Abstract
The SasA-RpaA two-component system constitutes a key output pathway of the cyanobacterial Kai circadian oscillator. To date, rhythm of phycobilisome associated (rpaA) is the only gene other than kaiA, kaiB, and kaiC, which encode the oscillator itself, whose mutation causes completely arrhythmic gene expression. Here we report a unique transposon insertion allele in a small ORF located immediately upstream of rpaA in Synechococcus elongatus PCC 7942 termed crm (for circadian rhythmicity modulator), which results in arrhythmic promoter activity but does not affect steady-state levels of RpaA. The crm ORF complements the defect when expressed in trans, but only if it can be translated, suggesting that crm encodes a small protein. The crm1 insertion allele phenotypes are distinct from those of an rpaA null; crm1 mutants are able to grow in a light:dark cycle and have no detectable oscillations of KaiC phosphorylation, whereas low-amplitude KaiC phosphorylation rhythms persist in the absence of RpaA. Levels of phosphorylated RpaA in vivo measured over time are significantly altered compared with WT in the crm1 mutant as well as in the absence of KaiC. Taken together, these results are consistent with the hypothesis that the Crm polypeptide modulates a circadian-specific activity of RpaA.
Keywords: chronobiology, transcription regulation
Acircadian clock controls rhythms of gene expression in nearly all organisms. In the model unicellular cyanobacterium Synechococcus elongatus PCC 7942, the majority of genes show circadian expression (1). The core oscillator of the cyanobacterial circadian clock is composed of the KaiA, KaiB, and KaiC proteins (2). The phosphorylation state, ATPase activity, and dynamic conformation of KaiC all cycle with an approximate 24-h period (3, 4). Interaction of output proteins with KaiC conveys timing information to downstream effectors to influence gene expression patterns (5), chromosome compaction (6), and timing of cell division (7, 8). The histidine kinase Synechococcus adaptive sensor A (SasA) interacts physically with KaiC (9) and forms a two-component system with its cognate response regulator RpaA, a transcription factor of the winged-helix family. Together, SasA and RpaA are thought to comprise the major regulatory system of circadian output. RpaA is a regulator of global gene expression, and disruption of the rpaA (regulator of phycobilisome associated) gene renders gene expression from essentially all promoters arrhythmic (10). The phosphorylated form of RpaA is presumed to constitute its active state in promotion of gene expression (10, 11). RpaA also was recently shown to be a cognate response regulator for the input protein CikA, which displays phosphatase activity on RpaA in vitro (11).
Here we report the identification of a previously undescribed clock-related genetic element, designated crm (for circadian rhythmicity modulator), located immediately upstream of rpaA. The crm ORF encodes a predicted 62-residue polypeptide with no recognized functional domains. A transposon insertion allele of this gene, crm1, causes arrhythmicity from different classes of promoters without affecting RpaA protein levels, results in alteration of the RpaA phosphorylation state and KaiC protein levels, and eliminates the KaiC phosphorylation cycle. We propose that Crm functions as an allosteric modulator of clock-related RpaA activity.
Results
A Specific Allele of crm Disrupts Gene Expression Rhythms.
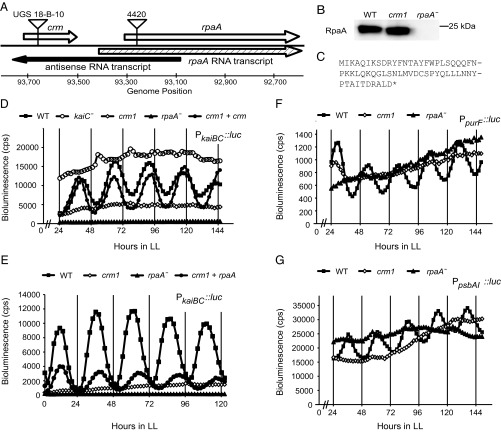
A mutant strain that exhibits arrhythmic expression of a PkaiBC::luc luciferase reporter, designated uni-gene set (UGS) mutant 18-B-10, was isolated from a transposon insertion library of S. elongatus (12) (Fig. 1D, crm1). The strain carries an insertion of a modified Tn5 transposon at 358 bp upstream of the start codon of the rpaA gene (Fig. 1A). Because rpaA deletion is known to cause arrhythmia, we analyzed UGS mutant 18-B-10 for possible disruption of RpaA expression. Immunoblot analysis demonstrated the presence of RpaA at WT levels in the UGS mutant 18-B-10 background, however (Fig. 1B, crm1). Sequence analysis of the S. elongatus genome indicated an ORF at the transposon insertion site (Fig. 1A) that encodes a predicted 62-aa protein with an estimated molecular weight of 7.25 kDa and no recognized functional domains (Fig. 1C).
