Abstract
Living cells communicate and cooperate to produce the emergent properties of tissues. Synthetic mimics of cells, such as liposomes, are typically incapable of cooperation and therefore cannot readily display sophisticated collective behavior. Here, we print tens of thousands of picoliter aqueous droplets that become joined by single lipid bilayers to form a cohesive material with cooperating microcompartments. 3D structures can be built with heterologous droplets in software-defined arrangements. The droplet networks can be functionalized with membrane proteins to allow rapid electrical communication along a specific path. The networks can also be programmed by osmolarity gradients to fold into otherwise unattainable designed structures. Printed droplet networks might be used as tissue engineering substrates, interfaced with tissues, or developed as mimics of living tissue.
Cells in living tissues communicate with each other in a precisely controlled manner. As a consequence, tissues display the emergent properties that distinguish them from collections of independently functioning cells. The reproduction of similarly sophisticated behavior in a synthetic system would be of scientific and technological value, but is not feasible with existing artificial membrane platforms. Of the various classes of structures containing artificial bilayers, liposomes resemble cells most closely, but the controlled insertion of specialized membrane proteins such as gap junctions (1) would be required to join liposomes and form a cohesive, cooperative system.
Lipid-coated aqueous droplets in oil adhere at their interfaces to form stable bilayers (2–5), which can be functionalized with membrane proteins. Small two-dimensional networks of droplets connected in this way have been shown to act cooperatively as light sensors (5), batteries (5) or simple electrical circuits (6). Further, the droplets can release their contents to bulk aqueous solution after a change in pH or temperature (7–9). Droplet networks are therefore a promising platform for the construction of complex functional devices. However, functional networks have been limited to small groups of droplets assembled by manual (5–7) or mechanical (10) manipulation, microfluidic means (3, 11, 12) or external fields (13–15). Larger assemblies have been constructed by packing droplets into microfluidic containers (16, 17), but their complexity is limited by the uncontrolled filling process.
Here, we automatically print tens of thousands of heterologous picoliter droplets in software-defined, three-dimensional mm-scale geometries. The resulting macroscopic material is cohesive and self-supporting, and consists of distinct aqueous microcompartments partitioned by single lipid bilayers. Printing can take place in bulk oil or within oil drops that reside in aqueous solution. The bilayers can be functionalized with membrane proteins to allow electrical communication along a specific route. Printed droplet networks can also be programmed by osmolarity gradients to fold after printing into various designed geometries not accessible by direct printing. Three characteristics distinguish printed networks from other shape-changing materials, such as the bimetallic strip or hydrogels patterned to undergo non-uniform volume changes under external stimuli (18, 19). Droplet networks are readily printed, consist of compartments that can communicate through membrane proteins, and their folding is driven by internal differences in osmolarity. The latter characteristics make the folding behavior closely analogous to the nastic movements exhibited by certain plants (20, 21). Printed picoliter droplet networks constitute a defined synthetic platform for sophisticated collective behaviors (Fig. 1A), and might be developed for medical applications (22).
Fig. 1.
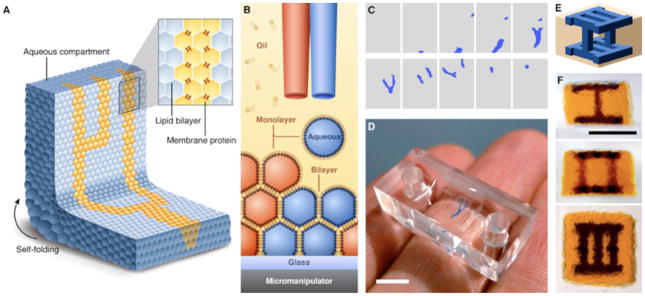
Printed droplet networks. (A) Illustration of a printed droplet network. (B) Schematic of the printing process. Two droplet generators eject droplets of different aqueous solutions into a solution of lipids in oil. The oil bath is mounted on a motorized micromanipulator. The droplets acquire a lipid monolayer, and form bilayers with droplets in the growing network. (C) Horizontal cross-sections of a design for a three-dimensional droplet network with a branching structure (blue) embedded in a cuboid (grey). The design comprises 20 layers of 50×35 droplets each; only alternate layers are shown. (D) Network printed according to the design in (C). Scale bar, 5 mm. (E) Schematic of a three-dimensional design that consists of 28 layers of 24×24 droplets each. (F) Three orthogonal views of a single network printed according to the design in (E). Scale bar, 1 mm.
