Abstract
APOBEC3B is one of seven human APOBEC3 DNA cytosine deaminases that function to inhibit the replication and persistence of retroelements and retroviruses. Human APOBEC3B restricts the replication of HIV-1 in HEK293 cells, while our laboratory clone of rhesus macaque APOBEC3B did not. We mapped the restriction determinant to a single amino acid difference that alters enzymatic activity. Human APOBEC3B D316 is catalytically active and capable of restricting HIV-1 while rhesus APOBEC3B N316 is not; swapping these residues alters the activity and restriction phenotypes respectively. Genotyping of primate center rhesus macaques revealed uniform homozygosity for aspartate at position 316. Considering the C-to-T nature of the underlying mutation, we suspect that our rhesus APOBEC3B cDNA was inactivated by its own gene product during subcloning in Escherichia coli. This region has been previously characterized for its role in substrate specificity, but these data indicate it also has a fundamental role in deaminase activity.
Keywords: APOBEC3B, DNA cytosine deamination, Human immunodeficiency virus-type 1, (HIV-1), Rhesus macaque, Restriction factors
Introduction
The human APOBEC3 proteins are a family of seven (APOBEC3A, B, C, D, F, G, and H) single-stranded DNA cytosine deaminases that function to protect the genome from parasitic genetic elements such as retroelements, retroviruses, and other forms of foreign DNA. APOBEC3A and APOBEC3B inhibit the replication and persistence of foreign DNA and endogenous retroelements, such as LINE-1 and Alu elements (Bogerd et al., 2006a, 2006b; Bulliard et al., 2011; Chen et al., 2006; Esnault et al., 2008; Muckenfuss et al., 2006; Stenglein et al., 2010; Stenglein and Harris, 2006; Wissing et al., 2011). APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H, on the other hand, inhibit the replication of retroviruses, such as HIV-1 (hereafter HIV), by incorporating into budding virions and catalyzing cytosine-to-uracil (C-to-U) mutations in the minus-strand DNA during reverse transcription (most recently (Hultquist et al., 2011; Refsland et al., 2012); reviewed by Harris et al. (2012), Malim and Emerman (2008)).
Consistent with a role in innate immune defense, the APOBEC3 locus has undergone sustained expansion during the evolutionary radiation of the mammalian lineage and most APOBEC3 proteins show evidence for positive selection (Conticello et al., 2005; Duggal et al., 2011; LaRue et al., 2009, 2008; Münk et al., 2012; Sawyer et al., 2004; Zhang and Webb, 2004). While mice have a single APOBEC3 gene, humans, chimpanzees, and rhesus macaques each have seven of similar domain architecture (Conticello et al., 2005; Hultquist et al., 2011; LaRue et al., 2009, 2008; Schmitt et al., 2011). Comparative analysis between different APOBEC3 family members and their homologs has thus been a powerful tool for mapping determinants of cellular localization, enzymatic activity, and protein-protein interactions (Albin et al., 2010; Bogerd et al., 2004; Dang et al., 2011; Lackey et al., 2012; Xu et al., 2004; Zhen et al., 2009). For example, comparative studies between the human and African Green monkey APOBEC3G proteins identified D128 as an important residue for interaction with the HIV Vif protein, an essential viral protein for neutralization of the APOBEC3 host defense (Bogerd et al., 2004; Mangeat et al., 2004; Schrofelbauer et al., 2004; Xu et al., 2004).
A recent study of the seven-member human and rhesus macaque APOBEC3 protein families reported that each rhesus APOBEC3 protein behaved similarly to its human homolog in HIV single cycle and spreading infection assays with the exception of APOBEC3B (Hultquist et al., 2011). While human APOBEC3B inhibited viral replication in HEK293T single cycle assays, rhesus APOBEC3B did not (Hultquist et al., 2011). Although this reflects prior reports indicating that human APOBEC3B is capable of restricting HIV (Bishop et al., 2004; Bogerd et al., 2007, 2006a, 2006b; Doehle et al., 2005; Hakata and Landau, 2006; Kinomoto et al., 2007; Rose et al., 2005; Yu et al., 2004), it contrasts with prior work demonstrating the capacity of rhesus APOBEC3B to restrict HIV in single cycle assays (Virgen and Hatziioannou, 2007).
To resolve this contradiction, we first used a series of comparative analyses to map the determinant for HIV restriction in human versus rhesus APOBEC3B. Taking advantage of the distinct antiviral phenotypes of our human and rhesus APOBEC3B laboratory clones, we constructed a series of chimeric proteins and single amino acid substitution mutants and tested them for antiviral and enzymatic activity using HIV single cycle and oligonucleotide cleavage deaminase assays, respectively. We found that a single amino acid in loop 7 of the C-terminal domain, D316, is essential for human APOBEC3B enzymatic and antiviral activity. Swapping this aspartic acid in human APOBEC3B for an asparagine, as in our rhesus APOBEC3B clone, completely ablates enzymatic activity and HIV restriction capacity. The corresponding N316D substitution, likewise, imparts rhesus APOBEC3B full enzymatic and antiviral activity. To determine if rhesus APOBEC3B naturally encodes an aspartic acid or asparagine at position 316, we genotyped a set of primate center rhesus macaques across this region. While these analyses revealed several nearby polymorphisms that may influence the activity and sequence specificity of rhesus APOBEC3B, all individuals were homozygous for D316. Thus, we infer our rhesus APOBEC3B D316N substitution arose during subcloning, quite possibly through the action of its own gene product. Thus, these results indicate that naturally occurring rhesus APOBEC3B (D316) is capable of HIV restriction in HEK293 cells and identify a previously unappreciated determinant for APOBEC3 intrinsic enzymatic and antiviral activity.
Results
APOBEC3B chimeras map the restriction determinant to the C-terminal domain
Clustal alignment of the human and rhesus macaque APOBEC3B protein sequences (derived from NM_004900 and NM_001246230, respectively) identified a total of 55 amino acid differences between the homologs. To first narrow in on the region responsible for the difference in restriction capacity, we created chimeric proteins between the human and rhesus APOBEC3B N-terminal and C-terminal domains. Human and rhesus APOBEC3B are each composed of two zinc-coordinating domains defined by a characteristic H-x-E-x25–31-C-x2−4-C motif (Conticello et al., 2005; Hultquist et al., 2011; Virgen and Hatziioannou, 2007). While the N-terminal domain of double-domain APOBEC3 proteins is thought to be important for nucleic acid binding, the C-terminal domain contains the active catalytic site (Haché et al., 2005; Hakata and Landau, 2006; Jónsson et al., 2006; Navarro et al., 2005; Stenglein and Harris, 2006). Human and rhesus APOBEC3B were then tested for HIV antiviral activity alongside these domain chimeras in a single-cycle infectivity assay. Increasing amounts of each protein were expressed alongside a constant amount of Vif-deficient HIV proviral plasmid. Virions produced from these cells were collected and used to infect a CEM-GFP reporter line to determine infectivity (Gervaix et al., 1997). Cell and viral-like particle (VLP) lysates were simultaneously collected and used for immunoblotting to detect protein expression and incorporation into viral particles. Supernatants from cells expressing only APOBEC3B (no virus) were concentrated in parallel as a negative control for virus-specific packaging and yielded no APOBEC3B by immunoblot (data not shown).
