response to foreign substances and produce secreted receptors that bind to these antigens. Jawless vertebrates use antigen receptors termed “variable lymphocyte receptors” that are somatically diversified by gene conversion,1 whereas jawed vertebrates use immunoglobulin (Ig) domain-based antigen receptors that are diversified by somatic rearrangement, somatic mutation, and gene conversion.2 The zebrafish has recently emerged as a complementary vertebrate model for the examination of immunity and disease, and many studies have used zebrafish to model human disease and enable drug discovery. An outstanding example is Mycobacteria marinum infection of zebrafish, which more closely mimics human tuberculosis than the mouse model.3 Importantly for immunologic studies, zebrafish possess the major blood cell lineages found in mammals, including neutrophils, eosinophils, mast cells, dendritic cells, monocytes/macrophages, and B and T lymphocytes.4 Several transgenic reporter lines, in which cell-specific enhancers drive the expression of fluorescent proteins, have been created to label specific blood cells, including T cells, erythrocytes, eosinophils, neutrophils, macrophages, and antigen-presenting cells.5-9 By combining fluorescent transgenesis with advanced imaging techniques, zebrafish offer unique advantages over other vertebrate models for visualizing the behavior of immune cells in living animals. Currently, no transgenic reporter lines exist that specifically label B lymphocytes in zebrafish, or in any other nonmammalian animal model.
The types of Igs produced by B cells in teleosts (bony fish) are more limited than those produced in mammals. Igs consist of a repeating structure of 2 identical heavy chains and 2 identical light chains, and the Ig isotype is defined by the heavy chain. Similar to that in mammals, the zebrafish locus contains exons that somatically rearrange to form an exon that is the variable antigen-binding portion of the heavy chain. Also similar to that in mammals, the constant region of the heavy chain is specified by either μ or δ isotypes, which are co-expressed by alternative splicing to produce IgM and IgD on naive B cells.10 As the immune response progresses in mammals, birds, reptiles, and amphibians, individual B cells delete the μ and δ exons as they switch the constant region of the heavy chain to other isotypes (γ, ε, α). Unlike mammals, all teleosts studied so far have neither downstream exons for other isotypes nor switch regions to mediate class switching.11 Instead, there are exons upstream of the μ and δ exons that encode another isotype (denoted ζ in zebrafish, Figure 1A), which has been found in most other teleosts.12,13 Studies in trout indicate that this alternative isotype is concentrated in B cells within the gut and that it convergently evolved to provide mucosal immunity, similar to IgA in mammals.13
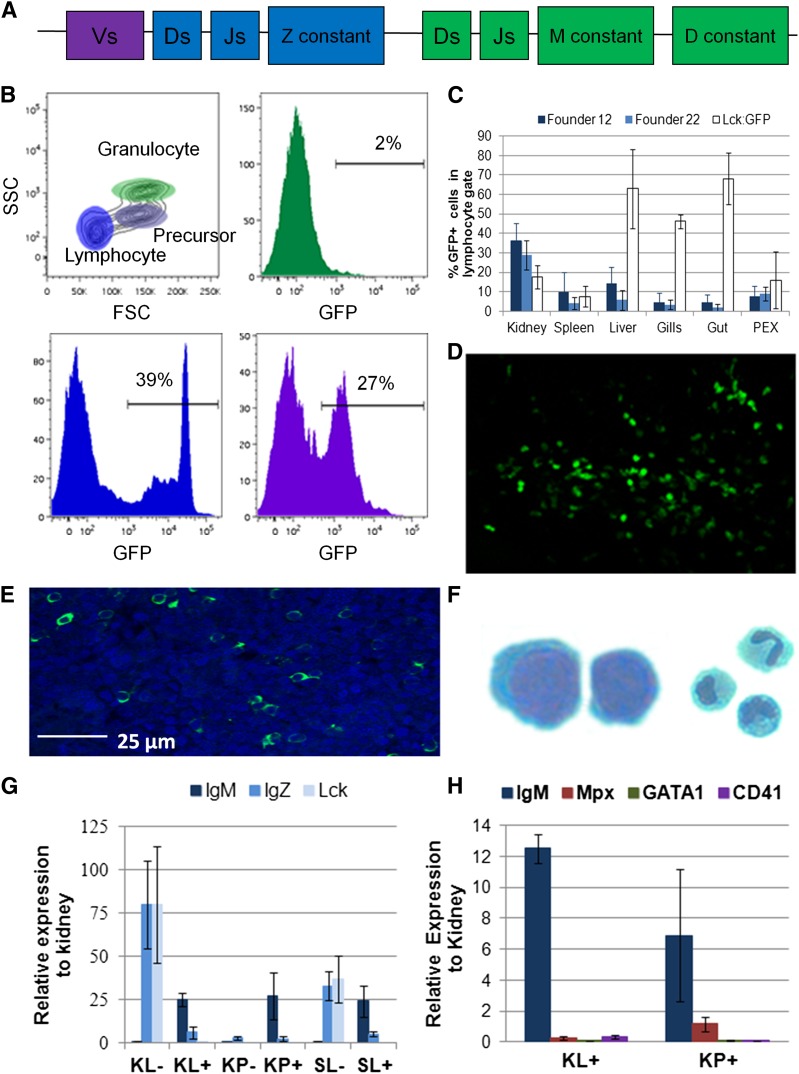
Figure 1.
Flow cytometric and gene expression analysis of IgM1:eGFP zebrafish. (A) Map of the zebrafish heavy chain locus with variable (V), diversity (D), joining (J), and constant region exons for IgZ, IgM, and IgD. (B) Organs were isolated from individual fish age 3 to 10 months and analyzed for GFP expression by flow cytometry. The forward scatter and side scatter (FSC/SSC) profile obtained from kidney is shown, with %GFP fluorescence obtained in the lymphocyte (bottom left), precursor (bottom right), and granulocyte (top right) gates. These results are representative of 16 to 22 individual fish from 10 independent experiments. (C) %GFP+ cells in the lymphocyte gate (average plus standard deviation [SD]) of various organs from 2 to 22 individual fish from 2 independent founders of the IgM1:eGFP line (founders 12 and 22) or from lck:GFP reporter fish for comparison. (D) Confocal imaging of GFP+ cells in the skin of the anterior portion of the trunk. (E) Merged image of 4,6 diamidino-2-phenylindole (DAPI) (blue) and anti-GFP (green) staining in the kidney. The image was taken with the ×25 objective of a Leica SP5 inverted confocal microscope. (F) GFP+ cells were sorted to at least 95% purity from the lymphocyte fraction of the kidney, stained with May-Grunwald/Giemsa stain, and photographed at ×1000 (left). More than 95% of the cells exhibited a lymphocytic morphology. Cells sorted from the granulocyte fraction of the kidney are shown for comparison (right, photographed at ×40). These results are a summary of 4 independent experiments. (G) GFP− and GFP+ cells were sorted from the lymphocyte (KL–, KL+) or precursor (KP−, KP+) fraction of the kidney or the lymphocyte fraction of the spleen (SL−, SL+) from 2 to 3 fish and analyzed by qRT-PCR. Average expression plus standard error of the mean (SEM) of IgM, IgZ, or lck relative to their expression in adult kidney (ΔΔCt) is shown. These results are a summary of 3 independent experiments. (H) GFP+ cells were sorted from the lymphocyte (KL+) or precursor (KP+) fraction of the kidneys from 2 to 3 fish. Average expression plus SD of IgM, mpx, gata1, and cd41 relative to their expression in adult kidney is shown. These results are a summary of 2 independent experiments.
