Abstract
The Siglec family of sialic acid-binding proteins are differentially expressed on white blood cells of the immune system and represent an attractive class of targets for cell-directed therapy. Nanoparticles decorated with high-affinity Siglec ligands show promise for delivering cargo to Siglec-bearing cells, but this approach has been limited by a lack of ligands with suitable affinity and selectivity. Building on previous work employing solution-phase sialoside library synthesis and subsequent microarray screening, we herein report a more streamlined ‘on-chip’ synthetic approach. By printing a small library of alkyne sialosides and subjecting these to ‘on-chip’ click reactions, the largest sialoside analog library to date was generated. Siglec-screening identified a selective Siglec-7 ligand, which when displayed on liposomal nanoparticles, allows for targeting of Siglec-7+ cells in peripheral human blood. In silico docking to the crystal structure of Siglec-7 provides a rational for the affinity gains observed for this novel sialic acid analog.
The 15-member Siglec family of sialic acid-binding proteins represent an attractive class of therapeutic targets due to their unique expression pattern on various white blood cell subsets and their capacity to deliver therapeutic cargo into the cell by endocytosis.1, 2,3, 4 For this reason, strategies for exploiting Siglecs for targeted-cell therapies are becoming increasingly pursued for diseases ranging from cancer to allergies. While first-generation strategies have employed antibodies,5 the use of liposomal nanoparticles decorated with Siglec ligands have shown promise as an alternative platform for targeting Siglecs due to the ease of loading and delivering a variety of therapeutic payloads.6, 7
In order to fully realize the potential of cell-specific therapy using ligand-targeted nanoparticles, unique high affinity ligands must be identified for each member of the Siglec family. As a strategy to achieving this goal, our group and others have utilized sialic acid as a “privileged scaffold” and have appended unnatural substituents to various positions on the sugar ring in an effort to identify ligand analogs with increased affinity and selectivity for individual Siglecs.8–13 Towards this end, we previously generated a library of analogs using sialoside scaffolds with azide and alkyne substituents at the C9 and C5 position of sialic acid and subjected these to high-throughput Cu(I)-catalyzed azide-alkyne cycloadditions14 (CuAAC) with corresponding libraries of alkynes and azides. Using microarray technology the library was then printed and screened with various Siglecs, leading to the identification of novel, high affinity ligands for Siglecs-9 and -10 suitable for targeting cells expressing these Siglecs in human peripheral blood.13
Despite the success of this approach, the size of the library was limited by the amounts of the sialoside scaffolds that could be prepared for construction of the library. We considered that this limitation could be removed if the synthesis of the library and screening could both be conducted ‘on-chip’. For example, while synthesis of a library of 1200 unique sialosides using solution-phase synthesis would require about 3 g of the desired sialoside scaffold,13 creating a library of similar diversity ‘on-chip’ would require microgram amounts of each scaffold, and submilligram amounts of each coupling partner. Thus, it was envisioned that a >1000 compound library could be readily generated by printing a small library of 10–15 alkyne derivatized sialoside scaffolds into each well of a multi-well (e.g. 48-well) glass slide, followed by CuAAC coupling to a small library of 60–100 different azide substituents.
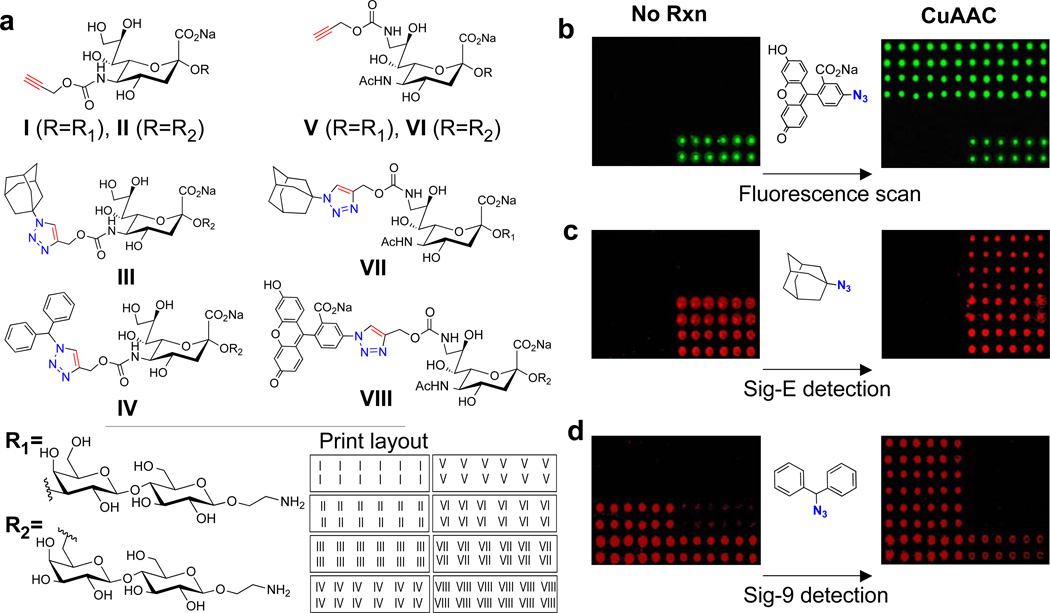
