Abstract
The proteasome is a cellular protease responsible for the selective degradation of the majority of the intracellular proteome. It recognizes, unfolds, and cleaves proteins that are destined for removal, usually by prior attachment to polymers of ubiquitin. This macromolecular machine is composed of two subcomplexes, the 19S regulatory particle (RP) and the 20S core particle (CP), which together contain at least 33 different and precisely positioned subunits. How these subunits assemble into functional complexes is an area of active exploration. Here we describe the current status of studies on the assembly of the 20S proteasome (CP). The 28-subunit CP is found in all three domains of life and its cylindrical stack of four heptameric rings is well conserved. Though several CP subunits possess self-assembly properties, a consistent theme in recent years has been the need for dedicated assembly chaperones that promote on-pathway assembly. To date, a minimum of three accessory factors have been implicated in aiding the construction of the 20S proteasome. These chaperones interact with different assembling proteasomal precursors and usher subunits into specific slots in the growing structure. This review will focus largely on chaperone-dependent CP assembly and its regulation.
1. Introduction
Proteolytic elimination of damaged proteins or regulatory proteins is essential for cellular homeostasis. The ubiquitin-proteasome system (UPS) is indispensable for degradation of most intracellular proteins, including many proteins critical to cellular regulatory pathways, such as cell-cycle progression, DNA damage repair, and antigen presentation [1-4]. After an enzymatic cascade covalently attaches a polymer(s) of ubiquitin to a substrate protein, the substrate is targeted for degradation by the proteasome [1]. The mechanisms of ubiquitylation have been well studied, and are reviewed elsewhere [1, 5-8].
Protein modifications analogous to ubiquitylation have been identified in actinobacteria and archaea. Enzymatic attachment of the small protein Pup (which is unrelated to ubiquitin) to substrate proteins in the actinomycete Mycobacterium tuberculosis [9] and addition of the ubiquitin-related SAMPs (small archaeal modifier proteins) to substrates in the archaeum Haloferax volcanii [10] show broad parallels to the eukaryotic UPS. In both cases, these modifications are believed to direct proteins for degradation by their compositionally simple proteasomes.
In eukaryotes, the 26S proteasome comprises a 20S proteasome or core particle (CP) capped by either one or two 19S regulatory particles (RPs) (Fig. 1A). The RP is responsible for the recognition, deubiquitylation and unfolding of polyubiquitylated substrates (and some non-ubiquitylated substrates), while the CP houses the proteasome’s proteolytic activities within its central chamber [11]. All 20S proteasomes are composed of two related types of subunits; α subunits, which form the outer two heptameric rings and are distinguished by highly conserved N-terminal extensions, and β subunits, which form the inner pair of heptameric rings and include the proteolytic active sites. The 20S cylinder has a central channel comprised of three separate chambers, as show in Fig. 1B. Archaeal and actinomycete (eubacterial) proteasomes have only one or two of each type of subunit, while eukaryotes are more complex, with seven different α subunits and seven different β subunits. In animals and plants, additional variants of the α and β subunits are found [12].
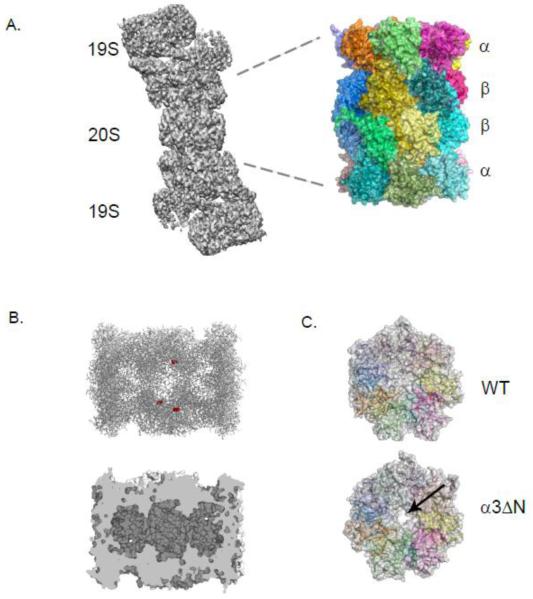
Figure 1. Structural features of the Eukaryotic 20S Proteasome.
A. An electron micrograph structure of the 26S proteasome (grey) from yeast with the 20S (CP) and 19S (RP) labeled. The yeast CP crystal structure is shown with each subunit colored.
B. In the top panel, a clipped structure of the CP on its side illustrates three of the six catalytically active sites shown as red spheres. The bottom panel highlights the three linked chambers within the proteasome.
C. The occluded pore of the CP (grey with individual subunits colored) from wild type cells is juxtaposed with the open pore from the α3ΔN strain indicated by an arrow.
All images were made using PyMol or Chimera using structures elucidated in Refs [15, 29].
Early electron microscopic (EM) studies allowed the first glimpse of the cylindrical shape and dimensions of the eukaryotic 20S proteasome [13]. Much of the key early EM work was performed on the compositionally simpler 20S proteasomes isolated from the archaeal species Thermoplasma acidophilum [13]. This culminated in the tour de force determination in 1995 of the T. acidophilum CP crystal structure in complex with a covalent active-site inhibitor [14]. The structure revealed the location of the active sites in a central chamber, the substrate entry ports at the ends of the cylinder, and the fact that proteasomes utilize an N-terminal threonine residue as the active-site nucleophile. Two years later, a crystal structure of a eukaryotic 20S proteasome was published [15]. This structure, from the yeast Saccharomyces cerevisiae, became the basis for many subsequent structure-function studies. Among other things, it confirmed that two copies each of 14 different but related subunits were incorporated into each CP, as expected from earlier genetic and biochemical studies [16, 17]. Most interestingly, the structure revealed that the pores at the ends of the proteasomal cylinder were blocked and therefore must somehow be opened (“gated”) to allow substrate entry (Fig. 1B, C).
The proteasome was the first identified threonine protease [18]. All catalytically active proteasome β subunits are synthesized with N-terminal propeptides. These propeptides are autocatalytically cleaved during assembly to reveal the active-site N-terminal threonine [19, 20]. Each of the identical β subunits in archaeal proteasomes is catalytically active; when assayed with short peptide substrates, the archaeal proteasome cleaves primarily after hydrophobic residues. In eukaryotic proteasomes, only three of the seven β subunits are catalytically active. The β5 subunit active site also preferentially cleaves after hydrophobic residues, but the β1 and β2 subunit active sites cleave after acidic and basic residues, respectively [21], enhancing the palette of peptides that can be produced. Vertebrates can synthesize alternative active β subunits that have protein cleavage-site preferences distinct from those of the constitutive subunits they replace [22-28] (see Section 6).
A major function for the proteasome α-subunit ring is to regulate entry into the central proteolytic chamber. The N-termini of these subunits assemble into the gate that occludes the entry pore into the CP (Fig. 1C). Deletion of an N-terminal peptide from the yeast α3 subunit is sufficient to open the gate [29]. Even in the open state, the pore is too narrow, 13 Å in diameter, to allow passage of folded protein domains; only single protein chains or extended loops can fit through this narrow opening, creating a molecular sieve that prevents the nonspecific degradation of folded cellular proteins.
The α subunits interact with proteasomal regulatory complexes that control the opening and closing of the gated pore. A crystal structure of the yeast CP with the PA26 (proteasome activator of 26 kDa) complex from trypanosomes revealed an open structure for the α-ring gate in which the seven N-termini of the α subunits were arrayed against the inner wall of the heptameric PA26 ring [30]. The C-terminal tails of the PA26 subunits insert into surface pockets between the α subunits. These inserted tails, together with an activator loop of the PA26 subunits, are thought to trigger conformational changes in the α ring and gate opening. Later studies have shown that multiple regulators and interacting proteins of the 20S proteasome possess a conserved C-terminal HbYX (hydrophobic-tyrosine-any amino acid) motif that inserts into these same surface pockets between the α subunits and can induce gate opening [31-33].
2. Overview of the assembly pathway of the 20S proteasome
Biogenesis of the eukaryotic 20S proteasome relies on the intrinsic ability of subunits to self-assemble, subunit-specific N-terminal and C-terminal extensions that help guide assembly, and dedicated extrinsic assembly chaperones. The simpler archaeal and bacterial 20S proteasomes might not have been expected to require assembly chaperones, but at least one archaeal complex implicated in proteasome assembly has been identified [31]. Like general molecular chaperones, proteasome-specific assembly chaperones do not form part of the final complex but associate with intermediate complexes or individual subunits. Eukaryotic 20S proteasome assembly appears to proceed through an ordered series of intermediates with step-wise addition of subunits (Fig. 3). As will be discussed in subsequent sections, assembly chaperones associate and dissociate at specific stages in this process.