To better understand the biology of zebrafish B cells, we generated novel transgenic reporter animals that mark the major IgM-expressing B cell subset. By analyzing the development and activation of B cells in the zebrafish, we hope to gain insight into the evolutionary development of adaptive immunity. Here we present the first characterization and analysis of zebrafish IgM+ B lymphocytes.
Methods
Zebrafish strains
Animals were maintained in accordance with University of California at San Diego (UCSD) Institutional Animal Care and Use Committee (IACUC) guidelines. Animal procedures were performed with the approval of the UCSD and Point Loma Nazarene University (PLNU) IACUCs. The transgenic lines used were rag2:DsRed14 = Tg(rag2:loxP-DsRed2-loxP-Hsa.KRAS_G12D), lck:eGFP6 = Tg(lck:lck-EGFP)cz1, insulin:GFP,15 fli1:DsRed16 = Tg(fli1a.ep:DsRedEx)um13, Tg(cd45:DsRed),17 IgM1:eGFP = Tg(Cau.Ighv-ighm:EGFP)sd19, blimp1:eGFP18 = TgPAC_F2219(prdm1a:EGFP)i106, and Tg(kdrl:cerulean)sd24.
Generation of transgenic animals
IgM1:eGFP animals were generated by cloning the 3-kb zebrafish Ig heavy chain enhancer and 0.2-kb goldfish V-region promoter19 upstream of green fluorescence protein (GFP) in a Tol2 transposon construct, which was injected with Tol2 transposase messenger RNA into 1-cell stage AB* embryos.20 Founders were identified by polymerase chain reaction (PCR) of genomic DNA from F1 embryos for GFP; transgenic F2 fish were identified by PCR of fin-clip DNA.
Tg(kdrl:cerulean) animals were generated by cloning 6.8 kb of the kdrl promoter/enhancer sequences21 upstream of the cerulean reporter construct. A total of 100 pg of kdrl:cerulean plasmid DNA was injected into 1-cell stage wild-type embryos with I-SceI meganuclease.22 Four founders were identified by screening for endothelial-specific fluorescent progeny, and Tg(kdrl:cerulean)sd24 exhibited the strongest expression.
Flow cytometry and cytology
Single-cell suspensions were prepared from zebrafish organs as described.9,23 Flow cytometry was performed with a fluorescence-activated cell sorter (FACS) AriaI or FACSCanto flow cytometer (Becton Dickinson). Cytospins and May-Grunwald/Giemsa stains were performed as described.24
Real-time qRT-PCR
RNA was purified from cells or homogenized tissues using RNeasy (Qiagen) and converted to complementary DNA (cDNA) by using SuperScript III First-Strand Synthesis System (Invitrogen) or qScript Flex cDNA Synthesis Kit (Quanta BioSciences) with oligo-dT (deoxy-thymine) primers. cDNA was amplified using BrilliantII SYBR Green QPCR Master Mix (Agilent) with 10 pmol of each primer. Quantitative reverse-transcription PCR (qRT-PCR) was performed with the Mx3000P System (Stratagene) or the Chromo4 System (BioRad). Elongation factor 1-alpha (ef1α) expression was scored for each sample in every experiment. Data were analyzed by the ΔCt method by using EF1α for normalization (2−(Ct,IgM – Ct,EF1α)) or the ΔΔCt method using EF1α and adult kidney for normalization (2[Ct(kidney,IgM)-Ct(organ,IgM)] × 2[Ct(organ,EF1α)-Ct(kidney,EF1α)]). The following primers were used:
BLIMP1: GGCGTCTTTACCTAGAAACC; ATCTTTGGGGACATTTTCTG
CD40: AGAGTTGCCGTTAAAGGTTC; TTCTCCGTACTCACATTTGG
CD41: CTGAAGGCAGTAACGTCAAC; TCCTTCTTCTGACCACACAC
EF1a: GAGAAGTTCGAGAAGGAAGC; CGTAGTATTTG TGGTCTCG
eGFP: GACCACATGAAGCAGCAC; TTGTCGGCCATGATATAGAC
GATA1: TGAATGTGTGAATTGTGGTG; ATTGCGTCTCCATAGTGTTG
IgM: AGCTTCTCTAGCTCCACCAG; ATTTTGGTGAAATGGAATTG
IgZ: AAAGCAACGATACCAAAGTG; AACAGCTTGCAAGACAATTC
IRF4: AACTGGAGAAAGAGCAGACC; AGCACAGAATGACCTGAGAG
Lck: TACGTAAACATGGGGAACTG; TCTTCTCCCCTTTCTCAAAC
MPX: TGATGTTTGGTTAGGAGGTG; GAGCTGTTTTCTGTTTGGTG
Pax5: CTGATTACAAACGCCAAAAC; CTAAATTATGCGCAGAA CG
Xbp1: TCAGATTCAGACTCCACCAC; TGTCTCTTGCTGTCTGTGC
Zebrafish immunization
Zebrafish were immunized by intraperitoneal injection with 5 μg keyhole limpet hemocyanin (KLH) (Sigma) emulsified in complete Freund’s adjuvant (1 mg/mL; Sigma), followed 2 to 3 weeks later by injection with 5 μg KLH emulsified in incomplete Freund’s adjuvant (IFA) (1 mg/mL; Sigma).
Immunohistochemistry
Frozen sections of zebrafish gut masses were fixed in 4% paraformaldehyde and permeabilized with methanol. For other organs, paraffin sections were prepared after fixation and decalcification of fish overnight at 4°C with Cal-ExII (Fisher). Sections were then incubated in Tris-EDTA at 85°C for 2 hours. Sections were stained with chicken anti-GFP (1:500; Aves Labs) overnight at 4°C, followed with AlexaFluor488-conjugated goat anti-chicken antibody (1:1000; Invitrogen) for 1 hour at 20°C, and a final 4′6 diamidino-2-phenylindole wash.
