Abstract
An emerging theme in medical microbiology is that extensive variation exists in gene content among strains of many pathogenic bacterial species. However, this topic has not been investigated on a genome scale with strains recovered from patients with well-defined clinical conditions. Staphylococcus aureus is a major human pathogen and also causes economically important infections in cows and sheep. A DNA microarray representing >90% of the S. aureus genome was used to characterize genomic diversity, evolutionary relationships, and virulence gene distribution among 36 strains of divergent clonal lineages, including methicillin-resistant strains and organisms causing toxic shock syndrome. Genetic variation in S. aureus is very extensive, with ≈22% of the genome comprised of dispensable genetic material. Eighteen large regions of difference were identified, and 10 of these regions have genes that encode putative virulence factors or proteins mediating antibiotic resistance. We find that lateral gene transfer has played a fundamental role in the evolution of S. aureus. The mec gene has been horizontally transferred into distinct S. aureus chromosomal backgrounds at least five times, demonstrating that methicillin-resistant strains have evolved multiple independent times, rather than from a single ancestral strain. This finding resolves a long-standing controversy in S. aureus research. The epidemic of toxic shock syndrome that occurred in the 1970s was caused by a change in the host environment, rather than rapid geographic dissemination of a new hypervirulent strain. DNA microarray analysis of large samples of clinically characterized strains provides broad insights into evolution, pathogenesis, and disease emergence.
Keywords: DNA microarray, evolution
Microbial pathogens have great versatility in their ability to colonize distinct ecological niches and hosts and cause infections at different anatomic sites. The molecular processes responsible for disease and host specificity are poorly understood but are presumed to be caused in part by differences in gene content and allelic variation between strains. Although analysis of variation on the basis of differences at a limited number of genetic loci has been useful for estimating population structure and evolution (1, 2), it is now possible to index diversity in overall chromosomal gene content in isolates of a bacterial species. For example, whole-genome DNA microarray analysis has been used to compare the genomes of 13 variants of the tuberculosis vaccine strain, B. bacillus Calmette–Guérin, and Mycobacterium tuberculosis H37Rv (3). The data provided new information about the evolution of bacillus Calmette–Guérin strains used globally for vaccinating humans against tuberculosis and, importantly, suggested rational approaches to the design of improved diagnostics and vaccines. Recently, analysis of 15 strains of Helicobacter pylori with a whole-genome microarray was used to identify chromosomal diversity and new candidate virulence genes (4).
Staphylococcus aureus is a serious human pathogen responsible for life-threatening septicemia, endocarditis, and toxic shock syndrome (TSS). It also causes economically important mastitis in cows and sheep. Multilocus enzyme electrophoresis (MLEE) has been used to estimate overall genetic relationships among S. aureus strains and establish a population genetic framework for analysis of pathogenesis, virulence gene distribution, and evolution (1). The data have shown that relatively few extant clones are responsible for the majority of infections, and some of the lineages are nonrandomly associated with distinct human infections and mammalian hosts. The observation that a limited number of lineages are responsible for a large proportion of S. aureus infections implies that interclone variance in relative virulence is large and raises the possibility that variation in genome content in natural populations plays an important role. To investigate the molecular population genetics of S. aureus on a genome scale and obtain information bearing on host and disease specificity, isolates representing the major lineages identified by MLEE were analyzed with a DNA microarray formulated on the basis of the genome of strain COL. Broad insights were obtained into the virulence, population genetics, and evolution of a critical human pathogen.
Materials and Methods
Bacterial Strains.
Thirty-six S. aureus strains isolated from different human disease types and bovine and ovine mastitis were studied (Table 1). The strains were selected to represent the most abundant lineages identified by a population genetic study of 2,077 S. aureus isolates from worldwide sources (1). The sample included 11 methicillin-resistant S. aureus (MRSA) strains from diverse phylogenetic lineages (5). In addition, 15 isolates of electrophoretic type (ET) 234 were studied, including 9 strains cultured from the urogenital tract of women with TSS, 2 MRSA isolates of unknown disease origin, 1 organism isolated from the nasopharynx of an asymptomatic individual, and 3 TSS toxin-1-producing strains isolated before the TSS epidemic in the 1970s. The bovine and ovine strains were selected to represent the most abundant S. aureus lineages associated with intramammary infection in diverse localities (1, 6, 7).