To determine the feasibility of the ‘on-chip’ synthetic approach, several 5-substituted and 9-substituted alkyne-containing sialoside scaffolds (I–II, V–VI) were printed onto NHS-activated slides together with a fluorescent sialoside (VIII), and CuAAC products originating from the above scaffolds which were previously identified as high affinity ligands of Siglecs-E (VII) and -9 (III–IV).13 To test the efficiency of the ‘on-chip’ CuAAC coupling, a solution of 5-azidofluorescein, 15 CuSO4, sodium ascorbate, and THPTA (tris(3-hydroxypropyltriazolylmethyl)amine, a Cu(I)-stabilizing ligand)16 was overlaid onto the array (Figure 1b) and after a 2 hr reaction, the array was washed and scanned for fluorescence. The results show that 5-azido-fluorescein was selectively and quantitatively clicked onto the array only where alkynes were present (I–II, V–VI) and that the reaction only proceeded in the presence of all necessary reaction components (Supplementary Figure 1). Since the eventual goal of this method is to synthesize high affinity Siglec ligands for subsequent detection, similar reactions were carried out with 1-azidoadamantane (Figure 1c) and (azidomethylene)dibenzene (Figure 1d) to synthesize high affinity ligands of Siglec-E and Siglec-9, respectively, and the arrays were probed with these Siglecs. Encouragingly, after the ‘on-chip’ reactions, Siglec-E readily detected the scaffolds V–VI and Siglec-9 detected I–II consistent with the fact that while both Siglecs show little preference for sialoside linkage (i.e. α2–3 or α2–6), Siglec-E typically binds sialoside analogs with C9 substituents, whereas Siglec-9 prefers analogs with C5 substituents.13 Interestingly, both Siglecs appeared to weakly detect the fluorescent sialoside, VIII, which is consistent with the C9-substituent preference of Siglec-E, however this was surprising for Siglec-9.
Figure 1. On-chip synthesis of sialoside analogs.
(A) The illustrated compounds, which include 5-substituted and 9-substituted alkyne-containing sialosides (I–II, V–VI), CuAAC products of the above scaffolds previously identified as high affinity ligands of Siglecs-E (VII) and -9 (III–V), and a fluorescent sialoside (VIII), were printed onto NHS-activated microarray slides in the pattern shown. (B) To show that azides can be selectively and quantitatively ‘clicked’ onto the microarray surface, 5-azido-fluorescein was used as a model substrate. (C) To demonstrate that high-affinity Siglec ligands can be synthesized on-chip and subsequently detected with Siglecs, 1-azidoadamantane was clicked onto the array to produce high affinity Siglec-E ligands on scaffolds V–VI and (D) (azidomethylene)dibenzene was clicked onto the array to produce known high-affinity ligands of Siglec-9 on scaffolds I–II. The arrays were then probed with the respective Siglecs. Images shown in green are from 488 nm wavelength scans (i.e. fluorescein readout), while those in red are from 555 nm (R-PE Siglec detection).
To ensure that the reaction conditions utilized above would yield quantitative or near-quantitative couplings for a diverse set of azides, a more systematic way to monitor reaction progress was needed. This was accomplished by carrying out the CuAAC reaction with the azide of interest first, and then performing a 2nd click reaction with 5-azido-fluorescein to visually detect any residual alkyne left on the slide. Using this procedure, it was found that (azidomethylene)dibenzene showed incredibly quick reaction kinetics with near-quantitative coupling in only five minutes, as evidenced by no subsequent reaction with 5-azido-fluorescein (Supplementary Figure 2). We then moved to one of the most challenging azides, the sterically hindered 1-azido-adamantane. To our delight, and consistent with the results above (Figure 1c), near-quantitative reaction of this azide was seen after a 2 hr reaction time (Supplementary Figure 2). Expanding the scope of these studies to a more diverse set of azides, showed that a 2 hr reaction time was sufficient (and typically far longer than necessary) to achieve quantitative or near-quantitative reactions in all cases (Supplementary Figure 3).