Fig. 1.
Transposon insertion in the crm gene affects circadian rhythmicity, but not rpaA expression. (A) Diagram of genomic region containing crm and rpaA genes. Arrows indicate direction of transcription. Positions of insertions are noted. (B) Immunoblots of whole-cell extracts of WT, crm1, and rpaA null (4420 insertion) strains (10 μg total protein/lane) probed with RpaA antiserum. The crm1 mutation did not alter levels of the RpaA protein. (C) Amino acid sequence of crm ORF. (D–G) Bioluminescence rhythms. After entrainment to a 12:12 LD cycle, bioluminescence traces were captured over 6 d under LL. crm1 causes arrhythmic expression from the kaiBC::luc reporter. rpaA− strains are arrhythmic and demonstrate only a barely detectable level of kaiBC reporter expression, whereas kaiC− strains display high-baseline arrhythmic expression. The crm1 mutant transformed with the crm coding sequence in neutral site I complements arrhythmic expression from kaiBC::luc in trans. Representative traces are shown. cps, counts per second. (E) Ectopic expression of an extra copy of rpaA in a crm1 background partially restores rhythmicity, displaying a low-amplitude, long-period rhythm compared with WT. Average bioluminescence levels of each strain are shown. (F and G) The crm1 allele abolishes rhythmic expression from the purF (F) and psbAI (G) promoters. Bioluminescence in crm1 strains is consistently near WT peak level as measured from the class II purF promoter, in contrast to WT trough level as measured from the class I promoters kaiBC and psbAI. Representative traces are shown.
Unexpectedly, however, substitution of the small ORF with an antibiotic resistance cassette did not result in arrhythmic kaiBC promoter activity (Fig. S1A), indicating that the circadian phenotype of UGS mutant 18-B-10 is allele-specific. The new ORF was named crm (for circadian rhythmicity modulator), and the Tn5 insertion allele was designated crm1. Recreation of the crm1 mutation in psbAI and purF reporter strains also caused arrhythmic expression from those promoters (Fig. 1 F and G). These genes and kaiBC have been assigned to different circadian output categories (13), suggesting that the crm1 allele exerts a global influence on gene expression rhythms.
Because an antisense RNA runs through the site of crm1 insertion (14) (Fig. 1A), we investigated whether the arrhythmic phenotype results from disruption of the crm ORF or of the antisense RNA. Quantification of the antisense transcript on either side of the crm1 insertion by quantitative RT-PCR (qRT-PCR) showed normal levels (Table S1). Moreover, circadian bioluminescence rhythms driven from the kaiBC promoter were restored when a WT crm gene was expressed in trans in the crm1 background, demonstrating complementation of arrhythmia (Fig. 1D). Additional experiments showed that the start codon of crm is necessary for complementation, suggesting the creation of a Crm protein (Fig. S1B). Addition of an affinity tag to the ectopically expressed allele abolished complementation, complicating confirmation of the protein product.
crm1 Phenotypes Are Distinct from Those of an rpaA Null Mutant.
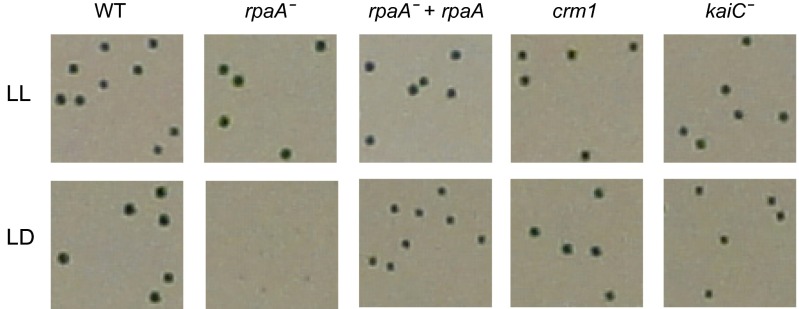
Several phenotypes distinguish a crm1 mutant from an RpaA-deficient strain. The overall level of expression from the PkaiBC::luc reporter is notably higher in a crm1 mutant background than in an rpaA null background (Fig. 1D). RpaA-deficient mutants do not form colonies under alternating 12-h light:dark (LD) cycles (10), but a crm1 mutant grew normally in LD (Fig. 2). The kinase CikA is a major sensor for the input pathway of the circadian clock and acts as a phosphatase for RpaA (11, 15, 16). In WT cells, ZsGreen-tagged CikA exhibits unipolar localization (17); in a subset of clones in which rpaA is disrupted, CikA is mislocalized and diffuse (Fig. S2). CikA remains localized to the pole in all cells of a crm1 background (Fig. S2), reinforcing the difference between this defect and that of an rpaA null.