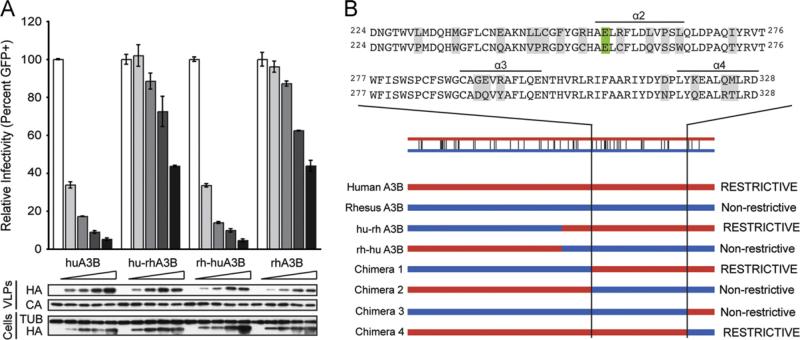
In the absence of APOBEC3B, viral infectivity is high (normalized to 100%, Fig. 1A). As an increasing amount of human APOBEC3B is expressed in the producer cells, more is packaged into the viral particles and infectivity rapidly decreases (Fig. 1A). Infectivity was not normalized to account for differences in virus release, so this decrease reflects the ability of APOBEC3B to elicit both foreign DNA and HIV restriction (Stenglein et al., 2010). The highest levels of human APOBEC3B expression reduced infectivity by greater than 90%, whereas even the highest levels of rhesus APOBEC3B expression failed to reduce infectivity below approximately 50% (Fig. 1A). The hu-rhAPOBEC3B (human N-terminal domain and rhesus C-terminal domain) chimera behaves pheno-typically similar to the rhesus protein, while the rh-huAPOBEC3B (rhesus N-terminal domain and human C-terminal domain) chimera phenocopies the human protein. Each protein was expressed and packaged into the viral particles at roughly equivalent levels (Fig. 1A). These results indicate that the restriction determinant tracks with the C-terminal domain of human APOBEC3B.
Fig. 1.
HIV-1 restriction potential maps to the human APOBEC3B C-terminal domain. (A) HIV-1 infectivity in the presence of increasing amounts of human APOBEC3B, rhesus APOBEC3B, or the human-rhesus/rhesus-human chimeras. A constant amount of Vif-deficient A200C HIV-1IIIB molecular clone was co-transfected into HEK293T cells with an increasing gradient of the indicated APOBEC3B vector. Percent infectivity of HIV-1IIIB is measured by infection of CEM-GFP in duplicate and flow cytometry, reported as the mean of the two technical replicates ± standard deviation. Representative immunoblots of HA-tagged APOBEC3B proteins (HA) in cell lysates and HIV-1 particles produced by those cells are shown with their respective tubulin (TUB) and p24 (CA) loading controls. (B) Schematic of human/rhesus APOBEC3B C-terminal domain chimeras and their HIV-1 restriction potential. HIV-1 single cycle assays were performed with each chimera as in (A) and found to be restrictive or non-restrictive depending on their similarity to the human or rhesus APOBEC3B parent, respectively. An alignment of human (top) and rhesus (bottom) APOBEC3B amino acid sequences to which the restriction determinant mapped is shown above. Differences between the human and rhesus sequences are highlighted in gray and the catalytic glutamate is highlighted in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further define the region of interest, we created chimeric proteins within the APOBEC3B C-terminal domain and tested them for antiviral activity. Chimeras that restricted infectivity of Vif-deficient HIV at a level similar to human APOBEC3B were deemed restrictive, while chimeras with restriction potential similar to rhesus APOBEC3B were considered non-restrictive (Fig. 1B). Chimera 1, containing rhesus amino acid residues 1–223 and human residues 224–382, remained restrictive while chimera 3, containing rhesus 1–333 and human 334–382, lacked restriction capacity (Fig. 1B). The reciprocal chimeras had opposite antiviral phenotypes (Fig. 1B). These results implicate human APOBEC3B residues 224–333 as important for restriction. Based on the homologous APOBEC3G C-terminal domain crystal structure (PDB: 3IR2, (Shandilya et al., 2010)), this region includes three predicted surface helices, alpha 2 through alpha 4.
Human APOBEC3B D316 is an important determinant of antiviral activity
Human and rhesus APOBEC3B differ in 20 amino acids between residues 224 and 333. Most of these differences cluster either near the active site or along the alpha 2, 3, and 4 helices (differences highlighted in gray, catalytic glutamate in green, Fig. 1B). We tested many of these amino acid differences for their impact on antiviral activity by changing the residues in human APOBEC3B to the corresponding residue in rhesus APOBEC3B. Many of these single or clustered amino acid changes (L230P, E241Q, L245V/L246P/C247R/F249D, R252C, R257C, R252C/R257C, L261Q, P263S, I272T, M325T, C354R) had no impact on antiviral activity or packaging in single-cycle assays (data not shown).