Table 1.
S. aureus strains used in this study
| Strain | Disease or characteristic | Lineage/electrophoretic type* | Source/date of isolation | Ref. |
|---|---|---|---|---|
| COL | MRSA | ND | Colindale, UK | 18 |
| MSA 820 | MRSA | F2/91 | Rhode Island | 5 |
| MSA 890 | MRSA | F2/93 | Texas | 5 |
| MSA 1601 | MRSA | D3/53 | Rhode Island/1980s | 5 |
| MSA 3400 | MRSA | F2/91 | Dublin, Ireland/1990 | 5 |
| MSA 3402 | MRSA | F2/89 | New York/1978 | 5 |
| MSA 3405 | MRSA | F2/91 | Edmonton, Canada/1980s | 5 |
| MSA 3407 | MRSA | H1/234 | New York/1980s | 5 |
| MSA 3410 | MRSA | F2/93 | London, UK/1960s | 5 |
| MSA 3412 | MRSA | H1/234 | New York/1980s | 5 |
| MSA 3418 | MRSA | F2/89 | Australia/1980s | 5 |
| MSA 3426 | MRSA | F2/89 | Dublin, Ireland/1980s | 5 |
| MSA 817 | Human origin | F4/114 | Rhode Island/1980s | 1 |
| MSA 1827 | TSST-1+ | H1/234 | Alabama/1964 | 1 |
| MSA 1832 | TSST-1+ | H1/234 | Georgia/1968 | 1 |
| MSA 1836 | TSST-1+ | H1/234 | Florida/1972 | 1 |
| MSA 2885 | Nasal commensal | H1/234 | Canada/1974 | 1 |
| MSA 537 | TSS | H1/234 | United States/1985 | 1 |
| MSA 700 | TSS | H1/234 | Canada | 1 |
| MSA 1205 | TSS | H1/234 | Arizona | 1 |
| MSA 2335 | TSS | H1/234 | Sweden | 1 |
| MSA 2346 | TSS | H1/234 | Sweden | 1 |
| MSA 2754 | TSS | H1/234 | Canada | 1 |
| MSA 2786 | TSS | H1/234 | Canada | 1 |
| MSA 3095 | TSS | H1/234 | Canada/1987 | 1 |
| MSA 1695 | Scalded skin syndrome | F2/93 | Japan | 1 |
| MSA 2020 | Scalded skin syndrome | F9/189 | France | 1 |
| MSA 2099 | Endocarditis | D2/32 | Denmark/1984 | 1 |
| MSA 2120 | Endocarditis | F4/146 | Denmark/1983 | 1 |
| MSA 2965 | Sepsis | F9/191 | Canada/1983 | 1 |
| MSA 2348 | Furunculosis | F9/189 | Sweden | 1 |
| MSA 2389 | Furunculosis | D2/39 | Sweden | 1 |
| RF122 | Bovine mastitis | F10/195 | Ireland/1993 | 7 |
| MSA 961 | Bovine mastitis | F4/146 | Louisiana | 1 |
| MSA 535 | Ovine mastitis | F1/66 | Germany | 1 |
| MSA 551 | Ovine mastitis | F1/70 | France | 1 |
MSA, Musser S. aureus strain number; TSS, toxic shock syndrome; TSST-1+, toxic shock syndrome toxin-1-positive; ND, not determined.
Phylogenetic lineage and electrophoretic type as designated by Musser and Selander (1).
DNA Microarray Construction, Hybridization, and Data Analysis.
DNA microarray experiments were carried out as described (published as supplemental data on the PNAS web site, www.pnas.org).