Fig. 2.
Phenotypes of crm1 mutants differ from those of an rpaA mutant. Growth of crm1 strain is not impaired in an LD cycle. WT, crm1, kaiC−, rpaA−, and rpaA-complemented strains were plated for single colonies and maintained in either 100 μmol m-2s-1 LL (Upper) or a 12:12 LD cycle (Lower) over 4 d. The crm1 mutant does not exhibit the growth impairment of an rpaA strain under the LD regimen.
KaiC Levels Are Reduced and Phosphorylation Cycling Is Impaired in crm1 Mutants.
Structural changes corresponding to ordered cycling of the phosphorylation state of KaiC are crucial for interactions with clock output effectors (18, 19). Disruption of rpaA has been reported to result in attenuated expression from the kaiBC promoter and to abolish cyclic phosphorylation of the core oscillator protein KaiC, which appears to be constitutively phosphorylated in an rpaA null strain under constant light (LL) conditions (10).
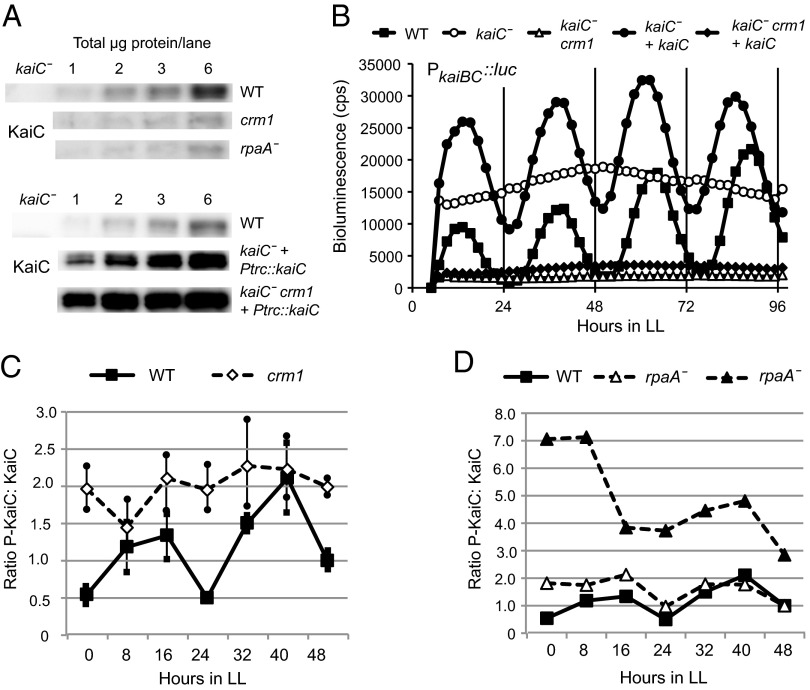
To assess whether the crm1 affects KaiC similarly to an rpaA disruption, we compared levels of KaiC in whole-cell extracts and examined in vivo phosphorylation cycling of KaiC in crm1 and an rpaA null strain over 48 h using Phos-tag gels and immunoblotting. Steady-state levels of KaiC were reduced by approximately two-thirds in both crm1 and rpaA null backgrounds compared with WT (Fig. 3A). Although both phosphorylated and nonphosphorylated KaiC were present in crm1 mutants at all time points, no cycling was detected (Fig. 3C). Surprisingly, although KaiC phosphorylation was sometimes constitutively elevated in an rpaA null background, rhythmic phosphorylation of KaiC was detected in replicate experiments (Fig. 3D).
Fig. 3.
