Significance
Phosphorylation of myosin regulatory light chain (RLC) alters actomyosin contractility and cell mobility. Finding that myosin IIA interacted directly with brefeldin A-inhibited guanine nucleotide-exchange factors (BIG)1 and BIG2, best known as activators of ADP ribosylation factor (Arf) GTPases for intracellular vesicular trafficking, we showed that BIG1 or -2 depletion increased RLC phosphorylation, which interfered with cell mobility independent of Arf activation. By acting noncatalytically as scaffolds for the assembly and operation of multimolecular phosphatase complexes that included phosphatase 1δ and myosin phosphatase-targeting subunit 1, BIG1 and BIG2 contribute to regulation of F-actin formation and cytoskeleton dynamics required for cell polarization and directed migration.
Abstract
Brefeldin A-inhibited guanine nucleotide-exchange factors BIG1 and BIG2 activate, through their Sec7 domains, ADP ribosylation factors (Arfs) by accelerating the replacement of Arf-bound GDP with GTP for initiation of vesicular transport or activation of specific enzymes that modify important phospholipids. They are also implicated in regulation of cell polarization and actin dynamics for directed migration. Reciprocal coimmunoprecipitation of endogenous HeLa cell BIG1 and BIG2 with myosin IIA was demonstrably independent of Arf guanine nucleotide-exchange factor activity, because effects of BIG1 and BIG2 depletion were reversed by overexpression of the cognate BIG molecule C-terminal sequence that follows the Arf activation site. Selective depletion of BIG1 or BIG2 enhanced specific phosphorylation of myosin regulatory light chain (T18/S19) and F-actin content, which impaired cell migration in Transwell assays. Our data are clear evidence of these newly recognized functions for BIG1 and BIG2 in transduction or integration of mechanical signals from integrin adhesions and myosin IIA-dependent actin dynamics. Thus, by anchoring or scaffolding the assembly, organization, and efficient operation of multimolecular myosin phosphatase complexes that include myosin IIA, protein phosphatase 1δ, and myosin phosphatase-targeting subunit 1, BIG1 and BIG2 serve to integrate diverse biophysical and biochemical events in cells.
Cell migration requires the coordinated spatiotemporal regulation of actomyosin function for alterations in cell shape and adhesion. Nonmuscle myosin II (NM II) is critical for regulation of structural remodeling and migration of nonmuscle cells. NM II comprises two heavy chains of 230 kDa, two 20-kDa regulatory light chains (RLCs), and two 17-kDa essential light chains that assemble into bipolar filaments with actin-stimulated ATPase activity. The resultant contractility and actin cross-linking drive assembly of actin filaments that form the actin cytoskeleton (1). In mammalian cells, NM II heavy chain proteins (NMHC) IIA, IIB, and IIC encoded, respectively, by three genes (Myh9, Myh10, and Myh14), are 60–80% identical. Three hexameric isoforms of NM II named NM IIA, IIB, and IIC, with both shared and unique properties, are designated by NMHC component, which accounts for their differences, and all function in events like cell polarization, migration, and adhesion that involve mechano-sensing and motility (2, 3).
Reversible phosphorylation of specific amino acids in the pair of RLCs and/or the heavy chains alters NM II activity. Without phosphorylation, NM II folds into a compact structure, in which one head interacts with the second head of the same molecule. The tails interact also with the heads to prevent ATP hydrolysis and thereby filament assembly. Phosphorylation of RLC on T18 and/or S19 disrupts head–head and head–tail interactions and promotes the formation of contractile actin bundles by enhancing the actin-activated ATPase of NM II (1). Rho-associated protein kinase (ROCK) (4), myotonic dystrophy kinase-related Cdc42-binding kinase (5), and myosin light chain kinase (MLCK) (6), the kinases most widely implicated in RLC T18 and/or S19 phosphorylation, are controlled by different signaling pathways. MLCK was activated by Ca2+-calmodulin (6), whereas the small GTPase RhoA activated ROCK (4). Dephosphorylation of RLC T18 and/or S19 was catalyzed by myosin phosphatase, a heterotrimeric enzyme comprising the 130-kDa myosin phosphatase-targeting subunit 1 (MYPT1), the 37-kDa catalytic subunit of protein phosphatase type 1δ (PP1cδ), and a 20-kDa subunit of unknown function (7). Myosin phosphatase and NM II are associated by direct interaction of the latter with both the PP1cδ and MYPT (8–11). Phosphorylation of MYPT was reported to alter the interaction of myosin phosphatase with NM II (7, 12) and myosin phosphatase catalytic activity (7, 13, 14).
Brefeldin A-inhibited guanine nucleotide-exchange proteins (BIG)1 (∼200 kDa) and BIG2 (∼190 kDa), which were initially purified together in a ∼670-kDa complex from bovine brain cytosol (15–17), like other members of the Sec7 family of ADP ribosylation factor (Arf) guanine nucleotide-exchange factors (GEFs), accelerate replacement of Arf-bound GDP with GTP to generate active Arf-GTP. Association of Arf-GTP with membranes, along with coat proteins and adaptors from cytosol, initiates vesicle formation for transport of cargo between Golgi and plasma membrane (PM) (15, 17–19). Microscopically, BIG1 was seen concentrated at trans-Golgi network, where it partially overlapped BIG2, which was associated also with recycling endosomes (19–21). Functions of BIG1 and BIG2 at the trans-Golgi network were described as redundant (22), although each protein clearly has specific roles that are not shared with the other. BIG1 was required for Golgi integrity, correct glycosylation of mature integrin β1 (23), and directed migration during wound healing (24). In contrast, BIG2 was involved in recycling of internalized transferrin receptors (21) and integrin β1 (25) to the cell surface, and in regulation of tumor necrosis factor receptor release in exosome-like vesicles (26). Besides the catalytic GEF activity of the Sec7 domain, there are additional highly conserved domains and sequences in BIG1 and BIG2 that remain functionally less well defined (27). They may act as scaffolds or platforms for assembly of multimolecular complexes required for integration of diverse signals that regulate several cellular processes. Both BIG1 and BIG2 molecules contain A kinase-anchoring protein sequences in their N-terminal region that bind specific regulatory subunits of protein kinase A (PKA) and act as scaffolds for assembly of multimolecular machines that limit intracellular PKA-cAMP signaling in time and space (28). PP1γ (29) and phosphodiesterase 3A (30) that, respectively, dephosphorylate PKA-modified BIG1 and BIG2 and terminate cAMP signals, have been identified in complexes with BIG1 and/or BIG2.
Depletion of BIG1 or BIG2 was reported to decrease cell motility as well as actin-based membrane protrusions (24, 25), although the mechanisms through which these proteins altered cytoskeleton dynamics remain unclear. Here, we provide evidence that NM IIA activity in HeLa cells, which is crucial for actin stress fiber formation, is also influenced by previously unrecognized interactions with the C termini of BIG1 and BIG2, independent of Arf GEF activity, in complexes that include PP1cδ, and MYPT1.
Results
Interaction of NMHC IIA with BIG1 and BIG2.