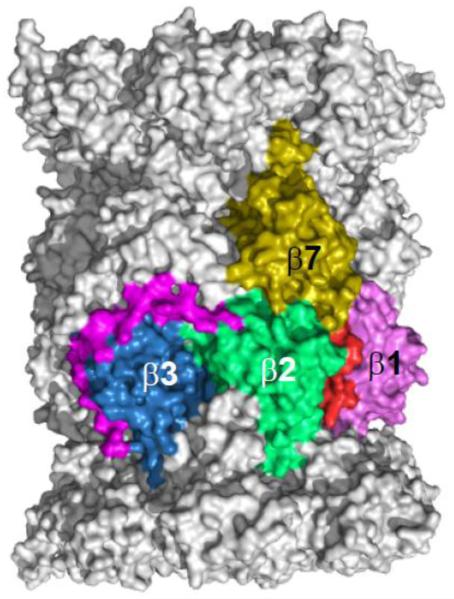
Figure 3. C-terminal extensions of the β2 and β7 subunits serve to stabilize the assembling CP.
The β2 subunit (green) forms extensive contacts with the β3 subunit through its C-terminal extension. The C-terminal 30 amino acid residues are shown here in magenta. The β7 subunit (dark yellow) inserts its C-terminal extension between the β2 and β1 subunit of the opposing β ring. The final 15 amino acids residues of β7 are shown in red. The image was made using PyMol using the structure elucidated in Refs [15].
E. coli-expressed actinomycete and archaeal proteasomes, which have a single α and a single β subunit (or one or two slightly different variants), are capable of full self-assembly without additional factors [34-37]. This ability to completely self-assemble is not shared by eukaryotic proteasomes. While deleting the genes for any of the known yeast assembly chaperones is not lethal, the mutants do show growth defects, and combining chaperone mutations with loss of the RPN4 gene is strongly deleterious or lethal [31, 38, 39]. Rpn4 is a transcription factor responsible for up-regulating proteasome gene expression when proteasome activity is compromised (see Section 5). Thus, yeast cells can compensate for inefficient proteasome assembly by overproducing its subunits.
2.1 Some α subunits can self-assemble into higher order structures
Archaeal α-subunits form heptameric rings when expressed without β subunits in E. coli. The ability to self-assemble requires a highly conserved N-terminal helix in the α subunit. The β subunits by themselves do not assemble into rings, but when co-expressed with the archaeal α subunits in E. coli, mature 20S proteasome particles are formed. The 8-residue β-subunit propeptide is dispensable for assembly in this system. From these data it was proposed that the α-subunit ring provides a template for assembly of the β subunits [34].
By contrast, α subunits from the actinomycete Rhodococcus erythropolis do not form rings on their own, likely because of the limited surface contacts between adjacent α subunits [37]. Instead, individual α subunits form α-β heterodimers that multimerize to make half-proteasome precursors [40]. The Rhodococcus β subunit has a long N-terminal propeptide that packs between the α subunits of neighboring α-β dimers, stabilizing their interaction and allowing assembly into a half-proteasome complex [37]. Finally, two half-proteasomes pair, and the propeptides are autocatalytically removed, creating a mature 20S proteasome [40]. Work on assembly of proteasomes from another actinomycete, Mycobacterium tuberculosis, revealed a distinct half-proteasome intermediate stage in which the β-subunit propeptides point outward and occlude the interface that must form when two half-proteasomes join [41]. These and other results reveal substantial conformational changes that must accompany the maturation of proteasome intermediates into 20S proteasomes.
Interestingly, higher order α–subunit complexes are also observed when specific eukaryotic α subunits are expressed in E. coli. Recombinant human α7 subunit can self-assemble into single or doubly stacked homoheptameric rings similar to the behavior of archaeal α subunits [42]. Human α7 also has the ability to incorporate either normal neighboring subunit into ring structures; these rings have a wide range of stoichiometries, indicating that additional information is needed for correct placement of the seven different α subunits in human cells [43]. The Trypanosoma brucei α5 subunit expressed in E. coli can also form heptameric rings [44]. These data imply that some but not all eukaryotic α subunits retain an ability to oligomerize into rings, which is likely to be important for the normal assembly pathway. However, in eukaryotes additional factors or more efficient binding of individual α subunits to the correct α-subunit neighbors (or both) is necessary to help ensure the correct order of subunits in each ring (see below).
2.2 Eukaryotic β-subunit extensions facilitate assembly
While the different α subunits in the eukaryotic proteasome are generally similar in length and positioning of secondary structure elements, many of the β subunits have unique N-terminal and C-terminal extensions that promote specific steps of assembly. Most of the N-terminal extensions are propeptides that are cleaved at the end of assembly and will be discussed in Section 3.1. The yeast proteasome crystal structure revealed that the β2 and β7 subunits have long C-terminal appendages in their mature structures that interact with adjacent subunits (Fig. 2) [15]. Similar interactions are seen in bovine and human proteasome structures [45, 46]. Deletion of the yeast β2 extension is lethal [47]. The β2 C-tail wraps around the neighboring β3 subunit in the same ring, making contacts with additional subunits in the same ring (β4) and the opposing β ring. Notably, the earliest stable assembly intermediate identifiable in yeast cells, the so-called 15S complex, contains a full α ring plus β2, β3, and β4, the three “early” β subunits [48]. In mammalian cells, systematic knockdowns of individual subunits suggested that β2 is the first β subunit added to the α ring en route to making a full half-mer intermediate [49].
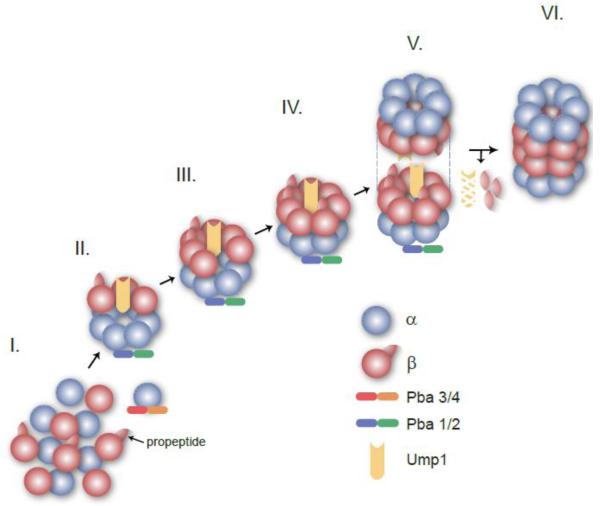
Figure 2. Model for the assembly of the Eukaryotic CP.
This cartoon depicts assembly precursors that were isolated, or shown to form, in yeast.
I. α (blue) and β subunits with propeptides (red) are synthesized as free polypeptides. An α ring is formed initially with the aid of the Pba3/4 (red and orange) chaperone that interacts specifically with the α5 subunit.
II. The isolated 15S intermediate contains a full α ring, the β2, β3 and β4 subunits and two assembly chaperones: Pba1/2 (blue and green) and Ump1 (yellow).
III. The –β7 intermediate is composed of a full complement of subunits except the β7 subunit and Pba1/2 and Ump1.
IV. The half-proteasome has a full α ring and full β ring, but is still associated with the assembly chaperones Pba1/2 and Ump1.
V. The dimerization of the half-proteasomes forms the preholoproteasome that is still immature.
VI. CP maturation is achieved by the autocatalytic processing of the β subunit propeptides and the degradation of the Ump1 chaperone. The Pba1/2 chaperone is also released upon maturation. This process yields a functional CP competent for protein degradation.
The β7 C-tail extends across the dyad axis of the proteasome and inserts into a groove formed on the outer surface between the β1 and β2 subunits in the opposing β-ring. A 19-residue deletion that includes the entire β7 tail causes the accumulation of specific assembly intermediates and active-site maturation defects [47, 48]. The β7 tail appears to stabilize the correct register between the two half-proteasomes and stimulate the autocatalytic processing of the propeptides during final proteasome maturation. When only the portion of the β7 tail that contacts the subunits in the dyad-related β-ring is truncated (15 residues), no growth defects were noted, but a strong drop in the post-acidic cleavage activity of the β1 subunit occurs [48, 50]. However, the β7 tail has a partially redundant role with the N-terminal β5 propeptide in half-mer dimerization (see Section 3.1 below).
3. Proteasomal intramolecular chaperones and dedicated assembly chaperones
Efficient, high-fidelity assembly of the eukaryotic 20S proteasome cannot be carried out without extrinsic accessory factors and intrinsic elements of proteasome precursors, in particular, the various N-terminal and C-terminal appendages of the β subunits.