Southern Hybridization and PCR Analysis.
To confirm the microarray data, Southern blotting and PCR analysis were performed as described in the supplemental data.
Results
Overall Genome Diversity in S. aureus.
DNA microarray analysis revealed extensive genome diversity within the S. aureus species. All 36 strains analyzed shared 2,198 ORFs, equivalent to ≈78% of all ORFs (n = 2,817). A maximum of 332 (12%) ORFs were absent in any one strain compared with the reference strain COL (Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). Many of these nonessential genes (n = 294) were located contiguously in regions 3–50 kb in size in strain COL. Eight of the regions contain genes encoding integrases or transposases.
The gene content of S. aureus strain COL was very similar to strains of ET 93, especially strain MSA3410 (Fig. 1); only 22 ORFs were absent in MSA3410 compared with strain COL. Eighteen of the 22 ORFs were phage-associated and were located in a single chromosomal region designated RD14 (see below).
Figure 1.
S. aureus genetic diversity. (A) Genome-wide distribution of absent ORFs among the 36 S. aureus strains studied. Red and black denote presence or absence of ORFs, respectively. RDs are marked by black arrows and were numbered arbitrarily, because the genome sequence of strain COL is not complete. (B) Dendrogram showing estimates of genomic relationships of the 36 strains constructed by complete linkage hierarchical cluster analysis. Strains were grouped with the cluster program on the basis of the presence or absence of ORFs with RDs treated as single loci, and the output was displayed with the computer program treeview. Phylogenetic lineage and ET of each strain are presented. Red text signifies MRSA strains. The scale represents the value of the Pearson correlation coefficient at each node. For a pairwise comparison, a coefficient of 1 would denote absolute identity, and zero would indicate complete independence.
Large Chromosomal Regions of Difference (RD).
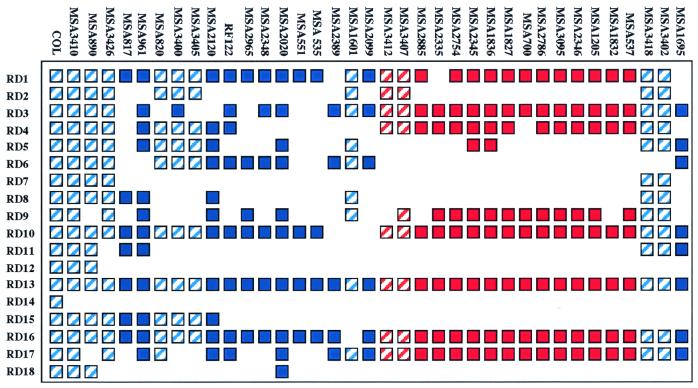
Seventeen chromosomal regions consisting of more than three contiguous ORFs were absent in one or more strains compared with the reference strain COL (Fig. 2). One chromosomal region (RD13) was not absent in entirety in any strain but varied considerably in gene content among the 36 strains examined. Each of these 18 regions will be referred to as a “RD,” a term used in studies of genome variation in M. tuberculosis (8). The RDs were numbered arbitrarily, and the size of each region given below refers to the length of the chromosomal segment in strain COL. RD8 and RD13 were characterized by PCR size variation, and RD1 and RD2 had large differences in gene content among the 36 strains examined (Fig. 3).
Figure 2.
Presence or absence of RDs in the 36 S. aureus isolates studied. Filled square denotes presence of an RD and an empty space, its absence. Hatched squares indicate presence of RDs in MRSA strains. Red signifies ET 234 strains.
Figure 3.