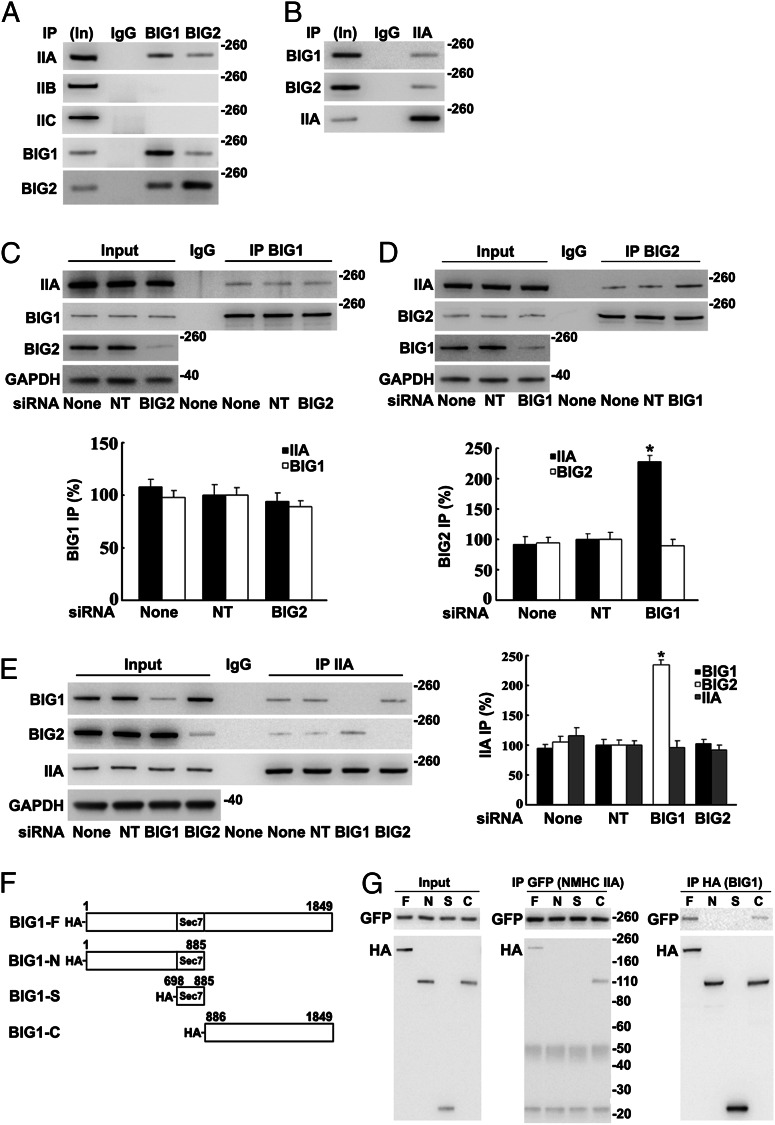
NMHC IIA was immunoprecipitated in multiprotein complexes from HeLa cell lysates with antibodies against BIG1 or BIG2 and identified via liquid chromatography MS/MS analysis of tryptic peptides from the collected proteins. To assess potential interactions of NMHC IIA with BIG1 and/or BIG2, we analyzed proteins coimmunoprecipitated from HeLa cells with antibodies against BIG1 or BIG2 (which often partially coprecipitate the other protein). Approximately 5% of the total cellular NMHC IIA was among proteins collected by BIG1 immunoprecipitation (IP), and ca. 1% by BIG2 IP, whereas no NMHC IIB or IIC was detected, although their presence in the cells was clear (Fig. 1A). IP of endogenous NMHC IIA also yielded BIG1 and BIG2 (Fig. 1B).
Fig. 1.
Co-IP of endogenous NMHC IIA with BIG1 and BIG2: co-IP of BIG2 with IIA increased after BIG1 depletion. Samples [5%, Input (In)] of HeLa cell proteins used for IP (100 µg) and immunoprecipitated proteins were analyzed by Western blotting with indicated antibodies and densitometric quantification. (A) Proteins from IP with control IgG (50%) or antibodies against BIG1 (25%) or BIG2 (50%) reacted with antibodies against NMHC IIA, -B, or -C or BIG1 or -2. (B) Proteins (50%) from IP with NMHC IIA (IIA) antibodies or control IgG reacted with BIG1 or -2 or IIA antibodies. (C) Effects of BIG2 siRNA depletion on IP of BIG1 (25%) or with control IgG (50%). (D) Effects of BIG1 depletion on IP with control IgG or BIG2 antibodies (50%). Data are reported as in C. *P < 0.005 vs. NT. (E) Effects of BIG1 or BIG2 depletion on IP of IIA (25%) or with control IgG (50%). Data are reported as in C, with representative blot on left. *P < 0.01 vs. NT. (F) Diagram of N-terminal-tagged full-length BIG1 and fragment sequences used in plasmids (pCMV-HA vector) plus EGFP-NMHC IIA to cotransfect cells 24 h before preparation of extracts. (G) Samples of extracts before IP (Input) and of proteins from IP (25%) with antibodies against GFP (IIA) or HA (BIG1) were reacted with tag antibodies on Western blots.
BIG1 and BIG2 interactions with NMHC IIA were further explored after selective depletion of BIG1 or BIG2 with siRNA. Although BIG2 depletion did not alter IP of NMHC IIA by BIG1 antibodies (Fig. 1C), IP of BIG2 after BIG1 depletion yielded approximately twice as much NMHC IIA (Fig. 1D), suggesting BIG1 priority in competition of the endogenous proteins for interaction with NMHC IIA. IP of NMHC IIA similarly yielded more than twice as much BIG2 from BIG1-depleted as from control (nontargeted, NT) cells (Fig. 1E).
To identify regions of the BIG1 molecule that interacted with NMHC IIA, HeLa cells were cotransfected with constructs encoding BIG1 amino acids 1–1849 (F, full-length), 1–885 (N), 698–885 (S), or 886–1849 (C) with N-terminal HA tags (Fig. 1F) and GFP-NMHC IIA (full-length) 24 h before preparation of lysates for IP with antibodies against GFP or HA. Neither HA-BIG1-N nor HA-BIG1-S was among proteins collected by IP of GFP-NMHC IIA, which did include HA-BIG1-F or HA-BIG1-C (Fig. 1G, Center). IP of HA-BIG1 or its fragments with HA antibodies yielded GFP-NMHC IIA only with the BIG1-F and BIG1-C proteins (Fig. 1G, Right), consistent with the conclusion that NMHC IIA interacted with structural elements in the C-terminal region of BIG1.
Effects of BIG1 or BIG2 Depletion on RLC Phosphorylation and Actin Organization.