3.1 β propeptides serve as intramolecular chaperones
Many proteases are synthesized with propeptides that are processed to allow formation of their catalytic active sites. All the active β subunits of the proteasome (β1, β2, and β5 in eukaryotes) are also translated with N-terminal propeptides that are autocatalytically cleaved during assembly [20, 51, 52]. A common function for these proteasomal propeptides is to protect the active site residues from Nα-acetylation [53]. The Nα-amino group of the active β subunits functions as a general base during substrate peptide-bond cleavage [52], so Nα-acetylation blocks active-site function. The propeptides are only removed after the two halves of the proteasome have joined, thus preventing access of Nα-acetyltransferases [20]. The N-termini of the β6 and β7 subunits are also proteolytically trimmed from their initial translation products, but this occurs within the matured 20S core through the action of nearby active site-containing subunits; the mature N-termini of β6 and β7 are repositioned following their processing [54]. The β-subunit propeptides also make contributions to 20S proteasome assembly. However, archaeal and M. tuberculosis β-subunit propeptides are fully dispensable for assembly, at least when expressed in E. coli [34, 55]. Interestingly, M. tuberculosis proteasome assembly is normally temperature dependent, with little or no β-subunit processing at low temperatures; however, if the β-subunit propeptide is deleted, assembly can occur at temperatures as low as 18°C [55]. This is consistent with the conformational repositioning of β-subunit propeptides implied from EM structural analysis (see above). In contrast, and as noted in Section 2.1, proteasomes from another actinomycete, Rhodococcus erythopolis, require their β propeptides for efficient assembly and proper folding of the subunit itself [56].
In eukaryotes, the different β-subunit propeptides play distinct roles in assembly. The β5 propeptide is necessary for efficient assembly in both yeast and mammalian cells. Deletion of this propeptide is normally lethal in yeast and causes the accumulation of precursor species in mammalian cells [20, 49, 57]. Separate expression of the 75-residue β5 propeptide rescues the lethality associated with the yeast β5 propeptide deletion allele and allows normal proteasome assembly. This ability of the propeptide to function in trans in assembly suggests that it normally functions as a type of intramolecular chaperone [20, 56]. The mammalian β5 propeptide appears to be necessary for incorporation of the neighboring β6 subunit into the growing β ring during assembly, though the propeptide may not be necessary for the incorporation of β5 itself [49, 57]. The 30-residue β2 propeptide also shows some capacity to function in trans [58].
Although the propeptides of β1 and β2 also contribute to efficient assembly and maturation of the proteasome, they are less important than the β5 propeptide [53]. Deletion of the β2 propeptide slows processing of the β5 propeptide as much as 2.5-fold and partially stabilizes model proteasome substrates in yeast cells. The propeptide of β1 is not essential for assembly, but its loss also causes β5 processing abnormalities. Simultaneous deletion of both the β1 and β2 propeptides reduces proteasome levels and causes a pronounced yeast growth defect not observed with either single mutant; this defect is far more pronounced than simply inactivating the β1 and β2 active sites, implying an overlapping role in proteasome assembly for the two propeptides [53]. Notably, the mature β6 subunit possesses a unique nine-residue N-terminal extension (relative to the aligned mature N-termini of all β subunits including those from prokaryotes); this NTE appears to function with the Ump1 assembly chaperone (see Section 3.2.1) in an assembly checkpoint prior to half-proteasome dimerization [48].
Determining the exact mechanisms by which the eukaryotic 20S proteasome propeptides facilitate assembly has been difficult, particularly since no in vitro assembly system has been developed. Unexpected insight into the contribution made by the β5 propeptide came from a high-copy suppressor screen using a yeast strain bearing β5 propeptide mutations that conferred temperature-sensitive growth [48]. The strongest full-length suppressor turned out to be PRE4, the gene encoding the β7 subunit. Suppression was lost if the β7 C-terminal tail was deleted or if point mutations were made in an invariant Trp residue in the C-tail or in residues in β1 from the opposing β ring that contact this Trp. Addition of the β7 subunit to the half-proteasome intermediate is a rate-limiting step in 20S proteasome assembly in vivo [48, 59]. The increase in β7 dosage allows the normally essential function of the β5 propeptide to be bypassed. Insofar as this suppression depends on the association of the β7 tail with the opposing β ring, these data strongly suggest that a key role of the β5 propeptide is also to align and stabilize two half-proteasome intermediates during formation of the pre-holoproteasome. Additional findings support this inference [48].
3.2 Efficient 20S proteasome assembly requires dedicated assembly chaperones
It is now clear that multiple accessory proteins help proteasome subunits fit together efficiently and in the correct arrangement [60, 61]. These proteins aid in specific steps of proteasome assembly, but they themselves are not part of the final mature structure (Fig. 3). The first 20S proteasome (CP) assembly chaperone to be identified was Ump1 in yeast (called hUMP1 or POMP1 in humans). Ump1 is conserved across eukaryotes and functions in later steps of assembly, although it may work slightly differently in yeast and mammals. The heterodimeric chaperones Pba1/Pba2 (human PAC1/PAC2) and Pba3/Pba4 (human PAC3/PAC4) participate in earlier assembly events. Two additional factors, Blm10 and Ecm29, have been proposed to have roles in 20S proteasome assembly, but they have also been assigned other functions, such as proteasome activation or inhibition.
3.2.1. Ump1 (hUMP1/ POMP/ Proteassemblin)
Ump1 was identified in a screen for yeast mutants that stabilized a short-lived model substrate protein [39]. Cells survive without Ump1 but grow poorly. Though they have low sequence similarity, both yeast Ump1 and its mammalian homolog have been shown to associate specifically with immature proteasomal precursors [39, 57]. Using an elegant antibody accessibility assay, it was shown that yeast Ump1 is encased in the proteasomal catalytic chamber when two half-proteasomes associate to form the preholoproteasome; upon autocatalytic cleavage of the β subunit propeptides, the trapped Ump1 is degraded by the nascent activated 20S proteasome [39].
Mutations in the β5 propeptide and Ump1 show a surprising genetic interaction. Normally, this propeptide is essential for proteasome assembly and cell viability; however, when the UMP1 gene is deleted, the β5 propeptide is no longer required. Ramos et al. [39] proposed that the β5 propeptide acts to reposition or conformationally alter Ump1 in a way that facilitates β5 autocleavage. Thus, if Ump1 were already deleted, the β5 propeptide would no longer be required. Ump1, in this model, is required to orchestrate β-subunit processing and proteasome maturation at a late stage of assembly. A different, possibly additional, function for Ump1 has been proposed from data, described earlier, showing that high levels of β7 can suppress defects associated with loss of the β5 propeptide and that this suppression requires the β7 C-tail to enhance proteasome half-mer dimerization [48]. Ump1 was proposed to have an assembly checkpoint function, inhibiting dimerization until the “late” β subunits, the last being β7, had incorporated into the half-mer. Premature dimerization would likely make it difficult to insert β7 and other late subunits into the complex. The β5 propeptide, which seems to promote dimerization, would become dispensable if Ump1 were no longer present to limit dimer formation, although overall assembly and maturation would be defective, leading to the continued growth defect associated with the UMP1 gene deletion.
3.2.2. Pba1/Pba2 (PAC1/PAC2; PbaA/PbaB)
It was some time before any additional 20S proteasome assembly factors were discovered. Hirano et al. [62] used a Flag-tagged β subunit to isolate proteasome-associated proteins from HEK293 cells, and identified a co-precipitating protein complex consisting of two proteins, which they named PAC1 and PAC2 (proteasome assembly chaperones 1 and 2). They went on to show that PAC1/PAC2 specifically associates with precursor proteasomes and that genetic knockdown of this complex impairs 20S proteasome assembly. Knockdown also leads to accumulation of aberrant α-ring dimers. Thus one of the functions of this chaperone pair may be to prevent the off-pathway interaction of two α-subunit rings [62]. PAC1 knockout mice are embryonic lethal, highlighting the importance of this assembly chaperone for mammalian development [63].
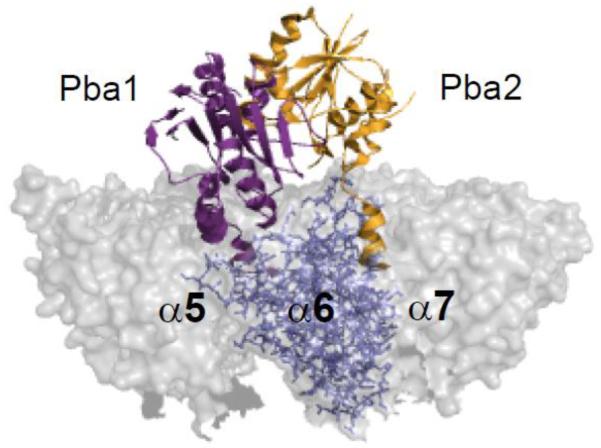
The yeast orthologs of PAC1 and PAC2, called Pba1 and Pba2 (proteasome biogenesis-associated factors 1 and 2), were initially identified by mass spectrometric analysis of 20S proteasome assembly intermediates but were not detected in mature proteasomes [48]. An earlier study had also identified Pba2 from a mutant that had a weak defect in the degradation of an abnormal endoplasmic reticulum-associated protein [64]. In another report, Pba1 and Pba2 (as well as Pba3 and Pba4) were identified from a screen for genetic suppressors of a hyperactive DNA damage-response mutant [65]. No growth defects are seen in pba1Δ or pba2Δ (or the double) mutants, but these mutations do exacerbate the phenotypic defects of proteasome mutants [48]. Interestingly, these chaperones possess C-terminal HbYX (hydrophobic-tyrosine-any amino acid) motifs. The HbYX motif has been found in a number of proteasome activators where it is required for activator function through binding to pockets formed between specific α subunits [32, 33, 66]. The Pba1 and Pba2 HbYX motifs both contribute to chaperone function and proteasome precursor binding [31]. A recent crystallographic study showed that these motifs do indeed associate with specific lysines in the intra-α subunit pockets [67]. The Pba1 C-terminus sits in the pocket between α5 and α6, while the Pba2 C-terminus interacts with the α6-α7 pocket (Fig. 4). In contrast to Ref. 31, these authors detected Pba1/Pba2 binding to mature proteasomes in vitro, although binding was very sensitive to salt, dropping over 225-fold (to a KD of 2.8 μM) when NaCl concentration was increased from 12.5 to 150 mM [67].