Phagocytosis assays
For in vivo phagocytosis assays, 2 μL pHrodo-Escherichia coli (Molecular Probes) was injected intraperitoneally into adult fish. Peritoneal exudate (PEX) was collected 1 to 10 hours after injection for flow cytometric analysis. For in vitro phagocytosis assays, 50 μL pHrodo-E coli was incubated with 200 μL of sorted cells in media at 32°C as described24 and then analyzed by flow cytometry.
Results
Generation and validation of IgM+ B-cell reporter fish
The regulatory modules that control expression of IgM have been well characterized. In mammals, an enhancer situated upstream of the first μ constant exon is required for B-cell–specific expression in transgenic animals.25 A similar enhancer situated between the μ and δ constant regions in zebrafish exhibited B-cell–specific expression when driving a goldfish V-region promoter in transient transfection assays.19 To create a transgenic reporter construct that would mark IgM+ B cells in zebrafish, these regulatory modules were cloned upstream of eGFP in a Tol2 vector, which was injected into 1-cell stage embryos with Tol2 transposase messenger RNA.20
We then validated two independent IgM1:eGFP transgenic founders. Transgenic F2 fish were assessed by flow cytometry and qRT-PCR to verify appropriate expression of the eGFP transgene. Studies in several teleosts have shown that hematopoiesis in adult animals occurs in the kidney.26,27 Additionally, mature blood cell lineages in the adult zebrafish kidney can be fractionated and resolved by flow cytometry based on forward scatter and side scatter differences, which delineate lymphoid, myeloid, erythroid, and precursor populations.4,5 The identity of these populations has been verified by Wright-Giemsa staining and qRT-PCR analysis of lineage-specific gene expression.5,23 As expected, GFP fluorescence from the IgM1:eGFP line was present in the lymphoid and precursor cell fractions in the kidney, which contain lymphocytes and hematopoietic stem cells (lymphoid fraction) and lymphoid and myeloid progenitor cells (precursor fraction).24 Importantly, GFP fluorescence was not detected in the granulocytic fraction, which primarily contains neutrophils and monocytes9 (Figure 1B). Flow cytometric analysis of other organs (Figure 1C) showed that B cells are present in the lymphocyte fractions of the spleen, liver, gills, gut, and PEX from two independent founders. Correspondingly, microscopic analysis and immunohistochemistry revealed GFP expression in cells in the skin, kidney, and spleen (Figure 1D-E; supplemental Figure 1).
To characterize the IgM1:eGFP+ cells, we sorted them from the lymphoid and precursor fractions of the kidney and from the lymphoid fraction of the spleen and analyzed them by qRT-PCR, comparing them to GFP− cells from the same fractions (Figure 1G). The GFP+ cells expressed IgM, but not IgZ or lck, a tyrosine kinase that is expressed in T cells,6 confirming that they were IgM-expressing B cells. In contrast, the GFP− cells expressed both lck and IgZ, indicating that this population contains a mixture of T cells and IgZ-expressing B cells. As expected, the GFP+ cells did not express cd41, gata1, or myeloperoxidase (mpx) (Figure 1H), which are expressed in cells from the thrombocytic, erythrocytic, and granulocytic lineages, respectively.5,7,8,28 Correspondingly, GFP+ lymphocytes from the kidney revealed a typical lymphocytic morphology (Figure 1F). These results indicate that GFP is appropriately expressed in IgM+ B lymphocytes in two independent IgM1:eGFP founder lines.
Analysis of IgM+ B cell development and ontogeny
Although hematopoiesis in adult zebrafish occurs in the kidney,27 in situ hybridization studies suggested that embryonic B cells might arise in the pancreas at 4 to 11 days postfertilization (dpf).29 To examine whether our transgenic lines recapitulated these findings, we first mated rag2:DsRed fish, which label both developing B and T cells,6,30 to insulin:GFP animals, which label β cells in the developing pancreas.15 Confocal imaging between 4 and 11 dpf revealed no rag2 expression near the pancreas, whereas high rag2 expression—corresponding to immature T cells—was observed only in the thymus, as previously described6 (supplemental Figure 2). Moreover, pancreatic B-cell development has not been observed in other teleosts.26
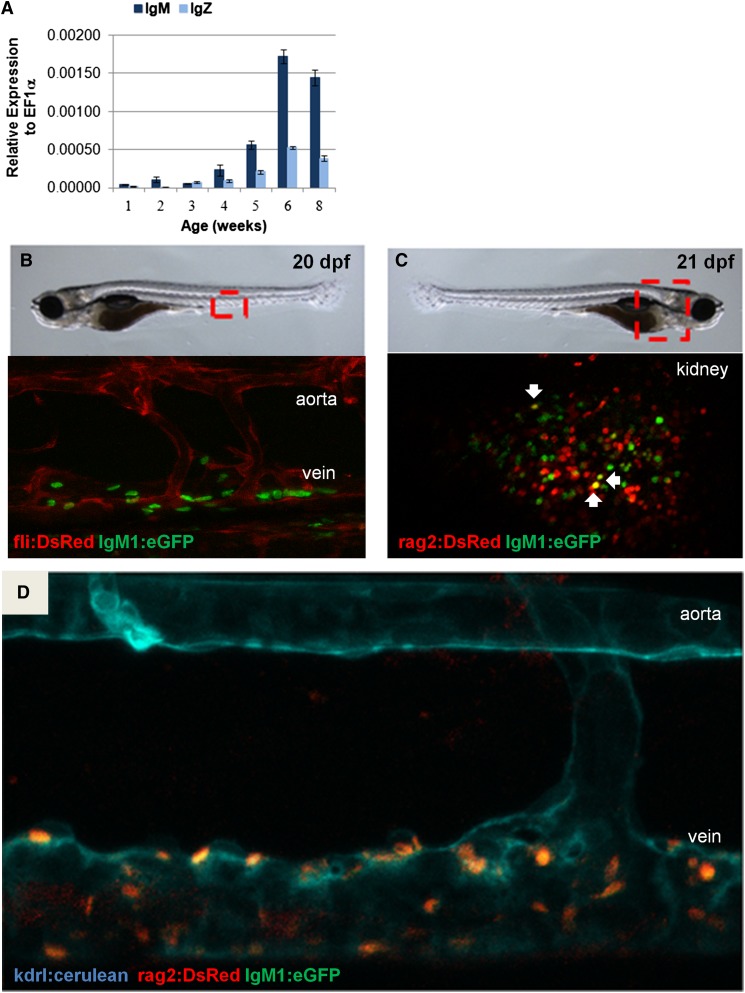
To further examine B-cell development, qRT-PCR analysis of whole larvae was performed, revealing that neither IgM nor IgZ expression was detectable until approximately 3 weeks postfertilization (wpf) (Figure 2A). Fish from several transgenic lines were next analyzed by confocal microscopy to determine the temporal and spatial location of the first IgM+ B cells in the developing zebrafish. Hematopoietic stem cells in zebrafish emerge along the dorsal aorta by 36 hours postfertilization and then colonize the thymus, caudal hematopoietic tissue, and kidney.17,31 To examine whether B cells were first detectable in these sites of hematopoietic development, we imaged double-transgenic fli1:DsRed;IgM1:eGFP fish to visualize developing IgM+ lymphocytes within the context of fli1+ vascular cells.32 Clusters of GFP+ cells were observed between the dorsal aorta and posterior cardinal vein around 20 dpf, suggesting that B cells could arise in this region (Figure 2B). This result was further corroborated by confocal imaging of rag2:DsRed;IgM1:eGFP fish, which revealed GFP+ B cells in the same area and in the kidney around 20 dpf (Figure 2C). A few of these cells were Rag2+IgM+ (yellow), as expected for the earliest IgM-expressing B cells. Because vascular cells also express flk (kdrl),21 we imaged triple-transgenic kdrl:cerulean;rag2:DsRed;IgM1:eGFP fish and again saw GFP+ cells appearing near the cardinal vein around 20 dpf (Figure 2D; supplemental Figure 3). Taken together, these results show that IgM+ B cells in zebrafish are found between the axial vessels of the trunk and in the kidney around 20 dpf.