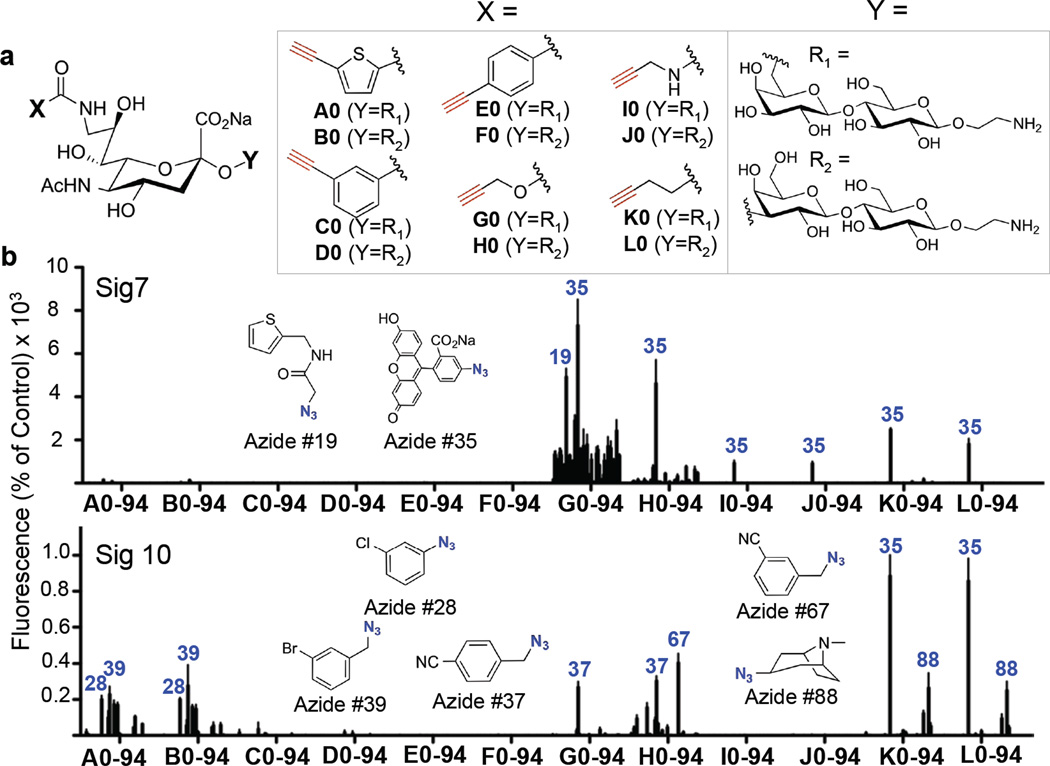
To synthesize a large library of sialoside analogs using the ‘on-chip’ approach, we first synthesized an expanded 12-member alkyne-sialoside scaffold library (A–L), comprising 6 different alkynes appended to the C9 position of sialic acid linked α2–3 or α2–6 to lactose (Figure 2a, Supplementary Schemes 1–3). Of these, scaffolds G and H had been previously documented to yield products recognized by a subset of Siglecs, including Siglec-7 and Siglec-E, for which ligands of sufficient avidity for in vivo targeting had not yet been identified.13 The sialoside scaffold library was then used to generate an 1140 compound sialoside analog library by printing each member of the library into every well of 48-well NHS-activated microarray slides. Subsequently, each well was then subjected to CuAAC with a different azide (Supplementary Figures 4–6). By using 94 different azides (Supplementary Figures 4–5), the full library of 1140 sialoside analogs was therefore constructed over 2 microarray slides using negligible amounts of materials.
Figure 2. Generation of a sialoside analog library by on-chip synthesis and subsequent screening with Siglecs-7 and -10.
(A) Twelve alkyne-bearing sialosides (A–H, Supporting Schemes 1–3) and Compound VII (Figure 1) were printed into each well of 48-well, NHS activated microarray slides. These were subsequently subjected to ‘on-chip’ CuAAC, ‘click’ reactions with 94 different azides (47 azides/slide, Supplementary Figures 4–6). The compound library nomenclature combines the letter of the parent scaffold with the number of the azide it was reacted with. (B) The resulting arrays were then screened with Siglec-7 and Siglec-10 Fc-chimeras to identify high affinity ligands. The binding intensity in each well was normalized to Compound VII (a weak ligand for Siglec-7 and a high affinity ligand for Siglec-10) and the structures of various hits are shown.
