Abstract
The mechanisms underpinning broad compatibility in root symbiosis are largely unexplored. The generalist root endophyte Piriformospora indica establishes long-lasting interactions with morphologically and biochemically different hosts, stimulating their growth, alleviating salt stress, and inducing local and systemic resistance to pathogens. Cytological studies and global investigations of fungal transcriptional responses to colonization of barley and Arabidopsis at different symbiotic stages identified host-dependent colonization strategies and host-specifically induced effector candidates. Here, we show that in Arabidopsis, P. indica establishes and maintains biotrophic nutrition within living epidermal cells, whereas in barley the symbiont undergoes a nutritional switch to saprotrophy that is associated with the production of secondary thinner hyphae in dead cortex cells. Consistent with a diversified trophic behavior and with the occurrence of nitrogen deficiency at the onset of saprotrophy in barley, fungal genes encoding hydrolytic enzymes and nutrient transporters were highly induced in this host but not in Arabidopsis. Silencing of the high-affinity ammonium transporter PiAMT1 gene, whose transcripts are accumulating during nitrogen starvation and in barley, resulted in enhanced colonization of this host, whereas it had no effect on the colonization of Arabidopsis. Increased levels of free amino acids and reduced enzymatic activity for the cell-death marker VPE (vacuolar-processing enzyme) in colonized barley roots coincided with an extended biotrophic lifestyle of P. indica upon silencing of PiAMT1. This suggests that PiAmt1 functions as a nitrogen sensor mediating the signal that triggers the in planta activation of the saprotrophic program. Thus, host-related metabolic cues affect the expression of P. indica’s alternative lifestyles.
Keywords: root cortical cell death, RCD, broad-host range, biotrophy, mutualism
Upon plant colonization, fungi adopt different strategies to gain access to host nutrients. Whereas necrotrophs kill plant cells with subsequent saprotrophic nutrition, other fungi maintain biotrophic relationships with their hosts either transiently (hemibiotrophs) or as lifelong interactions. The degree of specialization to a particular host and the host’s metabolic status may greatly influence plant colonization (1–4). Broad-host range root endophytes undergo long-term interactions with a large variety of plants, thereby playing a significant role in natural and managed ecosystems and in the evolution of land plants. To establish and maintain a compatible interaction with different hosts, these endophytes must respond and adapt to host-specific signals. Alternative lifestyles and colonization strategies in different host species thus may be a consequence of this adaptation to highly variable host environments. In this study, we addressed the question of whether endophytes adopt different strategies during colonization of distinct hosts or whether their success resides in a general colonization strategy. An interesting system to explore this issue is the mutualistic root endophyte Piriformospora indica (Basidiomycota, Sebacinales), with its large number of plant hosts. Among others, this generalist can establish a mutualistic interaction with roots of the agriculturally important monocot barley (Hordeum vulgare) and the dicot model plant Arabidopsis thaliana (5, 6), two biochemically and morphologically distinct plants. Cytological studies in both hosts have shown that P. indica has a biphasic colonization strategy (7–10). Initial root cell invasions are biotrophic where colonized host cells maintain membrane integrity and invasive hyphae of P. indica remain enveloped by the host plasma membrane and thus are not accessible to cell wall stains such as wheat germ agglutinin–Alexa Fluor 488 (WGA-AF488) conjugate (7, 8, 11). Later, P. indica is found more often in dead or dying host cells (9, 10), especially in the root cortex of barley. Colonization at later stages was shown to be reduced by overexpression of the negative cell death regulator BAX inhibitor 1 in barley and to be mediated by an endoplasmic reticulum stress-triggered caspase-dependent cell death in Arabidopsis (9, 10). Questions arise as to what extent hemibiotrophy in a root endophyte reflects a general colonization strategy of the symbiont to benefit from different plants and how the symbiotic lifestyle is influenced by the hosts. Our study reveals that broad compatibility in a root endophyte is associated with phenotypic plasticity of the symbiont and with the expression of alternative lifestyle strategies in a host-dependent way.
Results
Expression Patterns of P. indica Genes, Including Those Encoding Small Secreted Proteins, Support a Diversified Colonization Strategy for Barley and Arabidopsis.
