Abstract
In the central nervous system (CNS), neural stem cells (NSCs) differentiate into neurons, astrocytes, and oligodendrocytes - these cell lineages are considered unidirectional and irreversible under normal conditions. The introduction of a defined set of transcription factors has been shown to directly convert terminally differentiated cells into pluripotent stem cells, reinforcing the notion that preserving cellular identity is an active process. Indeed, recent studies highlight that tumor suppressor genes (TSGs) such as Ink4a/Arf and p53, control the barrier to efficient reprogramming, leaving open the question whether the same TSGs function to maintain the differentiated state. During malignancy or following brain injury, mature astrocytes have been reported to re-express neuronal genes and re-gain neurogenic potential to a certain degree, yet few studies have addressed the underlying mechanisms due to a limited number of cellular models or tools to probe this process. Here, we use a synthetic small-molecule (isoxazole) to demonstrate that highly malignant EGFRvIII-expressing Ink4a/Arf-/-; Pten-/- astrocytes down-regulated their astrocyte character, re-entered the cell cycle, and upregulated neuronal gene expression. As a collateral discovery, isoxazole small-molecules blocked tumor cell proliferation in vitro, a phenotype likely coupled to activation of neuronal gene expression. Similarly, histone deacetylase inhibitors induced neuronal gene expression and morphologic changes associated with the neuronal phenotype, suggesting the involvement of epigenetic-mediated gene activation. Our study assesses the contribution of specific genetic pathways underlying the de-differentiation potential of astrocytes and uncovers a novel pharmacological tool to explore astrocyte plasticity, which may bring insight to reprogramming and anti-tumor strategies.
Keywords: astrocyte plasticity, de-differentiation, epigenetics, glioblastoma, cancer stem cell
Introduction
It is well established that mature astrocytes lack neurogenic potential, especially during late postnatal stages and in adult brain [1]. Elegant work using genetic fate-mapping strategies confirmed that mature cortical astrocytes are largely quiescent and non-neurogenic, but retain the ability to proliferate and upregulate GFAP and other classical markers of reactive glia [2, 3]. Despite a few studies that examine the ability of glia to give rise to neuronal cells, this appears to only occur by forced expression of neuronal transcription factors in vitro, while the potential for this is drastically reduced in vivo, likely due to the non-neurogenic microenvironment [4-6]. Moreover, the finding that brain tumors often contain a mixture of neuronal and glial cell types has raised the notion that these tumors either contain multipotent or restricted stem/progenitors, or, arise from de-differentiated mature cell types, such as astrocytes [7]. Clearly, there is an urgent need to understand the cellular and molecular mechanisms underlying the proliferation and de-differentiation potential of mature astrocytes. Understanding this process, and especially, developing new strategies or tools to explore the extent of astrocyte plasticity may be relevant for designing neuroregenerative approaches and treating brain tumors.
Recent work from multiple labs indicates that reprogramming to pluripotent stem cells is markedly enhanced with the loss of tumor suppressor genes Ink4a or p53 [8, 9]. These data reinforce the connection between maintaining the differentiated state and initiating tumorigenesis. The Ink4a/Arf locus encodes two key tumor suppressor proteins (p16Ink4a and p19Arf) that, respectively, engage two critical anti-proliferative pathways, the retinoblastoma (Rb) and p53 pathways, both important for G1 checkpoint control [10, 11]. Ink4a (as well as its other Ink4 orthologs, Ink4b, Ink4c, and Ink4d) bind and inhibit the D-type cyclin-dependent kinases Cdk4 and Cdk6 that, in turn, relieve the cell-cycle inhibitory activity of Rb. On the other hand, Arf binds to and inactivates the Mdm2 protein, which is an E3 ubiquitin ligase that destabilizes p53. Both expression of p16Ink4a and p19ARF are critical for effective tumor suppression - including GBM, which frequently harbor homozygous deletions of the Ink4a/Arf locus [12-14]. Indeed, our previous studies indicate that Ink4a/Arf -/- astrocytes can undergo de-differentiation to a stem-like glioma cell and re-express progenitor markers such as Nestin and A2B5, retaining a capacity to become differentiated glial and neuronal progeny [15].