Our strategy was to eject aqueous droplets (Fig. S1 and Text S1) within a bath of lipid-containing oil that was mounted on a motorized micromanipulator, so that a droplet network was built up in horizontal layers (Fig. 1B). The construction of droplet networks raises challenges that preclude the use of a commercially available printer (Text S2), and which we addressed with a specially-designed system (23) (Figs. S2 to S8 and Table S1). A network is defined in horizontal cross-sections one droplet thick (Fig. 1C), and a custom computer program (23) accordingly synchronizes the motion of the oil bath with the ejection of droplets from two droplet generators to construct the desired network (Fig. 1D). We have printed precisely defined networks consisting of up to ~35,000 heterologous droplets ejected at a rate of ~1 s−1 (Fig. 1, C to F).
Printed droplet networks are self-supporting (Fig. 1, D and F), and a thermodynamic analysis of the system indicates that stable networks can be printed with at least several thousand layers (23). The lattice of lipid bilayers also allows droplet networks to retain their shape under gentle perturbation; each bilayer lends an effective spring constant of ~4 mN m−1 to connected droplets, with a tensile strength of ~25 Pa (23) (Figs. S9 and S10). We estimate that Young’s modulus of printed networks is of the order of ~100–200 Pa (23) (Fig. S11), which is comparable to the elastic moduli of brain, fat and other soft tissues (24).
We have shown (7) that droplet networks can be stabilized in bulk aqueous solution by encapsulation within small drops of oil, for prospective applications in synthetic biology and medicine. Whereas these networks were created manually and therefore were limited in complexity, here we demonstrate the printing of encapsulated networks consisting of thousands of droplets in designed three-dimensional patterns. This was achieved by printing inside an oil drop suspended in aqueous solution (23) (Fig. 2A and Text S3). Once printing is complete, excess oil can be removed by suction through one of the printing nozzles. Encapsulated printed networks (Fig. 2, B to D) were stable for at least several weeks, and will therefore serve to expand the functions previously demonstrated with simple encapsulated networks, including communication with the aqueous surroundings through membrane pores, and pH- or temperature-triggered release of contents (7).
Fig. 2.
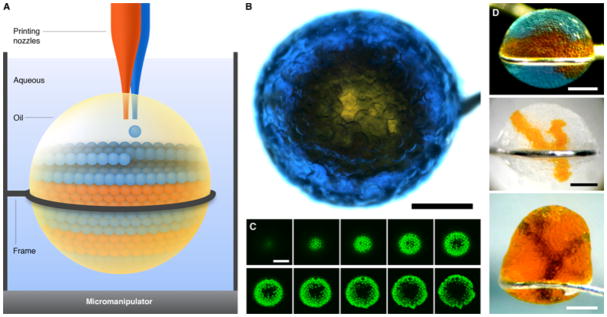
Droplet networks printed in bulk aqueous solution. (A) Schematic of printing in aqueous solution. Aqueous droplets are ejected into a drop of oil suspended in bulk aqueous solution. Excess oil can be removed after printing by suction through a printing nozzle. (B) Micrograph of a network printed in aqueous solution, viewed from above. A core of orange droplets is surrounded by a shell of blue droplets, which contain the fluorescent dye pyranine. Scale bar, 400 μm. (C) Horizontal sections of the network in (B) obtained by confocal microscopy, showing the fluorescent shell of droplets around the non-fluorescent core. The sections span approximately the bottom 150 μm of the network. Scale bar, 400 μm. (D) Micrographs of three other networks printed in bulk aqueous solution. Scale bars, 400 μm.