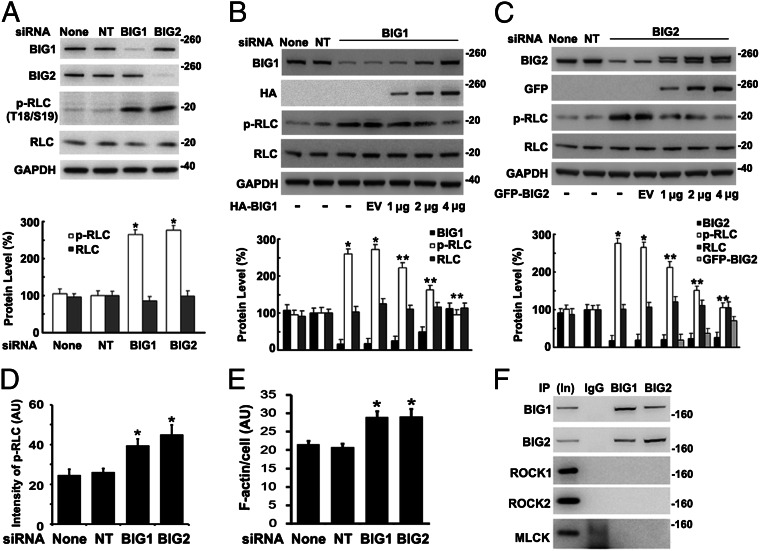
Reversible phosphorylation of RLC T18 and S19 determines MN IIA activity (1, 31). After BIG1 or BIG2 depletion (Fig. 2A), phosphorylation of RLC T18 and S19 was more than twice that in cells treated with the NT siRNA or vehicle alone. To evaluate specificity of these effects, their reversibility by overexpression of the appropriate BIG1 or BIG2 protein was assessed (Fig. 2 B and C) and confirmed participation of BIG1 and BIG2 in regulation of RLC phosphorylation.
Fig. 2.
Endogenous RLC phosphorylation and stress fibers increased after BIG1 or BIG2 depletion. (A) Cells were incubated with vehicle alone (None), or nontarget (NT), or specific BIG1 or BIG2 siRNA for 72 h before analysis of proteins by Western blotting and densitometric quantification. *P < 0.001 vs. NT. (B and C) After incubation with BIG1 (B) or BIG2 (C) siRNA as in A, cells were incubated with 4 μg of empty vector (EV) or 1, 2, or 4 μg of full-length HA-BIG1 or GFP-BIG2 DNA for 24 h before analysis of proteins. Data are presented as in A. *P < 0.001 vs. NT, **P < 0.005 vs. BIG1 siRNA + EV (B); *P < 0.002 vs. NT, **P < 0.005 vs. BIG2 siRNA + EV (C). (D and E) Cells, incubated for 72 h with vehicle (Mock) or with control (NT) or BIG1- or BIG2-specific siRNA were fixed and reacted with antibodies against phospho-RLC (p-RLC) (D) or NMHC IIA (E) plus Alexa Fluor 594-conjugated phalloidin (F-actin). Mean fluorescence intensity (arbitrary unit, AU) of phospho-RLC (D) or F-actin (E) in outlined area of each cell (>50 in each population) was measured in pixels by ImageJ software. *P < 0.005 vs. NT. Images of representative cells are in Fig. S1. (F) Samples (5%, In) of cell proteins (100 µg) used for IP or of those collected with control IgG or antibodies against BIG2 (50%) or BIG1 (50%) were subjected to Western blotting with anti-ROCK1, ROCK2, or MLCK antibodies.
Distributions of phosphorylated RLC in BIG1- or BIG2-depleted and control cells were compared microscopically (Fig. 2D and Fig. S1). BIG1 or BIG2 siRNA treatment increased significantly the mean fluorescence intensity of phospho-RLC (T18/S19) in each cell (Fig. 2D). Linear arrangements of phospho-RLC that colocalized with F-actin were clear in BIG1- or BIG2-depleted cells (Fig. S1). Because NM II activity is known to be related to F-actin organization, we looked for effects of BIG1 or BIG2 depletion on F-actin morphology. Stress fibers were significantly more prominent after BIG1 or BIG2 depletion than in control cells (Fig. 2E), consistent with the effects of BIG1 or BIG2 on RLC phosphorylation.
Because RLC phosphorylation is determined by opposing activities of kinase and phosphatases (1), we looked for association of BIG1 and BIG2 with three known NM II kinases, ROCK1, ROCK2, and MLCK, none of which was detected after BIG1 or BIG2 IP (Fig. 2F), providing no evidence of its association with BIG1 or BIG2.
Effects of BIG1 or BIG2 Depletion on RLC Phosphatase.
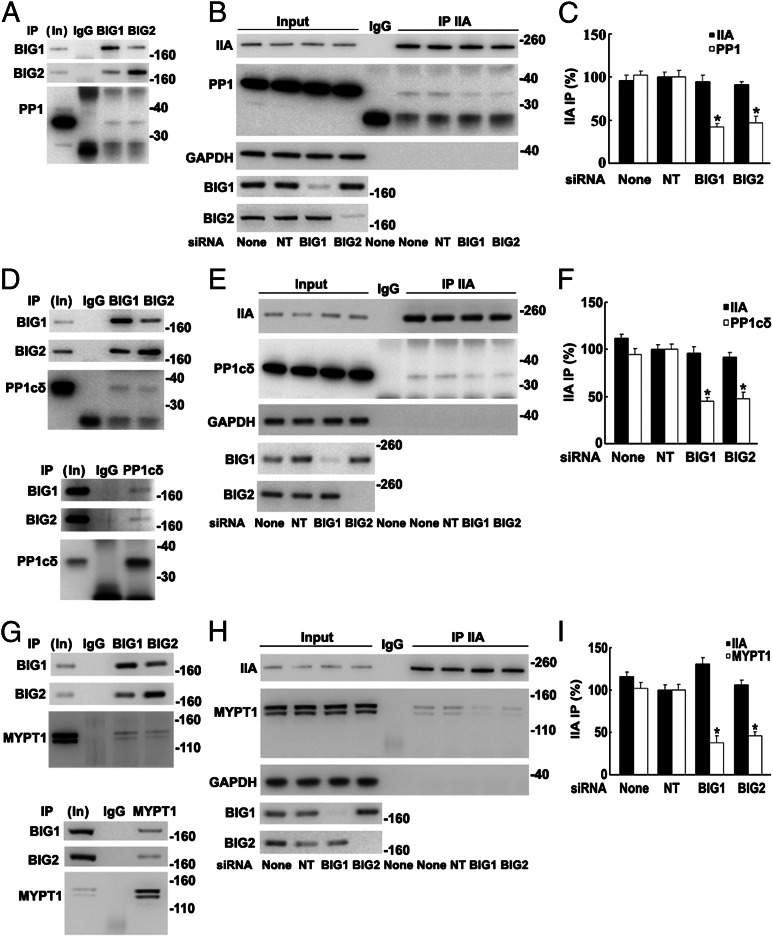
Endogenous PP1, reported to be responsible for dephosphorylation of myosin IIA (7), was among proteins collected by BIG1 and/or BIG2 IP (Fig. 3A). More notably, however, co-IP of PP1 with NMHC IIA was significantly decreased after BIG1 or BIG2 depletion (Fig. 3 B and C), consistent with BIG1 and BIG2 influences via PP1 on levels of RLC phosphorylation.
Fig. 3.
Co-IP of endogenous PP1cδ and MYPT1 with NMHC IIA decreased after BIG1 or BIG2 depletion. Samples of total proteins before (5%, In) or from IP with control IgG or indicated antibodies were analyzed by Western blotting. (A) Proteins from IP of BIG1 (25%) or BIG2 (50%) were analyzed by Western blotting with antibodies against BIG1, BIG2, or PP1. (B) Proteins (50%) from NMHC IIA IP of extracts of cells incubated 72 h without (None) or with indicated siRNA were reacted with antibodies against PP1, BIG1, BIG2, NMHC IIA, and GAPDH. (C) Data from three experiments like that in B. *P < 0.01 vs. NT. (D) Proteins from IP of BIG1 (25%), BIG2 (50%), or PP1cδ (50%) were reacted with antibodies against PP1cδ, BIG1, or BIG2. (E) Cells were incubated for 72 h without or with siRNA before IP of NMHC IIA from extracts. (F) Data from three experiments like that in E.*P < 0.005 vs. NT. (G) Proteins from IP with control IgG (50%) or antibodies against BIG1 (25%), BIG2 (50%), or MYPT1 were reacted with antibodies against BIG1, BIG2, or MYPT1. (H) Effect of BIG1 or BIG2 depletion on co-IP of MYPT1 with NMHC IIA. (I) Data from three experiments like that in H. *P < 0.01 vs. NT.