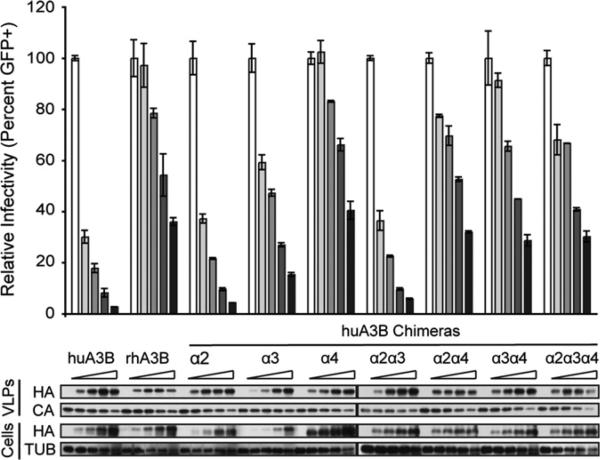
Many of the amino acid differences occur in clusters along alpha helices 2, 3, and 4 of the C-terminal domain. To see if the helices held the responsible determinant, we targeted clustered changes in the alpha helix 2 (R257C/L261Q/P263S/L265W), alpha helix 3 (G291D/E292Q/R294Y), and alpha helix 4 (D316N/K320Q/Q324R/M325T) regions. While human APOBEC3B, the human APOBEC3B alpha 2 chimera, and the human ABOPEC3B alpha 3 chimera all exhibited strong antiviral activity, the human APOBEC3B alpha 4 chimera showed a greatly diminished activity similar to that of rhesus APOBEC3B (Fig. 2). All human APOBEC3B constructs in which the alpha 4 helix region was altered to match the rhesus residues exhibited the same diminished activity, demonstrating a dominant effect of the alpha 4 substitutions on restriction activity (Fig. 2). The human APOBEC3B alpha 3 substitutions alone also yielded slightly reduced HIV restriction capacity, but this is likely due to lower expression levels of this protein. This reduced activity is not reproduced in the alpha2/alpha3 construct (Fig. 2). All of the above APOBEC3B helix chimeras incorporate into viral particles at roughly equal efficiencies.
Fig. 2.
HIV-1 restriction potential tracks with the putative alpha 4 helix region of human APOBEC3B. HIV-1 infectivity in the presence of increasing amounts of human APOBEC3B, rhesus APOBEC3B, or various human-rhesus alpha helix chimeras in the C-terminal domain. A constant amount of Vif-deficient A200C HIV-1IIIB molecular clone was co-transfected into HEK293T cells with an increasing gradient of the indicated APOBEC3B vector. Percent infectivity of HIV-1IIIB is measured by infection of CEM-GFP in duplicate and flow cytometry, reported as the mean of the two technical replicates±standard deviation. Representative immunoblots of HA-tagged APOBEC3B proteins (HA) in cell lysates and HIV-1 particles produced by those cells are shown with their respective tubulin (TUB) and p24 (CA) loading controls.
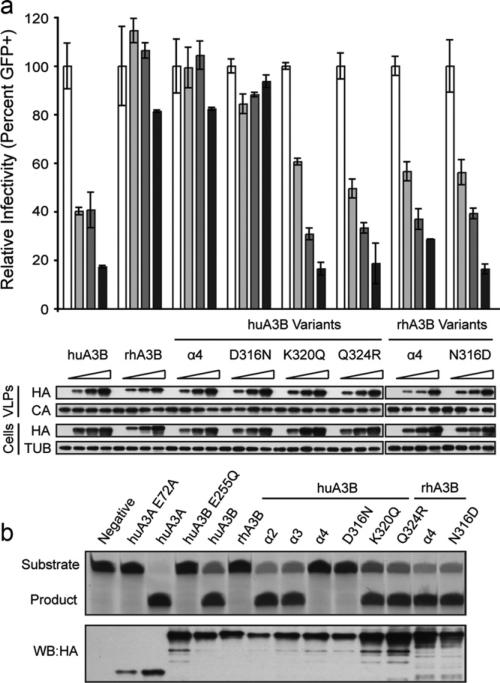
To define the specific amino acid change(s) responsible for the loss of restriction capacity in the human APOBEC3B alpha 4 chimera construct, we introduced each substitution singly (D316N, K320Q, or Q324R). Human APOBEC3B M325T was found to have no effect in our initial site-directed mutagenesis screen and so was not included here. Human APOBEC3B and the K320Q and Q324R constructs strongly restricted HIV replication, while human APOBEC3B D316N exhibited a dramatic loss of antiviral activity (Fig. 3A). All proteins were expressed and incorporated into viral particles equivalently. The corresponding rhesus-to-human amino acid substitutions were created in rhesus APOBEC3B to confirm the effect of residue 316. Rhesus APOBEC3B lacks strong antiviral activity, but substituting in the human alpha 4 region (N316D/Q320K/R324Q/T325M) or the N316D substitution alone greatly enhanced antiviral activity (Fig. 3A). Thus, an aspartic acid at residue 316 is necessary for antiviral activity in human and rhesus APOBEC3B.
Fig. 3.
An aspartic acid at APOBEC3B residue 316 is crucial for HIV-1 restriction and deaminase activity. (A) HIV-1 infectivity in the presence of increasing amounts of human APOBEC3B, rhesus APOBEC3B, or various helix or single amino acid substitution constructs. A constant amount of Vif-deficient A200C HIV-1IIIB molecular clone was co-transfected into HEK293T cells with an increasing gradient of the indicated APOBEC3B vector. Percent infectivity of HIV-1IIIB is measured by infection of CEM-GFP in duplicate and flow cytometry, reported as the mean of the two technical replicates ± standard deviation. Representative immunoblots of HA-tagged APOBEC3B proteins (HA) in cell lysates and HIV-1 particles produced by those cells are shown with their respective tubulin (TUB) and p24 (CA) loading controls. (B) Deaminase activity of human APOBEC3B, rhesus APOBEC3B, or various helix or single amino acid substitution constructs in cell lysates as measured by an oligo cleavage assay. A constant amount of each indicated APOBEC3 expression construct was transfected into HEK293T cells and lysates collected for oligo cleavage (top) or immunoblotting (bottom). Deaminase activity is indicated by cleavage of the oligo substrate to a smaller product. Expression of each deaminase was verified by immunoblotting for each of the HA-tagged APOBEC3 proteins as shown below.
D316 is required for the deaminase activity of APOBEC3B
To determine if the D316N substitution impacts APOBEC3B deaminase activity, we conducted an oligonucleotide cleavage assay in which a fluorescently labeled oligonucleotide containing the preferred 5′-TC-3′ APOBEC3B deamination target sequence is incubated with whole cell lysate containing the indicated APOBEC3 protein (Burns et al., 2013). After APOBEC3-mediated cytosine deamination, uracil is removed with uracil DNA glycosylase, and the phosphodiester backbone is cleaved at the abasic site by treatment with sodium hydroxide. Human APOBEC3A is a potent DNA deaminase with similar sequence specificity to APOBEC3B, and results in full deamination of the substrate and complete cleavage to product (Fig. 3B and (Carpenter et al., 2012; Love et al., 2012; Stenglein et al., 2010; Thielen et al., 2010; Wijesinghe and Bhagwat, 2012)). Human APOBEC3B likewise resulted in nearly full oligo cleavage. HEK293T lysate alone, the APOBEC3A E72A catalytic mutant, and the APOBEC3B E255Q catalytic mutant exhibited no deaminase activity. All human APOBEC3B constructs with an aspartic acid at position 316, including the alpha 2 chimera, alpha 3 chimera, K320Q, and Q324R, were all able to efficiently deaminate the substrate, resulting in near complete cleavage (Fig. 3B). However, all human APOBEC3B constructs with an asparagine residue at position 316, including the alpha 4 chimera and D316N, were inactive, comparable to the catalytic mutant. All rhesus APOBEC3B constructs were likewise inactive with the exception of N316D, which elicited strong deaminase activity (Fig. 3B).