Figure 4. Assembly chaperones Pba1 and Pba2 associate with the CP α ring.
Pba1 (purple) and Pba2 (orange) interact with intra-α subunit pockets of the 20S (grey) through their C-terminal HbYX motifs. The image was made using PyMol using the structure elucidated in Refs [67].
Since archaeal α and β subunits can form functional proteasomes by co-expression in E. coli in the absence of other factors, it was long thought that these proteasomes do not require chaperones. However, apparent homologs of the eukaryotic assembly chaperones, Pba1 and Pba2, have now been found. Biochemical analysis of the archaeal Methanococcus maripaludis PbaA and PbaB proteins demonstrated specific binding of PbaA (but not PbaB) to a preholoproteasome intermediate but not the mature M. maripaludis 20S proteasomes isolated by expression in E. coli [31]. PbaA, unlike PbaB, has a conserved C-terminal HbYX motif, and mutational analysis showed that the intact HbYX motif is necessary for preholoproteasome association; the specific lysine within the intra-α subunit pocket implicated in activator HbYX binding is also required. Interestingly, when the mature archaeal proteasome active sites are blocked by inhibitors, both PbaA and PbaB are found to associate with the inactive CP. These data imply that allosteric communication between the active sites in the central β-subunit chamber and the distant surface of the α ring can modulate PbaA and PbaB binding. Genetic studies are still needed to assess the exact physiological functions of PbaA and PbaB in archaeal 20S proteasome assembly and function.
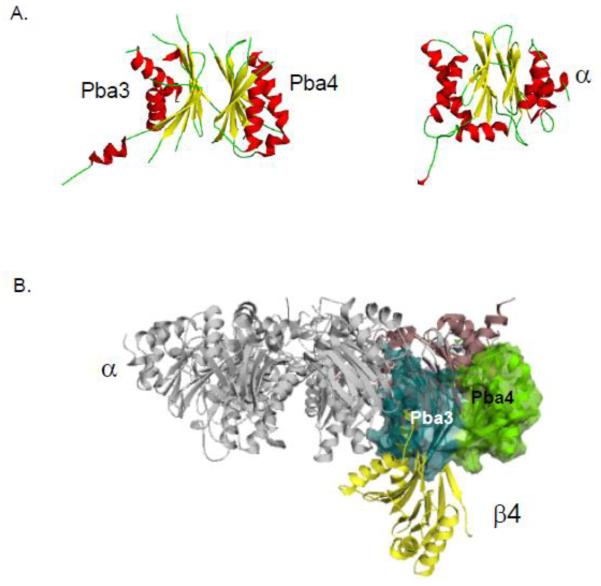
3.2.3 Pba3/Pba4 (PAC3/PAC4)
Using a stable cell line expressing Flag-tagged PAC1, Hirano et al. [68] affinity purified proteasome assembly intermediates and identified a novel factor, PAC3, by mass spectrometry. Knockdown of this protein caused proteasome assembly defects, lending credence to PAC3 being an assembly chaperone. Pba3, the S. cerevisiae homolog of PAC3, as well as an associated protein, Pba4, were identified by multiple research groups [38, 65, 69, 70]. It became clear that a mammalian homolog of Pba4, named PAC4, also exists. Loss of yeast Pba3 or Pba4 causes growth and proteasome assembly defects. The two proteins form a heterodimer, and the complex associates tightly with the α5 proteasome subunit. Structural studies of these chaperones revealed a surprising structural similarity of the dimer to proteasomal α and β subunits [69], as shown in Fig. 5A. However, the co-crystal structure of Pba3/Pba4-α5 indicated that the chaperone does not bind α5 in the same way as do either neighboring α or β subunits. Pba3/Pba4 appears to disassociate from the assembling intermediate when the β4 subunit is incorporated, accounting for its absence from the 15S intermediate, which includes the β4 subunit. This can be understood by modeling the chaperone-α5 complex onto the full 20S proteasome structure (Fig. 5B). The model shows a strong steric clash between the β4 subunit and Pba3/Pba4 [69].
Figure 5. Proteasome biogenesis associated (Pba) factors 3 and 4.
A. The structure of the Pba3/4 dimer is similar to the structure of proteasome subunits. A representative α subunit is shown for comparison. Helices are indicated in red, β strands in yellow and loops in green.
B. The Pba3 (blue) and Pba4 (green) chaperone associates with the α5 subunit (purple) in such a manner that there is a steric clash with the β4 subunit (yellow) unless Pba3/4 disassociates before this subunit is incorporated into the CP.
These images were made using PyMol using structures elucidated in Refs [15, 69].
An important function of Pba3/Pba4 is in controlling the incorporation of the α3 subunit into the proteasome [38]. The α3 subunit is unique in that it is the only dispensable subunit of the yeast 20S proteasome; its deletion causes only a minor growth defect under optimal conditions. In the absence of α3, a second copy the neighboring α4 subunit takes its place in each α ring [71]. Surprisingly, when yeast cells lack Pba3/Pba4, they also make proteasomes bearing neighboring α4 subunits despite the continued expression of α3; some 20-50% of proteasomes were estimated to have the α4-α4 configuration [38].
Pba3/Pba4 may form a scaffold on which α ring assembly is guided through a specific pathway ensuring that the α3 subunit is positioned between the α2 and α4 subunits in the ring [11, 38]. This model is consistent with the crystal structure of Pba3/Pba4 in complex with α5 and the modeling studies showing the possibility of extensive contacts between Pba3/Pba4 and the neighboring α subunits [69]. However, the exact mechanism by which α-ring assembly is regulated by the chaperone is not yet clear. Intriguingly, cells that make proteasomes with α4-α4-containing rings are more resistant to oxidative stresses [38]. Chronic oxidative stress may alter proteasome assembly such that more α4-α4 proteasomes are generated. Such proteasomes are expected to have a constitutively open α ring [72] and may interact differently with proteasome regulatory factors such as the RP.
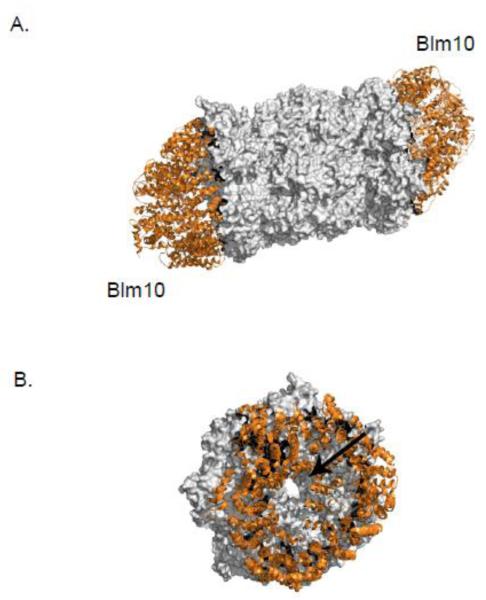
3.2.4. Blm10 (PA200)
A number of proteins and protein complexes have been described that bind the ends of the CP cylinder and stimulate its activity. The 19S regulatory particle caps the majority of 20S proteasomes, but other factors such as PA28α/β, PA28γ and PA200 in mammalian cells also bind and stimulate the CP. PA200, and its yeast ortholog Blm10, are large HEAT-repeat proteins that form a dome-shaped spiral atop the CP (Fig. 6A) [73]. Hybrid complexes, with a 19S RP on one end of the CP and Blm10/PA200 on the other, can also form [74]. Blm10/PA200 may have multiple functions as there are reports suggesting roles as a proteasome assembly factor, activator, or quality-control factor [32, 75-77].
Figure 6. The HEAT repeat protein, Blm10, caps the CP.
A. Blm10 (orange) occupies surfaces that are usually bound by regulators of the CP (grey). A top-down view of the Blm10 (orange) capped CP (grey) reveals a narrow opening through the proteasome, indicated here by an arrow. These images were made using PyMol using the structure elucidated in Refs [78].