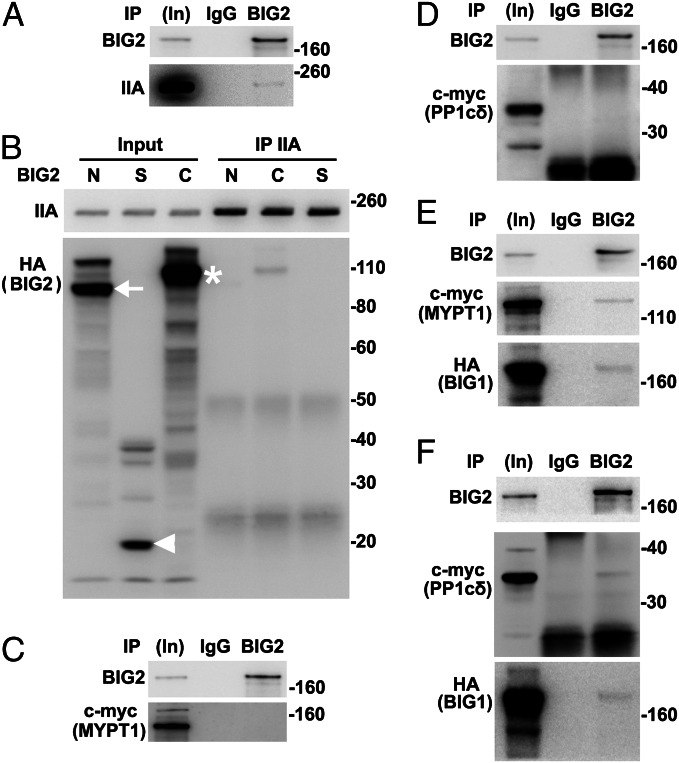
Mammalian cells contain α, δ (also called β), and γ forms of PP1c (32, 33). Endogenous PP1γ was identified after IP of BIG1 or BIG2 from microsomal fractions of HepG2 cells (29). Association of different PP1c forms with BIG1 or BIG2 in HeLa cells was compared, and PP1cδ was among proteins precipitated with BIG1 or BIG2 antibodies (Fig. 3D). Consistent with the conclusion that PP1cδ was the catalytic subunit of the NM II phosphatase (34, 35), IP with PP1cδ antibodies also yielded BIG1 and BIG2 (Fig. 3D). Similar to the findings in Fig. 3 B and C, co-IP of PP1cδ with NMHC IIA after BIG1 or BIG2 depletion was significantly less than that after treatment with control siRNA (Fig. 3 E and F).
Myosin phosphatase is a heterotrimer comprising the catalytic subunit of protein phosphatase type 1δ (PP1cδ), plus the ∼130-kDa myosin-binding MYPT1 and a smaller subunit (M20) of unknown function (9, 34). Phosphatase activity of the holoenzyme was reported to be greater than that of the free catalytic subunit, suggesting that myosin-binding by MYPT1 facilitated enzyme–substrate interaction (34, 35). Data in Fig. 3F are consistent with the notion that BIG1 and BIG2 could influence an NMHC IIA–PP1cδ interaction, perhaps involving MYPT1. To test this possibility, we looked for and found endogenous MYPT1 among proteins precipitated with antibodies against BIG1 or BIG2, but not control IgG (Fig. 3G). Reciprocal IP with antibodies against MYPT1 also collected BIG1 and BIG2 (Fig. 3G). Co-IP of MYPT1 with NMHC IIA was ca. 50% lower after BIG1 or BIG2 depletion (Fig. 3 H and I), consistent with their participation in assembly and/or stabilization of the large molecular complexes.
Phosphorylations of NM II RLC T18 and S19 enhance assembly and activity of NM II (1). To explore whether levels of RLC phosphorylation affected amounts of these multimolecular complexes, three RLC constructs with C-terminal GFP tags, wild-type RLC, nonphosphorylatable (RLC-T18A/S19A), or phosphomimetic (RLC-T18D/S19D), were overexpressed in HeLa cells. As had been reported (36), cells with RLC-T18D/S19D seemed to have more NM IIA and RLC in stress fiber-like arrangements than did those containing wild-type RLC or RLC-T18A/S19A (Fig. S2A). Associations of BIG1 and BIG2 with NM IIA, PP1cδ, and MYPT1 increased as the amount of overexpressed RLC-T18D/S19D, but not RLC-T18A/S19A, increased, consistent with a dependence on the negative charge conferred by phosphorylation of T18/S19 (Fig. S2B).
Direct Interaction of BIG1 with NMHC IIA, MYPT1, and PP1cδ.
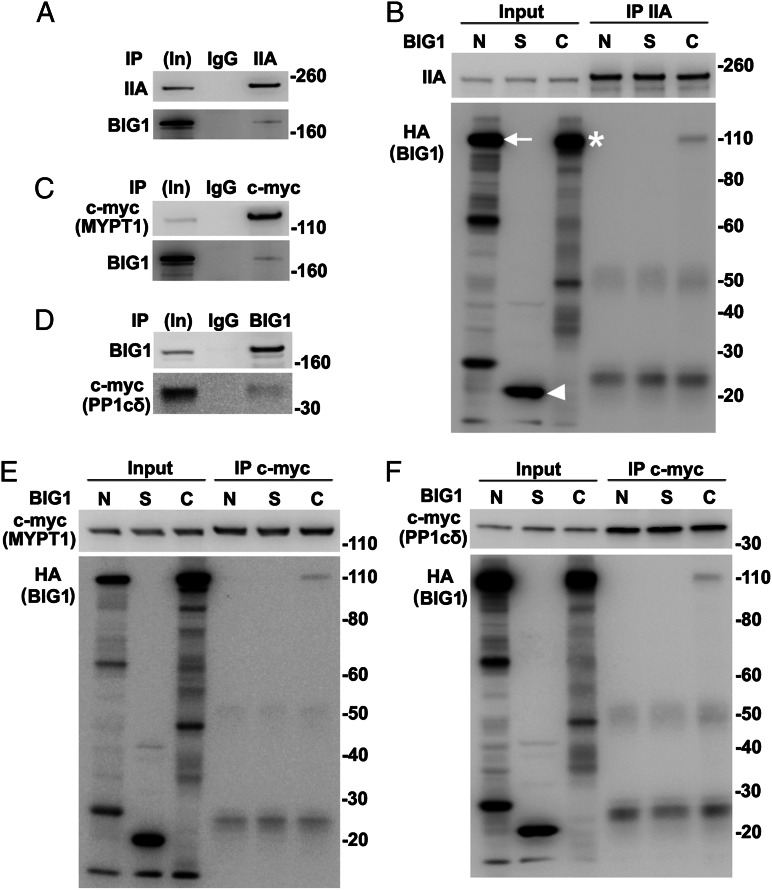
All of our data suggested the existence of endogenous BIG1, BIG2, NMHC IIA, MYPT1, PPc1δ, and perhaps additional proteins in macromolecular complexes. To evaluate possibly direct interactions of individual components, proteins synthesized in vitro (wheat germ extract) were used. BIG1, full-length or fragments, and NMHC IIA were synthesized singly and incubated together before IP. Approximately 2% BIG1-F and -C were found after IP of NMHC IIA (Fig. 4 A and B). Direct interactions of MYPT1 with NM IIA and PP1cδ had been reported (11). An absence of evidence that depletion of NM IIA affected co-IP of endogenous MYPT1 and PP1cδ with BIG1 (Fig. S3A) was consistent with direct interactions of BIG1 and those two proteins, which was confirmed by co-IP of ca. 2% in vitro-synthesized HA-BIG1-C with MYPT1 and PP1cδ (Fig. 4 C–F). Despite >70% sequence identity of the two C fragments, no interaction of BIG2-C with MYPT1 or PP1cδ was detected (Fig. S3 B and C).