Quantitation of in vivo KaiC levels and phosphorylation rhythm in crm1 and rpaA deletion mutants. (A) (Upper) Immunoblots of whole-cell extracts of WT, crm1, kaiC−, and rpaA− strains (1, 2, 3, and 6 μg total protein/lane) sampled at 9 h after onset of light and probed with KaiC antiserum. Steady-state levels of KaiC were reduced by approximately two-thirds in both crm1 and rpaA− mutants. (Lower) Immunoblots of whole-cell extracts of WT and strains expressing KaiC from uninduced trc promoter in kaiC− and kaiC− crm1 backgrounds. (B) Arrhythmic gene reporter expression in crm1 background is not due to low KaiC levels. Shown are representative bioluminescence traces of strains with KaiC expressed ectopically from a neutral site. Expression from kaiBC::luc reporter in a kaiC− crm1 background remains low and arrhythmic with ectopically expressed KaiC, whereas ectopic KaiC expression restores rhythms in a kaiC deletion background. (C and D) Densitometric analysis of immunoblots of whole-cell extracts from strains entrained to a 12:12 LD regime and sampled every 8 h in LL for 48 h. (C) Densitometric analysis showing average ± SD values of two blots of crm1 and two WT samples. The crm1 strain is characterized by constitutively elevated KaiC phosphorylation and no discernible overall rhythm over 48 h. (D) KaiC phosphorylation is elevated in rpaA− cells, but the rhythm of phosphorylation remains detectable. Quantitation of two separate blots of rpaA− samples is shown compared with the WT trace shown in C.
Phosphorylation Cycling of RpaA Is Perturbed in Arrhythmic crm1 and kaiC Strains.
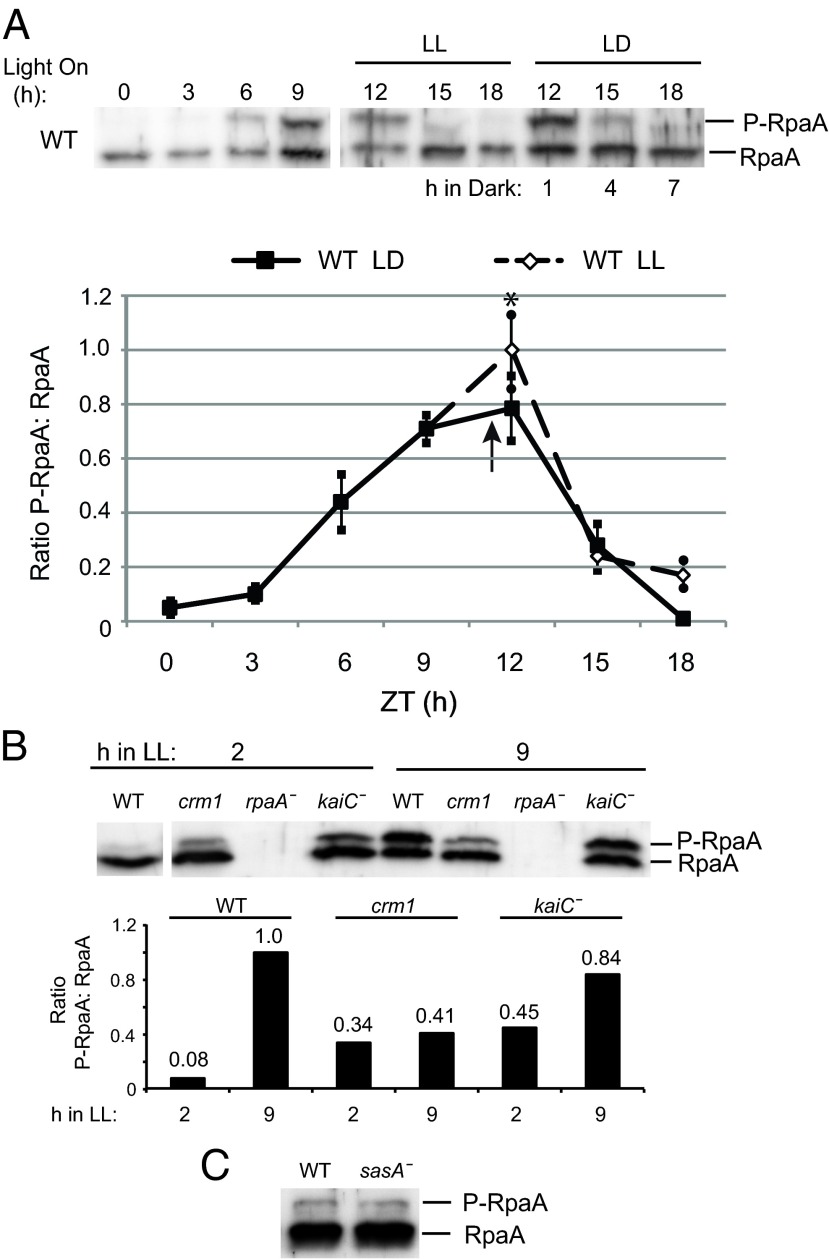
Because phosphorylated RpaA is postulated to reflect RpaA activity (10, 11), we investigated whether crm1 affects the in vivo phosphorylation state of RpaA in circadian and diurnal cycles. In WT cells, RpaA phosphorylation peaks at subjective dusk (8-12 h after light onset) and ebbs at subjective dawn (11, 20). To examine the influence of a dark cue on the RpaA phosphorylation pattern, we compared replicate WT samples that were entrained to a 12-h LD cycle (12:12 LD) and either left in the light or transferred to the dark at subjective dusk and sampled every 3 h after last light. The LD set was transferred to the dark at 11 h after lights on, so that the 12-h, 15-h, and 18-h samples from the two sets were from the same circadian times but experienced different LD cues. Samples were detected by separation on Phos-tag gels and immunoblotting with RpaA antiserum. Although exposure to darkness for 1 h slightly reduced the ratio of phosphorylated RpaA to nonphosphorylated RpaA at zeitgeber time (ZT) 12 (Fig. 4A), the overall pattern of phosphorylation was not substantially different under LL and LD, indicating that the RpaA phosphorylation cycle is largely under circadian clock control and is only mildly affected by a diurnal cycle.
Fig. 4.