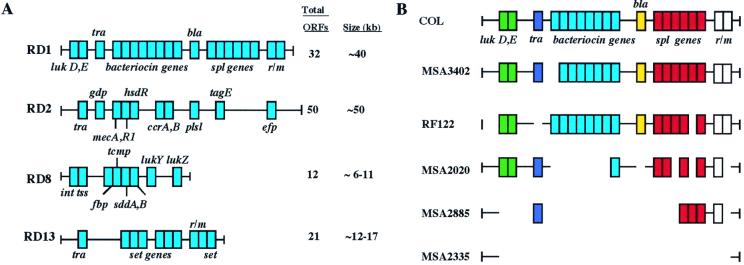
Variation in gene content of chromosomal RDs. (A) RDs with extensive variation in gene content or size among the 36 strains examined. Genes that encode proteins containing homology to proteins of known function are denoted by squares. Horizontal black lines represent regions containing ORFs that encode hypothetical proteins of unknown function. The hypothetical proteins encoded by the genes designated are as follows: lukD, E, leukotoxins D and E; tra, transposase; bla, β-lactamase; splA-F, serine protease A–F; r/m, restriction and modification proteins; gdp, glycerophosphoryl diester phosphodiesterase; mecA, penicillin-binding protein 2′; mecR1, signal transducer protein; hsdR, type 1 restriction endonuclease; ccrA, B, cassette chromosome recombinase A, B; plsl, Pls-like protein; tagE, poly(glycerol-phosphate)alpha-glucosyltransferase homologue; efp, efflux protein homologue; int, integrase; tss, phage terminase small subunit homologue; fbp, ferrichrome binding protein homologue; tcmp, tetracenomycin C synthesis protein homologue; sddA, B, succinyl-diaminopimelate desuccinylase-related proteins A, B; lukY, Z, leukotoxin F, S homologues; set genes, staphylococcal exotoxin protein homologues. (B) The major variants of RD1 identified among the 36 strains studied. In B, different colors are used to differentiate among genes encoding proteins with distinct functions.
RD1 (≈40 kb; 35 ORFs) has several noteworthy features, including a cluster of six genes encoding serine protease homologues with 49–93% identity to each other, a group of 8 genes encoding proteins involved in bacteriocin production and resistance, contiguous genes for leukotoxins D and E, and genes encoding proteins with 49% amino acid identity to restriction modification type Ic proteins made by Streptococcus thermophilus. The bacteriocin genes were present in 9 of 36 strains analyzed, and the genes for leukotoxins D and E were present in 17 strains. The entire serine protease gene cluster was absent in three strains. RD1 was highly variable in gene content among the 36 strains examined, with 5 major variants identified.
RD2 (≈50 kb; 50 ORFs) is the staphylococcal chromosomal cassette mec (SCCmec) and was present only in MRSA strains. There was considerable variation in the gene content of RD2 among the 11 MRSA strains (Fig. 3). However, all MRSA strains had 13 of the ORFs present in RD2 in strain COL, including the mecA gene encoding an altered penicillin-binding protein that confers resistance to methicillin and related antimicrobial agents, the mecRI gene encoding a signal transducer, a gene (gdp) encoding a glycerophosphoryl diester phosphodiesterase homologue, a transposase gene associated with the insertion sequence-like element IS431mec, a gene encoding a cassette chromosome recombinase homologue, and 8 genes that encode hypothetical proteins of unknown function. Each of the 11 MRSA strains had 14–50 of the ORFs present in RD2 in strain COL. Only one strain (MSA3410), an MRSA strain of ET 9 (1), had the same gene complement in RD2 as strain COL.
RD3 (≈4 kb; 5 ORFs) contains two genes encoding proteins with homology to transposases and two genes that encode proteins with no homology to known proteins. RD3 was present in 29 of 36 strains examined.
RD4 (≈5 kb; 4 ORFs) has a gene encoding a cobalt ABC transporter homologue. RD4 was present in 25 of the 36 strains analyzed.
RD5 (≈5 kb; 7 ORFs) has genes encoding a surface protein with homology to fibronectin-binding protein (FnbA) of S. aureus, two proteins with homology to the staphylococcal accessory regulator of virulence (SarA), and a protein with homology to a multidrug-resistance protein made by Pyrococcus abyssi. RD5 was present in 15 of the 36 strains.