Fig. 4.
Direct interactions of in vitro-synthesized full-length BIG1 and its C fragment with NMHC IIA, MYPT1, and PP1cδ. (A and B) BIG1, full-length and three fragments (BIG1-N, -S, and -C), with N-terminal HA tags and NMHC IIA (pCMV6-XL6-myosin IIA) were synthesized singly in wheat germ extract systems and incubated together as indicated before IP of NMHC IIA. Samples of in vitro-synthesized proteins before (5%, In) and from IP (50%) with antibodies against NMHC IIA were reacted with antibodies against (A) BIG1 or (B) HA or NMHC IIA. Arrow indicates BIG1-N, arrowhead BIG1-S, and asterisk BIG1-C. (C and D) Samples of in vitro-synthesized full-length BIG1 and c-myc-MYPT1 or c-myc-PP1cδ were mixed before IP with antibodies against c-myc and reaction of collected proteins (50%) with indicated antibodies. (E and F) In vitro-synthesized MYPT1 or PP1cδ and HA-tagged fragments of BIG1 were mixed as in B before IP of c-myc and reaction of collected proteins with antibodies against c-myc (MYPT1 or PP1cδ) or HA (BIG1).
Direct Interaction of BIG2 with BIG1 and NMHC IIA, but Not MYPT1 or PP1cδ.
After incubation of in vitro-synthesized BIG2 and NMHC IIA, BIG2 IP collected approximately 1% of added NMHC IIA (Fig. 5A). Similarly, ca. 1% BIG2-C fragment was present after IP of NMHC IIA (Fig. 5B), whereas co-IP of myc-tagged MYPT1 or PP1cδ with BIG2 was not detected (Fig. 5 C and D). Because BIG1 apparently interacted directly with MYPT1 and PP1cδ (Fig. 4), IP of BIG2 was expected to yield MYPT1 and PP1cδ with the presence of BIG1, as shown in Fig. 5 E and F. After depletion of endogenous BIG2, co-IP of BIG1 with MYPT1 and PP1cδ was ∼40% less than that from control cells (Fig. S4 A and B), suggesting a significant role for BIG2 in this complex, despite failure to show direct interactions with those two proteins.
Fig. 5.
Interaction of in vitro-synthesized BIG2 with BIG1 and NMHC IIA, but not MYPT1 or PP1cδ. BIG2, full-length (FL) and three fragments (BIG2-N, -S, and -C) with N-terminal HA tags and other proteins were synthesized singly in wheat germ extracts and mixed before IP and analyses of collected proteins. (A) After mixing BIG2 and NMHC IIA, samples before (In, 5%) and from IP with control IgG or anti-BIG2 antibodies (50%) were reacted with antibodies against BIG2 or NMHC IIA. (B) HA-tagged BIG2 fragments were mixed with NMHC IIA before IP with antibodies against NMHC IIA for analysis of collected proteins. Arrow indicates BIG2-N, arrowhead BIG2-S, and asterisk BIG2-C. (C and D) BIG2, MYPT1, and PP1cδ were mixed as indicated before IP with control IgG or antibodies against BIG2 that failed to collect MYPT1 (C) or PP1cδ (D). (E) BIG2, c-myc-MYPT1, and HA-BIG1 were mixed before IP with BIG2 antibodies that collected both MYPT1 and HA-BIG1. (F) IP of BIG2 after incubation of BIG2, PP1cδ, and BIG1 yielded PP1cδ with BIG2 and BIG1.
To explore further interactions among RLC and PP1cδ, BIG1, or BIG2, in vitro-synthesized molecules were used, with association of RLC and NMHC IIA as positive control (Fig. S4C). When BIG1 and BIG2, plus PP1cδ and RLC were synthesized singly and mixed together before IP, RLC was collected with IP of PP1cδ (i.e., both BIG1 and BIG2 apparently participated in the interactions of these proteins with RLC) (Fig. S4 D–F).
Overexpression of BIG1-C or BIG2-C Fragments Reversed Effects of BIG1 or BIG2 Depletion on Cell Migration and Actin Cytoskeleton.
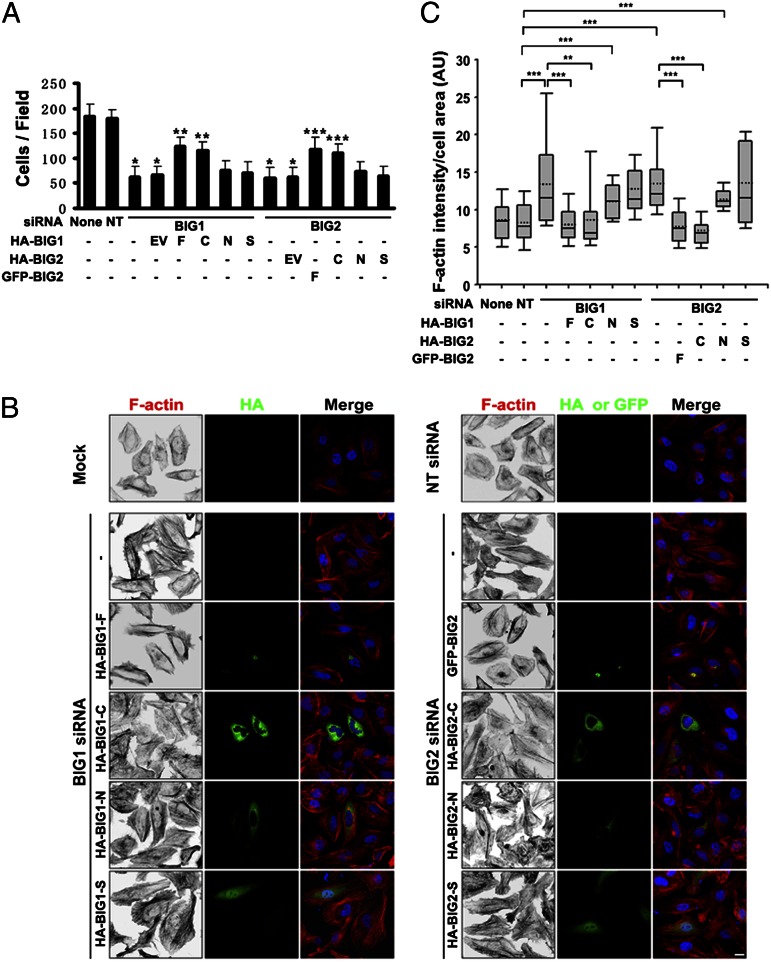
Because NM IIA is critical in cell motility, roles of BIG1 and BIG2 in cell movement were evaluated by Transwell migration assays, and findings were consistent with reports that depletion of BIG1 or BIG2 interfered with cell migration, albeit in different ways (23–25, 37). Motility of HeLa cells transfected with BIG1 or BIG2 siRNA was significantly impaired relative to that of cells transfected with NT siRNA, and overexpression of BIG1 or BIG2, full-length or C fragment, but not N or S fragments, reversed those effects (Fig. 6A and Fig. S5). Effects on stress fibers were similar, perhaps reflecting the changes in RLC phosphorylation (Fig. 6 B and C). Reversal of effects of BIG1 or BIG2 depletion on co-IP of PP1δ and MYPT1 with NMHC IIA (Fig. S6 A–D) was associated with dephosphorylation of RLC (Fig. S6E). Because C fragments effectively replaced the full-length proteins in these experiments, a dependence on Arf GEF activity was excluded.