The in vivo RpaA phosphorylation cycle is minimally sensitive to a dark cue and is perturbed in arrhythmic strains. (A) Time points from representative immunoblots (Upper) and densitometric analysis showing average ± SD values from multiple immunoblots (Lower) demonstrating overall levels of RpaA phosphorylation in WT cells entrained to a 12:12 LD cycle and sampled every 3 h over 18 h. Samples were either maintained in LL or collected after entering dark at 11 h after onset of light. The overall RpaA phosphorylation level in LD is significantly lower after 1 h in dark compared with the 12-h LL sample, but the overall phosphorylation cycle remains substantially the same under both LL and LD conditions. The arrow indicates onset of darkness (at ZT = 11 h) for LD samples. (B) Representative immunoblot (Upper) and quantification (Lower) of the P-RpaA:RpaA ratio of whole-cell extracts (10 μg total protein/lane) sampled at 2 h and 9 h after onset of light after entrainment to a 12:12 LD cycle and probed with RpaA antiserum. A high proportion of RpaA is phosphorylated in the WT strain at 9 h after entering light, whereas the majority of RpaA appears nonphosphorylated at 2 h. The P-RpaA:RpaA ratio does not change substantially in crm1 cells sampled at 2 h compared with 9 h. In a kaiC deletion strain, P-RpaA:RpaA remains at intermediate levels compared with WT at both time points. (C) Immunoblot of nonentrained WT and sasA deletion strain probed with RpaA antiserum. P-RpaA is detectable in the absence of SasA in vivo.
In entrained cultures, phosphorylated RpaA was present at higher levels in crm1 samples than in WT near subjective dawn (LL2), and did not show the marked increase at subjective dusk (LL9) characteristic of WT (Fig. 4B). These results support a mechanism in which the crm1 allele affects rhythmic gene transcription via modulation of RpaA activity. Unexpectedly, substantial levels of phosphorylated RpaA were detected in the absence of KaiC at both subjective dawn (L 2 h) and dusk (L 9 h) (Fig. 4B), demonstrating that a functional clock is not required for RpaA phosphorylation, but suggesting that regulation of RpaA dephosphorylation requires the core clock. Surprisingly, phospho-RpaA was detected in a sasA null background (Fig. 4C), indicating that other kinases are able to substitute for SasA in vivo for phospho-transfer to RpaA.
Discussion
The timing information from the circadian oscillator in cyanobacteria is proposed to be conveyed through at least three output pathways, one of which is the two-component SasA-RpaA system (20). According to recent models, CikA and LabA act as negative effectors and converge on RpaA, which functions as the master circadian regulator of transcription (20), and circadian regulation of RpaA activity is the result of opposing kinase activity of SasA and phosphatase activity of CikA (11). Deletion of rpaA has pleiotropic effects on transcript levels and physiology in S. elongatus. Overall transcript levels are dramatically reduced, and cells cannot survive multiple LD cycles (10). In contrast, crm1 or kaiC mutants grow robustly in LD, and reporter levels suggest transcripts within the normal range, although closer to trough levels when Crm is disrupted as a crm1 allele. These data suggest that Crm has more specific effects on circadian aspects of gene expression compared with RpaA. As such, a crm1 background provides an arrhythmic but otherwise fit strain in which to assess the roles of circadian gene expression.