To establish whether membrane proteins could be included in specific bilayers, we printed a network in which only the droplets along a defined pathway contained staphylococcal α-hemolysin (αHL), in order to create an ionically conductive route across an otherwise insulating network (Fig. 3, A and B). To probe the network electrically, a drop of buffer of diameter ~500 μm containing αHL pores was manually pipetted onto each of two Ag/AgCl electrodes with agarose-coated ends. The drops were then brought into contact with different parts of the network, so that they formed bilayers with the droplets on the network surface (Fig. 3A). When the two large drops were placed on either end of the αHL-containing pathway (Fig. 3B), we measured a stepwise increase in ionic current under an applied potential (Fig. 3C). After one of the drops was separated and brought back into contact with the network away from the αHL-containing pathway (Fig. 3D), only transient currents were observed (Fig. 3E). When this drop was separated from the network again and replaced in its original position, a stepwise increase in current was again observed (Fig. S12). Droplet networks in which no droplets contained αHL showed negligible current flow, whereas the current measured across droplet networks in which every droplet contained αHL was similar to that shown in Fig. 3C (Fig. S12).
Fig. 3.
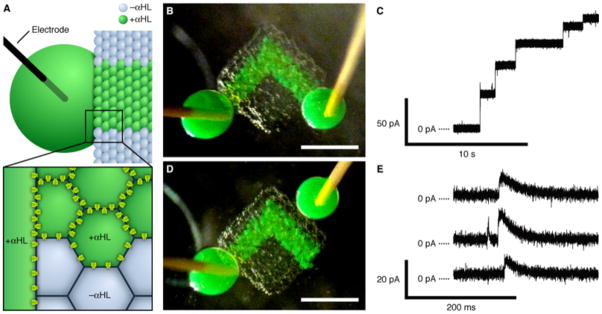
Electrically conductive pathway. (A) Schematic of part of a network printed with an ionically conductive pathway. Only the green droplets and the large drop contain αHL pores. The large drop is impaled with an Ag/AgCl electrode. The magnified section illustrates the αHL pores in the bilayers around the αHL-containing droplets. (B) Photograph of a printed network with electrode-impaled drops placed on either end of the conductive pathway. The green droplets contain αHL, while the other droplets contain no protein. Scale bar, 500 μm. (C) Stepwise increase in the ionic current as measured in the configuration in (B), at 50 mV in 1 M KCl at pH 8.0. (D) Photograph of the network in (B), after separating one of the large drops and rejoining it onto the network away from the conductive pathway. Scale bar, 500 μm. (E) Selected portions of a single recording as measured in the configuration in (D) at 50 mV, showing transient increases in ionic current.
To interpret these results, we performed computational simulations of the electrical behavior of printed droplet networks (23) (Fig. S13). The electrical model was consistent with the measured currents exemplified in Fig. 3, C and E, provided most of the bilayers along the αHL-containing pathway contained several pores, and the other bilayers in the network contained none (Text S4 and Fig. S14), so that the pathway presented an already established conductive route through the otherwise insulating network. The stepwise increase in current in Fig. 3C was therefore most likely caused by pore insertions into the bilayers between the large electrode drops and the pathway droplets, rather than into bilayers within the printed network (Text S4). The current spikes in Fig. 3E correspond to pore insertions into the bilayers between the electrode drop newly placed away from the pathway and insulating droplets in the network, which transmit transient capacitive currents but do not permit a steady resistive current (Text S4). Based on these findings, we maintain that droplet networks can be printed with protein pores in specific bilayers. Further, printed networks can be made tolerant of slight variations in structure and in the number of proteins that insert in each bilayer, recalling the robustness of living tissues to minor flaws. The printed network presented here is functionally analogous to a nerve axon in enabling rapid, long-distance electrical communication along a defined path, but does not mimic an axon’s mechanism of signal propagation.