Several key questions are raised by these studies: (1) Are there specific tumor suppressor genes and/or oncogenes that govern the differentiation potential of malignant astrocytes, and (2) What is the extent of phenotypic plasticity of malignant astrocytes and is it reversible? In this report, we use a synthetic small-molecule 3,5-disubstituted isoxazole (compound 1), identified in a previous high-throughput chemical compound screen for inducers of differentiation of P19 embryonal carcinoma cells [16, 17], to interrogate the molecular pathways that control the lineage plasticity of malignant astrocytes. We demonstrate that Ink4a/Arf, Pten, and EGFRvIII pathways interact to maintain the differentiated state of astrocytes, and that in this context isoxazole acts as a stem cell modulator (SCM) to trigger neuronal gene expression and block tumor cell proliferation. Our findings provide novel insights into Ink4a/Arf-mediated de-differentiation of malignant astrocytes and identify a potential starting point for future glioma therapeutic drug design. Most importantly, we demonstrate the use of a novel pharmacological tool to explore the phenotypic plasticity of astrocytes, which is relevant in the context of cellular de-differentiation, reprogramming, and malignancy.
Materials and Methods
Astrocyte cell culture
SS05 cells or primary astrocytes were isolated from cerebral cortices of 5-day-old wild-type, Ink4a/Arf/f, Ptenf/f; Ink4a/Arf-/-; Ptenf/f, or p53-/-; Ptenf/f pups according to previous methods [15]. The floxed Ink4a/Arf or Pten allele was deleted using an adenovirus expressing Cre. Infection of astrocytes with lentiviruses expressing constitutively active EGFR(EGFRvIII) has previously been described [15]. Cells were cultured in 10% FBS in DMEM:F12 media (Omega Scientific). For differentiation experiments, astrocytes harboring glioma-relevant mutations were switched to serum-free (or indicated concentrations of FBS) DMEM:F12 media supplemented with N2 and B27 (Invitrogen) and treated with vehicle (DMSO) or different concentrations of 1 or 2 at indicated time points. In some experiments, cells were treated with N2/B27 media containing LIF and BMP-2 (50 ng/ml each) and 1% FBS for 4 days to promote astrocyte differentiation [17, 44]. For TSA or VPA-induction experiments, cells were plated in 10% FBS, switched to N2/B27 media the next day and treated with either 50 or 100 nM TSA (Millipore) or 300 nM or 1 mM VPA (Sigma) for 4 days.
Immunostaining and cell quantification
Cells were fixed with 4% paraformaldehyde, followed by immunocytochemical staining as previously described [44, 45]. Labeled cells were visualized using a Nikon TE2000-U inverted microscope (Nikon, Inc.) and a CoolSnap digital camera (Photometrics, Inc.). Quantification of cell phenotype from 3 independent experiments was done by sampling 6-8 random fields in each well and counting a total of 250-500 cells at 20× magnification. Throughout the study, definitive neuronal or glial cells were scored on the basis of morphological criteria (elaboration of processes), as well as immunoreactivity with various markers (e.g. Tuj1 or GFAP staining). 4′,6-diamidino-2-phenylindole (DAPI) was used to identify individual cells. In some cultures, propidium iodide (1 mg/ml, Molecular Probes) or Hoechst 33342 (1 mg/ml, Sigma) were added to label dead or all cells, respectively. Staining of live (rather than fixed) cultures was used to avoid underestimating cell death because of possible detachment of dying and/or dead cells from culture substrates. The following primary antibodies were used: rabbit anti-Tuj1 (1:2500; Covance) or mouse anti-Tuj1 (1:500; Sigma), guinea pig anti-GFAP (1:2500; Advanced Immunochemical, Inc.), mouse anti-Olig2 (1:500; Chemicon), or rabbit anti-Ki67 (1:400; Accurate). Secondary antibodies were all from Jackson ImmunoResearch and used at 1:250 dilution.
Clonal assay
SS05 cells were dissociated and plated at single cell density (20 cells/l) in clear 96-well plates in a final volume of 100 μl of serum-free DMEM media supplemented with N2 and B27 and treated with either vehicle or 1. After 12-24 hours, healthy, live single cells were identified and marked and at least 50-100 clones per treatment were scored for Ki67 or Tuj1 staining after 4 days.
Reporter gene assay
All reporter gene assays were done in 96-well format, and each data point represents the average of 12 replicates. Reporter genes included GluR2- and NR1-luciferase and each well contained ∼25,000 cells in a volume of 100 μL. 5×106 cells SS05 cells were transfected with 5 μg DNA by electroporation (Amaxa) and plated in growth media overnight. Compounds were added the day following transfection and luciferase assays were performed 24 hour later.