In light of the drastic effects of crm1 on promoter rhythmicity and complementation of the mutant phenotype with the crm ORF expressed in trans, the lack of a discernible phenotype in a crm deletion strain is puzzling. The Tn5 insertion may produce a “poison” product that interferes with protein–protein interactions key to generating circadian rhythms; complementation suggests that even a small amount of WT Crm is sufficient to restore normal regulation, and, similarly, expression of an extra copy of rpaA in a crm1 background partially restores rhythmic expression from the kaiBC promoter (Fig. 1E). Lack of a detectable circadian phenotype in a deletion strain also has been observed for the circadian feedback regulator lalA, in which effects on rhythmic gene expression and cell growth are seen only as a consequence of overexpression (21).
The inability to complement the mutant phenotype with a tagged allele hindered our efforts to confirm overexpression in strains that carry inducible constructs and show no mutant phenotype. Notably, however, expression of native crm from the strong class I psbAI promoter in WT and crm deletion backgrounds resulted in substantial damping of kaiBC promoter rhythms over several days in free run (Fig. S3A), suggesting that Crm has a modulating effect on circadian rhythms.
In kaiC deletion strains, elevated RpaA phosphorylation correlates with high expression from the kaiBC promoter (Figs. 1 and 4), suggesting that P-RpaA is a prerequisite for activity of that promoter. In a previous study, the crm1 allele was erroneously used as an rpaA deletion strain, under the assumption that the insertion blocked rpaA expression, consistent with arrhythmicity of reporter gene expression (8). The crm1 allele, like a true rpaA or sasA null, suppresses the cell-division defect of ΔcikA mutant (8); in addition, we found that the combination of an rpaA null allele with either crm1 or crm deletion alleles resulted in PkaiBC reporter expression and light:dark sensitivity characteristic of an rpaA single mutant (Fig. S3B). These findings support the hypothesis that the crm1 mutation disrupts RpaA activity. The observation that crm1 suppresses high expression from the kaiBC promoter even in a kaiC deletion background (Fig. 3B) also supports this hypothesis. Moreover, the location of the crm gene just upstream of rpaA raises the possibility that transcription of crm itself is coupled to RpaA function. Few transcripts were identified through this region in deep-sequencing datasets, however (14).
Despite similar attenuation of KaiC levels, an rpaA null strain retains low-amplitude KaiC phosphorylation rhythms, whereas a crm1 mutant does not. The crm1 allele may influence kaiBC feedback regulation and synchrony and/or the mechanism of KaiC cycling. The lack of bioluminescence reporter rhythms in an rpaA null background suggests a decoupling or insensitivity to the KaiC oscillations. RpaA activity is critical for sustaining the transcription/translation feedback loop of KaiBC expression (20); however, a recent study suggested that the response regulator RpaB, not RpaA, binds directly to the promoters of rhythmically regulated target genes, including kaiBC, and represses them (22). The authors suggested that RpaA functions cooperatively with RpaB in transcriptional regulation and postulated that phosphorylated RpaA dissociates RpaB from promoters. An interfering effect of a translation product from the crm1 allele on RpaA’s ability to associate with RpaB also would be consistent with this model. Alternatively, the arrhythmia observed in crm1 mutant cells could be related to a defective mechanism of entrainment wherein cells cannot respond to LD cues and are asynchronous in the population.
We propose that the native form of Crm acts as an allosteric modulator of RpaA, although direct interaction with the oscillator cannot be ruled out. Crm likely forms a disordered coil, given that secondary structure prediction of the Crm amino acid sequence did not reveal recognized structural motifs. The genome of the multicellular nitrogen-fixing cyanobacterium Anabaena sp. PCC 7120 contains a short putative ORF, including 19 contiguous bases identical to those of S. elongatus crm, upstream of a two-component histidine kinase (all4261), but no crm-like sequence upstream of the Anabaena homolog of rpaA, suggesting that crm-like proteins exist in other cyanobacteria and perhaps function in other signaling pathways. A precedent for allosteric regulation of transcription factors by small proteins in cyanobacteria is provided by PipX, an 89-residue protein that interacts either with the carbon/nitrogen balance sensor PII or the transcription factor NtcA depending on nutrient conditions (23).