To control for the possibility that one or more of these substitutions could have changed the substrate specificity of human or rhesus APOBEC3B, this assay was repeated using a 5′-TCC-3′ fluorescently labeled oligonucleotide (data not shown). As before, all APOBEC3B constructs with an aspartic acid at position 316, including human APOBEC3B and rhesus APOBEC3B N316D, were all able to efficiently deaminate the substrate. Conversely, all APOBEC3B constructs with an asparagine residue at position 316, including rhesus APOBEC3B and human APOBEC3B D316N, were inactive. These results demonstrate that an aspartic acid at position 316 is an important determinant for the intrinsic DNA deaminase activity of APOBEC3B.
Genotyping of rhesus APOBEC3B exon 6 reveals several polymorphic residues
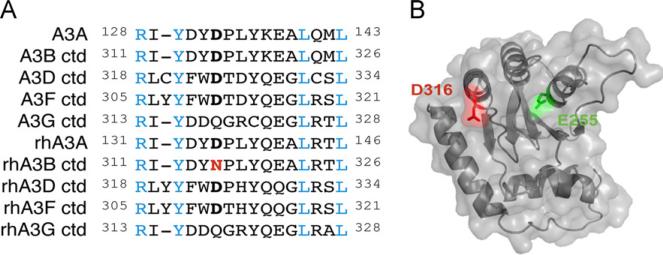
Phylogenetic comparisons of several catalytic human and rhesus APOBEC3 deaminase domains indicate conservation of an aspartic acid residue at position 316 with the exception of the APOBEC3G homologs and, notably, our rhesus APOBEC3B clone (Fig. 4A). Structural modeling of the human APOBEC3B C-terminal domain positions residue 316 in close proximity to the catalytic glutamate, suggesting it may be important for substrate binding or local active site architecture (Fig. 4B). In contrast to the rhesus APOBEC3B mRNA sequence in Genbank (NM_001246230), the APOBEC3B gene in the rhesus genome assembly indicates an aspartic acid at residue 316 as does at least one prior report of APOBEC3B cDNA from rhesus macaque PBMCs (assembly AANU00000000.1 and (Virgen and Hatziioannou, 2007)). Taken together with the fact that APOBEC3 proteins are under positive selection and therefore have extensive variation within species, it is possible that this residue is polymorphic within the natural population. However, it is also possible that the D316N substitution emerged during our initial subcloning of rhesus APOBEC3B. Mutation of a GAC (aspartic acid) to an AAC (asparagine) codon could be catalyzed by APOBEC3B expression in E. coli, resulting in a less toxic and therefore self-selecting mutant.
Fig. 4.
Conservation and predicted structural context of APOBEC3B D316. (A) Amino acid sequence alignment of the predicted loop 7 regions of human and rhesus APOBEC3B and their related family members. Blue residues indicate perfect conservation. APOBEC3B residue 316 and its predicted homologs are bolded in black (aspartic acid) or red (asparagine). (B) Predicted structure of human APOBEC3B C-terminal domain based on the human APOBEC3G C-terminal domain crystal structure (PDB: 3IR2). D316 is highlighted in red in close proximity to the catalytic E255 in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
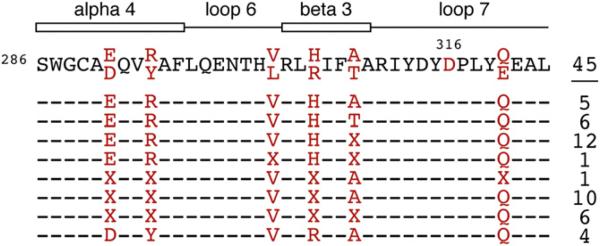
To determine if rhesus APOBEC3B is naturally polymorphic at position 316, we sequenced a portion of exon 6 in genomic DNA isolated from 45 independent rhesus macaques. While we found extensive variation in this region and identified six polymorphic residues, all individuals were homozygous for alleles encoding an aspartic acid at position 316 (Fig. 5). It should be noted that all genomic DNA samples were collected from primates in captivity, and that these polymorphism frequencies may not be representative of frequencies in the wild. Nevertheless, we consider it unlikely that N316 is a naturally occurring variant and, therefore, believe it to be the result of a mutation that arose during subcloning in E. coli.
Fig. 5.
Naturally occurring APOBEC3B variants in rhesus macaques. An amino acid alignment displaying a portion of rhesus APOBEC3B near residue 316 and polymorphisms observed among 45 individual rhesus macaques. A dash ‘–’ indicates that all animals surveyed were homozygous for alleles encoding the residue in the above consensus. At positions in red, some individuals were homozygous for alleles encoding alternate amino acids, as indicated. An ‘X’ indicates that the individual is heterozygous for alleles encoding the two possible amino acids observed at this site. The numbers of individuals found to have each of the indicated genotypes are indicated to the right. A schematic of the predicted structural motifs based on the human APOBEC3G C-terminal domain crystal structure is depicted above (PDB: 3IR2). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
We used a series of comparative analyses to map the determinant for HIV restriction present in human APOBEC3B, but not in our laboratory clone of rhesus macaque APOBEC3B. In testing the antiviral activity of APOBEC3B chimeras and single amino acid substitution mutants, we identified a single residue, D316, necessary for the HIV restriction capacity of APOBEC3B. Swapping this residue for the corresponding N316 of our rhesus APOBEC3B clone rendered human APOBEC3B inactive in both single cycle and oligonucleotide cleavage assays. Conversely, rhesus APOBEC3B N316D displayed full catalytic activity and HIV restriction capacity. Genotyping of primate center rhesus macaques yielded uniform homozygosity for an aspartic acid at position 316, making it likely that our N316 mutant arose during subcloning ex vivo. While the presence of N316 could not be verified as naturally occurring variant in rhesus APOBEC3B, these results implicate residue 316 as a key determinant of enzymatic and antiviral activity. Furthermore, the existence of multiple polymorphisms in this region may indicate naturally circulating rhesus APOBEC3B variants with altered activities or substrate specificities.