The arrays were screened with Siglecs-7, -10, and -E revealing hits for each of the three Siglecs (Figure 2b and Supplementary Figure 7). As observed previously, all Siglecs bound to analogs originating from scaffolds G and H, but exhibiting differential recognition of the substituents. Siglec-7 did not recognize any sialosides of series A–F, but recognized one or more analogs in the G–L series. The most promising substituent for Siglec-7 was the product of azide 35, 5-azido-fluorescein, which was picked up as a hit as a substituent in the G–L alkyne sialoside series, with the most promising hit being G35. Although, Siglec-10 also bound to products with the azide 35, Siglec-10 exhibited highest avidity for K35 and L35 and little binding to G35. Notably, Siglec-7 recognizes G35 with an increase in affinity over compound VII, the highest affinity analog analog for this siglec identified to date (Supporting Tables 1–2). Importantly, the 5-azido-fluorescein substituent (azide 35) by itself does not bind to either siglec as evidenced by lack of binding to compounds A35, B35, C35, D35, and F35 . .
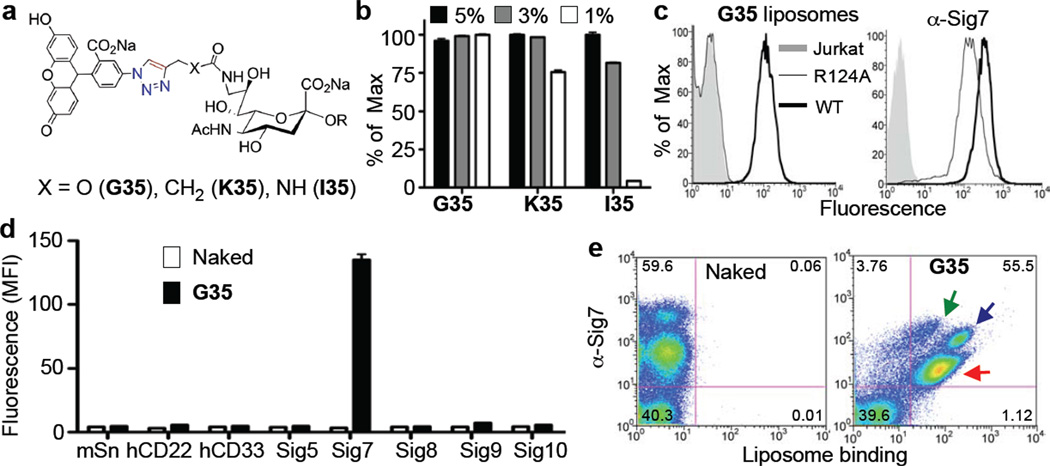
Since the overall goal is to identify sialoside analogs capable of selectively targeting ligand-bearing liposomes to Siglec-expressing cells, the most promising Siglec-7 hits containing the 5-azido-fluorescein substituent` (G35, I35, and K35) were resynthesized, coupled to PEGylated lipids (Figure 3a and Supplementary Scheme 4), and formulated into liposomal nanoparticles to assess binding to Siglec-7 expressing cells. While all ligand-bearing liposomes were able to bind to Siglec-7 expressing cells, G35-liposomes bound with greatest functional avidity (Figure 3b) and increased selectivity (Supplementary Figure 9). Further evidence that the sialic acid scaffold of G35 was critical for binding to Siglec-7 was documented by comparison of the binding of liposomes to Jurkat cells expressing Siglec-7 and a mutant missing the essential arginine (R124A),17 a mutation known to fully ablate sialic acid binding18 (Figure 3c). The specificity of these liposomes for Siglec-7 over other Siglecs was then assessed with a panel of recombinant Siglec-expressing cell lines. The G35-liposomes bound only Siglec-7 expressing cells and not any of the other 7 Siglec-expressing cell lines examined (Figure 3d), including Siglec-9, which exhibited some weak cross reactivity in the microarray studies (Figure 1d and Supporting Figure 10), and Siglec-10, which recognizes the fluorescein substituent on other sialic acid scaffolds (Supplementary Figure 9). Moreover, the G35-liposomes also showed excellent selectively in a complex cellular mixture. Upon isolation of white blood cells from peripheral human blood, the G35 liposomes bound selectively to Siglec-7 positive cells, while cells that were Siglec-7 negative showed no binding (Figure 3e). Consistent with the documented expression pattern of Siglec-7, G35-liposomes bound to monocytes, granulocytes, and a subset of lymphocytes.19 Together, these results validate G35 as selective, high affinity Siglec-7 ligand with potential utility for understanding the function of this receptor, and for the selective targeting of Siglec-7 positive cells for possible therapeutic applications.
Figure 3. Avidity and selectivity of G35-liposomes for Siglec-7 expressing cells.