Figure 2.
IgM expression during development. (A) qRT-PCR analysis was performed on individual whole fish. Average expression plus SEM of IgM or IgZ relative to ef1α (ΔCt) for 3 to 4 fish per time point is shown. (B) Confocal imaging of the area between the dorsal aorta and the posterior cardinal vein of fli1:DsRed;IgM1:eGFP fish at 20 to 21 dpf. (C) Confocal imaging of the kidney of rag2:DsRed;IgM1:eGFP fish at 21 dpf. DsRed+eGFP+ cells are indicated with arrows. (D) Confocal imaging of the area between the dorsal aorta and the posterior cardinal vein of kdrl:cerulean;rag2:DsRed;IgM1:eGFP fish at 22 dpf. All of the cells near the vein are DsRed+eGFP+. These results are representative of results obtained with at least 3 individual fish of each type. All embryos were imaged in water at room temperature with an ×25 objective using a Leica SP5 inverted confocal microscope with Leica Application Suite Advanced Fluorescence (LAS AF) acquisition and analysis software.
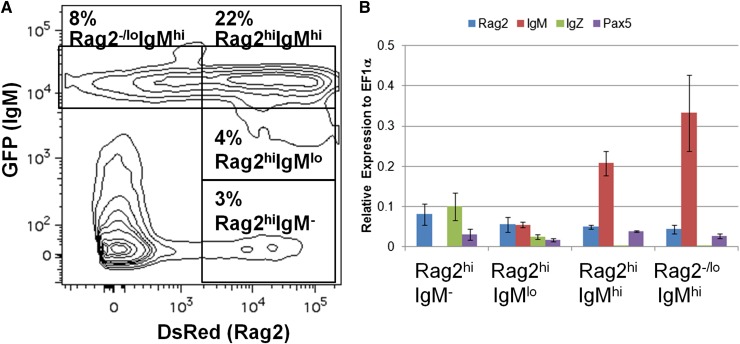
To investigate B-cell ontogeny in the adult, we analyzed cell populations in the kidneys of rag2:DsRed;IgM1:eGFP fish. Flow cytometric analysis of the lymphocyte and precursor fractions in the kidney revealed four prominent populations: Rag2+IgM−, Rag2+IgMlo, Rag2+IgMhi, and Rag2−/loIgMhi (Figure 3A). To examine gene expression in these populations, the cells were sorted and analyzed by qRT-PCR (Figure 3B). As expected, increasing levels of IgM expression correlated with levels of GFP fluorescence. All of the populations expressed pax5, a transcription factor that is expressed in most stages of B-cell development,33 whereas only the IgM− and IgMlo populations expressed IgZ. Each population also expressed rag2, although its expression was lower in the IgM-expressing populations. These results show patterns of gene expression similar to those observed during mammalian B-cell ontogeny from pro-B cells (Pax5+Rag2+IgM−) to pre-B cells (Pax5+Rag2+IgM+) to immature/mature B cells (Pax5+Rag2−/loIgM+), indicating that IgM1:eGFP transgenic fish closely recapitulate mammalian B-cell ontogeny.33,34
Figure 3.
B-cell ontogeny in IgM1:eGFP fish. (A) Kidneys were isolated from adult rag2:DsRed;IgM1:eGFP fish and analyzed by flow cytometry. The GFP/DsRed profile of cells in the combined lymphocyte and precursor gates is shown. The indicated populations were sorted from the fish and analyzed by qRT-PCR. (B) Average expression plus SD of rag2, IgM, IgZ, or pax5 relative to ef1α (ΔCt) for the indicated cell populations from 3 individual fish is shown.