Finally, we explored a means for droplets in a printed network to produce a designed, macroscopic change in the network geometry through cooperative action. Water permeates readily through droplet interface bilayers even in the absence of protein channels or pores, with a permeability coefficient of 27 ± 5 μm s−1 (mean ± SD, n = 6) under the conditions of this study (Fig. S15), consistent with other permeability measurements of droplet interface bilayers (25–27) and other lipid bilayer systems (28). Consequently, two droplets of higher and lower osmolarity joined by an interface bilayer will respectively swell and shrink until their osmolarities are equal (Fig. 4A). By extension, water transfer between droplets in a network composed of droplets of different osmolarities will cause spontaneous deformation of the network as long as adhesion between droplets is maintained (Fig. 4B).
Fig. 4.
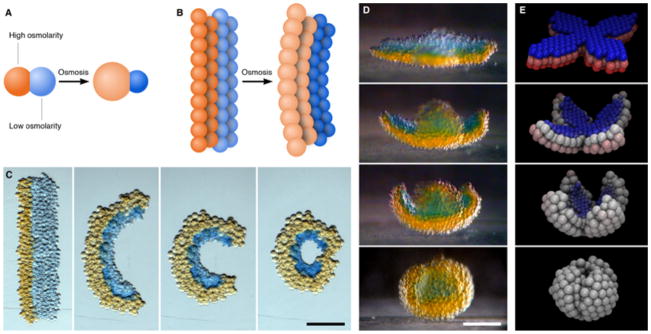
Self-folding droplet networks. (A) Schematic of two droplets of different osmolarities joined by a lipid bilayer. The flow of water through the bilayer causes the droplets to swell or shrink. (B) Schematic of a droplet network that comprises two strips of droplets of different osmolarities. The transfer of water between the droplets induces an overall deformation of the network. (C) Photographs of a rectangular network folding into a circle over ~3 h. The orange and blue droplets initially contain 250 mM KCl and 16 mM KCl, respectively. Scale bar, 250 μm. (D) Photographs of a flower-shaped network folding spontaneously into a hollow sphere. The orange and blue droplets initially contain 80 mM KCl and 8 mM KCl, respectively. The photographs cover a period of 8 h. Scale bar, 200 μm. (E) Frames from a folding simulation of a network with a similar initial geometry to the network in (D). Blue and red represent the lowest and highest initial osmolarities, respectively, and white indicates the average of the two.
We found several prerequisites for droplet networks to fold in a predictable way (see also Text S5). First, to prevent droplets from being printed onto incorrect positions in the network, the network must fold slowly compared to the printing time. Second, the swelling and shrinking of the two droplet types induces a stress between regions of connected droplets that can cause the network to buckle in an uncontrolled manner. This is analogous to the buckling instability in tissues that grow at inhomogeneous rates, such as certain leaves (29) and flower petals (30). In certain cases, the stress may instead cause connected droplets to separate and thereby prevent the network from folding further around the fracture zone (Fig. S16). These various problems can be solved through judicious choices of printing rate, salt concentrations and droplet size, and adjustments to the network geometry (Text S5). The final geometry of the network is then determined in a well-defined way by its initial geometry, the distribution of the two types of droplets, and the ratio of their osmolarities. Using a simple computational model (23) (Table S2) that allows us to predict the folding behavior of a given droplet network, we designed droplet networks that folded successfully.