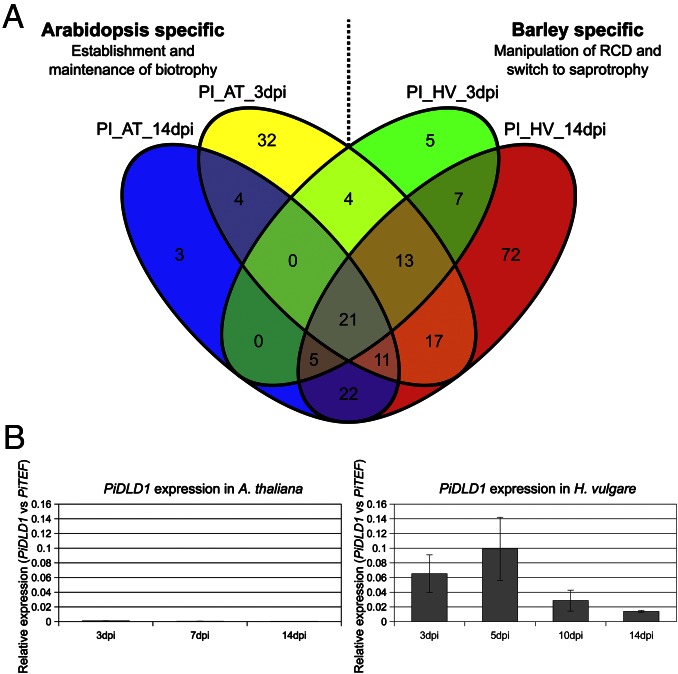
We hypothesized that successful root colonization of different hosts would require host-related colonization strategies, and we studied these differences by a global characterization of fungal transcriptional responses to barley and Arabidopsis at different developmental stages. A customized Agilent microarray was designed to monitor P. indica’s gene expression during root colonization of plants grown on sugar-free minimal medium (PNM) at 3 (early biotrophic phase) and 14 (late saprotrophic phase) days post inoculation (dpi). Fungal material grown on PNM was used as a control. From the 11,463 P. indica genes represented on the microarray chip, about 2,400 genes were differentially regulated at the early biotrophic phase in barley compared with the control. At this time point, 3,162 fungal genes were differentially regulated in colonized Arabidopsis compared with the control. At the late colonization stage, far more genes (4,482 genes) were differentially regulated in colonized barley than in colonized Arabidopsis roots (1,948 genes; Dataset S1). In total, about 70% (2,023) of the in planta-induced genes (2,861 genes) encoding intracellular proteins and 60% (277 of 463 induced genes) of those encoding putative secreted proteins had host-specific expression profiles (SI Appendix, Fig. S1). In particular, 123 of the 216 induced genes encoding small secreted proteins (SSPs; <300 amino acids), also known as putative effectors (12), were either Arabidopsis or barley responsive (Fig. 1A), suggesting that colonization of different hosts may require exploitation of distinct effectors that can interact with elements characteristic to each host. SSPs recently were shown to facilitate colonization by manipulating host defense and reprogramming plant metabolism during symbiosis (13, 14). Many of these proteins are cysteine rich (15) or possess distinctive features, such as a regular pattern of histidine and alanine residues found in all members of the P. indica-specific DELD family (7, 8). Eighteen of the 29 genes encoding P. indica DELD proteins were plant responsive and largely induced in barley but to a lesser extent in Arabidopsis (Fig. 1B and Dataset S2). Additionally, a small set of 21 SSPs were identified that are expressed in both hosts at comparable symbiotic stages (Fig. 1A). These SSPs may represent general determinants that target conserved recognition and signaling pathways in roots. Congruent with the broad definition of effectors (12), at the early symbiotic stage, 16 and 14 of P. indica’s top 20 up-regulated SSPs in Arabidopsis and in barley, respectively, encoded proteins with no known functional domains. A different situation was found at 14 dpi, when 50% of P. indica’s SSPs induced during colonization of barley, but not of Arabidopsis, encoded putative hydrolases (mainly glycoside hydrolase families GH10, GH11, and GH61; Dataset S3). Taken together, these data are consistent with a diversified colonization strategy for barley and for Arabidopsis, especially at 14 dpi, and prompted us to further characterize the two interactions at this stage.
Fig. 1.
Host-dependent expression profiles of in planta-induced P. indica genes encoding SSPs (<300 amino acids). (A) Number of SSPs significantly induced at 3 and 14 dpi during Arabidopsis (PI_AT) and barley (PI_HV) colonization, calculated vs. PNM control. (B) RT-qPCR analysis of PiDLD1 (PIIN_05872), a member of the P. indica-specific putative effector family DELD, during colonization of Arabidopsis (Left) and barley (Right) at different time points from three independent experiments.
P. indica Undergoes Major Trophic, Phenotypic, and Transcriptional Rearrangements During Colonization of Barley, Whereas It Maintains a Predominant Biotrophic Nutrition in Arabidopsis.
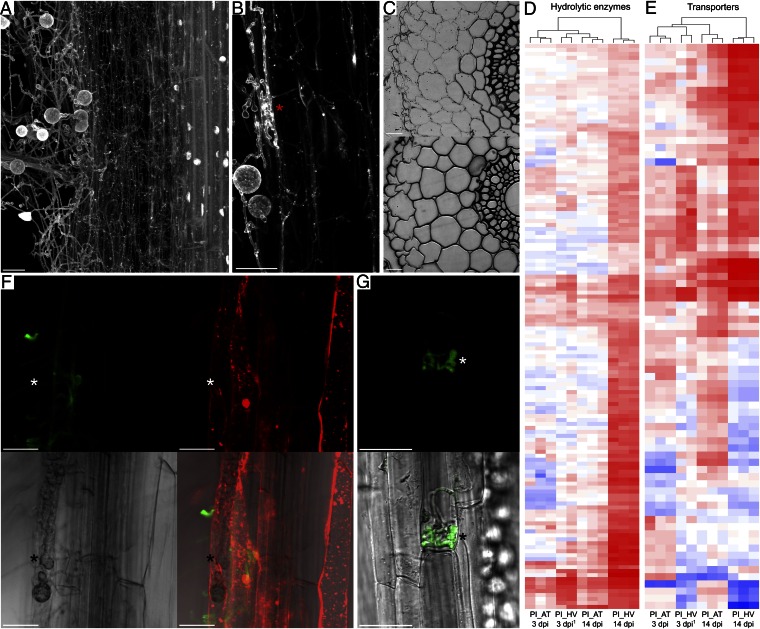
Transcriptional data, together with cytological analyses, show that in barley, in response to a progressively increasing natural root cortical cell death (RCD) in older/basal root zones (16), P. indica undergoes a distinct nutritional shift from biotrophic to saprotrophic nutrition associated with the production of secondary thinner hyphae and with the secretion of hydrolytic enzymes (Fig. 2 A–D and SI Appendix, Fig. S2). Thus, after biotrophic colonization of the outermost cell layer (3–5 dpi), on its route toward the endodermis, the fungus progresses inter- and intracellularly by digestion of barley cortical cell walls, which became most evident at 30 dpi (Fig. 2C), but without visible root necrosis at the macroscopic level (10, 17). Host cell wall appositions, named papillae, often are visible beneath the site of fungal penetration attempts of living host barley cells (8), and at later colonization stages, hyphal contact with the endodermis, which is not penetrated (10), results in a strong autofluorescence of the host cell wall, indicating activation of plant defense upon attempted penetration of the root vasculature (SI Appendix, Fig. S3). Despite a predominant saprotrophic lifestyle, the presence of P. indica in barley leads to beneficial effects (6, 17).