(A) The Siglec-7 hits G35, K35, and I35 were resynthesized, coupled to PEGylated lipids (Supporting Scheme 4), and formulated into liposomal nanoparticles at various ligand percentages. (B) These were then assessed for binding to Jurkat Siglec-7 expressing cells in triplicate. (C) The best ligand liposomes, G35-liposomes (1% ligand), were then assessed for binding to Jurkat (grey shaded), wild-type Jurkat Siglec-7 (dark black line), and R124A Jurkat Siglec-7 (thin black line) cells (top) to show that the essential Arg is critical for binding. As a control, an anti-Siglec-7 antibody was used (bottom). (D) The specificity of these liposomes (1% ligand) was then assessed against a panel of Siglec-expressing cell lines in triplicate. Naked (no ligand) liposomes were used as a negative control. (E) White blood cells from peripheral human blood were obtained and incubated with Naked or G-35 liposomes (5% ligand) followed by staining with an anti-Siglec 7 antibody showing that G-35 liposomes bind to all Siglec-7 positive populations (green arrow: lymphocyte subset, blue arrow: monocytes, red arrow: granulocytes), but not cells that are Siglec-7 negative.
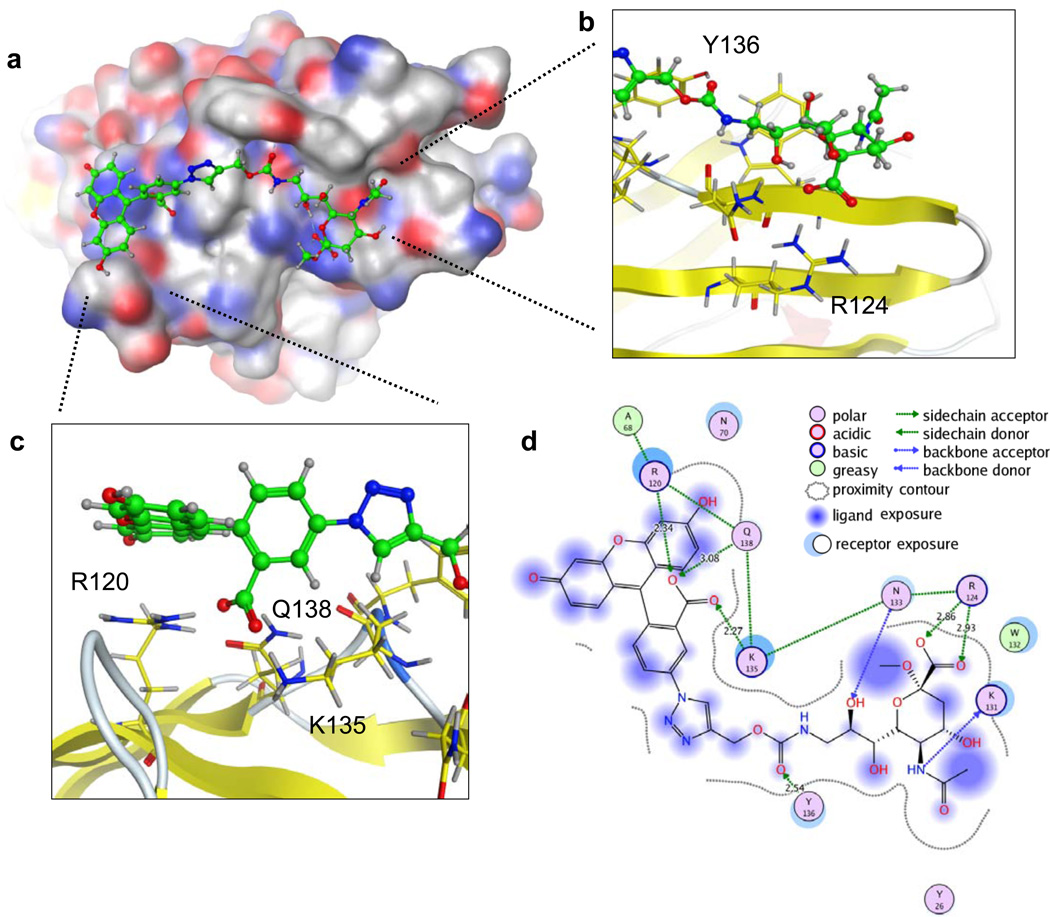
To gain insights into the ability of the fluorescein substituent to increase affinity, we used available Siglec-7 crystal structures20–22 to construct a model in complex with G35 (Figure 4). The reported crystal structures to date document that siglec-7 contains the conserved features of the sialic acid binding site found in other siglecs, including an arginine (R124) required to coordinate with the C-1 carboxyl group of sialic acid. Although, Siglec-7 exhibits preference for binding glycans with a terminal disialyl sequence, NeuAcα2-8NeuAcα2-3Gal, a co-crystal structure with the glycan moiety of the ganglioside GT1b, NeuAcα2-8NeuAcα2-3(NeuAcα2-3GalNAcβ1-3)Galβ1-4Glc-R, shows that it is the non-reducing terminal sialic acid that is bound to the conserved sialic acid binding site, analogous to the binding of ligands with a single sialic acid in other siglecs20–22. Since we had already established that binding is abolished by the R124A mutation (Figure 3d), we fixed the sialic acid moiety of G35 to the conserved binding site, and investigated the possible modes of interaction of the triazole and fluorescein moiety extending from the C-9 position. Accordingly, the area extending from C-9 that would be contacted by the fluorescein moiety were analyzed to identify potentially flexible amino acids as evidenced by differential side chain positions amongst the various structures reported to date. This showed that, in the context of potential fluorescein recognition, side chains of residues in the GG’ linker and CC’ loop of Siglec-7 are considerably flexible. Keeping the sialic acid binding site and the sialic acid scaffold fixed (Figure 4b), the identified residues (Arg120, Lys 135, Tyr 136) and the sialic acid substituent were then subjected to a low-mode molecular dynamics (LMMD) simulation. This allowed us to probe