Fig. 6.
Overexpression of BIG1-C or BIG2-C reversed effects of BIG1 or BIG2 depletion on cell migration and stress fiber content. (A) After 72-h depletion of BIG1 or BIG2, cells were incubated for 24 h with EV, or BIG1- or BIG2-full length (F) or indicated fragments, before Transwell migration assays. Cells on undersides of filters after 4 h at 37 °C were fixed, stained with crystal violet, and counted as described in Materials and Methods. Data are means ± SEM of values from three experiments. *P < 0.005 vs. NT, **P < 0.01 vs. BIG1 siRNA + EV, ***P < 0.05 vs. BIG2 siRNA + EV. (B) Cells were incubated with vehicle (mock) or siRNA, either nonspecific (NT) or specific for BIG1 or BIG2, before transfection with indicated N-terminal HA- or GFP-tagged BIG1 or BIG2 constructs. After 24 h, cells were fixed, reacted with antibodies against HA, and stained for F-actin by Alexa Fluor-594-phalloidin before microscopy and imaging. (Scale bar, 10 µm.) (C) Box-blot analysis of integrated phalloidin intensity per cell area for each group (dotted line, mean; box, 25th, 50th, and 75th percentile; whiskers, maximum and minimum). Data for 40 cells from each of three experiments were analyzed. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
NM II action is fundamental in cell migration (1) through regulation of localized contraction of the actin cytoskeleton, maintainence of cell polarity, modulation of cell adhesions, and retraction of trailing edges. Here we showed that endogenous NMHC IIA, RLC, MYPT1, and PP1cδ were coimmunoprecipitated from HeLa cells with BIG1 and BIG2. Depletion of BIG1 or BIG2 decreased co-IP of MYPT1 and PP1cδ with NMHC IIA, enhanced phosphorylation of RLC T18 and S19, and led to increased content of F-actin fibers that impaired cell movement. Effects of BIG1 or BIG2 depletion on RLC phosphorylation, actomyosin filament remodeling, and cell migration were reversed, at least in part, by overexpression of BIG1 or BIG2, respectively, or the C-terminal fragments of those molecules that did not include the catalytic Arf GEF Sec7 domain. On the basis of these data, we propose a newly recognized role for BIG1 and BIG2, namely their contributions to critical interactions of myosin phosphatase and NMHC IIA that modulate NM IIA activity.
Increased RLC phosphorylation could result from reduced myosin phosphatase amount or activity (7, 38), or elevation of kinase levels or action (6, 10, 39). BIG1 and BIG2 in HeLa cells were apparently associated with NMHC IIA, and RLC phosphorylation was significantly increased after BIG1 or BIG2 depletion, consistent with a role for BIG1/BIG2 in multimolecular complex that regulates RLC activity. We found no evidence of RLC kinase, MLCK, ROCK1, or ROCK2 among proteins precipitated with BIG1 or BIG2 antibodies. Co-IP of catalytic PP1cδ with BIG1 or BIG2 was clear, however, and co-IP of PP1cδ or MYPT1 with NMHC IIA was significantly decreased after BIG1 or BIG2 depletion. It seemed that BIG1 or BIG2 could affect RLC phosphorylation via interactions with myosin phosphatase that included PP1cδ and MYPT1. BIG1 or BIG2 depletion reduced co-IP of myosin phosphatase with NM IIA, consistent with loss of the phosphatase from actomyosin filaments (stress fibers), leading to accumulation of phospho-RLC, but other mechanisms through which BIG1 or BIG2 depletion might affect the phosphatase activity or localization could not be excluded.
In fibroblasts, myosin phosphatase was localized to actin-myosin stress fibers (40, 41), and MYPT1 association with stress fibers in HeLa cells suggests a potential role for it in cellular localization of the heterotrimeric phosphatase. Regulation and intracellular targeting of myosin phosphatase are, however, complex and incompletely understood. Myosin phosphatase was reported also in cytosol and at the PM (40, 42), as was the presence of MYPT1 at several other intracellular sites. Myosin phosphatase is known to dephosphorylate several substrates, including adducin, moesin, tau, MAP, and a transcriptional repressor HDAC7, perhaps reflecting functions in addition to myosin regulation (7). Our co-IP and other in vitro experiments suggested the existence of multimolecular complexes comprising BIG1, BIG2, NM IIA, MYPT1, and PP1δ (i.e., including both substrate and phosphatase). In such assemblies, BIG1 might interact directly with the other four components through its C-terminal region, serving as the core of a macromolecular “machine.” By apparently interacting directly with BIG1 and NM IIA, but not MYPT1 and PP1cδ, BIG2 could contribute to stabilization and/or coordination of function components in a regulatory molecular structure based on interactions of BIG1 with MYPT1 and PP1cδ. Although the mechanistic details of this BIG1 role remain unclear, via interactions with NM IIA and myosin phosphatase, it might be required, at least in part, for targeting and action of the enzyme on phospho-RLC or could influence phosphorylation of MYPT1 and thus phosphatase activity (43–47). We do not know how BIG1/2 is recruited to or retained at appropriate sites, or, in fact, the intracellular location of the complexes that were immunoprecipitated, but they do seem to be related to RLC phosphorylation.
In vitro experiments had shown that phosphorylation of RLC interfered with the assembly of NM II filaments (48). In cells, bipolar myosin filaments comprising 14–20 molecules can interact with actin filaments in thick bundles to form structures like stress fibers (1). HeLa cell RLC phosphorylation was significantly increased after depletion of BIG1 or BIG2, and the levels of actin stress fibers were >30% higher than those in control cells. These findings are consistent with reported promotion and stabilization of stress fibers by phospho-RLC (49, 50), as well as the increased HeLa cell stress fibers after silencing MYPT1 (50). MYPT1 binding to PP1cδ altered conformation of the catalytic cleft, increasing enzyme activity and specificity (51). MYPT1/PP1cδ phosphatase activity is itself controlled by phosphorylation catalyzed by several kinases, e.g., ROCK (52–54). Future work will be directed toward understanding how the association of MYPT1–BIG1–BIG2 serves in the spatiotemporal regulation of myosin during cell migration.