Fig. 2.
Colonization patterns and P. indica gene expression polymorphisms correlate with extended biotrophy in Arabidopsis (AT) and with a switch from biotrophic to saprotrophic nutrition during colonization of barley (HV). (A) Maximum projection of a barley root colonized by P. indica at 30 dpi. Broad extraradical hyphae are visible at the boundary of the epidermis, whereas thin secondary hyphae are filling the cortical cells. Host nuclei are absent in the cortex cells while the cylinder is undamaged and preserves intact nuclei. The root was stained with acid fuchsine. (B) Closer view of biotrophic broad invasive hyphae within a barley epidermal cell and secondary hyphae in cortical cells (*). (C) Transverse 4-μm sections of barley roots inoculated with P. indica (30 dpi), stained with toluidine blue. (Upper) Heavily colonized cortex cells. (Lower) A noncolonized part of the root. (D and E) Heat map showing log2-fold expression changes of P. indica genes encoding (D) hydrolytic enzymes and (E) transporters. Significant (t test, P < 0.05) log2-fold expression changes were calculated vs. PNM control. A consistent divergence is observed at 14 dpi, where a strong induction is visible for most of these genes during colonization of barley only (for a detailed overview, see Dataset S1). 1Raw expression data for barley 3 dpi were retrieved from ref. 8. Color coding indicates up-regulation (red) and down-regulation (blue) of genes. (F) Biotrophic broad invasive hyphae (white *) in Arabidopsis epidermal cell at 14 dpi. In contrast to extracellular hyphae, invasive hyphae are not stainable with WGA-AF488 (green) because of the presence of a plant-derived membrane. Endomembrane structures stained by FM4-64 (red) are visible inside the plant cells, indicating cell viability. (G) Arabidopsis epidermal cell with biotrophic invasive hyphae (white *) of P. indica GFP strain (14 dpi). The scale bars represent 25 µm.
Colonization of Arabidopsis by P. indica results in growth promotion (5) (SI Appendix, Fig. S4) and is characterized by a long-term feeding relationship with living host cells via the production of thicker bulbous invasive hyphae in epidermal cells and no sign for papillae formation upon penetration (Fig. 2 F and G and SI Appendix, Fig. S5). We show here that these intracellular, non–WGA-stainable multilobed invasive hyphae are present throughout the Arabidopsis colonization process, leading to a nondestructive progression within the epidermis and cortex layers, as demonstrated by colonized living root cells capable of endocytosis also at later time points (Fig. 2F). Consistent with this observation is the reduced expression of P. indica genes involved in host cell wall and lipid degradation at 3 and 14 dpi in this host compared with the situation in barley (Fig. 2D and SI Appendix, Fig. S2). The occurrence of a long-term biotrophic nutrition in Arabidopsis and of a switch to a saprotrophic nutrition in barley is supported further by comparative expression analyses of fungal genes involved in primary metabolism and nutrient transport. Whereas in colonized Arabidopsis fungal transcripts for amino acid biosynthetic processes and glycolysis are abundant, they show lower expression values in colonized barley at 14 dpi (SI Appendix, Fig. S6 and Dataset S4A). Conversely, at this time point, transcripts encoding fungal carbohydrate and nitrogen transporters are strongly induced during colonization of barley (Fig. 2E and SI Appendix, Fig. S7 and Dataset S4B). In particular, the high-affinity ammonium transporter PiAmt1 (PIIN_02036), whose transcripts are accumulating upon nitrogen starvation in axenic culture (SI Appendix, Fig. S8A), is induced during the late saprotrophic phase in colonized barley but to a much lower extent in colonized Arabidopsis (SI Appendix, Fig. S9 A and B), strongly indicating that in barley, but not in Arabidopsis, a status of nitrogen depletion is reached.
Transcripts for P. indica ABC transporters and other transporters most probably implicated in detoxification processes are well represented in both hosts, but to a greater extent in colonized Arabidopsis at 14 dpi (SI Appendix, Figs. S2 and S10). ABC transporters are efflux pumps that have been implicated in resistance to antifungal compounds in various host–fungal interactions, but little is known about their substrate specificities (18–20). Their host-dependent expression profiles therefore might represent a fungal stress response matched to distinct phytoalexins and other antifungal compounds produced by different hosts (21).