Unlike the RP, Blm10/PA200 does not bind or hydrolyze ATP, but its interaction with the CP disorders the α-ring gate and promotes entry of peptide substrates [32, 78]. A recent crystal structure shows that a tyrosine residue in the conserved C-terminal HbYX motif of this protein interacts with the α ring via α6-Lys66 in the pocket formed between the α5 and α6 subunits [78]. This interaction displaces the α5 N-terminal segment, effectively disordering the gate as judged by activation of peptide hydrolysis. A blm10Δ mutant does not exhibit general proteasomal proteolytic defects; Blm10 may target a specific subset of proteins, such as ones that are intrinsically disordered [74]. The dome structure of Blm10 completely covers the CP end, with only a narrow orifice at the top of the dome, implying that only peptides or unfolded substrates would be able to gain entry into the proteasome (Fig. 6B).
Participation of Blm10 in proteasome assembly was initially postulated when it was found to co-elute from a gel filtration column with an Ump1-containing precursor complex [79]. The authors reported that in a blm10Δ null mutant, the rate of β5 subunit-propeptide processing and Ump1 degradation is accelerated, implying that Blm10 normally delays assembly. Blm10 is found associated with several CP assembly intermediates, which would in principle allow a Blm10 contribution to the proteasome maturation process [48, 59]. Notably, when the blm10Δ allele is combined with a mutation in an RP subunit that impairs RP-CP binding, enhanced defects in CP maturation are observed [59]. These data argue for a positive role for Blm10 in CP assembly by a mechanism that can be compensated in part by the RP.
Blm10 has also been proposed to bind preferentially to 20S proteasomes with an open or disordered axial gate. Blm10 binding to the CP is increased in CP mutants with one or more α subunit N-termini deleted or in cells with the ump1Δ mutation [80]. Atomic-force microscopy has shown that the gated axial pores of the CP open and close stochastically, with approximately 25% of proteasomes adopting the open conformation at steady-state [72]. One proposed role for Blm10 is to sequester 20S proteasomes that have their gates open an aberrantly high proportion of the time, thereby preventing uncontrolled substrate access to the catalytic chamber [80].
3.2.5. Ecm29
Ecm29 was initially identified by its binding to yeast 26S proteasomes under low salt conditions [81]. Intact 26S proteasomes can be purified without ATP when Ecm29 is present, but ATP is required for maintaining a stable RP-CP interaction in the absence of this factor. This suggests that Ecm29 can stabilize the 26S proteasome complex. Like Blm10, Ecm29 is a HEAT-repeat protein and is conserved in humans, where it localizes to the nucleus and cytoplasmic membrane structures [82, 83].
There is evidence that Ecm29 may function in proteasome quality control by either sequestering aberrantly formed proteasomes or repairing them. In one report, Ecm29 co-purified with aberrant 26S proteasomes that apparently lacked the β3 subunit and possessed immature versions of the β5 subunit but associated with RP [84]. Reconstitution assays suggested that exogenous addition of Ecm29 was sufficient to drive the maturation of the β5 subunit in these immature or aberrant RP-CP complexes [84]. The loss of β3 subunits from the Ecm29-26S proteasome complexes was not observed in several subsequent studies, but Ecm29 association with otherwise aberrant or misassembled proteasomes was confirmed [85, 86].
Proteasomes disassemble into their RP and CP components in response to oxidative stress; Ecm29 plays a pivotal role in this disassembly by associating specifically with the RP [87]. This occurs in both yeast and human cells. Ecm29-dependent disassembly of proteasomes is necessary for growth of yeast under oxidative stress conditions. Despite the many intriguing observations linking Ecm29 to proteasomes, the exact mechanism(s) of Ecm29 action remains uncertain. There is general agreement that the ability to identify atypical or abnormal proteasomes is most likely a core function of Ecm29.
4. Assembly of the proteasome and its subcellular localization may be coordinated
Proteasomes in yeast are mainly nuclear [88, 89], while mammalian proteasomes are distributed both in the nucleus and cytoplasm but are often concentrated in the nucleus as well [90, 91]. Whether de novo proteasome assembly steps occur at distinct subcellular sites is unknown, as are many details of the dynamics of proteasome localization in general. In an early study, a GFP-tagged version of the β1i CP subunit was shown to be quantitatively incorporated into proteasome particles and was used to follow proteasome dynamics in cultured mammalian cells. Photobleaching experiments demonstrated that the nuclear and cytoplasmic pools of proteasomes are in very slow exchange except when the nuclear envelope breaks down during mitosis. Within each of these compartments, however, proteasomes diffuse rapidly [90].
Several proteasome subunits have nuclear localization sequences (NLSs), and a possible way to regulate localization of either mature proteasomes or assembly intermediates may be to mask or unmask these sequences [92, 93]. Only a subset of proteasome subunits bear an NLS, suggesting that, for most of the subunits, transport into the nucleus requires prior formation of either mature proteasomes or proteasomal subparticles. Some 20S proteasome assembly chaperones, such as Pba1/Pba2 and Pba3/Pba4, may be primarily in the cytoplasm [65, 94], which supports the idea that at least early stages of CP assembly occur in the cytoplasm. Proteasomal precursors may be imported into the nucleus and then assembled into mature CPs within the nucleus where Ump1 is concentrated [95]. Interestingly, in yeast cells lacking Ump1, Pba3 and Pba4 accumulate in the nucleus [65]. This supports the possibility that later steps of 20S proteasome maturation occur in the nucleus, releasing the early-acting Pba3/Pba4 chaperone, whereupon it exits the nucleus.
Data are also available in support of the ability of fully assembled proteasomes to cross the nuclear membrane. Employing a nuclear reconstitution assay based on Xenopus egg extracts, Savulescu et al. [96] showed that particles containing the fully assembled CP, two RP subunits (Rpn1 and Rpn2), Hsp90 and importin β can enter the reformed nuclei. Purified CP by itself collected at the nuclear periphery but did not appear to enter nuclei in this in vitro system, but the actively transported proteasomal species in vivo remain to be catalogued. Interestingly, in yeast cells that enter a quiescent state (stationary phase), proteasomes exit the nucleus and collect in one or two large cytoplasmic structures called proteasome storage granules (PSGs). Within minutes of addition of fresh medium, the PSGs break down and proteasomes re-enter the nucleus. Therefore fully mature 20S and/or 26S proteasomal particles are likely to be able to traffic across the nuclear membrane, possibly by a mechanism distinct from the transport of precursor complexes [97].
5. Regulation of 20S proteasome biogenesis
Besides maintaining basal levels of proteasomes under optimal growth conditions, cells must be able to regulate proteasome activity in response to various stimuli. Both basal and induced levels of proteasomes are set in yeast by the transcription factor Rpn4 [98]. Recent studies have identified a functional equivalent in humans, Nrf1, which is also responsible for the upregulation of proteasome subunits during stresses known to induce an excess of aberrant, potentially toxic proteins [99]. Another recent report suggests that the CCAAT box-binding protein NF-Y is responsible for maintaining basal proteasome levels in mammalian cells [100]. Rpn4 and Nrf1 stimulate the expression of proteasome subunits and other genes involved in the UPS [101].
In S. cerevisiae, a nonameric DNA consensus sequence in the promoter regions of nearly all proteasome subunits called the proteasome-associated control element (PACE) was identified. This element was shown to bind Rpn4, a 60 kDa protein with a C2H2 type zinc finger motif [98]. Rpn4 is an extremely short-lived protein, with a half-life of about two minutes, and is degraded by both ubiquitin-dependent and ubiquitin-independent mechanisms [102, 103]. Both mechanisms require the proteasome, providing a negative-feedback control on Rpn4 levels: When proteasome activity becomes limiting, Rpn4 levels rise, increasing proteasome gene transcription and driving proteasome assembly, and when proteasome activity increases, Rpn4 will be degraded more rapidly, reducing its levels. The first evidence for the existence of such a feedback mechanism came from studies of proteasome assembly [20]. When the processing or activity of β5 was impaired, large increases in the levels of β5 were observed, leading to the suggestion of a positive regulator of β5 expression that was itself a substrate of the proteasome.
Genetic evidence highlights the importance of the Rpn4 pathway in ensuring adequate basal levels of proteasomes and in response to proteotoxic stresses such as high temperature. Yeast mutants with the RPN4 gene deleted are temperature sensitive for growth, and the rpn4Δ mutation is synthetically lethal with many other proteasome-pathway mutations [31, 65, 104]. It is of note that none of the yeast 20S proteasome assembly chaperones, Ump1 and Pba1-4, possesses a PACE in its promoter, although Blm10 and Ecm29 do. It is possible that the basal levels of assembly chaperones are sufficient for maximal assembly rates regardless of conditions. They are normally present at much lower levels than total proteasomes. Conceivably, some environmental stimuli might cause one or another assembly chaperone to become limiting and as a result, channel assembly through a pathway yielding proteasomes of distinct composition. Such a hypothesis has been proposed to account for the α4-α4 proteasome found in pba4Δ mutants, with the assumption that wild-type cells might make these same alternative forms under some conditions [38].