The relatively recently recognized effect of BIG1 depletion on directional persistence of cell migration in wound-healing assays (24), revealed unanticipated functions of BIG1 in regulation of cell polarization and directed migration. In moving cells, protrusions that direct and drive translocation contain two kinds of actin-based structures, the lamellipodium and lamellum (55, 56). NM II in the lamellum controls retrograde actin flow and promotes adhesion maturation, thereby limiting protrusion, suggesting a negative role of NM II in cell migration (57–60). In our migration assays, HeLa cell motility was significant impaired after BIG1 and/or BIG2 depletion, perhaps because RLC phosphorylation caused cross-linking with actin, and the actin-bundling function of NM IIA is critical to adhesion maturation (60). Activation of Arf1, a substrate of BIG1 and BIG2 GEFs, was reported to affect migration of MDA-MB-231 and HeLa cells (61, 62). Although effects of BIG1 or BIG2 depletion on HeLa cell migration might well have been due, at least in part, to interference with Arf activation, we showed that regions of BIG1 or BIG2, which when overexpressed reversed effects, respectively, of BIG1 or BIG2 depletion, did not include the Arf-activating Sec7 domain.
On the basis of the data presented here, we propose that BIG1, BIG2, NMHC IIA, MYPT1, and PP1δ contribute to HeLa cell function via concerted actions in macromolecular machines, variously assembled and situated to accomplish diverse signaling and regulatory actions that coordinate and integrate mechanical events with multiple signaling and metabolic aspects of cell life. Through one important segment of those interactions, BIG1 and BIG2 can regulate RLC phosphorylation and abundance of actin stress fibers by contributing to the interactions of NM IIA and myosin phosphatase.
Materials and Methods
Sources of antibodies and plasmids are reported in SI Materials and Methods. Details of IP and procedures for in vitro protein synthesis in Wheat Germ Extract System are also reported in SI Materials and Methods. HeLa cells were grown, and Transwell assays for analyses of cell migration as well as confocal immunofluorescence microscopy experiments were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Robert S. Adelstein and Mary Ann Conti [National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH)] for their generous provision of expert advice and consultation, in addition to antibodies against NMHC IIB and IIC; Dr. Zissis C. Chroneos (The Pennsylvania State University) for his important gift of NMHC IIA plasmids that enabled us to demonstrate direct interactions of in vitro-synthesized proteins; and Drs. Daniela Malide and Christian Combs (Light Microscopy Core Facility, NHLBI) for their invaluable help in confocal microscopy. This research was supported by the Intramural Research Program of the NIH, NHLBI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312531110/-/DCSupplemental.
References
- 1.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10(11):778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang A, Ma X, Conti MA, Adelstein RS. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochem Soc Trans. 2011;39(5):1131–1135. doi: 10.1042/BST0391131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vicente-Manzanares M, Newell-Litwa K, Bachir AI, Whitmore LA, Horwitz AR. Myosin IIA/IIB restrict adhesive and protrusive signaling to generate front-back polarity in migrating cells. J Cell Biol. 2011;193(2):381–396. doi: 10.1083/jcb.201012159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riento K, Ridley AJ. Rocks: Multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4(6):446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 5.Tan I, Yong J, Dong JM, Lim L, Leung T. A tripartite complex containing MRCK modulates lamellar actomyosin retrograde flow. Cell. 2008;135(1):123–136. doi: 10.1016/j.cell.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 6.Somlyo AP, Somlyo AV. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: Modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev. 2003;83(4):1325–1358. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- 7.Matsumura F, Hartshorne DJ. Myosin phosphatase target subunit: Many roles in cell function. Biochem Biophys Res Commun. 2008;369(1):149–156. doi: 10.1016/j.bbrc.2007.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sellers JR, Pato MD. The binding of smooth muscle myosin light chain kinase and phosphatases to actin and myosin. J Biol Chem. 1984;259(12):7740–7746. [PubMed] [Google Scholar]
- 9.Hartshorne DJ, Ito M, Erdödi F. Myosin light chain phosphatase: Subunit composition, interactions and regulation. J Muscle Res Cell Motil. 1998;19(4):325–341. doi: 10.1023/a:1005385302064. [DOI] [PubMed] [Google Scholar]
- 10.Tan I, Ng CH, Lim L, Leung T. Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J Biol Chem. 2001;276(24):21209–21216. doi: 10.1074/jbc.M102615200. [DOI] [PubMed] [Google Scholar]
- 11.Koga Y, Ikebe M. A novel regulatory mechanism of myosin light chain phosphorylation via binding of 14-3-3 to myosin phosphatase. Mol Biol Cell. 2008;19(3):1062–1071. doi: 10.1091/mbc.E07-07-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murányi A, et al. Phosphorylation of Thr695 and Thr850 on the myosin phosphatase target subunit: Inhibitory effects and occurrence in A7r5 cells. FEBS Lett. 2005;579(29):6611–6615. doi: 10.1016/j.febslet.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 13.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116(2):167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson S, Paterson HF, Marshall CJ. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat Cell Biol. 2005;7(3):255–261. doi: 10.1038/ncb1230. [DOI] [PubMed] [Google Scholar]
- 15.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274(18):12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 16.Morinaga N, Adamik R, Moss J, Vaughan M. Brefeldin A inhibited activity of the sec7 domain of p200, a mammalian guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274(25):17417–17423. doi: 10.1074/jbc.274.25.17417. [DOI] [PubMed] [Google Scholar]
- 17.Yamaji R, et al. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc Natl Acad Sci USA. 2000;97(6):2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinotsuka C, Yoshida Y, Kawamoto K, Takatsu H, Nakayama K. Overexpression of an ADP-ribosylation factor-guanine nucleotide exchange factor, BIG2, uncouples brefeldin A-induced adaptor protein-1 coat dissociation and membrane tubulation. J Biol Chem. 2002;277(11):9468–9473. doi: 10.1074/jbc.M112427200. [DOI] [PubMed] [Google Scholar]
- 19.Shin HW, Morinaga N, Noda M, Nakayama K. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: Its localization to recycling endosomes and implication in the endosome integrity. Mol Biol Cell. 2004;15(12):5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Lasell TK, Melançon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: Evidence for distinct functions in protein traffic. Mol Biol Cell. 2002;13(1):119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen X, et al. Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc Natl Acad Sci USA. 2006;103(8):2635–2640. doi: 10.1073/pnas.0510599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishizaki R, Shin HW, Mitsuhashi H, Nakayama K. Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol Biol Cell. 2008;19(6):2650–2660. doi: 10.1091/mbc.E07-10-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen X, Hong MS, Moss J, Vaughan M. BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein, is required for correct glycosylation and function of integrin beta1. Proc Natl Acad Sci USA. 2007;104(4):1230–1235. doi: 10.1073/pnas.0610535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li CC, et al. Effects of brefeldin A-inhibited guanine nucleotide-exchange (BIG) 1 and KANK1 proteins on cell polarity and directed migration during wound healing. Proc Natl Acad Sci USA. 2011;108(48):19228–19233. doi: 10.1073/pnas.1117011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen X, et al. Brefeldin A-inhibited ADP-ribosylation factor activator BIG2 regulates cell migration via integrin β1 cycling and actin remodeling. Proc Natl Acad Sci USA. 2012;109(36):14464–14469. doi: 10.1073/pnas.1211877109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam A, et al. The brefeldin A-inhibited guanine nucleotide-exchange protein, BIG2, regulates the constitutive release of TNFR1 exosome-like vesicles. J Biol Chem. 2007;282(13):9591–9599. doi: 10.1074/jbc.M607122200. [DOI] [PubMed] [Google Scholar]