The Switch to Saprotrophic Nutrition in Barley Is Affected by Nitrogen Availability in a PiAMT1-Dependent Manner.
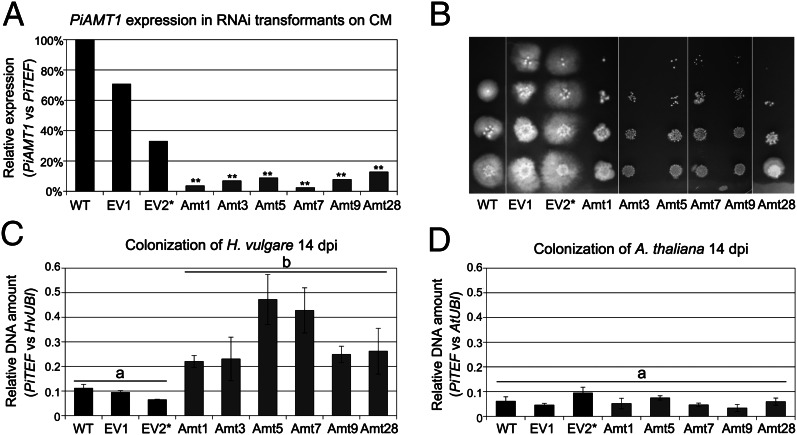
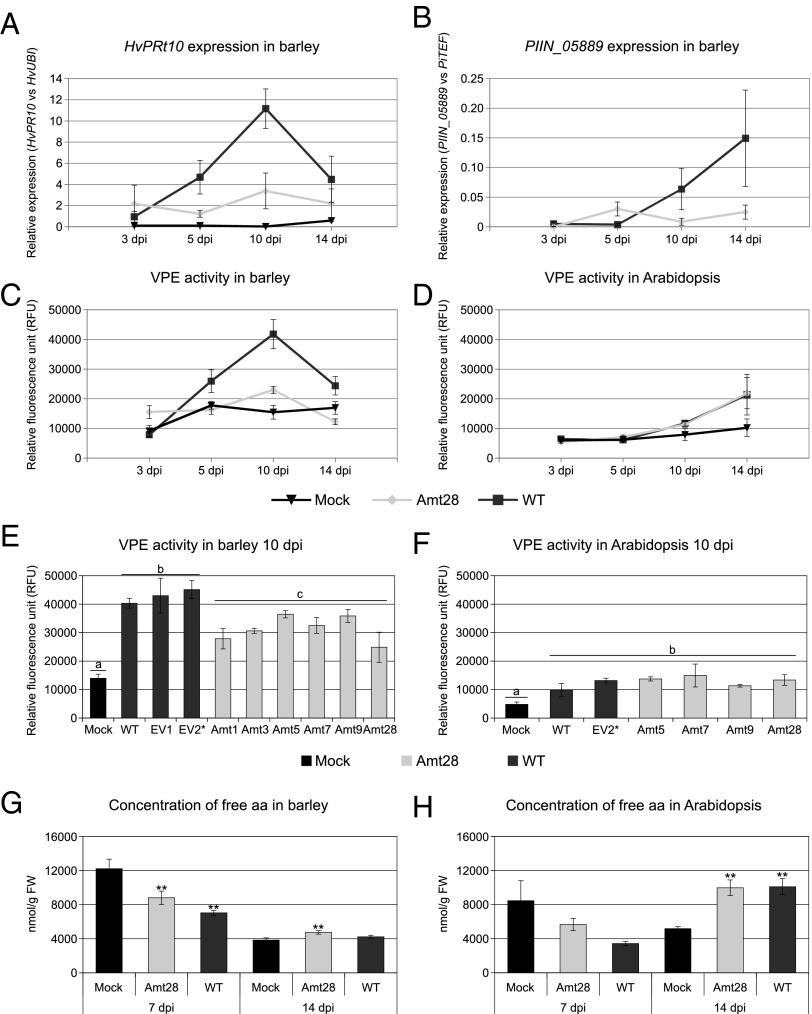
To address whether nitrogen availability is one of the key switches to saprotrophic nutrition in planta, P. indica RNAi strains carrying a silencing construct targeting the high-affinity ammonium transporter PiAMT1 were generated. The success of transformation for selected P. indica RNAi strains was confirmed by Southern blot, and the efficiency of silencing was verified by quantitative PCR (qPCR) experiments (Fig. 3A and SI Appendix, Fig. S11A). PiAmt1 displays strong homology (75% amino acid residues identity) to the high-affinity ammonium transporters AMT1 and AMT2 from Hebeloma cylindrosporum and to the high-affinity ammonium permease MEP2 from Saccharomyces cerevisiae (SI Appendix, Fig. S8B). These transporters were proposed to function as ammonium sensors, generating downstream signals in response to nitrogen starvation (22, 23). In yeast, MEP2 is required for pseudohyphal growth, which is the outgrowth of nuclear-free hyphae to better forage the medium at low N availability (22, 24, 25). The ammonium import function of PiAmt1 was verified by yeast complementation (SI Appendix, Fig. S8C), and the predicted topological structure of the P. indica Amt1 polypeptide demonstrated the presence of a long cytoplasmic tail at the C-terminus, which might be involved in downstream signaling (SI Appendix, Fig. S8D). To verify that ammonium uptake by the P. indica RNAi strains is reduced relative to wild-type (WT) and empty-vector (EV) controls, these strains were analyzed for growth on minimal medium with a low concentration of ammonium as the sole nitrogen source. Their growth on complex media and media supplied with large amounts of ammonium was not affected (SI Appendix, Fig. S11B), whereas hyphal growth at low N was suppressed (Fig. 3B). The RNAi strains were investigated further for altered root colonization. Despite a substantial reduction in growth under a low supply of ammonium as the sole nitrogen source and the absence of any evidence for a compensatory up-regulation of the second ammonium transporter PiAMT2 (SI Appendix, Fig. S9C), the RNAi strains displayed a significantly increased colonization of barley roots at 14 dpi compared with the WT and EV controls, calculated as the ratio of fungal DNA to plant DNA (elogation factor PiTEF/ubiquitin HvUBI) (Fig. 3C) or as the ratio of PiTEF to HvUBI transcripts (SI Appendix, Fig. S9D). The temporal expression pattern for transcripts encoding the barley pathogenesis-related gene PR10 was examined to determine the plant’s response to colonization by the P. indica WT and the RNAi strain Amt28. A decreased transcript accumulation for the PR gene in roots colonized by the RNAi strain compared with the WT strain at the onset of saprotrophy was observed (Fig. 4A). Consistent with this, expression of the P. indica saprotrophic marker gene, PIIN_05889 (encoding a putative xylanase), was reduced in the RNAi strain compared with P. indica WT (Fig. 4B). This indicates an extended biotrophic phase of the RNAi strain Amt28, which likely contributes to the enhanced colonization of barley roots at 14 dpi. To verify this