Analysis of mature IgM+ B cells and plasma B cells
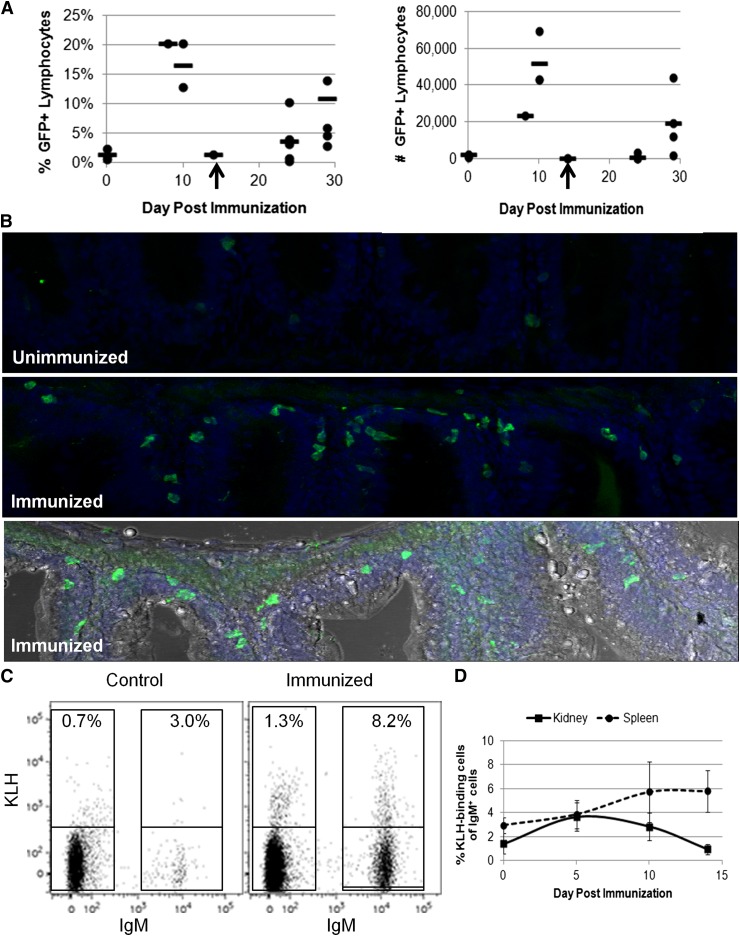
With the ability to now specifically track B lymphocytes, we used IgM1:eGFP animals to study B-cell responses. Previous studies examined B-cell responses in trout or zebrafish via IgM secretion, which was detected 4 weeks after intraperitoneal injection with T-cell–dependent antigens.35,36 To examine IgM+ B-cell function in our transgenic animals, IgM1:eGFP fish were immunized with the T-cell–dependent antigen KLH emulsified in complete Freund’s adjuvant. After several weeks, the fish were boosted with KLH in IFA. The percentage and number of IgM+ B cells from various organs were examined by flow cytometry at several time points. Although lymph nodes have not been detected in zebrafish, lymphocytes are found in the kidney, spleen, skin, and peritoneal cavity, as well as in lymphoid tissue associated with the gut and gills5,6,37 (Figure 1). Correspondingly, we observed increases in the percentage and number of IgM+ B cells in the kidney, spleen, gut, and PEX; however, there was substantial variability from fish to fish (Figure 4A; supplemental Figure 4). Immunohistochemistry of gut sections also revealed a threefold increase in the number of GFP-expressing B cells in animals boosted with KLH in IFA 14 days previously (Figure 4B). To more accurately measure the immune response, we conjugated KLH to AlexaFluor647 and measured the number of antigen-binding B cells in various organs (Figure 4C). The kidney, spleen, gut, and PEX from individual fish were analyzed after the booster immunization. The highest percentages of KLH-binding B cells were observed in the spleen 14 days after the booster immunization (Figure 4D) with 2% to 8% of the IgM+ B cells binding KLH compared with only 1% to 3% in control fish. These results suggest that significant numbers of antigen-binding B cells in zebrafish are not present until 10 to 14 days after immunization.
Figure 4.
Analysis of IgM+ B cells during an immune response. Adult IgM1:eGFP fish were injected intraperitoneally with KLH in complete Freund’s adjuvant and then boosted 14 to 21 days later with KLH in IFA. (A) Spleens were isolated from individual fish at the indicated times and analyzed by flow cytometry for the percentage and number of GFP+ lymphocytes. Each point represents 1 fish, and the line represents the average response at that time point. Two independent experiments are represented in these data. The booster immunization is indicated with an arrow. (B) The gut mass was isolated from individual fish 14 days after the booster immunization and sectioned. DAPI (blue) /anti-GFP (green) immunohistochemical analyses from an unimmunized (top) and immunized (middle) fish are shown. Also shown is a compilation of the DAPI and anti-GFP stains in an immunized fish with light microscopy for context (bottom). There was an average of 10 ± 4 (SD) GFP+ cells in unimmunized gut sections vs 35 ± 14 (SD) GFP+ cells in immunized gut sections. These data are representative of results from 3 unimmunized and 4 immunized fish. The images were taken with a ×25 objective using a Leica SP5 inverted confocal microscope with LAS AF acquisition and analysis software. (C) Organs were isolated from individual adult IgM1:eGFP fish and analyzed by flow cytometry for KLH-binding cells. The percentages of KLH-binding cells from a control and immunized spleen 14 days after the booster immunization are shown. (D) Average percentages of KLH-binding cells among the IgM+ B cells + SD at several time points after the booster immunization (2 to 5 fish per time point). These results are representative of 2 independent experiments.
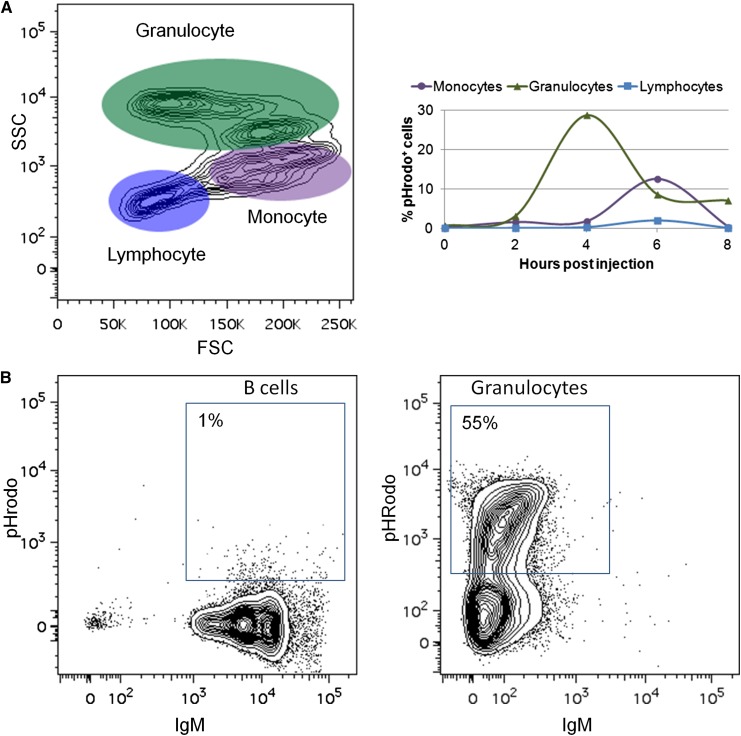
To examine another hallmark of teleost B cells,38 we analyzed the ability of zebrafish IgM+ B cells to phagocytose pathogens. Zebrafish were injected with pHrodo-labeled E coli, which contain a rhodamine-based dye that emits fluorescence in the acidic environment of the phagolysosome.39 In contrast with previous studies,38 zebrafish B cells were only marginally phagocytic, with no more than 2% of zebrafish B cells phagocytosing pathogens (Figure 5A). This assay was repeated using latex beads38 with similar results (data not shown). Finally, we purified B cells and granulocytes from the kidneys of IgM1:eGFP fish by flow cytometric sorting and incubated them in vitro with pHrodo-labeled E coli. More than half the granulocytes were phagocytic, but only 1% of the B cells phagocytosed the E coli (Figure 5B).
Figure 5.