Residue 316 is predicted to be in the loop 7 region of the APOBEC3B C-terminal domain between beta-strand 4 and alpha-helix 4 (Fig. 4B). This region has been the focus of several other studies seeking to map determinants in the APOBEC3 proteins. A comparison of human APOBEC3D and APOBEC3F revealed a key determinant for HIV restriction in the same loop 7 region of the C-terminal domain of APOBEC3F. When cysteine 320 of APOBEC3D was swapped for the corresponding tyrosine residue of APOBEC3F, antiviral activity of APOBEC3D was enhanced several fold (Dang et al., 2011). This same loop region has also been studied for its role in substrate specificity and DNA binding (Carpenter et al., 2010; Kohli et al., 2009; Rausch et al., 2009; Wang et al., 2010). Chimeric AID/APOBEC3G proteins in which loop 7 of APOBEC3G and the corresponding region of AID were swapped, mirrored the substrate specificity of the protein from which the loop region originated (Carpenter et al., 2010; Kohli et al., 2009; Wang et al., 2010). Furthermore, a screen of potential APOBEC3G inhibitors identified two classes of small molecules that inhibit deaminase activity by covalently binding APOBEC3G C321, which is also located within loop 7 (Li et al., 2011; Olson et al., 2013). These findings, combined with this study, highlight the broad importance of this loop region for both APOBEC3 substrate specificity and deaminase activity. One possibility is that aspartic acid 316 is required for substrate binding as it lies directly adjacent to the predicted nucleic acid binding cleft (Fig. 4B).
Phylogenetic comparison of several catalytic human and rhesus APOBEC3 deaminase domains indicate conservation of an aspartic acid residue at position 316 (Fig. 4A and Fig. 5). Our attempts failed to verify the presence of N316 as a naturally occurring variant within the rhesus population. While these results do not rule out the possibility that residue 316 is polymorphic, we believe that the D316N substitution emerged during our initial cloning as such a mutant would undoubtedly be less toxic to E. coli. This and other experiences in our laboratory have since justified the inclusion of an intron in all APOBEC3A and APOBEC3B expression constructs to ensure safe propagation in bacteria. In independent work several years ago, a similar human APOBEC3B mutant was identified during attempts to clone the human APOBEC3B cDNA (W228L and D316N (Stenglein and Harris, 2006)). We must update our prior findings, therefore, to report that naturally occurring rhesus APOBEC3B D316 can restrict HIV-1 in HEK293T cells.
While N316 was not verified as a naturally occurring polymorphism in rhesus APOBEC3B, our results indicated the occurrence of six other polymorphic residues nearby (Fig. 5). As all these residues lie in a region shown to be important for enzyme activity and substrate specificity, it is possible that there are naturally occurring variants that differ in one or both of these regards. The existence of naturally occurring variations that inactivate rhesus APOBEC3B would parallel the documented deletion of APOBEC3B in certain human populations (Kidd et al., 2007). Such a scenario would strongly implicate a high cost-to-benefit ratio for the maintenance of APOBEC3B. The strong DNA deamination activity and nuclear localization of APOBEC3B may combine to present a significant risk to genomic integrity (e.g. (Burns et al., 2013)).
While the possible risks of maintaining an active APOBEC3B allele are clear, the physiologic role of APOBEC3B remains ambiguous. While several studies have demonstrated the capacity for APOBEC3B to restrict HIV in HEK293T based single cycle assays (Bishop et al., 2004; Bogerd et al., 2007, 2006a, 2006b; Doehle et al., 2005; Hakata and Landau, 2006; Kinomoto et al., 2007; Rose et al., 2005; Virgen and Hatziioannou, 2007; Yu et al., 2004), neither human nor rhesus APOBEC3B are appreciably expressed in human or rhesus CD4+ T lymphocytes and neither are capable of packaging into or inhibiting the replication of HIV when expressed stably in T cells (Haché et al., 2008; Hultquist et al., 2011; Refsland et al., 2010; Schmitt et al., 2011). Furthermore, human APOBEC3B is not targeted for degradation by HIV Vif (Bishop et al., 2004; Doehle et al., 2005; Hultquist et al., 2011; Rose et al., 2005; Yu et al., 2004), though rhesus APOBEC3B is partially sensitive to degradation by SIVmac239 and SIVagm Vif (Hultquist et al., 2011; Virgen and Hatziioannou, 2007). Therefore, the role of APOBEC3B in retrovirus restriction remains unclear. Alternate roles in retroelement restriction and foreign DNA restriction have also been proposed (Bogerd et al., 2006a, 2006b; Chen et al., 2006; Esnault et al., 2008; Muckenfuss et al., 2006; Stenglein et al., 2010; Stenglein and Harris, 2006; Wissing et al., 2011). Regardless, the benefit may not always outweigh the cost of encoding such a potent DNA mutator, a result that may be reflected in the selective loss of APOBEC3B function in certain primate subpopulations.
In conclusion, we have characterized the restriction determinant that accounts for the distinct antiviral phenotypes previously observed for human and rhesus APOBEC3B (Hultquist et al., 2011). While rhesus APOBEC3B N316 may not be a naturally occurring variant, the data presented here provide new information regarding DNA deaminase domain active site architecture and local amino acid requirements. Extension of these findings to the other APOBEC3 family members will likely aid in the understanding of the general APOBEC3 deaminase and HIV restriction mechanism (Haché et al., 2005; Miyagi et al., 2007; Navarro et al., 2005; Schumacher et al., 2008). Furthermore, identification of several polymorphisms in a region of rhesus APOBEC3B known to play a role in deaminase activity and substrate specificity suggest the existence of naturally occurring functional variants that may shed further light on the physiologic roles and potential pathological risks of APOBEC3B.
Materials and methods
Expression constructs
The Vif-deficient (X26×27) HIV-1IIIB A200C proviral expression construct has been reported (Haché et al., 2008). The carboxy(C)-terminally 3xHA-tagged, human APOBEC3A, intron-containing human APOBEC3B, and intron-containing rhesus macaque APOBEC3A expression constructs have also been reported (Hultquist et al., 2011). The derivative human APOBEC3A E72A and human APOBEC3B E255Q catalytic mutants were generated via site-directed mutagenesis using primers 5′-GGC CGC CAT GCG GCG CTG CGC TTC TTG-3′/5′-CAA GAA GCG CAG CGC CGC ATG GCG GCC-3′ and 5′-GCC GCC ATG CGC AGC TGC GCT TC-3′/5′-GAA GCG CAG CTG CGC ATG GCG GC-3′, respectively.