Similarly, Crm may interact transiently with RpaA, possibly in response to a state of the KaiABC oscillator, and also may interact with other clock components in the circadian regulatory network. However, the WT oscillations of gene expression in a crm null background indicate a more complex network in which redundancy is present; thus, the identity of crm is made possible by the negative effect of the neomorphic crm1 allele. Other data support a wider network than has been described previously, with detection of RpaA phosphorylation in a sasA null background (Fig. 4C). Biochemical or proteomic approaches may reveal the mechanism of Crm activity and identify potential interaction partners.
Taken together, our data are largely consistent with current models in which phosphorylated RpaA represents an active state corresponding to promotion of circadian gene expression from most genes. However, we found that in the absence of KaiC, a substantial ratio of phospho-RpaA is continuously present in the cell, suggesting a more critical role for the oscillator in regulating RpaA dephosphorylation. Although a recent study did not detect RpaA phosphorylation in the absence of SasA, those experiments were conducted using a strain induced to restore native levels of KaiC (11). Our observation that phosphorylated RpaA is present in the absence of SasA in vivo suggests that an alternative interaction partner of RpaA may help maintain residual gene expression rhythms from the kaiBC promoter in an sasA null (20), a hypothesis that is consistent with a network organization model for two-component systems in S. elongatus (24).
In addition, the detectable sensitivity of RpaA phosphorylation level to an LD transition may imply a connection between the redox-sensing input system and the output acting in parallel with the central oscillator. In this model, association of RpaA with a previously described dynamic multiprotein circadian complex, termed the periodosome, consisting of input proteins, output factors, and the Kai oscillator (25, 26), best explains the interplay between traditionally defined output proteins and the core of the clock, such as the negative regulation of RpaA by CikA (10, 11). Such an arrangement would facilitate the integration of environmental and time signals and ensure robust circadian cellular responses.
Materials and Methods
Bacterial Strains, Media, and Culture.
S. elongatus PCC 7942 and transformed derivatives (Table S2) were maintained in BG-11 liquid or solid medium with antibiotics as needed for selection at standard concentrations (27).
Bioluminescence Monitoring.
Bioluminescence of PkaiBC-luc firefly luciferase fusion reporter strains was monitored at 30 °C under LL conditions as described previously (27). Data were analyzed with the Biological Rhythms Analysis Software System (http://millar.bio.ed.ac.uk/pebrown/brass/brasspage.htm) import and analysis program using Microsoft Excel. Results shown are representative of at least three independent experiments.
RT-PCR.
Cyanobacterial strains in liquid medium were grown for 3 d under a 12:12 LD regimen, and 10–15 mL of culture was collected at 4 h after lights on. cDNA was prepared from total RNA using the SuperScript III Kit (Life Technologies). qRT-PCR was performed with the Applied Biosystems StepOne Plus system (Life Technologies) and SybrGold Master Mix (Life Technologies) with three biological replicates per strain. Abundance was calculated via the ΔΔCT method and normalized to rpoA as a control.
Construction of Plasmids.
Plasmids (Table S3) were constructed using the GeneArt Seamless Cloning and Assembly Kit (Life Technologies) or GeneArt Synechococcus pSyn1_D/TOPO Vector Kit (Life Technologies) without the addition of nickel.
Site-Directed Mutagenesis.
Site-directed mutagenesis designed to substitute two stop codons for the start codon of the crm ORF was performed on plasmid pAM4666 (Table S3) following the Quickchange method (Stratagene) using complementary primers 5′-CCTGATTACTTGAGTAGTAGATCAAGGCTCAG-3′ and 5′-CTGAGCCTTGATCTACTACTCAAGTAATCAGG-3′.
Immunoblotting.
SDS/PAGE was performed according to standard methods with the following exceptions. Phosphorylation of RpaA and KaiC was detected using 10% SDS-polyacrylamide gels supplemented with Phos-tag ligand (Wako Chemicals USA) at a final concentration of 25 μM and manganese chloride at a final concentration of 50 μM. Gels were incubated once for 10 min in transfer buffer supplemented with 100 mM EDTA, followed by a 10-min incubation in transfer buffer without EDTA before standard wet transfer. For phospho-RpaA detection, protein extracts and electrophoretic apparatus were kept chilled to minimize hydrolysis of heat-labile phospho-Asp. Protein extracts for use in Phos-tag gels were prepared in Tris-buffered saline, and extracts for standard SDS/PAGE were prepared in PBS. RpaA antiserum (a gift of E. O’Shea, Harvard University, Cambridge, MA) was used at a dilution of 1:1,000. KaiC immunoblotting was performed as described previously (28) with modifications described elsewhere (8). Densitometric analyses were performed using National Institutes of Health ImageJ software.