Zebrafish B cells are marginally phagocytic. (A) Adult zebrafish were injected intraperitoneally with fluorescent E coli (pHrodo). After several hours, PEX was collected and analyzed by flow cytometry. Left: FSC/SSC profile of cells from the PEX. Right: Total percentage of pHrodo+ cells in each cell population at various time points. (B) IgM+ B cells and granulocytes were purified by flow cytometric sorting from adult IgM1:eGFP fish and cultured in vitro with pHrodo. Flow cytometric profiles of B cells and granulocytes after 4 hours of culture are shown. These results are representative of at least 3 independent experiments.
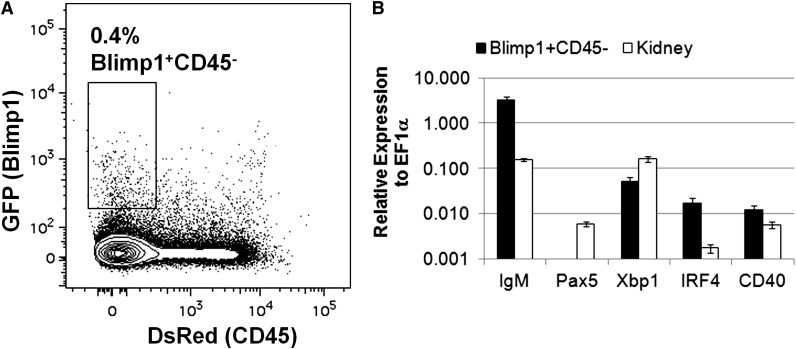
With the ability to study B lymphocytes in the zebrafish, we set out to identify and characterize plasma cells. The transcription factor Blimp1 is required for plasma B-cell development in mammals,40 and blimp1:GFP fish18 possess GFP+ cells in the lymphocyte and precursor fractions from adult kidneys (supplemental Figure 5). To prospectively isolate zebrafish plasma cells, we examined double transgenic cd45:DsRed;blimp1:GFP fish by flow cytometry. Whereas the endogenous cd45 gene is expressed in all zebrafish leukocytes, we previously showed that a cd45:DsRed transgene marks T cells but not B cells.9 Flow cytometric analysis of lymphocytes and precursors from the kidneys of cd45:DsRed;blimp1:GFP fish revealed a small CD45−Blimp1+ population, as well as larger populations of CD45−Blimp1− and CD45+Blimp1− cells (Figure 6A). Each fraction was sorted and analyzed by qRT-PCR for markers of plasma B cells. The CD45−Blimp1+ population expressed large amounts of IgM, but no pax5 (Figure 6B), in agreement with results for mammalian plasma B cells.33 Further qRT-PCR analysis of this population revealed that these cells also expressed other markers of plasma B cells,41 including cd40 and the transcription factor X-box binding protein 1 (xbp1) and interferon regulatory factor 4 (irf4) (Figure 6B). These data suggest that cd45:DsRed;blimp1:GFP reporter fish can be used to isolate and characterize zebrafish plasma B cells.
Figure 6.
Isolation and analysis of plasma B cells in zebrafish. (A) Kidneys were isolated from adult cd45:DsRed;blimp1:eGFP fish and analyzed by flow cytometry. The GFP/DsRed profile of cells in the combined lymphocyte and precursor fractions is shown. The indicated population of cells was sorted from individual fish and analyzed by qRT-PCR. (B) Average expression (plus range) of IgM, pax5, irf4, xbp1, or cd40 relative to ef1α (ΔCt) for cells from the Blimp1+CD45− fraction from two individual fish using whole-kidney cDNA as a comparison. These results are representative of 3 independent experiments.
Discussion
We have generated a novel transgenic zebrafish line to characterize the temporal and spatial development of B cells in the early embryo, as well as to study the function of these cells in the adult organism. Specifically, the IgM1:eGFP transgenic line allows precise tracking of IgM+ B cells. This line represents the first nonmammalian B-cell reporter animal, and it is highly specific, since it marks IgM- but not IgZ-expressing B cells. Moreover, we illustrate the usefulness of combining this transgenic line with several other previously generated reporter fish to study B-cell development and function. In particular, IgM1:eGFP;rag2:DsRed animals can be used to define pro-B, pre-B, and immature/mature B-cell populations via their differential expression of IgM, rag2, and pax5. We also show that crossing the IgM1:eGFP fish with our previously described cd45:DsRed fish can be used to visualize and isolate B cells, T cells, and monocytes. We note that this is because of a unique feature of this CD45- transgenic line whereby DsRed is expressed in all leukocytes except B cells.9 Thus, flow cytometric analysis of IgM1:eGFP;cd45:DsRed animals yields GFP+IgM+ B cells and DsRed+ T cells within the lymphoid fraction. Moreover, the cd45:DsRed;blimp1:GFP double transgenic line can be used to visualize and isolate presumptive plasma B cells based on their CD45−Blimp1+ phenotype. Taken together, the IgM1:eGFP reporter line can be used alone or in combination with other lineage-specific reporter lines to further immunologic studies in the zebrafish, including immune cell development and activation, host-pathogen interactions, and forward genetic screens.
We also show that the first B cells in zebrafish arise at 20 to 21 dpf. We did not observe B cells in or around the pancreas at earlier time points, as previously suggested.29 Instead, we first detected them between the dorsal aorta and posterior cardinal vein. We also show that Rag2+ and/or IgM+ cells are present in the kidney around this time. Although we expected to observe developing B cells in the kidney, which is the site of hematopoiesis in adults, their appearance between the axial vessels of the trunk is a novel finding. These results suggest that B-cell development is dynamic and features shifting sites during development, similar to a developmental shift in mammals whereby stem cells from the fetal liver engraft in the bone marrow for the remainder of the organism’s life.42 A similar region near the dorsal aorta in chickens, termed “para-aortic foci,” shows clusters of CD45+ cells that express genes characteristic of lymphoid and myeloid progenitors at 6 to 7 days of embryonic development.43 It will be interesting to determine the precise relationships of the zebrafish sites of B-cell development via precursor-progeny and lineage-tracing experiments, since our imaging-based approaches did not uniformly detect initial B-cell emergence in either site. Our results in zebrafish, however, suggest that shifting sites of B-cell development are a feature of all jawed vertebrates.
It was initially surprising to us that the earliest B cells in zebrafish were not detected until 3 wpf, since very low levels of IgM, IgD, and IgZ transcripts were detected at 2 dpf in previous studies.10 However, the transcript levels of these genes were increased 10-fold at 14 dpf.10 There are several possible reasons for the differences between our findings, including the size of the fish, which can vary greatly depending on caloric intake, temperature, and the strain of the fish. In particular, our experiments were performed by using larvae fed artificial artemia, which grow more slowly than rotifer-fed animals.