6. Assembly of Alternative Proteasome Isoforms
The canonical 20S proteasome consists of a constitutive set of 14 different subunits usually encoded by 14 different genes. In vertebrates, interferon γ-inducible proteasome subunit genes (β1i, β2i and β5i) are expressed at high levels in immune tissues such as the spleen and by stimulated class I antigen-presenting cells [22-26, 105, 106]. This results in the formation of the immunoproteasome. In the thymus, a specialized proteasome called the thymoproteasome is made, which includes β1i and β2i but replaces β5i with another specialized subunit, β5t [27, 28, 107, 108]. In Drosophila, testis-specific proteasomes have been documented. These include an alternative α6 subunit (α6T) that is essential for fertility [109]. However, it appears that α6T is not qualitatively different from its constitutive counterpart since the fertility of an α6T null mutant can be rescued by ectopic α6 expression; high α6T expression in the testis is presumably needed to maintain sufficient proteasome levels during sperm formation [110]. Plants also express a large set of 20S proteasome subunit paralogs, which could in principle have distinct functions [111]. Finally, an alternative proteasome with two α4 subunits in each α ring was identified in mutant S. cerevisiae, but whether such proteasomal forms are important for wild-type yeast function remains uncertain [71] (Section 3.2.3).
Much of what is known about the assembly of alternative proteasomes comes from studies on the immunoproteasome [60, 107]. The immunoproteasome contains catalytic β subunits that share ~60% amino-acid identity with their constitutive counterparts but differ in their active-site specificities; these different protein cleavage preferences generate an alternative repertoire of peptides, including ones suitable for antigen presentation [107]. Importantly, the immunoproteasome-specific subunits do not incorporate into the proteasome independently of one another. Most proteasomes from interferon-stimulated cells, for example, will have all three βi subunits in each β ring of each particle [112]. β1i is the first to enter the assembly pathway, and it influences the preferred incorporation of the β2i and β5i subunits into the assembling proteasome [112, 113]. This assembly pathway appears to be different from constitutive proteasome β-subunit incorporation, where the β2 subunit enters the ring first and the rest then enter sequentially [49]. The influence of βi subunits on subunit selection resides primarily in their divergent propeptides. Chimeric subunits that have immunoproteasome βi subunit-derived propeptides fused to constitutive β subunit mature domains behave like the respective βi subunit. The apparent cooperative assembly behavior of interferon-induced catalytic β subunits ensures that immunoproteasomes comprise the majority of proteasomes in inflamed tissues [114, 115]. Nevertheless, proteasomes isolated from infected β5i -/- mice contain the immunosubunits β1i and β2i but a constitutive β5, indicating that in the absence of the preferred β5i, the constitutive β5 can be incorporated [116].
The assembly chaperone hUmp1 is induced upon interferon stimulation and associates strongly with the β5i subunit [113]. Immunoproteasome formation is ~4-fold faster compared to constitutive proteasome assembly and requires hUmp1. The tighter apparent binding of hUmp1 to β5i relative to β5 may further bias assembly toward immunoproteasome formation. Mature immunoproteasomes are also degraded about 5-fold faster than the constitutive particle (half-life of ~1 d versus ~5 d), consistent with a rapid and transient adaptive immune response [113]. Crystal structures of the immunoproteasome in complex with inhibitors have suggested possible mechanisms for the differences in substrate processing between constitutive proteasomes and immunoproteasomes, but additional structural work will be required for a deeper understanding of the distinctive biogenesis and dynamics of the immunoproteasome [117].
The proteasome peptidase activator PA28α/β has also been implicated in the efficient assembly of immunoproteasomes [118]. PA28α/β binds immunoproteasome precursors, and an early study of mice lacking PA28α/β implicated the activator in immunoproteasome assembly [119]. However, in a subsequent study, immunoproteasomes were present at nearly wild-type levels in the spleens of mice missing PA28α/β, calling into question the necessity of these activators for assembly and instead suggesting a more specialized role for PA28α/β in the processing of certain antigens [120, 121].
6. Concluding remarks
Efficient and high-fidelity assembly of the 28-subunit 20S proteasome, including correct assembly of alternative forms, is a challenging task to carry out in the crowded confines of the cell. Maintaining the fidelity of proteasome assembly is essential to proper cell function, particularly under conditions of proteotoxic stress, and it has been shown that altering proteasome composition can change its interactions with inhibitors [117, 122]. Certain steps of the assembly process rely on intrinsic features of the subunits or subunit subcomplexes, while others require the help of dedicated, conserved accessory factors. These proteasome assembly chaperones can function as guides, checkpoints, and perhaps even as remodeling agents for aberrant intermediates. Though several proteins involved in the 20S core particle assembly pathway have been biochemically and even structurally characterized, a mechanistic understanding of exactly what these chaperones do is lacking.
Among the open questions remaining, a prominent one is the question of which aspects of proteasome assembly occur by stochastic self-assembly and which require chaperones. Computational studies indicate that very high intersubunit affinities can delay assembly of generic ring structures; very weak interactions yield the same results [123]. Intermediate affinities are optimal for efficient assembly of rings, especially those containing three or more subunits. Archaeal subunits can assemble into stable proteasomes without the aid of additional factors, while eukaryotic subunits probably cannot. The more complicated compositions of eukaryotic proteasomes have evidently necessitated the evolution of assembly chaperones. These may play a role in modulating subunit-subunit interactions to build stable ring structures. The possibility of restricted subcellular localization of assembly steps and assembly chaperone action and whether this has any bearing on the types of proteasomes that are formed are also still largely unknown. Uncovering the function of these chaperones remains as a major challenge for the field as does understanding how these factors might be regulated to build different types of proteasomes.
Highlights.
The 28-subunit 20S proteasome complex is found in all three domains of life
Propeptides of specific β subunits function as intramolecular chaperones
Proteasome assembly requires specialized appendages of specific β subunits
Multiple dedicated assembly chaperones are required for the proper assembly
Acknowledgements
We would like to thank Robert Tomko Jr. and Yanjie Li for critical reading of this manuscript. Work on the proteasome from our laboratory has been supported by the NIH R01 grants GM046904 and GM083050 to M.H. M.J.K. was supported in part by training grant GM007223 and an NRSA predoctoral fellowship from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hochstrasser M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- [2].Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ciechanover A, Schwartz AL. The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim. Biophys. Acta. 2004;1695:3–17. doi: 10.1016/j.bbamcr.2004.09.018. [DOI] [PubMed] [Google Scholar]
- [4].Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- [5].Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- [6].Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- [7].Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- [8].Hochstrasser M, Deng M, Kusmierczyk AR, Li X, Kreft SG, Ravid T, Funakoshi M, Kunjappu M, Xie Y. Molecular genetics of the ubiquitin-proteasome system: lessons from yeast. Ernst Schering found. Symp. Proc. 2008;1:41–66. doi: 10.1007/2789_2008_100. [DOI] [PubMed] [Google Scholar]
- [9].Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol. Chem. 2008;389:1143–1151. doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Maupin-Furlow J. Proteasomes and protein conjugation across domains of life. Nat. Rev. Microbiol. 2011;10:100–111. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Baumeister W, Dahlmann B, Hegerl R, Kopp F, Kuehn L, Pfeifer G. Electron microscopy and image analysis of the multicatalytic proteinase. FEBS Lett. 1988;241:239–245. doi: 10.1016/0014-5793(88)81069-x. [DOI] [PubMed] [Google Scholar]
- [14].Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- [15].Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- [16].Chen P, Hochstrasser M. Biogenesis, structure and function of the yeast 20S proteasome. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Heinemeyer W, Trondle N, Albrecht G, Wolf DH. PRE5 and PRE6, the last missing genes encoding 20S proteasome subunits from yeast? Indication for a set of 14 different subunits in the eukaryotic proteasome core. Biochemistry. 1994;33:12229–12237. doi: 10.1021/bi00206a028. [DOI] [PubMed] [Google Scholar]