hypothesis, we measured the activity of the vacuolar processing enzyme (VPE). This is a cysteine protease with caspase-like activity responsible for the maturation of various vacuolar proteins in higher plants and reported to be induced in dying cells (9, 26). VPE-like enzymatic activity increased significantly in barley roots colonized by P. indica WT compared with barley mock treated and barley roots colonized by the RNAi strain Amt28, especially at the onset of saprotrophy (Fig. 4C). Analyses of the VPE-like enzymatic activity performed at 10 dpi with the other RNAi strains and EV controls confirmed this finding (Fig. 4E). Remarkably, in the RNAi strains, elicitation of growth promotion was not affected compared with the WT situation (SI Appendix, Fig. S12). This indicates that an extended biotrophic phase does not seem to influence P. indica’s beneficial effects on barley.
Fig. 3.
Silencing of PiAMT1 affects P. indica colonization of barley but not of Arabidopsis. (A) Relative expression of PiAMT in P. indica WT and EV controls and RNAi strains grown in liquid complete medium (CM) for 7 d. **ANOVA, P < 0.01. (B) Colony phenotype of P. indica WT and transformants on yeast nitrogen base medium supplemented with 2 mM NH4Cl as the sole nitrogen source, 14 dpi. (C and D) Relative amount of fungal DNA in (C) barley and (D) in Arabidopsis roots colonized by P. indica WT or transformants at 14 dpi. Plants were grown on 1/10 PNM. Error bars represent SE of the mean from three independent biological repetitions. Grouping was done by ANOVA. *Southern blot analyses showed that all transformed strains had one to two integrations of the plasmids, with the exception of EV2, which had multiple integrations (SI Appendix, Fig. S11).
Fig. 4.
Gene expression profiles, VPE-like enzymatic activity, and amino acid levels in barley roots suggest extended biotrophy in P. indica AMT1-RNAi strains compared with WT. (A) Relative expression of the plant defense-related gene PR10 and (B) of the P. indica gene PIIN_05889 encoding a putative xylanase, during colonization of barley by the P. indica WT and RNAi strain Amt28 at different time points. Expression data are standardized relative to HvUBI or to PiTEF. SEs are calculated from three independent biological repetitions. (C) VPE-like enzymatic activities during biotrophic (≤5 dpi) and cell death-associated colonization (≥10 dpi) of barley roots colonized by P. indica WT and RNAi strain Amt28 or mock treated. For the assay, the fluorescent VPE-specific substrate Ac-ESEN-MCA was added to the root extracts for spectrophotometric determination of enzymatic activities (for details, see SI Appendix). (D) VPE-like enzymatic activities during early (3 and 5 dpi) and late (10 and 14 dpi) colonization of Arabidopsis roots. (E) VPE-like enzymatic activities at the onset of saprotrophy (10 dpi) in barley roots and (F) in Arabidopsis roots. (G) Concentrations of free amino acids in the roots of barley noncolonized or colonized by P. indica WT and RNAi strain Amt28 at 7 and 14 dpi. (H) Concentrations of free amino acids in the roots of Arabidopsis at 7 and 14 dpi. Error bars represent SEM from three to four independent biological repetitions. **ANOVA, P < 0.01. Columns not sharing a letter are significantly different (ANOVA, P < 0.01).