- 27.Bui QT, Golinelli-Cohen MP, Jackson CL. Large Arf1 guanine nucleotide exchange factors: Evolution, domain structure, and roles in membrane trafficking and human disease. Mol Genet Genomics. 2009;282(4):329–350. doi: 10.1007/s00438-009-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Adamik R, Pacheco-Rodriguez G, Moss J, Vaughan M. Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) Proc Natl Acad Sci USA. 2003;100(4):1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroda F, Moss J, Vaughan M. Regulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) and BIG2 activity via PKA and protein phosphatase 1gamma. Proc Natl Acad Sci USA. 2007;104(9):3201–3206. doi: 10.1073/pnas.0611696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puxeddu E, et al. Interaction of phosphodiesterase 3A with brefeldin A-inhibited guanine nucleotide-exchange proteins BIG1 and BIG2 and effect on ARF1 activity. Proc Natl Acad Sci USA. 2009;106(15):6158–6163. doi: 10.1073/pnas.0901558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirata N, Takahashi M, Yazawa M. Diphosphorylation of regulatory light chain of myosin IIA is responsible for proper cell spreading. Biochem Biophys Res Commun. 2009;381(4):682–687. doi: 10.1016/j.bbrc.2009.02.121. [DOI] [PubMed] [Google Scholar]
- 32.Cohen PT. Protein phosphatase 1—targeted in many directions. J Cell Sci. 2002;115(Pt 2):241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 33.Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84(1):1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 34.Alessi D, MacDougall LK, Sola MM, Ikebe M, Cohen P. The control of protein phosphatase-1 by targetting subunits. The major myosin phosphatase in avian smooth muscle is a novel form of protein phosphatase-1. Eur J Biochem. 1992;210(3):1023–1035. doi: 10.1111/j.1432-1033.1992.tb17508.x. [DOI] [PubMed] [Google Scholar]
- 35.Shirazi A, et al. Purification and characterization of the mammalian myosin light chain phosphatase holoenzyme. The differential effects of the holoenzyme and its subunits on smooth muscle. J Biol Chem. 1994;269(50):31598–31606. [PubMed] [Google Scholar]
- 36.Beach JR, Licate LS, Crish JF, Egelhoff TT. Analysis of the role of Ser1/Ser2/Thr9 phosphorylation on myosin II assembly and function in live cells. BMC Cell Biol. 2011;12:52. doi: 10.1186/1471-2121-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. Brefeldin A-inhibited guanine exchange factor 2 regulates filamin A phosphorylation and neuronal migration. J Neurosci. 2012;32(36):12619–12629. doi: 10.1523/JNEUROSCI.1063-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartshorne DJ, Ito M, Erdödi F. Role of protein phosphatase type 1 in contractile functions: myosin phosphatase. J Biol Chem. 2004;279(36):37211–37214. doi: 10.1074/jbc.R400018200. [DOI] [PubMed] [Google Scholar]
- 39.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15(7):371–377. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Murata K, Hirano K, Villa-Moruzzi E, Hartshorne DJ, Brautigan DL. Differential localization of myosin and myosin phosphatase subunits in smooth muscle cells and migrating fibroblasts. Mol Biol Cell. 1997;8(4):663–673. doi: 10.1091/mbc.8.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surks HK, et al. Regulation of myosin phosphatase by a specific interaction with cGMP- dependent protein kinase Ialpha. Science. 1999;286(5444):1583–1587. doi: 10.1126/science.286.5444.1583. [DOI] [PubMed] [Google Scholar]
- 42.Ito M, et al. Interaction of smooth muscle myosin phosphatase with phospholipids. Biochemistry. 1997;36(24):7607–7614. doi: 10.1021/bi9702647. [DOI] [PubMed] [Google Scholar]
- 43.Kimura K, et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273(5272):245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 44.Kaibuchi K, Kuroda S, Amano M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu Rev Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- 45.Feng J, et al. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem. 1999;274(52):37385–37390. doi: 10.1074/jbc.274.52.37385. [DOI] [PubMed] [Google Scholar]
- 46.MacDonald JA, et al. Identification of the endogenous smooth muscle myosin phosphatase-associated kinase. Proc Natl Acad Sci USA. 2001;98(5):2419–2424. doi: 10.1073/pnas.041331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borman MA, MacDonald JA, Murányi A, Hartshorne DJ, Haystead TA. Smooth muscle myosin phosphatase-associated kinase induces Ca2+ sensitization via myosin phosphatase inhibition. J Biol Chem. 2002;277(26):23441–23446. doi: 10.1074/jbc.M201597200. [DOI] [PubMed] [Google Scholar]
- 48.Scholey JM, Taylor KA, Kendrick-Jones J. Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature. 1980;287(5779):233–235. doi: 10.1038/287233a0. [DOI] [PubMed] [Google Scholar]
- 49.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133(6):1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia D, Stull JT, Kamm KE. Myosin phosphatase targeting subunit 1 affects cell migration by regulating myosin phosphorylation and actin assembly. Exp Cell Res. 2005;304(2):506–517. doi: 10.1016/j.yexcr.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 51.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429(6993):780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 52.Birukova AA, et al. Role of Rho GTPases in thrombin-induced lung vascular endothelial cells barrier dysfunction. Microvasc Res. 2004;67(1):64–77. doi: 10.1016/j.mvr.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 53.Birukova AA, et al. Microtubule disassembly induces cytoskeletal remodeling and lung vascular barrier dysfunction: Role of Rho-dependent mechanisms. J Cell Physiol. 2004;201(1):55–70. doi: 10.1002/jcp.20055. [DOI] [PubMed] [Google Scholar]
- 54.Grassie ME, Moffat LD, Walsh MP, MacDonald JA. The myosin phosphatase targeting protein (MYPT) family: A regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1δ. Arch Biochem Biophys. 2011;510(2):147–159. doi: 10.1016/j.abb.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 55.Heath JP, Holifield BF. Cell locomotion: New research tests old ideas on membrane and cytoskeletal flow. Cell Motil Cytoskeleton. 1991;18(4):245–257. doi: 10.1002/cm.970180402. [DOI] [PubMed] [Google Scholar]
- 56.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 57.Cai Y, et al. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91(10):3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Even-Ram S, et al. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9(3):299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 59.Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128(3):561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi CK, et al. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat Cell Biol. 2008;10(9):1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boulay PL, Cotton M, Melançon P, Claing A. ADP-ribosylation factor 1 controls the activation of the phosphatidylinositol 3-kinase pathway to regulate epidermal growth factor-dependent growth and migration of breast cancer cells. J Biol Chem. 2008;283(52):36425–36434. doi: 10.1074/jbc.M803603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krndija D, et al. The phosphatase of regenerating liver 3 (PRL-3) promotes cell migration through Arf-activity-dependent stimulation of integrin α5 recycling. J Cell Sci. 2012;125(Pt 16):3883–3892. doi: 10.1242/jcs.104885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.