- [18].Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- [19].Witt S, Kwon YD, Sharon M, Felderer K, Beuttler M, Robinson CV, Baumeister W, Jap BK. Proteasome assembly triggers a switch required for active-site maturation. Structure. 2006;14:1179–1188. doi: 10.1016/j.str.2006.05.019. [DOI] [PubMed] [Google Scholar]
- [20].Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- [21].Arendt CS, Hochstrasser M. Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7156–7161. doi: 10.1073/pnas.94.14.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T, Tanahashi N, Yoshimura T, Tanaka K, Ichihara A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994;115:257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- [23].Glynne R, Powis SH, Beck S, Kelly A, Kerr LA, Trowsdale J. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature. 1991;353:357–360. doi: 10.1038/353357a0. [DOI] [PubMed] [Google Scholar]
- [24].Groettrup M, Kraft R, Kostka S, Standera S, Stohwasser R, Kloetzel PM. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur. J. Immunol. 1996;26:863–869. doi: 10.1002/eji.1830260421. [DOI] [PubMed] [Google Scholar]
- [25].Kelly A, Powis SH, Glynne R, Radley E, Beck S, Trowsdale J. Second proteasome-related gene in the human MHC class II region. Nature. 1991;353:667–668. doi: 10.1038/353667a0. [DOI] [PubMed] [Google Scholar]
- [26].Nandi D, Jiang H, Monaco JJ. Identification of MECL-1 (LMP-10) as the third IFN-gamma-inducible proteasome subunit. J. Immunol. 1996;156:2361–2364. [PubMed] [Google Scholar]
- [27].Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- [28].Tomaru U, Ishizu A, Murata S, Miyatake Y, Suzuki S, Takahashi S, Kazamaki T, Ohara J, Baba T, Iwasaki S, Fugo K, Otsuka N, Tanaka K, Kasahara M. Exclusive expression of proteasome subunit {beta}5t in the human thymic cortex. Blood. 2009;113:5186–5191. doi: 10.1182/blood-2008-11-187633. [DOI] [PubMed] [Google Scholar]
- [29].Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- [30].Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- [31].Kusmierczyk AR, Kunjappu MJ, Kim RY, Hochstrasser M. A conserved 20S proteasome assembly factor requires a C-terminal HbYX motif for proteasomal precursor binding. Nat. Struct. Mol. Biol. 2011;18:622–629. doi: 10.1038/nsmb.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dange T, Smith D, Noy T, Rommel PC, Jurzitza L, Cordero RJ, Legendre A, Finley D, Goldberg AL, Schmidt M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J. Biol. Chem. 2011;286:42830–42839. doi: 10.1074/jbc.M111.300178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y. Interactions of PAN’s C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 2010;29:692–702. doi: 10.1038/emboj.2009.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat. Struct. Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- [35].Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J. Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pouch MN, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete Frankia. Mol. Microbiol. 2000;35:368–377. doi: 10.1046/j.1365-2958.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- [37].Kwon YD, Nagy I, Adams PD, Baumeister W, Jap BK. Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly. J. Mol. Biol. 2004;335:233–245. doi: 10.1016/j.jmb.2003.08.029. [DOI] [PubMed] [Google Scholar]
- [38].Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- [39].Ramos PC, Hockendorff J, Johnson ES, Varshavsky A, Dohmen RJ. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- [40].Sharon M, Witt S, Glasmacher E, Baumeister W, Robinson CV. Mass spectrometry reveals the missing links in the assembly pathway of the bacterial 20 S proteasome. J. Biol. Chem. 2007;282:18448–18457. doi: 10.1074/jbc.M701534200. [DOI] [PubMed] [Google Scholar]
- [41].Li D, Li H, Wang T, Pan H, Lin G, Li H. Structural basis for the assembly and gate closure mechanisms of the Mycobacterium tuberculosis 20S proteasome. EMBO J. 2010;29:2037–2047. doi: 10.1038/emboj.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gerards WL, Enzlin J, Haner M, Hendriks IL, Aebi U, Bloemendal H, Boelens W. The human alpha-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the beta-type subunits HsDelta or HsBPROS26. J. Biol. Chem. 1997;272:10080–10086. doi: 10.1074/jbc.272.15.10080. [DOI] [PubMed] [Google Scholar]
- [43].Gerards WL, de Jong WW, Bloemendal H, Boelens W. The human proteasomal subunit HsC8 induces ring formation of other alpha-type subunits. J. Mol. Biol. 1998;275:113–121. doi: 10.1006/jmbi.1997.1429. [DOI] [PubMed] [Google Scholar]
- [44].Yao Y, Toth CR, Huang L, Wong ML, Dias P, Burlingame AL, Coffino P, Wang CC. alpha5 subunit in Trypanosoma brucei proteasome can self-assemble to form a cylinder of four stacked heptamer rings. Biochem. J. 1999;344(Pt 2):349–358. doi: 10.1042/0264-6021:3440349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol. Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- [46].Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- [47].Ramos PC, Marques AJ, London MK, Dohmen RJ. Role of C-terminal extensions of subunits beta2 and beta7 in assembly and activity of eukaryotic proteasomes. J. Biol. Chem. 2004;279:14323–14330. doi: 10.1074/jbc.M308757200. [DOI] [PubMed] [Google Scholar]
- [48].Li X, Kusmierczyk AR, Wong P, Emili A, Hochstrasser M. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 2007;26:2339–2349. doi: 10.1038/sj.emboj.7601681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hirano Y, Kaneko T, Okamoto K, Bai M, Yashiroda H, Furuyama K, Kato K, Tanaka K, Murata S. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008;27:2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hilt W, Enenkel C, Gruhler A, Singer T, Wolf DH. The PRE4 gene codes for a subunit of the yeast proteasome necessary for peptidylglutamyl-peptide-hydrolyzing activity. Mutations link the proteasome to stress- and ubiquitin-dependent proteolysis. J. Biol. Chem. 1993;268:3479–3486. [PubMed] [Google Scholar]
- [51].Schmidtke G, Kraft R, Kostka S, Henklein P, Frommel C, Lowe J, Huber R, Kloetzel PM, Schmidt M. Analysis of mammalian 20S proteasome biogenesis: the maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J. 1996;15:6887–6898. [PMC free article] [PubMed] [Google Scholar]
- [52].Seemuller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–471. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- [53].Arendt CS, Hochstrasser M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J. 1999;18:3575–3585. doi: 10.1093/emboj/18.13.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Groll M, Heinemeyer W, Jager S, Ullrich T, Bochtler M, Wolf DH, Huber R. The catalytic sites of 20S proteasomes and their role in subunit maturation: a mutational and crystallographic study. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10976–10983. doi: 10.1073/pnas.96.20.10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, Parsons T, Li P, Chen Z, Zwickl P, Weich N, Nathan C. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol. Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- [56].Zuhl F, Seemuller E, Golbik R, Baumeister W. Dissecting the assembly pathway of the 20S proteasome. FEBS Lett. 1997;418:189–194. doi: 10.1016/s0014-5793(97)01370-7. [DOI] [PubMed] [Google Scholar]
- [57].Witt E, Zantopf D, Schmidt M, Kraft R, Kloetzel PM, Kruger E. Characterisation of the newly identified human Ump1 homologue POMP and analysis of LMP7(beta 5i) incorporation into 20 S proteasomes. J. Mol. Biol. 2000;301:1–9. doi: 10.1006/jmbi.2000.3959. [DOI] [PubMed] [Google Scholar]
- [58].Jager S, Groll M, Huber R, Wolf DH, Heinemeyer W. Proteasome beta-type subunits: unequal roles of propeptides in core particle maturation and a hierarchy of active site function. J. Mol. Biol. 1999;291:997–1013. doi: 10.1006/jmbi.1999.2995. [DOI] [PubMed] [Google Scholar]
- [59].Marques AJ, Glanemann C, Ramos PC, Dohmen RJ. The C-terminal extension of the beta 7 subunit and activator complexes stabilize nascent 20s proteasomes and promote their maturation. J. Biol. Chem. 2007 doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- [60].Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- [61].Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem. Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- [62].Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, Natsume T, Tanaka K, Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- [63].Sasaki K, Hamazaki J, Koike M, Hirano Y, Komatsu M, Uchiyama Y, Tanaka K, Murata S. PAC1 gene knockout reveals an essential role of chaperone-mediated 20S proteasome biogenesis and latent 20S proteasomes in cellular homeostasis. Mol. Cell. Biol. 2010;30:3864–3874. doi: 10.1128/MCB.00216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Scott CM, Kruse KB, Schmidt BZ, Perlmutter DH, McCracken AA, Brodsky JL. ADD66, a gene involved in the endoplasmic reticulum-associated degradation of alpha-1-antitrypsin-Z in yeast, facilitates proteasome activity and assembly. Mol. Biol. Cell. 2007;18:3776–3787. doi: 10.1091/mbc.E07-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Le Tallec B, Barrault MB, Courbeyrette R, Guerois R, Marsolier-Kergoat MC, Peyroche A. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- [66].Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Stadtmueller BM, Kish-Trier E, Ferrell K, Petersen CN, Robinson H, Myszka DG, Eckert DM, Formosa T, Hill CP. Structure of a Proteasome Pba1-Pba2 Complex: IMPLICATIONS FOR PROTEASOME ASSEMBLY, ACTIVATION, AND BIOLOGICAL FUNCTION. J. Biol. Chem. 2012;287:37371–37382. doi: 10.1074/jbc.M112.367003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Murata S. Multiple chaperone-assisted formation of mammalian 20S proteasomes. IUBMB Life. 2006;58:344–348. doi: 10.1080/15216540600733144. [DOI] [PubMed] [Google Scholar]