the extended binding site for structural plasticity under the limitations of a large ligand structure, and an ensemble of low energy conformations were identified. The resulting model suggests a number of potential interactions between Siglec-7 and the triazole-linked fluorescein moiety of G35. First, the Tyr136 hydroxyl is proposed to make a hydrogen bonding contact with the carbamate carbonyl (Figure 4b,d). Second, the side chains of Lys135, Gln138, and Arg120 are in hydrogen bonding contact with the carboxylic acid of the fluorescein moiety (Figure 4c,d). Lastly, the interactions of Arg120 and Gln138 with each other and the above-mentioned carboxylate orient the Arg120 to make a favorable stacking interaction with the xanthene ring (Figure 4c,d). The latter two sets of interactions, namely the acid coordination and the Arg stacking, have been previously observed in another fluorescein-protein interaction23 (Supplementary Figure 11), indicating the feasibility of these interactions in the proposed Siglec-7-G35 model.
Figure 4. Molecular modeling of G35 in the Siglec-7 binding site.
(A) G35 was docked into a composite model of existing Siglec-7 crystal structures by keeping the sialic acid and sialic acid binding site fixed, and carrying out a low mode MD simulation on surrounding flexible residues and the sialic acid substituent. (B) Expansion of the interactions between Siglec-7 and the sialic acid scaffold. (C) A close-up view of the proposed interactions between Siglec-7 and the triazole-linked fluorescein moiety of G35. (D) Summary of all of the proposed interactions, and the nature of these interactions, between Siglec-7 and G35.
As illustrated from the identification of G35 as a selective, high-affinity Siglec-7 ligand, the ‘on-chip’ approach represents a promising and attractive platform for high-throughput synthesis and screening. The ability to construct and screen large libraries of ligand analogs using minimal amounts of material allows synthetic efforts to be considerably reduced, and therefore these efforts can be focused on diversity rather than scale, an important factor for complex classes of molecules. Although this report documented the utility of on-chip synthesis of a sialoside analog library for identification of ligands for Siglecs, this approach should be applicable for generating and identifying high-affinity ligands for other families of glycan-binding proteins and likely for even more diverse protein-ligand systems. Moreover, while CuAAC was employed as the workhorse for library generation in this work, one can envision using other robust, orthogonal chemistries for ‘on-chip’ library construction, allowing for different areas of chemical space to be rapidly surveyed. Utilizing high-throughput approaches such as these and others,9, 11, 13 the identification of specific, high-affinity ligands for every Siglec will likely be possible in the near future, allowing for the promise of ligand-targeted nanoparticles for Siglec-mediated therapeutics to be fully realized.