Consistent with low expression of the PiAMT1 gene in Arabidopsis at 14 dpi, analyses of the P. indica RNAi strains impaired in ammonium uptake and possibly ammonium sensing displayed no difference in their colonization patterns compared with the WT and EV controls (Fig. 3D and SI Appendix, Fig. S9E). Similarly, no differences in the VPE-like enzymatic activity were observed in Arabidopsis roots colonized by P. indica WT or RNAi strain Amt28, in which both displayed moderate increased activity compared with mock control at later stages (Fig. 4 D and F). We therefore concluded that PiAmt1 is not required for biotrophic growth but is involved in the lifestyle switch of P. indica to saprotrophic growth as observed in barley.
Quantification of free amino acid levels in barley roots by ultra-pressure reversed-phase chromatography (see SI Appendix for details on this method) showed that amino acid concentrations decreased significantly in colonized roots by P. indica WT compared with roots colonized by the RNAi strain Amt28 and control roots at the onset of the saprotrophic stage/late biotrophic stage (Fig. 4G and SI Appendix, Fig. S13). At 14 dpi, the concentrations of free amino acids were remarkably lower than at 7 dpi in the older/basal root zone of barley, irrespective of colonization (Fig. 4G and SI Appendix, Fig. S13). In particular, asparagine and glutamine decreased by approximately five- and sevenfold, respectively, in control roots (SI Appendix, Fig. S13). This might be the result of reallocation of nitrogen to younger root zones in response to developmental RCD (16) and is in agreement with the microarray data, which indicate limited nitrogen availability to P. indica in the basal root zone of barley at 14 dpi. In Arabidopsis, colonization by P. indica WT decreased amino acid concentrations 7 dpi (Fig. 4H and SI Appendix, Fig. S13), whereas the level of free amino acids increased significantly 14 dpi (Fig. 4H and SI Appendix, Fig. S13). The altered organic nitrogen allocation upon P. indica colonization at 14 dpi was mainly the result of changes in asparagine and glutamine (SI Appendix, Fig. S13) and suggests that nitrogen supply to the fungus is not limited in this phase. Biotrophic and hemibiotrophic fungi have been shown to induce plant nitrogen mobilization and accumulation at the site of infection. In particular, nitrogen-rich amino acids such as glutamine and asparagine have been identified as the major forms of nitrogen reallocation during infection of different plant hosts (27–29). Changes in free amino acid pools during biotrophy have been speculated to reflect the demand for organic nitrogen by the fungus or to be required to launch defense responses by the host (27, 30). Asparagine and glutamine are the preferred nitrogen source of P. indica in axenic culture, and when grown on these amino acids as the sole nitrogen source, P. indica produces enlarged hyphae that resemble the multilobed biotrophic hyphae in planta (SI Appendix, Fig. S14). On nitrate-containing medium, known to be a poor nitrogen source for P. indica (8), or on medium without nitrogen, P. indica hyphae are thin and less branched, similar to the secondary hyphae found in barley at later colonization stages (SI Appendix, Fig. S14). Asparagine and glutamine therefore may represent a ready source of organic nitrogen during biotrophy.
Taken together, these results strongly suggest that host-related nutritional cues affect P. indica’s lifestyle and that the switch from biotrophic to saprotrophic nutrition during colonization of barley is affected by PiAmt1 and by nitrogen availability.
Discussion
Establishment of biotrophy during colonization of Arabidopsis and barley by P. indica is an important feature of the symbioses with both plant hosts, and it implies a strong interdependence between host metabolism and fungal nutrient uptake in this endophyte. Transcriptional profiling revealed that far fewer genes need to be induced to maintain biotrophy compared with those necessary for coordinating the switch to saprotrophy and the manipulation of RCD. Evolution of obligate biotrophy was shown to correlate, among others, with the loss of genes involved in nitrate metabolism, and it is speculated that host plants provide a ready source of organic nitrogen in the form of amino acids (15). In the draft genome of P. indica, no nitrate transporters or reductases were found (8), indicating that nitrate is not essential for this symbiont. P. indica can grow on ammonium and on glutamine and asparagine, which are its favorite nitrogen sources. The low expression of the P. indica high-affinity ammonium transporter in Arabidopsis at later colonization stages suggests that an adequate source of nitrogen is provided by this host during colonization. Increased concentrations of free amino acids, principally of glutamine and asparagine upon P. indica colonization of this host at 14 dpi, strongly support this conclusion. The induction of PiAMT1 and other fungal nitrogen transporters in barley leads to the assumption that this plant cannot provide P. indica with sufficient organic nitrogen during the onset of the RCD program and that nitrogen depletion therefore may function as a trigger for the in planta expression of fungal genes encoding hydrolytic enzymes and for the activation of the saprotrophic program. Silencing of the high-affinity ammonium transporter PiAMT1 by RNAi resulted in reduced expression of fungal xylanase, barley defense response, and VPE activity in colonized roots, supporting the hypothesis that PiAmt1 is involved in sensing the N status and in downstream signaling upon nitrogen depletion in P. indica. Because biotrophic growth of P. indica in Arabidopsis was independent of PiAmt1, we conclude that expression and a signaling function of PiAmt1 are needed for the switch of P. indica’s lifestyle to saprotrophy. Loss or suppression of the expression of PiAMT1 would represent a step toward a progressively more intimate biotrophic association with its hosts. The maintenance of this gene at the expense of biotrophy might benefit P. indica during prolonged saprotrophic growth on decaying plant material, making this fungus also able to survive in the absence of living hosts or on dying host cells.