Live-Cell Fluorescence Microscopy.
Strains were patched on BG-11 solid medium supplemented with appropriate antibiotics and then inoculated into liquid BG-11 medium. Immediately before imaging, 1 μL of culture was placed onto 1% agarose dissolved in BG-11 medium, coverslipped, and imaged on a DeltaVision RT inverted epifluorescence microscope (Applied Precision) with an Olympus Plan Apochromat 100× objective at 24 °C using appropriate filters. Images were captured using a CoolSnap HQ CCD camera (Photometrics) and deconvolved using the SoftWorx imaging program (Applied Precision). Micrographs were saved as 16-bit gray-scale images and then converted to 8-bit images, and channels were combined in ImageJ.
Supplementary Material
Acknowledgments
We thank current and former members of the S.S.G. laboratory; A. Taton for assisting with plasmid design and construction; and S. Diamond, B. Flaherty, S. Cohen, and M. Paddock for providing experimental support and helpful discussion. We also thank E. O’Shea for kindly providing the RpaA antiserum. This work was supported by National Institutes of Health Grant R01GM062419 (to S.S.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312793110/-/DCSupplemental.
References
- 1.Mackey SR, Golden SS, Ditty JL. The itty-bitty time machine: Genetics of the cyanobacterial circadian clock. Adv Genet. 2011;74:13–53. doi: 10.1016/B978-0-12-387690-4.00002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281(5382):1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 3.Nishiwaki T, et al. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA. 2004;101(38):13927–13932. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terauchi K, et al. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104(41):16377–16381. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Golden SS, Kondo T, Ishiura M, Johnson CH. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J Bacteriol. 1995;177(8):2080–2086. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103(22):8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93(19):10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong G, et al. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140(4):529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki H, et al. A kaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101(2):223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 10.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103(32):12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutu A, O’Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol Cell. 2013;50(2):288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holtman CK, et al. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 2005;12(2):103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 13.Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181(11):3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vijayan V, Jain IH, O’Shea EK. A high-resolution map of a cyanobacterial transcriptome. Genome Biol. 2011;12(5):R47. doi: 10.1186/gb-2011-12-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289(5480):765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 16.Mackey SR, et al. Proteins found in a CikA interaction assay link the circadian clock, metabolism, and cell division in Synechococcus elongatus. J Bacteriol. 2008;190(10):3738–3746. doi: 10.1128/JB.01721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol Microbiol. 2006;60(3):658–668. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]
- 18.Chang YG, Kuo NW, Tseng R, LiWang A. Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2011;108(35):14431–14436. doi: 10.1073/pnas.1104221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valencia S J, et al. Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells. 2012;17(5):398–419. doi: 10.1111/j.1365-2443.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- 20.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci USA. 2010;107(7):3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniguchi Y, Nishikawa T, Kondo T, Oyama T. Overexpression of lalA, a paralog of labA, is capable of affecting both circadian gene expression and cell growth in the cyanobacterium Synechococcus elongatus PCC 7942. FEBS Lett. 2012;586(6):753–759. doi: 10.1016/j.febslet.2012.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Hanaoka M, et al. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J Biol Chem. 2012;287(31):26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espinosa J, Forchhammer K, Burillo S, Contreras A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol Microbiol. 2006;61(2):457–469. doi: 10.1111/j.1365-2958.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 24.Kato H, et al. Exploration of a possible partnership among orphan two-component system proteins in cyanobacterium Synechococcus elongatus PCC 7942. Biosci Biotechnol Biochem. 2012;76(8):1484–1491. doi: 10.1271/bbb.120172. [DOI] [PubMed] [Google Scholar]
- 25.Kageyama H, Kondo T, Iwasaki H. Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J Biol Chem. 2003;278(4):2388–2395. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- 26.Golden SS. Meshing the gears of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2004;101(38):13697–13698. doi: 10.1073/pnas.0405623101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackey SR, Ditty JL, Clerico EM, Golden SS. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol. 2007;362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
- 28.Ivleva NB, Golden SS. Protein extraction, fractionation, and purification from cyanobacteria. Methods Mol Biol. 2007;362:365–373. doi: 10.1007/978-1-59745-257-1_26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.