RT-PCR, Q-PCR and protein blotting
Total RNA was isolated by Trizol reagent (Invitrogen) and RT-PCR was carried out as previously described [46]. For Q-PCR, genomic DNA contamination was digested with RNase-free DNase when isolating total RNA. First strand cDNA was synthesized from 1.0 μg of total RNA in 20 μl volume with an iScript cDNA Synthesis Kit (Bio-Rad), diluted to 40 μl by DNase-free water, and 0.5 μl is used as template to measure the transcripts level by Q-PCR. Q-PCR was performed by using SYBR green dye (Bio-RAD) to measure duplex DNA formation with the Bio-RAD CFX384™ Real-Time PCR Detection System and normalized to the expression Gapdh. Primer sequences are available upon request. For protein blotting, whole cell lysates were prepared from astrocytes cells cultured in undifferentiated conditions (DMSO vehicle control) or from differentiating conditions (1 or 2). For protein blotting analysis, whole cell lysates were prepared from SS05 cells using RIPA buffer (Tris-HCl, 50 mM, pH 7.4; NP-40, 1%; Na-deoxycholate, 0.25%; NaCl, 150 mM; EDTA, 1 mM; PMSF, 1 mM; aprotinin, leupeptin, pepstatin, 1 mg/ml; Na3VO4, 1 mM; NaF, 1 mM). Protein concentration in centrifugation-clarified cell lysates were measured by the BCA Protein Assay Kit (Pierce) and equal amounts of protein were separated on a 4-12% SDS-PAGE and transferred to Hybond PVDF (Amersham Biosciences). Protein blots were done using the NuPage gel and transfer system with 4-20% Tris-Bis gels (Invitrogen). Primary Abs for protein blotting included: mouse anti-cyclin D1 and mouse anti-cyclin E1 (1:500; Cell Signaling); mouse anti-p21 (1:500; Santa Cruz Biotechnology), mouse anti-βTubIII (1:500; Sigma), and mouse anti-GAPDH (1:5000; Chemicon). Immunoreactive bands were visualized using AP-conjugated secondary antibodies, followed by BCIP/NBT detection (KPL).
Cell cycle analysis
SS05 cells were collected, centrifuged, washed, and resuspended in PBS and 70% (v/v) ETOH, then washed with PBS followed by incubation with 200 μg/ml RNase A and 40μg/ml of propidium iodide (Sigma). Cell cycle was monitored using a FACScalibur flow cytometer (Becton-Dickson) with 10,000 events/determination and analyzed by using FLOWJO analysis software (Tree Star, Inc.). Control (DMSO- and FBS-treated) experiments were performed in parallel. Cells that have 2N DNA content are G1 and cells with greater than 2N DNA content are S, G2, and M. The cell cycle analysis was repeated in at least 3 independent experiments and the data presented are from one representative experiment.
Statistics
Experiments with two groups were analyzed for statistical significance using unpaired two-tailed Student's t-test, and all error bars are expressed as ± standard deviation (s.d.), unless otherwise stated. Values of p < 0.05 or p < 0.001 were considered statistically significant.
Results
Isoxazole SCMs block cell proliferation and induce neuronal gene expression in malignant astrocytes
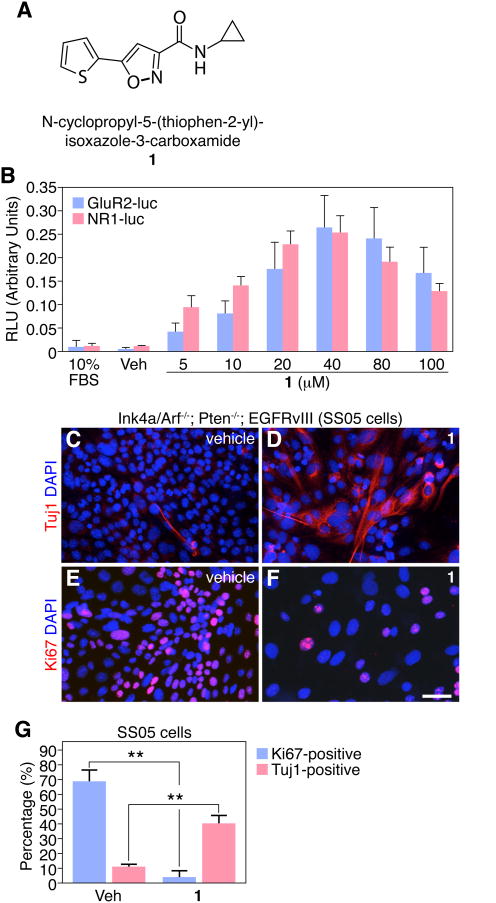