In contrast to B-cell development, T cells arise early in zebrafish, with lck+ cells in the thymus at 4 dpf and then in circulation by 8 dpf.6 In mice, γδ-T-cell antigen receptor (TCR) –expressing cells appear first, followed several days later by the main population of αβ-TCR–expressing cells.44 It will be interesting to discover when the αβ-TCR–expressing cells arise in zebrafish now that we know the first B cells arise by 20 to 21 dpf. TCRβ chain expression in zebrafish was detected at 5 dpf,45 but TCRα chain expression has not been examined. Interestingly, it was recently shown that the adult pattern of globin gene expression does not begin until 22 dpf in zebrafish.46 This group also found that hematopoietic stem cells, which emerge at 36 hours postfertilization, did not contribute to the mature erythroid pool until 10 to 14 dpf. This suggests that a shift from larval to adult programming occurs around 3 wpf. This observation correlates closely with our observations of B-cell development, suggesting that the adaptive immune system in zebrafish may not be fully functional until this larval-to-adult shift occurs. In support of this hypothesis, sequencing of the complete zebrafish antibody repertoire reveals limited usage of VDJ gene segment combinations in 2-week-old fish with more diversification as the zebrafish age.47
Although B cells from several teleost species are capable of phagocytosis,38 we did not observe significant phagocytosis of E coli or latex beads by zebrafish B cells, either in vivo or in vitro. In multiple assays, we never observed more than 2% of zebrafish B cells to be phagocytic. Interestingly, up to 60% of the B cells in other bony fish exhibited phagocytic activity to latex beads, whereas only 11% to 14% of the mouse B1-cell population exhibited phagocytosis when injected with E coli,39 which is the same assay that we used. It is possible that zebrafish B cells are different from those of other teleosts; alternatively, some unknown component of the culture system could be contributing to the discrepancy.
Interestingly, the immune response in zebrafish appears to be delayed relative to mammals, where germinal center B cells appear in lymphoid tissue only 2 to 4 days after immunization and plasma B cells appear in the bone marrow by 7 to 10 days.48 In contrast, we did not observe significant numbers of total B cells or antigen-binding B cells until 10 to 14 days postimmunization. These results explain why IgM secretion is not detected until 4 weeks after intraperitoneal immunization of zebrafish or trout.35,36 We found increases in B cell numbers in the kidney, spleen, gut, and PEX after intraperitoneal immunization, although the most dramatic increases in antigen-binding B cells were in the spleen and gut. Correspondingly, these sites were reported to be likely sites of immune responses to bacterial infections in zebrafish.49 Although neither classical germinal centers nor lymph nodes have been described in fish, germinal center-like structures were found in the spleen of catfish.50 With our new transgenic animals, we have generated the appropriate reagents to more precisely address the architecture of the immune response in fish.
We also show that zebrafish plasma B cells can be isolated by using combined cd45:DsRed;blimp1:GFP transgenic lines. The CD45−Blimp1+ lymphoid population displayed a gene expression profile highly characteristic of mammalian plasma cells. In particular, the lack of pax5 and the expression of irf4, xbp1, and cd40 are expected of plasma B cells.33,41 These transgenic animals, along with the IgM1:eGFP animals, will allow for a detailed analysis of the time course and location of plasma B-cell development during various types of immune responses in zebrafish.
In conclusion, we have generated IgM1:eGFP reporter zebrafish, which in combination with other available reporter lines, will be an essential tool for studies of B-cell development and activation, as well as for an integrated study of how each immune cell subset interacts in vivo, thus adding to our understanding of the evolution of the adaptive immune response.
Supplementary Material
Acknowledgments
We thank David Stachura for critical review of the manuscript and assistance with zebrafish cell culture and cell sorting and Albert Kim for assistance with confocal microscopy.
This work was supported by National Science Foundation grant IOS-1052561 (D.M.P. and D.T.), National Science Foundation grant MRI-0619057 (D.M.P.), a Natural Sciences and Engineering Research Council of Canada grant (B.G.M.), a Howard Hughes Medical Institute Science Education Grant (to PLNU, and by PLNU Research Associates.
Footnotes
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.M.P. wrote the paper, designed and performed research, and analyzed data; V.W. and J.Y.B. designed and performed research and analyzed data; K.L.L., D.N.P., N.D., S.E.S., C.M., B.H.J., A.D., and Y.P. performed research and analyzed data; H.Y. and N.C.C. made the kdrl:cerulean fish; B.G.M. made the IgM1:eGFP DNA construct; and D.T. designed research, assisted in writing the paper, and provided crucial reagents and laboratory space.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Corresponding author: Dawne M. Page, Department of Biology, Point Loma Nazarene University, 3900 Lomaland Dr, RS102B, San Diego, CA 92106; e-mail: dapage@pointloma.edu.
References
- 1.Boehm T, Iwanami N, Hess I. Evolution of the immune system in the lower vertebrates. Annu Rev Genomics Hum Genet. 2012;13:127–149. doi: 10.1146/annurev-genom-090711-163747. [DOI] [PubMed] [Google Scholar]
- 2.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124(4):815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10(5):1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- 4.Traver D. Cellular dissection of zebrafish hematopoiesis. Methods Cell Biol. 2004;76:127–149. doi: 10.1016/s0091-679x(04)76008-2. [DOI] [PubMed] [Google Scholar]
- 5.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4(12):1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 6.Langenau DM, Ferrando AA, Traver D, et al. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci USA. 2004;101(19):7369–7374. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol. 2006;80(6):1281–1288. doi: 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- 8.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 9.Wittamer V, Bertrand JY, Gutschow PW, Traver D. Characterization of the mononuclear phagocyte system in zebrafish. Blood. 2011;117(26):7126–7135. doi: 10.1182/blood-2010-11-321448. [DOI] [PubMed] [Google Scholar]
- 10.Zimmerman AM, Moustafa FM, Romanowski KE, Steiner LA. Zebrafish immunoglobulin IgD: unusual exon usage and quantitative expression profiles with IgM and IgZ/T heavy chain isotypes. Mol Immunol. 2011;48(15-16):2220–2223. doi: 10.1016/j.molimm.2011.06.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16(4):257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Danilova N, Bussmann J, Jekosch K, Steiner LA. The immunoglobulin heavy-chain locus in zebrafish: identification and expression of a previously unknown isotype, immunoglobulin Z. Nat Immunol. 2005;6(3):295–302. doi: 10.1038/ni1166. [DOI] [PubMed] [Google Scholar]