Plant-associated fungi are either specialists, which are adapted to one or a few distinct hosts, or generalists that can thrive in highly variable host environments. Specialists and their hosts are in an evolutionary arms race that leads to the development of fungal tools and colonization strategies that are efficiently tailored to the respective host. Conversely, broad-host range species must evolve adaptations to cope with a plethora of different host-associated signals and host-specific defense mechanisms. The evolutionary force, in this case, possibly drives the expansion and diversification of the fungal toolkit and the host-adapted gene expression to better suit different plants. Recently it was shown that the obligate biotrophic ascomycete powdery mildew pathogen, Blumeria graminis f. sp. hordei, which can grow and reproduce only on living cells of its natural host, barley, displays a conserved transcriptional program during early pathogenesis on barley and on immunocompromised Arabidopsis (31). Although we cannot exclude that at early time points (<2 dpi, prepenetration stage) a certain conservation of the transcriptional program may be present, our data show that the broad-host range fungal root symbiont P. indica responds differently to divergent hosts, especially at later time points during establishment and maintenance of the intracellular biotrophic interaction.
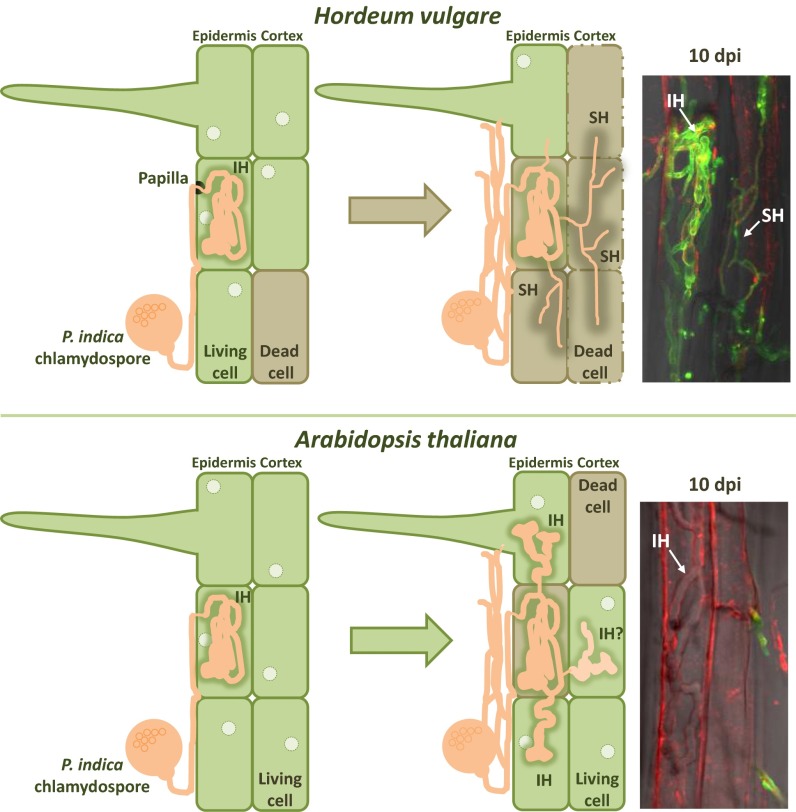
In conclusion, the transcriptional and phenotypic plasticity of P. indica during symbioses (summarized in Fig. 5) establishes a highly adaptive capacity in a root endophyte with broad compatibility that can reconfigure itself and its lifestyle in response to different environmental and host signals.
Fig. 5.
Schematic representation of P. indica colonization strategies at different symbiotic stages in barley and in Arabidopsis. (Upper) Invasive hyphae (IH) and secondary thin hyphae (SH) of P. indica in barley dead cells (10 dpi). (Lower) P. indica non−WGA-stainable biotrophic broad invasive hyphae in an Arabidopsis epidermal cell (10 dpi). Fungal structures were stained with WGA-AF488 (green); membranes were stained with FM4-64 (red). Staining of fungal structures by WGA-AF488 is enhanced in dead host cells. In H. vulgare roots, cell death is initiated a few days after germination by developmental RCD (16) and is increased by P. indica colonization.
Material and Methods
Microarray Analyses.
Microarray experiments were performed with total RNA extracted from P. indica-inoculated barley and Arabidopsis roots. As a control, total RNA from P. indica grown on 1/10 PNM–agar was used. Root samples from three independent biological replicates were labeled and hybridized according to Agilent’s One-Color Microarray-Based Gene Expression Analysis Low Input Quick Amp Labeling protocol. For details, see SI Appendix.
RNAi Vector Construction and P. indica Transformation.
A 570-bp fragment of the PiAMT1 gene was amplified by PCR (Dataset S5) from cDNA and inserted in the EcoRV site of the convergent dual-promoter vector pPiRNAi (17). P. indica was transformed with vector pPiRNAi-AMT1 and the EV control as described in ref. 17. For details, see SI Appendix.
Supplementary Material
Acknowledgments
We thank Elmar Meyer for technical support and Regine Kahmann and Gregor Langen are gratefully acknowledged for reading the manuscript prior to submission. A.Z. acknowledges support from Max-Planck-Gesellschaft and Deutsche Forschungsgemeinschaft Grant ZU263/2-1. U.L. acknowledges support from the International Max Planck Research School for Environmental, Cellular and Molecular Microbiology (IMPRS-MIC) Marburg.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE47775).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301653110/-/DCSupplemental.