RD6 (≈6 kb; 7 ORFs) has genes that encode proteins that lack significant homology to known proteins. Fifteen of the 36 strains examined had RD6.
RD7 (≈6 kb; 8 ORFs) has one gene encoding a protein with homology to a hypothetical protein encoded by S. thermophilus insertion element IS1191 and 7 genes that encode proteins of unknown function. Only 5 of 36 strains examined had RD7.
RD8 (≈11 kb; 12 ORFs) is highly variable in size and gene content among the 36 strains examined (Fig. 3). Depending on the strain, RD8 has genes encoding an integrase-like protein, a putative ferrichrome-binding protein (febp), and proteins with homology to leukotoxin subunits F and S. PCR analysis found that RD8 varied from 6 to 11 kb among the 36 strains examined.
RD9 (≈14 kb; 15 ORFs) is a putative transposable element. RD9 has a transposase gene and a gene encoding a protein with 40% identity to TrsG, the transfer complex protein of S. aureus plasmid pG01. The other 13 ORFs in RD9 encode hypothetical proteins with no significant identity to proteins of known function. RD9 was present in 21 of the 36 strains investigated. Interestingly, microarray and Southern blot analysis indicated that strain MSA1601 contains more than one copy of RD9. Moreover, PCR amplification of the putative insertion site of RD9 indicated that this element has more than one chromosomal integration site among the 36 strains examined (data not shown).
RD10 (≈6 kb; 4 ORFs) contains genes that encode a putative oligopeptide transporter complex. This region was present in 33 of the 36 strains studied.
RD11 (≈3 kb; 5 ORFs) contains genes encoding proteins that lack significant homology to known proteins. RD11 was present in 7 of the 36 strains examined.
RD12 (≈16 kb; 25 ORFs) is a putative pathogenicity island that has genes for staphylococcal enterotoxins b (seb), k (sek), a novel enterotoxin; an integrase; and β-lactamase (bla). RD12 was present in only 2 of the 36 strains studied. However, 4–21 of the RD12 ORFs were present somewhere in the chromosome in each of the 36 strains, indicating that related elements may exist in some strains.
RD13 (≈12 kb; 14 ORFs) contains 7 genes that encode proteins with homology to recently described staphylococcal exotoxin-like proteins (9), and 2 genes encoding a putative restriction and modification system. There was considerable variation among strains in the structure of RD13. Different combinations of the 7 exotoxin genes were identified (Fig. 3), and long-range PCR analysis found that RD13 varied in size from 12 to 17 kb among the 36 strains analyzed.
RD14 (≈44 kb; 72 ORFs) may represent a previously undescribed bacteriophage. PCR analysis of the putative integration site of RD14 confirmed that all test strains lacked the element at this chromosomal location. However, each of the 36 strains analyzed contained 8–63 of the ORFs present in RD14 in strain COL, indicating that related elements may exist elsewhere in the genome of some strains.
RD15 (≈4 kb; 4 genes) consists of 4 genes (cap5g, cap5h, cap5i, and cap5j) involved in synthesis of serotype 5 capsule. RD15 was present in 9 of 36 strains investigated, suggesting that these organisms produce serotype 5 capsule.
RD16 (≈12 kb; 12 ORFs) has a gene encoding a protein with 56% identity to a 67-kDa antigen made by Streptococcus pyogenes that crossreacts with myosin. RD16 was present in 35 of the 36 strains studied.
RD17 (≈10 kb; 10 ORFs), present in 28 of the 36 strains, contains genes that encode hypothetical proteins of unknown function.
RD18 (≈5 kb; 5 ORFs) is a plasmid related to pT181 of S. aureus. RD18 has genes that encode a tetracycline-resistance protein, a recombination protein, and a replication initiation protein. Microarray data indicated that RD18 was present as a multicopy plasmid in 3 of 36 strains examined.
Southern Hybridization and PCR Analysis.