- [69].Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, Niwa S, Kasahara M, Kurimoto E, Sakata E, Takagi K, Suzuki A, Hirano Y, Murata S, Kato K, Yamane T, Tanaka K. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- [70].Hoyt MA, McDonough S, Pimpl SA, Scheel H, Hofmann K, Coffino P. A genetic screen for Saccharomyces cerevisiae mutants affecting proteasome function, using a ubiquitin-independent substrate. Yeast. 2008;25:199–217. doi: 10.1002/yea.1579. [DOI] [PubMed] [Google Scholar]
- [71].Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J. 2004;23:500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Osmulski PA, Hochstrasser M, Gaczynska M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure. 2009;17:1137–1147. doi: 10.1016/j.str.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Savulescu AF, Glickman MH. Proteasome activator 200: the heat is on. Mol. Cell. Proteomics. 2011;10:R110.006890. doi: 10.1074/mcp.M110.006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- [75].Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi LB, Jr, White JM, Walker LM, Carnes K, Hess RA, Sleckman BP. Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Lopez AD, Tar K, Krugel U, Dange T, Ros IG, Schmidt M. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol. Biol. Cell. 2011;22:528–540. doi: 10.1091/mbc.E10-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell. 2010;37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Fehlker M, Wendler P, Lehmann A, Enenkel C. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 2003;4:959–963. doi: 10.1038/sj.embor.embor938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lehmann A, Jechow K, Enenkel C. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 2008;9:1237–1243. doi: 10.1038/embor.2008.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- [82].Gorbea C, Goellner GM, Teter K, Holmes RK, Rechsteiner M. Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J. Biol. Chem. 2004;279:54849–54861. doi: 10.1074/jbc.M410444200. [DOI] [PubMed] [Google Scholar]
- [83].Kajava AV, Gorbea C, Ortega J, Rechsteiner M, Steven AC. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J. Struct. Biol. 2004;146:425–430. doi: 10.1016/j.jsb.2004.01.013. [DOI] [PubMed] [Google Scholar]
- [84].Lehmann A, Niewienda A, Jechow K, Janek K, Enenkel C. Ecm29 fulfils quality control functions in proteasome assembly. Mol. Cell. 2010;38:879–888. doi: 10.1016/j.molcel.2010.06.016. [DOI] [PubMed] [Google Scholar]
- [85].Lee SY, De la Mota-Peynado A, Roelofs J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J. Biol. Chem. 2011;286:36641–36651. doi: 10.1074/jbc.M111.280875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Park S, Kim W, Tian G, Gygi SP, Finley D. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J. Biol. Chem. 2011;286:36652–36666. doi: 10.1074/jbc.M111.285924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci. Signal. 2010;3:ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wilkinson CR, Wallace M, Morphew M, Perry P, Allshire R, Javerzat JP, McIntosh JR, Gordon C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- [90].Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Amsterdam A, Pitzer F, Baumeister W. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc. Natl. Acad. Sci. U. S. A. 1993;90:99–103. doi: 10.1073/pnas.90.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tanaka K, Yoshimura T, Tamura T, Fujiwara T, Kumatori A, Ichihara A. Possible mechanism of nuclear translocation of proteasomes. FEBS Lett. 1990;271:41–46. doi: 10.1016/0014-5793(90)80367-r. [DOI] [PubMed] [Google Scholar]
- [93].Nederlof PM, Wang HR, Baumeister W. Nuclear localization signals of human and Thermoplasma proteasomal alpha subunits are functional in vitro. Proc. Natl. Acad. Sci. U. S. A. 1995;92:12060–12064. doi: 10.1073/pnas.92.26.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- [95].Lehmann A, Janek K, Braun B, Kloetzel PM, Enenkel C. 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J. Mol. Biol. 2002;317:401–413. doi: 10.1006/jmbi.2002.5443. [DOI] [PubMed] [Google Scholar]
- [96].Savulescu AF, Rotem A, Harel A. Proteasomes crossing the nuclear border. Nucleus. 2011;2 doi: 10.4161/nucl.2.4.16267. [DOI] [PubMed] [Google Scholar]
- [97].Laporte D, Salin B, Daignan-Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mannhaupt G, Schnall R, Karpov V, Vetter I, Feldmann H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999;450:27–34. doi: 10.1016/s0014-5793(99)00467-6. [DOI] [PubMed] [Google Scholar]
- [99].Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Xu H, Fu J, Ha SW, Ju D, Zheng J, Li L, Xie Y. The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochim. Biophys. Acta. 2012;1823:818–825. doi: 10.1016/j.bbamcr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- [101].Xie Y. Structure, assembly and homeostatic regulation of the 26S proteasome. J. Mol. Cell. Biol. 2010;2:308–317. doi: 10.1093/jmcb/mjq030. [DOI] [PubMed] [Google Scholar]
- [102].Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Ju D, Xie Y. Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and -independent. J. Biol. Chem. 2004;279:23851–23854. doi: 10.1074/jbc.C400111200. [DOI] [PubMed] [Google Scholar]
- [104].Ju D, Wang L, Mao X, Xie Y. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 2004;321:51–57. doi: 10.1016/j.bbrc.2004.06.105. [DOI] [PubMed] [Google Scholar]
- [105].Brown MG, Driscoll J, Monaco JJ. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991;353:355–357. doi: 10.1038/353355a0. [DOI] [PubMed] [Google Scholar]
- [106].Tanaka K. Role of proteasomes modified by interferon-gamma in antigen processing. J. Leukoc. Biol. 1994;56:571–575. doi: 10.1002/jlb.56.5.571. [DOI] [PubMed] [Google Scholar]
- [107].Kruger E, Kloetzel PM. Immunoproteasomes at the interface of innate and adaptive immune responses: two faces of one enzyme. Curr. Opin. Immunol. 2012;24:77–83. doi: 10.1016/j.coi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- [108].Groettrup M, Kirk CJ, Basler M. Proteasomes in immune cells: more than peptide producers? Nat. Rev. Immunol. 2010;10:73–78. doi: 10.1038/nri2687. [DOI] [PubMed] [Google Scholar]
- [109].Belote JM, Miller M, Smyth KA. Evolutionary conservation of a testes-specific proteasome subunit gene in Drosophila. Gene. 1998;215:93–100. doi: 10.1016/s0378-1119(98)00256-x. [DOI] [PubMed] [Google Scholar]
- [110].Zhong L, Belote JM. The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development. 2007;134:3517–3525. doi: 10.1242/dev.004770. [DOI] [PubMed] [Google Scholar]
- [111].Fu H, Doelling JH, Arendt CS, Hochstrasser M, Vierstra RD. Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics. 1998;149:677–692. doi: 10.1093/genetics/149.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J. Exp. Med. 1998;187:97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Heink S, Ludwig D, Kloetzel PM, Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].De M, Jayarapu K, Elenich L, Monaco JJ, Colbert RA, Griffin TA. Beta 2 subunit propeptides influence cooperative proteasome assembly. J. Biol. Chem. 2003;278:6153–6159. doi: 10.1074/jbc.M209292200. [DOI] [PubMed] [Google Scholar]
- [115].Kingsbury DJ, Griffin TA, Colbert RA. Novel propeptide function in 20 S proteasome assembly influences beta subunit composition. J. Biol. Chem. 2000;275:24156–24162. doi: 10.1074/jbc.M001742200. [DOI] [PubMed] [Google Scholar]
- [116].Joeris T, Schmidt N, Ermert D, Krienke P, Visekruna A, Kuckelkorn U, Kaufmann SH, Steinhoff U. The Proteasome System in Infection: Impact of beta5 and LMP7 on Composition, Maturation and Quantity of Active Proteasome Complexes. PLoS One. 2012;7:e39827. doi: 10.1371/journal.pone.0039827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, Groll M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- [118].Hill CP, Masters EI, Whitby FG. The 11S regulators of 20S proteasome activity. Curr. Top. Microbiol. Immunol. 2002;268:73–89. doi: 10.1007/978-3-642-59414-4_4. [DOI] [PubMed] [Google Scholar]
- [119].Preckel T, Fung-Leung WP, Cai Z, Vitiello A, Salter-Cid L, Winqvist O, Wolfe TG, Von Herrath M, Angulo A, Ghazal P, Lee JD, Fourie AM, Wu Y, Pang J, Ngo K, Peterson PA, Fruh K, Yang Y. Impaired immunoproteasome assembly and immune responses in PA28-/- mice. Science. 1999;286:2162–2165. doi: 10.1126/science.286.5447.2162. [DOI] [PubMed] [Google Scholar]
- [120].Murata S, Udono H, Tanahashi N, Hamada N, Watanabe K, Adachi K, Yamano T, Yui K, Kobayashi N, Kasahara M, Tanaka K, Chiba T. Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J. 2001;20:5898–5907. doi: 10.1093/emboj/20.21.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Macagno A, Kuehn L, de Giuli R, Groettrup M. Pronounced up-regulation of the PA28alpha/beta proteasome regulator but little increase in the steady-state content of immunoproteasome during dendritic cell maturation. Eur. J. Immunol. 2001;31:3271–3280. doi: 10.1002/1521-4141(200111)31:11<3271::aid-immu3271>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- [122].Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem. Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- [123].Deeds EJ, Bachman JA, Fontana W. Optimizing ring assembly reveals the strength of weak interactions. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2348–2353. doi: 10.1073/pnas.1113095109. [DOI] [PMC free article] [PubMed] [Google Scholar]