References
- 1.López-Berges MS, Rispail N, Prados-Rosales RC, Di Pietro A. A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell. 2010;22(7):2459–2475. doi: 10.1105/tpc.110.075937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuttmann J, et al. Perturbation of Arabidopsis amino acid metabolism causes incompatibility with the adapted biotrophic pathogen Hyaloperonospora arabidopsidis. Plant Cell. 2011;23(7):2788–2803. doi: 10.1105/tpc.111.087684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kämper J, et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444(7115):97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- 4.Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 5.Peskan-Berghofer T, et al. Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol Plant. 2004;122(4):465–477. [Google Scholar]
- 6.Waller F, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA. 2005;102(38):13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lahrmann U, Zuccaro A. Opprimo ergo sum—evasion and suppression in the root endophytic fungus Piriformospora indica. Mol Plant Microbe Interact. 2012;25(6):727–737. doi: 10.1094/MPMI-11-11-0291. [DOI] [PubMed] [Google Scholar]
- 8.Zuccaro A, et al. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 2011;7(10):e1002290. doi: 10.1371/journal.ppat.1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiang X, Zechmann B, Reitz MU, Kogel KH, Schäfer P. The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress-triggered caspase-dependent cell death. Plant Cell. 2012;24(2):794–809. doi: 10.1105/tpc.111.093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshmukh S, et al. The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc Natl Acad Sci USA. 2006;103(49):18450–18457. doi: 10.1073/pnas.0605697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs S, et al. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 2011;156(2):726–740. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stergiopoulos I, de Wit PJ. Fungal effector proteins. Annu Rev Phytopathol. 2009;47:233–263. doi: 10.1146/annurev.phyto.112408.132637. [DOI] [PubMed] [Google Scholar]
- 13.Plett JM, et al. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol. 2011;21(14):1197–1203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 14.Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21(14):1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330(6010):1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 16.Liljeroth E, Bryngelsson T. DNA fragmentation in cereal roots indicative of programmed root cortical cell death. Physiol Plant. 2001;111(3):365–372. doi: 10.1034/j.1399-3054.2001.1110314.x. [DOI] [PubMed] [Google Scholar]
- 17.Hilbert M, et al. Indole derivative production by the root endophyte Piriformospora indica is not required for growth promotion but for biotrophic colonization of barley roots. New Phytol. 2012;196(2):520–534. doi: 10.1111/j.1469-8137.2012.04275.x. [DOI] [PubMed] [Google Scholar]
- 18.Schoonbeek H, Del Sorbo G, De Waard MA. The ABC transporter BcatrB affects the sensitivity of Botrytis cinerea to the phytoalexin resveratrol and the fungicide fenpiclonil. Mol Plant Microbe Interact. 2001;14(4):562–571. doi: 10.1094/MPMI.2001.14.4.562. [DOI] [PubMed] [Google Scholar]
- 19.Fleissner A, Sopalla C, Weltring KM. An ATP-binding cassette multidrug-resistance transporter is necessary for tolerance of Gibberella pulicaris to phytoalexins and virulence on potato tubers. Mol Plant Microbe Interact. 2002;15(2):102–108. doi: 10.1094/MPMI.2002.15.2.102. [DOI] [PubMed] [Google Scholar]
- 20.Urban M, Bhargava T, Hamer JE. An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 1999;18(3):512–521. doi: 10.1093/emboj/18.3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahuja I, Kissen R, Bones AM. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012;17(2):73–90. doi: 10.1016/j.tplants.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Lorenz MC, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17(5):1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Javelle A, et al. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol Microbiol. 2003;47(2):411–430. doi: 10.1046/j.1365-2958.2003.03303.x. [DOI] [PubMed] [Google Scholar]
- 24.Marini AM, André B. In vivo N-glycosylation of the mep2 high-affinity ammonium transporter of Saccharomyces cerevisiae reveals an extracytosolic N-terminus. Mol Microbiol. 2000;38(3):552–564. doi: 10.1046/j.1365-2958.2000.02151.x. [DOI] [PubMed] [Google Scholar]
- 25.Marini AM, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17(8):4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara-Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M. Vacuolar processing enzyme: An executor of plant cell death. Curr Opin Plant Biol. 2005;8(4):404–408. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Horst RJ, et al. Ustilago maydis infection strongly alters organic nitrogen allocation in maize and stimulates productivity of systemic source leaves. Plant Physiol. 2010;152(1):293–308. doi: 10.1104/pp.109.147702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon PS, Oliver RP. The nitrogen content of the tomato leaf apoplast increases during infection by Cladosporium fulvum. Planta. 2001;213(2):241–249. doi: 10.1007/s004250000500. [DOI] [PubMed] [Google Scholar]
- 29.Tavernier V, et al. The plant nitrogen mobilization promoted by Colletotrichum lindemuthianum in Phaseolus leaves depends on fungus pathogenicity. J Exp Bot. 2007;58(12):3351–3360. doi: 10.1093/jxb/erm182. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, et al. Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell. 2010;22(11):3845–3863. doi: 10.1105/tpc.110.079392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hacquard S, et al. Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc Natl Acad Sci USA. 2013;110(24):E2219–E2228. doi: 10.1073/pnas.1306807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.