All microarray data were confirmed by Southern blot analysis with one or more probes specific for ORFs in each RD. Two RDs with large size variation were identified by PCR among the 36 strains studied. Specifically, RD8 had size variants of 6–11 kb, and RD13 varied from 12 to 17 kb (data not shown).
Overall Chromosomal Relatedness of S. aureus Strains.
We used hierarchical clustering (4) to investigate chromosomal relationships among the 36 S. aureus strains on the basis of their gene content. The strains were clustered into distinct groups that correlate strongly with estimates of chromosomal relationships revealed by MLEE (1) (Fig. 3). Virtually all strains of closely related ETs identified by MLEE were grouped together (Fig. 1).
Diversity of S. aureus Strains Containing the mec Element.
The mec element (RD2) was present in all 11 MRSA strains as assessed by microarray and Southern blot analysis, although extensive diversity was present in the gene content of this region. The mec gene was present in at least five distinct S. aureus lineages that are highly differentiated in overall chromosomal gene content (Fig. 1).
Diversity of Isolates of Multilocus Electrophoretic Type 234, the Predominant Clone Causing TSS.
A previous study of 315 S. aureus isolates recovered from the urogenital tract of women with TSS by MLEE provided evidence that 90% of the organisms were clonally related by descent (10). To further assess the possibility of a close genetic relationship among urogenital TSS isolates, 15 isolates of ET 234 (cluster H1) (1) cultured from diverse geographic localities were studied. The isolates were similar in gene content, indicating close genetic affiliation of all 15 strains (Fig. 1). However, the RD content varied among the strains. For example, TSS isolate MSA2335 lacked RD1, and some of the 15 strains differed in the exotoxin gene complement in RD13. In addition, MSA1832, MSA2885, and MSA3412 lacked RD9, and 8 of the 15 strains contained 20–26 ORFs present in a contiguous region of RD14 in strain COL. These RD14 ORFs were absent in the other strains of ET 234 examined. Analysis of ORFs not contained within RDs revealed that a total of 208 were variably absent among the 15 ET 234 S. aureus isolates examined (supplemental Table 2). Hence, although ET 234 organisms were closely related, there was considerable heterogeneity in genome composition, indicating that the organisms have not evolved very recently from a common ancestor.
Diversity of Bovine and Ovine S. aureus Strains.
Molecular population genetic analysis has suggested the existence of bovine- and ovine-specialist strains of S. aureus (1, 6). To investigate genetic diversity among S. aureus isolates cultured from cows and determine whether these organisms had a distinct genome composition, strains were studied that represent the two most abundant lineages causing bovine mastitis (1, 6). Bovine strains RF122 (ET cluster F10) and MSA961 (ET cluster F4) were assigned to different clonal lineages by MLEE analysis (1). The two ovine mastitis strains studied (MSA535 and MSA551) were selected to represent the major clonal lineage (ET cluster F1) causing this disease (1). We found that the ovine strains and bovine strain RF122 were genetically allied (Fig. 1). In addition, bovine strain MSA961 was similar in gene content to human strain MSA817. Together, these data strongly support MLEE results, indicating that many bovine isolates are more closely allied to human isolates than to other bovine isolates (1, 6).
Discussion
Species Genome Diversity.
We identified extensive genetic variation among strains representing the most abundant disease-causing clonal lineages of S. aureus (1), with as many as 332 ORFs absent in one strain compared with the reference strain COL. We also discovered that 2,198 (78%) of the 2,817 ORFs represented on the DNA microarray were present in all S. aureus strains studied, a result suggesting that these genes are essential for S. aureus cell maintenance. Some of these 2,198 genes encode known virulence factors, suggesting a critical role for these proteins in staphylococcal survival and pathogenesis. Conversely, ≈22% of S. aureus genes represent nonessential genetic information that is strain-specific. Some of these genes encode factors that facilitate colonization of specialized host or environmental niches. We note that a recent comparative genomics study of 15 H. pylori strains also reported that 22% of genes were not essential for core cell functions (4). In Escherichia coli, as many as 25% of genes have been reported to be strain specific (11). Taken together, the data suggest an emerging theme that a very large number of genes in the chromosome of several common human bacterial pathogens are devoted to contingency functions.