Methods
Proof-of-Principle Array
The array described in Figure 1 was assembled by printing the noted compounds13 (100 µM) onto Schott-Nexterion Slide-H microarray slides24 into 16 subarrays which were then physically separated using a 16-well adhesive superstructure (Schott cat. No. 1178061). The on-chip click reactions were performed in DMF/H2O (3:1 v/v) by adding the desired azide (50 mM) to precomplexed THPTA/CuSO4 (5 mM/1 mM), followed by sodium ascorbate (10 mM) and applying this solution to the chip surface. Reactions were carried out for the indicated times, the solution was removed, and the arrays were washed by manual pipetting 3× with DMF, PBS + 0.05% Tween-20, H2O, and centrifuged to dry. For Siglec detections, Siglec-Fc chimeras were precomplexed with an R-PE secondary antibody and applied to the arrays as previously described.13
Large Scale On-Chip Synthesis and Screening
A library of amine-terminated alkyne sialosides (Figure 2a, Supplementary Methods, Supplementary Schemes 1–3) along with VII, were printed (100 µM, 4 spots/compound) onto Slide-H MPX-48 slides (Schott).24 The on-chip reactions with 94 azides (Supplementary Figures 4–5, 5 µl/well) were carried out for 2 hrs, as described above, with the exception that DMF/H2O (6.5:3.5 v/v) was used as the solvent to maintain integrity of the Teflon mask. The minor change in solvent did not slow the ‘on-chip’ reactions of a variety of diverse azides (data not shown), with the notable exception of 1-azidoadamantane whose solubility, and therefore reaction rate, was considerably slowed. The library was constructed over 2 slides with one unreacted subarray per slide (for a schematic see Supplementary Figure 6). Arrays were then immersed in PBS-Tween, vigorously washed with 10% SDS/PBS to remove any insoluble precipitate deposited on the array surface, washed extensively with water, centrifuged to dry, and probed with various Siglecs.13 As above, all Siglecs were precomplexed with an R-PE secondary antibody. Image analysis was preformed using IMAGENE, and the signals within each of the 96 subarrays were normalized to compound VII as 100%.
Liposome Preparation and Cell Binding Studies
Siglec-7 hits G35, K35, and I35 were resynthesized and coupled to PEGylated lipids (Supplementary Methods and Supplementary Scheme 4). Fluorescent liposomes (~100 nm diameter) containing various percentages of Siglec-7 hits or a “naked” PEGylated lipid were made essentially as previously described6, 7, 9, 13 with the following composition: 0.1 mol% Alexa-Fluor-647 lipid,6 5 mol% PEGylated lipid (= Siglec-7 Hit + “naked” lipid), 38 mol% cholesterol, and 57% distearoyl phosphatidylcholine. Binding experiments were done as previously described13 using 10–50 µM liposomes (final lipid concentration) for recombinant and 3 µM liposomes for primary cells, reading out the AF-647 channel in each case. The WT and R124A Jurkat Siglec-7 cells were prepared previously17 and the R-PE anti-Siglec 7 antibody was obtained from eBiosciences (12-5759-42).
Construction of the Siglec-7-G35 Model
The X-ray crystallographic structure of Siglec-7 (PDB ID: 2HRL) was used to derive a structural model of G35 in the binding site.21 To allow access to the extended binding site, the side chain of Lys135 had to be placed into an alternative low energy conformation as found for other Siglec-7 structures. The MOE rotamer library was employed. A preliminary ligand structure was then built in the binding site. Due to the high number of rotatable bonds in G35, finding low energy conformations is particularly challenging and low mode molecular dynamics simulations were employed using the following parameters25: rejection limit 100, iteration limit 1000, RMS gradient 0.01, maximum minimization iteration limit 500, enforcement of chair conformations was enabled, energy window 7, conformation limit 10000, RMSD limit 0.25. The MMFF94x force field was used. All molecular modeling was performed in MOE (Molecular Operating Environment, Chemical Computing Group, version 2011.10) on a regular desktop computer.
Supplementary Material
Acknowledgements
This work was supported by the NIH (P01HL107151 and AI050143 to J.C.P., T32AI007606 to C.D.R., and GM087620 to V.V.F), a Schering-Plough Research Institute postdoctoral Fellowship (to E.S.), a Rubicon fellowship from the Netherlands Organization For Scientific Research (NWO) (to E.S.), an Emmy Noether fellowship from the German Research Foundation (RA911/2-1 to C.R.), and the Max Planck Society (C.R.).