- 13.Zhang YA, Salinas I, Li J, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11(9):827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langenau DM, Keefe MD, Storer NY, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21(11):1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H, Vogel SS, Liu N, Melton DA, Lin S. Analysis of pancreatic development in living transgenic zebrafish embryos. Mol Cell Endocrinol. 2001;177(1-2):117–124. doi: 10.1016/s0303-7207(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 16.Covassin LD, Siekmann AF, Kacergis MC, et al. A genetic screen for vascular mutants in zebrafish reveals dynamic roles for Vegf/Plcg1 signaling during artery development. Dev Biol. 2009;329(2):212–226. doi: 10.1016/j.ydbio.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135(10):1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elworthy S, Hargrave M, Knight R, Mebus K, Ingham PW. Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development. 2008;135(12):2115–2126. doi: 10.1242/dev.015719. [DOI] [PubMed] [Google Scholar]
- 19.Ellestad KK, Magor BG. Evolution of transcriptional enhancers in the immunoglobulin heavy-chain gene: functional characteristics of the zebrafish Emu3′ enhancer. Immunogenetics. 2005;57(1-2):129–139. doi: 10.1007/s00251-005-0785-3. [DOI] [PubMed] [Google Scholar]
- 20.Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234(2):244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 21.Beis D, Bartman T, Jin SW, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132(18):4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 22.Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118(1-2):91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 23.Lugo-Villarino G, Balla KM, Stachura DL, Bañuelos K, Werneck MB, Traver D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc Natl Acad Sci USA. 2010;107(36):15850–15855. doi: 10.1073/pnas.1000494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stachura DL, Svoboda O, Lau RP, Balla KM, Zon LI, Bartunek P, Traver D. Clonal analysis of hematopoietic progenitor cells in the zebrafish. Blood. 2011;118(5):1274–1282. doi: 10.1182/blood-2011-01-331199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 26.Hansen JD. Characterization of rainbow trout terminal deoxynucleotidyl transferase structure and expression. TdT and RAG1 co-expression define the trout primary lymphoid tissues. Immunogenetics. 1997;46(5):367–375. doi: 10.1007/s002510050290. [DOI] [PubMed] [Google Scholar]
- 27.Willett CE, Cortes A, Zuasti A, Zapata AG. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev Dyn. 1999;214(4):323–336. doi: 10.1002/(SICI)1097-0177(199904)214:4<323::AID-AJA5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106(12):3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danilova N, Steiner LA. B cells develop in the zebrafish pancreas. Proc Natl Acad Sci USA. 2002;99(21):13711–13716. doi: 10.1073/pnas.212515999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langenau DM, Traver D, Ferrando AA, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299(5608):887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 31.Murayama E, Kissa K, Zapata A, et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25(6):963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Brown LA, Rodaway AR, Schilling TF, Jowett T, Ingham PW, Patient RK, Sharrocks AD. Insights into early vasculogenesis revealed by expression of the ETS-domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech Dev. 2000;90(2):237–252. doi: 10.1016/s0925-4773(99)00256-7. [DOI] [PubMed] [Google Scholar]
- 33.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27(1):49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 34.Roessler S, Grosschedl R. Role of transcription factors in commitment and differentiation of early B lymphoid cells. Semin Immunol. 2006;18(1):12–19. doi: 10.1016/j.smim.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Gong YF, Xiang LX, Shao JZ. CD154-CD40 interactions are essential for thymus-dependent antibody production in zebrafish: insights into the origin of costimulatory pathway in helper T cell-regulated adaptive immunity in early vertebrates. J Immunol. 2009;182(12):7749–7762. doi: 10.4049/jimmunol.0804370. [DOI] [PubMed] [Google Scholar]
- 36.Swan CM, Lindstrom NM, Cain KD. Identification of a localized mucosal immune response in rainbow trout, Oncorhynchus mykiss (Walbaum), following immunization with a protein-hapten antigen. J Fish Dis. 2008;31(5):383–393. doi: 10.1111/j.1365-2761.2008.00918.x. [DOI] [PubMed] [Google Scholar]
- 37.Bernard D, Six A, Rigottier-Gois L, et al. Phenotypic and functional similarity of gut intraepithelial and systemic T cells in a teleost fish. J Immunol. 2006;176(7):3942–3949. doi: 10.4049/jimmunol.176.7.3942. [DOI] [PubMed] [Google Scholar]
- 38.Li J, Barreda DR, Zhang YA, et al. B lymphocytes from early vertebrates have potent phagocytic and microbicidal abilities. Nat Immunol. 2006;7(10):1116–1124. doi: 10.1038/ni1389. [DOI] [PubMed] [Google Scholar]
- 39.Parra D, Rieger AM, Li J, et al. Pivotal advance: peritoneal cavity B-1 B cells have phagocytic and microbicidal capacities and present phagocytosed antigen to CD4+ T cells. J Leukoc Biol. 2012;91(4):525–536. doi: 10.1189/jlb.0711372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19(4):607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 41.Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J., Jr Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102(2):592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- 42.Douagi I, Vieira P, Cumano A. Lymphocyte commitment during embryonic development, in the mouse. Semin Immunol. 2002;14(6):361–369. doi: 10.1016/s1044532302000702. [DOI] [PubMed] [Google Scholar]
- 43.Säynäjäkangas R, Uchida T, Vainio O. Differential gene expression in CD45 cells at para-aortic foci stage of chicken haematopoiesis. Scand J Immunol. 2009;70(3):288–294. doi: 10.1111/j.1365-3083.2009.02304.x. [DOI] [PubMed] [Google Scholar]
- 44.Havran WL, Carbone A, Allison JP. Murine T cells with invariant gamma delta antigen receptors: origin, repertoire, and specificity. Semin Immunol. 1991;3(2):89–97. [PubMed] [Google Scholar]
- 45.Schorpp M, Bialecki M, Diekhoff D, et al. Conserved functions of Ikaros in vertebrate lymphocyte development: genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J Immunol. 2006;177(4):2463–2476. doi: 10.4049/jimmunol.177.4.2463. [DOI] [PubMed] [Google Scholar]
- 46.Ganis JJ, Hsia N, Trompouki E, et al. Zebrafish globin switching occurs in two developmental stages and is controlled by the LCR. Dev Biol. 2012;366(2):185–194. doi: 10.1016/j.ydbio.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang N, Weinstein JA, Penland L, White RA, III, Fisher DS, Quake SR. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc Natl Acad Sci USA. 2011;108(13):5348–5353. doi: 10.1073/pnas.1014277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarlinton DM, Smith KG. Dissecting affinity maturation: a model explaining selection of antibody-forming cells and memory B cells in the germinal centre. Immunol Today. 2000;21(9):436–441. doi: 10.1016/s0167-5699(00)01687-x. [DOI] [PubMed] [Google Scholar]
- 49.Pressley ME, Phelan PE, III, Witten PE, Mellon MT, Kim CH. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev Comp Immunol. 2005;29(6):501–513. doi: 10.1016/j.dci.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Saunders HL, Oko AL, Scott AN, Fan CW, Magor BG. The cellular context of AID expressing cells in fish lymphoid tissues. Dev Comp Immunol. 2010;34(6):669–676. doi: 10.1016/j.dci.2010.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.