In one experiment, we printed a network that comprised two strips of droplets of different salt concentrations, connected along their lengths (Fig. 4C). The network folded spontaneously in the horizontal plane over ~3 h, until droplets at opposing ends of the network formed new bilayers in a closed ring. We also programmed a network to fold spontaneously out of the horizontal plane to attain a geometry that would be difficult to print directly. We printed a flower-shaped network with four petals, in which the lower layers had higher osmolarity than the upper layers. The permeation of water from the upper into the lower layers induced a curvature that raised the petals and folded them inwards (Fig. 4D and Movie S1). The folded network was self-supporting and approximated the geometry of a hollow sphere, with the originally upper layer contained within a shell formed by the originally lower layer (Fig. 4E and Fig. S17). The evolution of the geometry of the network is in good agreement with that of a simulated folding network with similar initial conditions (Fig. 4F and Movie S2). We estimate the energy available from the osmosis-driven flow of water to be of the order of 1 μJ for this network (23), which is comparable to the total energy of bilayer formation in the network, and several orders of magnitude greater than the energy required to lift the droplets against gravity (23).
We have devised a means of printing, in designed three-dimensional geometries, a soft material composed of aqueous microcompartments bound together by lipid bilayers. The bilayers allow the compartments to interact directly through membrane proteins or osmotic flows of water, and thereby enable the engineering of collective properties such as long-range electrical communication or macroscopic deformation.
Here we used the well-characterized αHL pore and a simple salt solution to achieve cooperative action between droplets in a printed network. Additional membrane proteins and their engineered forms (5, 6) should allow printed networks to transduce a wider range of signals, and stimulus-responsive osmolytes might offer greater control of folding. New means of communication and cooperation will enable printed networks to more closely reproduce the functions of tissues. Another interesting challenge is the integration of droplet networks with living organisms. The outer surface of a printed network might be engineered to interact in a designed way with the biological environment, for example to deliver drugs upon a specific physiological signal (7–9). More sophisticated networks might be interfaced with failing tissues to support their functions. Alternatively, cells could be included inside the droplets during printing for various applications, such as to immobilize cells within a defined three-dimensional scaffold for tissue engineering (22).
Supplementary Material
Acknowledgments
The authors thank Q. Li for the αHL protein, and A. Wainman for assistance with confocal microscopy. This work was supported by grants from the National Institutes of Health and the European Commission’s Seventh Framework Programme Revolutionary Approaches and Devices for Nucleic Acid Analysis Consortium. G.V. was supported by an Engineering and Physical Sciences Research Council Life Sciences Interface Doctoral Training Centre studentship. A.D.G. was supported by a Biotechnology and Biological Sciences Research Council Doctoral Training Programme in Molecular Biochemistry and Chemical Biology studentship. The authors have submitted a patent application related to this work.
Footnotes
The authors declare no competing financial interests.
G.V., A.D.G. and H.B. planned the research. G.V. and A.D.G. designed the experimental system and performed the experiments. G.V. wrote the software, performed the modeling and analyzed the data. G.V. and H.B. wrote the paper.
Materials and Methods
References and Notes