To match the intron placement in the human APOBEC3B expression vector, a β-globin intron was inserted into the rhesus macaque APOBEC3B coding region between exons 5 and 6. The 5′ end and 3′ ends of rhesus APOBEC3B were amplified from a cDNA construct previously provided by T. Hatziioannou using primer pairs 5′-NNN NGG TAC CAC CAT GAA TCC ACA GAT CAG-3′/5′-ATC TCC TGG ACT CAC CTG GTT GCA TAG AAA-3′ and 5′-CTT TCA TCT CAA CAG GCT AAG AAT GTT CCC-3′/5′-NNN NGC GGC CGC CCG TTT CCC TGA TTC TG-3′, respectively. The β-globin intron was amplified using primers 5′-GTG AGT CCA GGA GAT GTT TCA GCA CTG TTG CC-3′ and 5′-CTG TTG AGA TGA AAG GAG ACA ATA AAG ATG AC-3′. The fragments were gel purified (GeneJET gel extraction kit; Fermentas) and connected by overlapping PCR. The full-length, intron-containing segment was PCR amplified, gel purified, digested with KpnI/NotI, and ligated into the similarly digested pcDNA3.1(+) vector with a C-terminal 3xHA tag.
The human-rhesus APOBEC3B domain chimeras were generated by digestion of the human and rhesus APOBEC3B expression constructs described above with BamHI, which cuts once in the intron and once in the 3′ flanking region within the pcDNA3.1(+) vector sequence. The fragment containing part of the intron and exons 6 through 8 of human APOBEC3B was ligated into the similarly digested rhesus APOBEC3B expression construct and vice versa. The resultant chimeric expression constructs were sequence verified.
The other human-rhesus APOBEC3B chimeras were generated by overlapping PCR between fragments generated from the human and rhesus expression constructs. For chimeric proteins 1 and 2 (human APOBEC3B residues 1–223 with rhesus APOBEC3B residues 224–382 and vice versa) the N-terminal fragments of human and rhesus APOBEC3B were PCR amplified using primers 5′-NNN NAA GCT TGG TAC CAC CAT GAA TCC ACA GAT CAG-3′/5′-GAC CCA GGT GCC ATT GTC-3′ and the C-terminal fragments were amplified using primers 5′-GAC AAT GGC ACC TGG GTC-3′/5′-GTC GAC GTT TCC CTG ATT CTG GAG AAT GGC CC-3′. For chimeric proteins 3 and 4 (human APOBEC3B residues 1–333 with rhesus APOBEC3B residues 334–382 and vice versa) the amino(N)-terminal fragments of human and rhesus APOBEC3B were PCR amplified using primers 5′-NNN NAA GCT TGG TAC CAC CAT GAA TCC ACA GAT CAG-3′/5′-AGA CTT GGG CCC CAG CAT C-3′ and the C-terminal fragments were amplified using primers 5′-GAT GCT GGG GCC CAA GTC T-3′/5′-GTC GAC GTT TCC CTG ATT CTG GAG AAT GGC CC-3′. Reciprocal fragments were stitched together into full-length, intron-containing chimeric complementary (c)DNAs by overlapping PCR. These were gel purified, digested with KpnI/NotI, and ligated into a similarly digested pcDNA3.1(+) vector with a C-terminal 3xHA tag.
Additional APOBEC3B amino acid substitution constructs were created using site-directed mutagenesis on the human or rhesus APOBEC3B expression plasmids. See Table 1 for a full list of constructs and deoxyoligonucleotide primer sequences used in this study.
Table 1.
Deoxyoligonucleotides used in this study for mutagenesis.
| Construct | Primers |
|---|---|
| hsAPOBEC3B L230P | 5′-ggcacctgggtcccgatggaccagcac-3′ |
| 5′-gtgctggtccatcgggacccaggtgcc-3′ | |
| hsAPOBEC3B E241Q | 5′-ggctttctatgcaaccaggtgagtccaggag-3′ |
| 5′-ctcctggactcacctggttgcatagaaagcc-3′ | |
| hsAPOBEC3B L245V/L246P/C247R/F249D | 5′-cctttcatctcaacaggctaagaatgttccccgtggcgattacggccgccatg-3′ |
| 5′-catggcggccgtaatcgccacggggaacattcttagcctgttgagatgaaag-3′ | |
| hsAPOBEC3B R252C | 5′-gtggcttttacggctgccatgcggagctg-3′ |
| 5′-cagctccgcatggcagccgtaaaagccac-3′ | |
| hsAPOBEC3B R257C | 5′-gccatgcggagctgtgcttcttggacctg-3′ |
| 5′-caggtccaagaagcacagctccgcatggc-3′ | |
| hsAPOBEC3B R252C/R257C | 5′-gtggcttttacggctgccatgcggagctg-3′ |
| 5′-cagctccgcatggcagccgtaaaagccac-3′ | |
| 5′-gccatgcggagctgtgcttcttggacctg-3′ | |
| 5′-caggtccaagaagcacagctccgcatggc-3′ | |
| hsAPOBEC3B L261Q | 5′-ctgcgcttcttggaccaggttccttctttgcag-3′ |
| 5′-ctgcaaagaaggaacctggtccaagaagcgcag-3′ | |
| hsAPOBEC3B P263S | 5′-cgcttcttggacctggttagttctttgcagttggacc-3′ |
| 5′-ggtccaactgcaaagaactaaccaggtccaagaagcg-3′ | |
| hsAPOBEC3B I272T | 5′-gacccggcccagacctacagggtcact-3′ |
| 5′-agtgaccctgtaggtctgggccgggtc-3′ | |
| hsAPOBEC3B M325T | 5′-atataaggaggcgctgcaaacgctgcgggatg-3′ |
| 5′-catcccgcagcgtttgcagcgcctccttatat-3′ | |
| hsAPOBEC3B C354R | 5′-gtaccgccagggacgtcccttccagcc-3′ |
| 5′-ggctggaagggacgtccctggcggtac-3′ | |
| hsAPOBEC3B R257C/L261Q/P263S/L265W | 5′-tgcgcttcttggaccaggttagttcttggcagttggacccggc-3′ |
| 5′-gccgggtccaactgccaagaactaacctggtccaagaagcgca-3′ | |
| 5′-gccatgcggagctgtgcttcttggaccag-3′ | |
| 5′-ctggtccaagaagcacagctccgcatggc-3′ | |
| hsAPOBEC3B G291D/E292Q/R294Y | 5′-gcttctcctggggctgtgccgatcaggtgtatgcgttccttcaggagaacac-3′ |
| 5′-gtgttctcctgaaggaacgcatacacctgatcggcacagccccaggagaagc-3′ | |
| hsAPOBEC3B D316N/K320Q/Q324R/M325T | 5′-gcccgcatctatgattacaaccccctatatcaggaggcgct-3′ |
| 5′-gcatttgcagcgcctcctgatatagggggttgtaatcatagatgcgggc-3′ | |