Importance of Horizontal Gene Transfer in the Evolution of S. aureus.
Many of the strain-variable ORFs identified in this study form 18 discrete RDs, including pathogenicity island-, phage-, plasmid-, transposon-, and insertion sequence-related elements. Only RD2 and RD15 have been previously characterized (12, 13). Our analysis found that several RDs have extensive variation in gene content and size. Of particular note are RD1 and RD13, which contain many putative virulence factor genes and genes for transposases. In this regard, a recently described operon containing enterotoxin genes (14) is present in the RD1 homologue of MRSA strain 252 (http://www.sanger.ac.uk). In addition, a pathogenicity island (SaPIbov) described in bovine S. aureus strain RF122 is present in the chromosome of RF122, adjacent to RD13 (15). It is clear that multiple deletion, integration, and recombination events have contributed to the variation in the RDs and the genome overall. At least 19 genes that would encode proteins with homology to mediators of lateral gene transfer (such as integrases and transposases) are present in strain COL; 14 of these 19 genes were located in RDs. Moreover, most RDs were widely distributed among strains of divergent clonal lineages that have not shared a recent ancestor. Taken together, it is clear that horizontal gene transfer has played a fundamental role in the evolution of pathogenic S. aureus, especially by assortive recombination of RDs containing virulence and antibiotic-resistance genes. It is also evident that the overall genomic relationships revealed by MLEE and whole-genome microarray analysis are largely cognate, indicating the presence of phylogenetic signal in the data. This means that chromosomal relationships inferred by these techniques are useful for study of many biomedically relevant characteristics.
Chromosomal Relatedness of S. aureus Strains from Different Host Species.
Previous studies have indicated that successful cross-species transfer of pathogenic S. aureus is infrequent (6). We found that a strain representing the most abundant clonal lineage causing bovine mastitis (RF122) was closely related to strains of the predominant ovine mastitis clonal type. This result suggests that a similar gene complement may be required to cause bovine and ovine intramammary infection. However, DNA microarray analysis allows comparison only of the gene content of strains to genes represented on the array. Hence, it is unlikely that we identified all strain-specific genes present in the strains examined. Identification of genes present in the 36 test strains but absent in strain COL is likely to provide new insight into the molecular mechanisms of pathogenic clone adaptation to specialized niches such as the mammary gland.
Evolution of MRSA Strains.
MRSA strains were first reported in 1961, soon after methicillin entered clinical use. Outbreaks of infections caused by MRSA were reported soon thereafter in Europe, and by the mid-1970s, MRSA had become a significant problem in the United States. MRSA are critical public health threats because they cause hospital-acquired infections that can be difficult and expensive to treat. Many MRSA strains are susceptible only to vancomycin, and this antibiotic is used extensively to treat patients infected with these organisms. Hence, there is also concern that MRSA may serve as a reservoir of organisms that may give rise to vancomycin-resistant strains that could not be killed with available antibiotics.
Two very different hypotheses have been put forth to explain the origin of MRSA strains (5, 16). The association of the mec gene with genetically diverse lineages of S. aureus, together with data indicating that the mec gene was horizontally transferable in the laboratory, led to the hypothesis that MRSA strains had evolved many independent times by lateral gene transfer of the mec element into phylogenetically distinct methicillin-susceptible precursor cell lines (5). In contrast, data obtained from study of MRSA by restriction fragment length polymorphism analysis with probes for mecA and Tn554 were interpreted to mean that MRSA organisms evolved from a susceptible clone that acquired the mec element and subsequently generated substantial chromosomal diversity (16). We discovered that MRSA strains were assigned to at least five distinct chromosomal genotypic groups that, relative to one another, are highly divergent, in some cases differing by greater than several hundred genes. Hence, the only reasonable interpretation of the data is that the MRSA strains have arisen multiple independent times by lateral transfer of the mec element into methicillin-susceptible precursors. Taken together, the microarray data unambiguously settle a longstanding controversy in the S. aureus field.