References
- 1.O'Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci. 2009;30:240–248. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jandus C, Simon HU, von Gunten S. Targeting siglecs--a novel pharmacological strategy for immuno- and glycotherapy. Biochem Pharmacol. 2011;82:323–332. doi: 10.1016/j.bcp.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 4.Pillai S, Netravali IA, Cariappa A, Mattoo H. Siglecs and immune regulation. Annu Rev Immunol. 2012;30:357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricart AD. Antibody-drug conjugates of calicheamicin derivative: gemtuzumab ozogamicin and inotuzumab ozogamicin. Clin Cancer Res. 2011;17:6417–6427. doi: 10.1158/1078-0432.CCR-11-0486. [DOI] [PubMed] [Google Scholar]
- 6.Chen WC, Kawasaki N, Nycholat CM, Han S, Pilotte J, Crocker PR, Paulson JC. Antigen Delivery to Macrophages Using Liposomal Nanoparticles Targeting Sialoadhesin/CD169. PLoS One. 2012;7:e39039. doi: 10.1371/journal.pone.0039039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen WC, Completo GC, Sigal DS, Crocker PR, Saven A, Paulson JC. In vivo targeting of B-cell lymphoma with glycan ligands of CD22. Blood. 2010;115:4778–4786. doi: 10.1182/blood-2009-12-257386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng Y, Rademacher C, Nycholat CM, Futakawa S, Lemme K, Ernst B, Paulson JC. High affinity sialoside ligands of myelin associated glycoprotein. Bioorg Med Chem Lett. 2011 doi: 10.1016/j.bmcl.2011.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nycholat CM, Rademacher C, Kawasaki N, Paulson JC. In silico-aided design of a glycan ligand of sialoadhesin for in vivo targeting of macrophages. J Am Chem Soc. 2012;134:15696–15699. doi: 10.1021/ja307501e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaccai NR, Maenaka K, Maenaka T, Crocker PR, Brossmer R, Kelm S, Jones EY. Structure-guided design of sialic acid-based Siglec inhibitors and crystallographic analysis in complex with sialoadhesin. Structure. 2003;11:557–567. doi: 10.1016/s0969-2126(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 11.Shelke SV, Cutting B, Jiang X, Koliwer-Brandl H, Strasser DS, Schwardt O, Kelm S, Ernst B. A fragment-based in situ combinatorial approach to identify high-affinity ligands for unknown binding sites. Angew Chem Int Ed Engl. 2010;49:5721–5725. doi: 10.1002/anie.200907254. [DOI] [PubMed] [Google Scholar]
- 12.Mesch S, Lemme K, Wittwer M, Koliwer-Brandl H, Schwardt O, Kelm S, Ernst B. From a library of MAG antagonists to nanomolar CD22 ligands. ChemMedChem. 2012;7:134–143. doi: 10.1002/cmdc.201100407. [DOI] [PubMed] [Google Scholar]
- 13.Rillahan CD, Schwartz E, McBride R, Fokin VV, Paulson JC. Click and pick: identification of sialoside analogues for siglec-based cell targeting. Angew Chem Int Ed Engl. 2012;51:11014–11018. doi: 10.1002/anie.201205831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: coppeRI)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong V, Presolski SI, Ma C, Finn MG. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew Chem Int Ed Engl. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikehara Y, Ikehara SK, Paulson JC. Negative regulation of T cell receptor signaling by Siglec-7 (p70/AIRM) and Siglec-9. J Biol Chem. 2004;279:43117–43125. doi: 10.1074/jbc.M403538200. [DOI] [PubMed] [Google Scholar]
- 18.Angata T, Varki A. Siglec-7: a sialic acid-binding lectin of the immunoglobulin superfamily. Glycobiology. 2000;10:431–438. doi: 10.1093/glycob/10.4.431. [DOI] [PubMed] [Google Scholar]
- 19.Nicoll G, Ni J, Liu D, Klenerman P, Munday J, Dubock S, Mattei MG, Crocker PR. Identification and characterization of a novel siglec, siglec-7, expressed by human natural killer cells and monocytes. J Biol Chem. 1999;274:34089–34095. doi: 10.1074/jbc.274.48.34089. [DOI] [PubMed] [Google Scholar]
- 20.Alphey MS, Attrill H, Crocker PR, van Aalten DM. High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J Biol Chem. 2003;278:3372–3377. doi: 10.1074/jbc.M210602200. [DOI] [PubMed] [Google Scholar]
- 21.Attrill H, Imamura A, Sharma RS, Kiso M, Crocker PR, van Aalten DM. Siglec-7 undergoes a major conformational change when complexed with the alpha(2,8)-disialylganglioside GT1b. J Biol Chem. 2006;281:32774–32783. doi: 10.1074/jbc.M601714200. [DOI] [PubMed] [Google Scholar]
- 22.Attrill H, Takazawa H, Witt S, Kelm S, Isecke R, Brossmer R, Ando T, Ishida H, Kiso M, Crocker PR, van Aalten DM. The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design. Biochem J. 2006;397:271–278. doi: 10.1042/BJ20060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korndörfer IP, Beste G, Skerra A. Crystallographic analysis of an "anticalin" with tailored specificity for fluorescein reveals high structural plasticity of the lipocalin loop region. Proteins. 2003;53:121–129. doi: 10.1002/prot.10497. [DOI] [PubMed] [Google Scholar]
- 24.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labute P. LowModeMD--implicit low-mode velocity filtering applied to conformational search of macrocycles and protein loops. J Chem Inf Model. 2010;50:792–800. doi: 10.1021/ci900508k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.