| 5′-atatcaggaggcgctgagaacgctgcgggatgctggg-3′ | |
| 5′-cccagcatcccgcagcgttctcagcgcctcctgatat-3′ | |
| hsAPOBEC3B D316N | 5′-gctgcccgcatctatgattacaaccccctatataag-3′ |
| 5′-cttatatagggggttgtaatcatagatgcgggcagc-3′ | |
| hsAPOBEC3B K320Q | 5′-tctatgattacgaccccctatatcaggaggcgctgc-3′ |
| 5′-gcagcgcctcctgatatagggggtcgtaatcataga-3′ | |
| hsAPOBEC3B Q324R | 5′-ctatataaggaggcgctgagaatgctgcgggatgctgg-3′ |
| 5′-ccagcatcccgcagcattctcagcgcctccttatatag-3′ | |
| rhAPOBEC3B N316D/Q320K/R324Q/T325M | 5′-ctgcccgcatctatgattacgaccccctgtatcag-3′ |
| 5′-ctgatacagggggtcgtaatcatagatgcgggcag-3′ | |
| 5′-tctatgattacgaccccctgtataaggaggcactgc-3′ | |
| 5′-gcagtgcctccttatacagggggtcgtaatcataga-3′ | |
| 5′-ataaggaggcactgcaaatgctgcgggatgctgg-3′ | |
| 5′-ccagcatcccgcagcatttgcagtgcctccttat-3′ | |
| rhAPOBEC3B N316D | 5′-ctgcccgcatctatgattacgaccccctgtatcag-3′ |
| 5′-ctgatacagggggtcgtaatcatagatgcgggcag-3′ | |
| rhAPOBEC3B Q320K | 5′-tctatgattacaaccccctgtataaggaggcactgc-3′ |
| 5′-gcagtgcctccttatacagggggttgtaatcataga-3′ | |
| rhAPOBEC3B R324Q | 5′-caggaggcactgcaaacgctgcgggat-3′ |
| 5′-atcccgcagcgtttgcagtgcctcctg-3′ |
Cell lines
Human embryonic kidney (HEK)293T cells were maintained in Dulbecco's modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 0.5% penicillin/streptomycin (P/S). CEM-GFP cells, obtained from the AIDS Research and Reference Reagent Program, were maintained in Roswell Park Memorial Institute (RPMI) media containing 10% FBS and 0.5% P/S.
HIV single cycle assay with replication proficient virus
At 50% confluency, HEK293T cells were transfected (TransIt, Mirus) with 1 μg Vif-deficient (X26×27) HIV-1IIIB A200C proviral expression construct and either 25, 50, 100, 200, or 400 ng of the appropriate APOBEC3B-3xHA expression construct. After 48 h, the virus-containing supernatants were harvested, filter purified, and used to infect the CEM-GFP reporter cell line to quantify infectivity. Cell and viral particle lysates were collected for immunoblotting.
Immunoblotting
HEK293T cells were pelleted by centrifugation at 8000 rpm for 2 min, washed in 1X PBS, and lysed directly in 2.5× Laemmli Sample Buffer (25 mM Tris pH 6.8, 8% glycerol, 0.8% SDS, 2% 2-mercaptoethanol, 0.02% bromophenol blue). Virus particles from the culture supernatants were purified using 0.45 μm PVDF filters (Millipore), pelleted by centrifugation at 13,000 rpm for 2 h through a 20% sucrose 1X PBS buffer, and lysed directly in 2.5× Laemmli Sample Buffer. All samples were homogenized at 98C for 30 min prior to loading. Samples were run on 12.5% Tris–HCl SDS-PAGE resolving gels with 4% stacking gels both with a 37.5 acrylamide:1 bis-acrylamide ratio (BioRad Criterion) at 90 V for 30 min followed by 150 V for 1 h. Proteins were transferred to PVDF membranes by methanol-based electrotransfer (BioRad Criterion Blotter) at 90 V for 2 h. Membranes were blocked in 4% Milk in 1X PBS with 0.1% Tween-20 overnight. Proteins were detected using primary antibodies against HA to detect HA-tagged APOBEC3B (HA.11; Covance), TUB (tubulin; Covance), or p24 (NIH ARRRP 3537 courtesy of B. Chesebro and K. Wehrly). Anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson ImmunoResearch) was detected using Hyglo HRP detection reagents (Denville Scientific). Blots were incubated in stripping buffer (1xPBS, 0.2M glycine, 1.0% SDS, 1.0% Tween-20, pH 2.2) before reprobing.
Flow cytometry
At 48 h post infection, CEM-GFP cells were fixed in 4% paraformaldehyde in 1× PBS. GFP fluorescence was measured using a Becton Dickinson FACS Canto II flow cytometer. HIV infectivity was quantified by calculating the percent of live cells that were positive for GFP signal using FlowJo Flow Cytometry Analysis Software (Version 8.8.6). Data was normalized to the 0 ng APOBEC3B control as 100% infectivity.
Oligonucleotide cleavage assay
At 50% confluency, HEK293T cells were transfected (TransIt, Mirus) with 100 ng of the indicated APOBEC3 expression construct. After 48 h, the cells were washed and split into two samples, one half for immunoblotting and the other for an oligonucleotide cleavage assay. Transfected cells were resuspended in 1X HED buffer (25 mM HEPES, 5 mM EDTA, 10% Glycerol, 1mM DTT, proteasome inhibitor, pH 7.8) and homogenized with three short (5 s) pulses of sonication (Misonix XL-2000). The deamination reaction was prepared by mixing 16.5 μL cleared lysate, 0.25 μL RNaseA (20 Units, QIAgen), 1 μL fluorescein-labelled oligonucleotide (4 pmol, 5′-ATT ATT ATT ATT CTA ATG GAT TTA TTT ATT TAT TTA TTT ATT T-fluorescein-3′), 2 μL 10× UDG buffer, and 0.25 μL UDG (1.25 Units). After 2 h incubation at 37 °C, 2 μL of 1.0 M sodium hydroxide were added to each reaction to induce cleavage at abasic sites. Samples were mixed with 2X Formamide Buffer (80% Formamide, 1× TBE, 0.05% Bromophenol blue, 0.01% Xylene cyanol) and run on a 15% Urea-TBE gel. Fluorescein tagged oligonucleotide products were detected using a Fugi FLA-5000 scanner.