Origin of the TSS Epidemic.
Ninety percent of S. aureus strains recovered from women with urogenital-associated TSS are a distinct multilocus enzyme genotype, designated ET 234 (1, 10). Several lines of evidence (summarized in refs. 10 and 17) led to the hypothesis that the epidemic of TSS cases was caused by modification of the host population. However, the alternative hypothesis—an episode of periodic selection involving a fitness mutation occurring in a cell representing ET 234—could not be ruled out. Although the DNA microarray data confirmed that ET 234 organisms are genetically related and have shared a common ancestor, clearly they are not genetically identical, and the last ancestor has not been very recent in evolutionary time. Thus, the data indicate that the epidemic was caused by a change in the host environment (preadapted clone hypothesis) (10) and rule out the alternative explanation that rapid intercontinental dissemination of a new hypervirulent clone of S. aureus was responsible.
Concluding Comment.
Our studies provide an overview of genome diversity present in natural populations of S. aureus, an important human and animal pathogen worldwide. Genome-scale DNA microarray analysis contributed improved understanding about the evolution and virulence gene distribution in this bacterium. Importantly, the data resolved the controversy pertaining to the evolutionary origin of MRSA strains and strongly supported the hypothesis that the TSS epidemic was caused by a change in the host cervicovaginal environment. Rapid indexing of genome-scale variation present in large samples of microbial isolates that are well characterized clinically will provide many new insights into pathogen virulence, host and disease specificity, and disease emergence and reemergence.
Supplementary Material
Acknowledgments
We thank the laboratory of S. Falkow for assistance with microarray procedures, and A. Elias, G. Somerville, and R. Belland for critical review of the manuscript.
Abbreviations
- ET
electrophoretic type
- MLEE
multilocus enzyme electrophoresis
- MRSA
methicillin-resistant S. aureus
- RD
region of difference
- TSS
toxic shock syndrome
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Musser J M, Selander R K. In: Molecular Biology of the Staphylococci. Novick R P, editor. New York: VCH; 1990. pp. 59–67. [Google Scholar]
- 2.Enright M C, Day N P, Davies C E, Peacock S J, Spratt B G. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 4.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musser J M, Kapur V. J Clin Microbiol. 1992;30:2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapur V, Sischo W M, Greer R S, Whittam T S, Musser J M. J Clin Microbiol. 1995;33:376–380. doi: 10.1128/jcm.33.2.376-380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald J R, Meaney W J, Hartigan P J, Smyth C J, Kapur V. Epidemiol Infect. 1997;119:261–269. doi: 10.1017/s0950268897007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahairas G, G, Sabo P J, Hickey M J, Singh D C, Stover C K. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams R J, Ward J M, Henderson B, Poole S, O'Hara B P, Wilson M, Nair S P. Infect Immun. 2000;68:4407–4415. doi: 10.1128/iai.68.8.4407-4415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musser J M, Schlievert P M, Chow A W, Ewan P, Kreiswirth B N, Rosdahl V T, Naidu A S, Witte W, Selander R K. Proc Natl Acad Sci USA. 1990;87:225–229. doi: 10.1073/pnas.87.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochman H, Jones I B. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, Katayama Y, Hiramatsu K. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarraud S, Peyrat M A, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. J Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald J R, Monday S R, Foster T J, Bohach G A, Hartigan P J, Meaney W J, Smyth C J. J Bacteriol. 2001;183:63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 17.Musser J M, Krause R M. In: Emerging Infections. Krause R M, editor. New York: Academic; 1998. pp. 185–218. [Google Scholar]
- 18.Shafer W M, Iandolo J J. Infect Immun. 1979;25:902–911. doi: 10.1128/iai.25.3.902-911.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.