- 1.Nakagawa S, Maeda S, Tsukihara T. Curr Opin Struct Biol. 2010;20:423. doi: 10.1016/j.sbi.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Poulin P, Bibette J. Langmuir. 1998;14:6341. [Google Scholar]
- 3.Funakoshi K, Suzuki H, Takeuchi S. Anal Chem. 2006;78:8169. doi: 10.1021/ac0613479. [DOI] [PubMed] [Google Scholar]
- 4.Malmstadt N, Nash MA, Purnell RF, Schmidt JJ. Nano Lett. 2006;6:1961. doi: 10.1021/nl0611034. [DOI] [PubMed] [Google Scholar]
- 5.Holden MA, Needham D, Bayley H. J Am Chem Soc. 2007;129:8650. doi: 10.1021/ja072292a. [DOI] [PubMed] [Google Scholar]
- 6.Maglia G, et al. Nature Nanotech. 2009;4:437. doi: 10.1038/nnano.2009.121. [DOI] [PubMed] [Google Scholar]
- 7.Villar G, Heron AJ, Bayley H. Nature Nanotech. 2011;6:803. doi: 10.1038/nnano.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Needham D. Nature Nanotech. 2011;6:761. doi: 10.1038/nnano.2011.218. [DOI] [PubMed] [Google Scholar]
- 9.Eisenstein M. Nat Methods. 2012;9:13. [Google Scholar]
- 10.Sarles SA, Leo DJ. Anal Chem. 2010;82:959. doi: 10.1021/ac902555z. [DOI] [PubMed] [Google Scholar]
- 11.Bai Y, et al. Lab Chip. 2010;10:1281. doi: 10.1039/b925133b. [DOI] [PubMed] [Google Scholar]
- 12.Zagnoni M, Cooper JM. Lab Chip. 2010;10:3069. doi: 10.1039/c0lc00219d. [DOI] [PubMed] [Google Scholar]
- 13.Aghdaei S, Sandison ME, Zagnoni M, Green NG, Morgan H. Lab Chip. 2008;8:1617. doi: 10.1039/b807374k. [DOI] [PubMed] [Google Scholar]
- 14.Poulos JL, Nelson WC, Jeon TJ, Kim CJ, Schmidt JJ. Appl Phys Lett. 2009;95:013706. [Google Scholar]
- 15.Dixit SS, Kim H, Vasilyev A, Eid A, Faris GW. Langmuir. 2010;26:6193. doi: 10.1021/la1010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanley CE, et al. Chem Commun. 2010;46:1620. doi: 10.1039/b924897h. [DOI] [PubMed] [Google Scholar]
- 17.Thutupalli S, Herminghaus S, Seemann R. Soft Matter. 2011;7:1312. [Google Scholar]
- 18.Hu ZB, Zhang XM, Li Y. Science. 1995;269:525. doi: 10.1126/science.269.5223.525. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Hanna JA, Byun M, Santangelo CD, Hayward RC. Science. 2012;335:1201. doi: 10.1126/science.1215309. [DOI] [PubMed] [Google Scholar]
- 20.Forterre Y, Skotheim JM, Dumais J, Mahadevan L. Nature. 2005;433:421. doi: 10.1038/nature03185. [DOI] [PubMed] [Google Scholar]
- 21.Skotheim JM, Mahadevan L. Science. 2005;308:1308. doi: 10.1126/science.1107976. [DOI] [PubMed] [Google Scholar]
- 22.Vunjak-Novakovic G, Scadden DT. Cell Stem Cell. 2011;8:252. doi: 10.1016/j.stem.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Materials and methods are available as supplementary material on Science Online.
- 24.Levental I, Georges PC, Janmey PA. Soft Matter. 2007;3:299. doi: 10.1039/b610522j. [DOI] [PubMed] [Google Scholar]
- 25.Thiam AR, Bremond N, Bibette J. Langmuir. 2012;28:6291. doi: 10.1021/la3003349. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Sigworth FJ, LaVan DA. Adv Mater. 2010;22:120. doi: 10.1002/adma.200901945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixit SS, Pincus A, Guo B, Faris GW. Langmuir. 2012;28:7442. doi: 10.1021/la3005739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boroske E, Elwenspoek M, Helfrich W. Biophys J. 1981;34:95. doi: 10.1016/S0006-3495(81)84839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nath U, Crawford BCW, Carpenter R, Coen E. Science. 2003;299:1404. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- 30.Liang HY, Mahadevan L. Proc Natl Acad Sci USA. 2011;108:5516. doi: 10.1073/pnas.1007808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maglia G, et al. Nano Lett. 2009;9:3831. doi: 10.1021/nl9020232. [DOI] [PubMed] [Google Scholar]
- 32.Oesterle A. P-1000 & P-97 Pipette Cookbook (rev G) Sutter Instrument Co; Novato, CA: 2011. [Google Scholar]
- 33.Aronson MP, Princen HM. Nature. 1980;286:370. [Google Scholar]
- 34.Hamer WJ, Wu YC. J Phys Chem Ref Data. 1972;1:1047. [Google Scholar]
- 35.Humphrey W, Dalke A, Schulten K. J Molec Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 36.Kankare J, Vinokurov IA. Langmuir. 1999;15:5591. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