Sequencing of rhesus APOBEC3B exon 6
Blood samples or transformed B cell lines were obtained from 45 rhesus macaque (Macaca mulatta) individuals housed at either the Keeling Center for Comparative Medicine and Research (42/45) or at the New England Primate Research Center (3/45). B cell lines were expanded in RPMI, 20% FBS, P/S, l-glutamine, HEPES, and AZT. Genomic DNA from blood and B cells was isolated using the All Prep DNA/RNA Mini Kit (Qiagen #80204). A 506 base-pair fragment was amplified by PCR using Supermix High Fidelity Polymerase (Invitrogen #10790-020), forward primer 5′-GTT TCC TTT TCT AGG CTA AGA ATG TTC TCC-3′ and reverse primer 5′-CAC TTC CAG CCA CCC TCC-3′. PCR products were sequenced directly using the forward primer. Chromatogram analysis was carried out using Sequencher software (Gene Codes). Work with primate materials was conducted at UT Austin under an approved biosafety protocol (#2008-03-0049).
Acknowledgments
We thank Drs. Greg Wilkerson and Welkin Johnson for providing rhesus macaque samples and the NIH AIDS Research and Reference Reagent Program for materials. This research was funded by NIH R01 AI064046 and P01 GM091743 to RSH and NIH R01 GM093086 to SLS. JFH was supported in part by a UMN Graduate Student Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Albin JS, LaRue RS, Weaver JA, Brown WL, Shindo K, Harjes E, Matsuo H, Harris RS. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J. Biol. Chem. 2010;285(52):40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14(15):1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. A single amino acid difference in the host APOBEC3G protein controls the primate species specifi-city of HIV type 1 virion infectivity factor. Proc. Nat. Acad. Sci. U.S.A. 2004;101(11):3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006a;34(1):89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Nat. Acad. Sci. U.S.A. 2006b;103(23):8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Cullen BR. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364(2):486–493. doi: 10.1016/j.virol.2007.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, Zoete V, Castro- Diaz N, Turelli P, Telenti A, Michielin O, Weitzman MD, Trono D. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 2011;85(4):1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013 doi: 10.1038/nature11881. http://dx.doi.org/10.1038/nature11881, February 6th, advance online publication. [DOI] [PMC free article] [PubMed]
- Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amst) 2010;9(5):579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, Brown WL, Harris RS. Methyland normal-cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 2012;287(41):34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22(2):367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Dang Y, Abudu A, Son S, Harjes E, Spearman P, Matsuo H, Zheng YH. Identification of a single amino acid required for APOBEC3 antiretroviral cytidine deaminase activity. J. Virol. 2011;85(11):5691–5695. doi: 10.1128/JVI.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehle BP, Schafer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339(2):281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Duggal NK, Malik HS, Emerman M. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J Virol. 2011;85(21):11361–11371. doi: 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Priet S, Ribet D, Heidmann O, Heidmann T. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo “traces” on the murine IAPE and human HERV-K elements. Retrovirology. 2008;5:75. doi: 10.1186/1742-4690-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervaix A, West D, Leoni LM, Richman DD, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Nat. Acad. Sci. U.S.A. 1997;94(9):4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haché G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 2005;280(12):10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- Haché G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 2008;18(11):819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281(48):36624–36631. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 2012 doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 2011;85(21):11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson SR, Haché G, Stenglein MD, Fahrenkrug SC, Andrésdóttir V, Harris RS. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006;34(19):5683–5694. doi: 10.1093/nar/gkl721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3(4):e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35(9):2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 2009;284(34):22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Jónsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, Hötzel I, Andrésdóttir V, Smith TP, Harris RS. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue RS, Andrésdóttir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jónsson SR, Landau NR, Löchelt M, Malik HS, Malim MH, Münk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 2009;83(2):494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L, Demorest ZL, Land AM, Hultquist JF, Brown WL, Harris RS. APOBEC3B and AID have similar nuclear import mechanisms. J. Mol. Biol. 2012;419(5):301–314. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem. Biol. 2011;7(3):506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love RP, Xu H, Chelico L. Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A. J. Biol. Chem. 2012;287(36):30812–30822. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microb. 2008;3(6):388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Liao S, Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 2004;279(15):14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 2007;81(24):13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Münk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Münk C, Willemsen A, Bravo IG. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol. 2012;12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333(2):374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Olson ME, Li M, Harris RS, Harki DA. Small-molecule APOBEC3G DNA cytosine deaminase inhibitors based on a 4-amino-1,2,4-triazole-3-thiol scaffold. Chem. Med. Chem. 2013;8(1):112–117. doi: 10.1002/cmdc.201200411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch JW, Chelico L, Goodman MF, Le Grice SF. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J. Biol. Chem. 2009;284(11):7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38(13):4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8(7):e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose KM, Marin M, Kozak SL, Kabat D. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retroviruses. 2005;21(7):611–619. doi: 10.1089/aid.2005.21.611. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2(9):E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt K, Guo K, Algaier M, Ruiz A, Cheng F, Qiu J, Wissing S, Santiago ML, Stephens EB. Differential virus restriction patterns of rhesus macaque and human APOBEC3A: implications for lentivirus evolution. Virology. 2011;419(1):24–42. doi: 10.1016/j.virol.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Nat. Acad. Sci. U.S.A. 2004;101(11):3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Haché G, MacDuff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 2008;82(6):2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18(1):28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retro-transposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 2010;17(2):222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielen BK, McNevin JP, McElrath MJ, Hunt BV, Klein KC, Lingappa JR. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J. Biol. Chem. 2010;285(36):27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgen CA, Hatziioannou T. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J. Virol. 2007;81(24):13932–13937. doi: 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J. Exp. Med. 2010;207(1):141–153. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinghe P, Bhagwat AS. Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res. 2012;40(18):9206–9217. doi: 10.1093/nar/gks685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J. Biol. Chem. 2011;286(42):36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Nat. Acad. Sci. U.S.A. 2004;101(15):5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 2004;279(51):53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Webb DM. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 2004;13(16):1785–1791. doi: 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]
- Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. A single amino aciddifference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J. Virol. 2009;84(4):1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]