To explore the lineage potential of malignant astrocyte cultures, we used a continuous line of cultured astrocytes (SS05) derived from genetically engineered mice with homozygous deletion of both Ink4a/Arf and Pten tumor suppressor genes. SS05 cells initially expressed the astrocyte marker (GFAP), but downregulated their astrocyte phenotype and increased proliferation during in vitro cell culture with 10% FBS (data not shown). SS05 cells also harbor constitutively active epidermal growth factor receptor variant III (EGFRvIII), the most commonly amplified/mutated oncogene in GBM [18-20]. Phenotypic plasticity was tested with 1 (Figure 1A), a second-generation analog of a SCM identified in our original screen [16] and previously shown to induce neuronal differentiation in wild-type NSCs from adult rat hippocampus [17]. To establish an effective drug concentration, serum was withdrawn and SS05 cells were treated in adherent cultures with 1 (0-100 μM) for 4 days. We observed a concentration-dependent activation of GluR2- and NR1-luciferase reporter genes, markers of neuronal differentiation, with 1 treatment in SS05 cells, compared with vehicle-treated control cells or cells grown in 10% FBS (Figure 1B). We also found that 1 (40 μM) increases the number of cells of a neuronal phenotype (Tuj 1 +) and decreases the number of proliferating cells (Ki67+) (Figure 1C-G). Higher concentrations of 1 (>40 μM), however, resulted in significant cell death compared with vehicle-treated cells (Figure 4B), hence we used 1 at 40 μM in the majority of our studies since this concentration conferred maximal Tuj1+ cells with minimal toxicity. In addition, treated cells rapidly (<24 hours) flattened and exhibited enlarged nuclei and extended morphologic processes (Figure 1D). Although the Tuj1-induction is robust, 1-treated SS05 cells do not exhibit typical neuronal morphology and still retain astrocyte-like features. Furthermore, the neuronal marker Map2ab is not induced in malignant astrocytes after 1 treatment (data not shown), suggesting that 1 is able to active some neuronal genes, but not the entire lineage program.
Figure 1.
1 activates neuronal genes in Ink4a/Arf-/-; PTEN-/-; EGFRvIII astrocytes (SS05 cells). (A) Chemical structure of lead isoxazole (1). (B) 1 induces a concentration-dependent increase in both GluR2- and NR1-luciferase (luc) reporters. (C-D) 1 promotes neuronal gene expression (Tuj 1) in SS05 cells compared to vehicle control (DMSO) in 4-day cultures. (E-F) 1 blocks proliferation of SS05 cells, compared to vehicle control (DMSO) in 4-day cultures. Scale bar: 25 μm. (G) Quantification of Ki67+ and Tuj1+ cells after 4 days of vehicle or 1 treatment is shown. (**p < 0.001, t-test, data in graphs represent ± s.d. from three independent experiments).
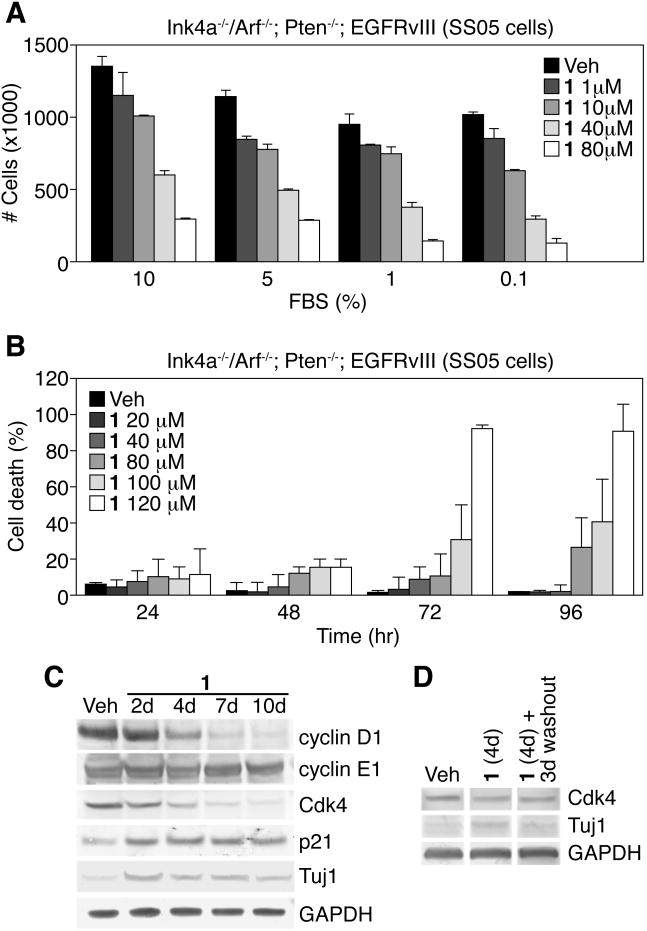
Figure 4.
1 prevents proliferation, but not survival, of Ink4a/Arf-/-; PTEN-/-; EGFRvIII astrocytes (SS05 cells). (A) Total number of cells/plate treated with different concentrations of 1 and FBS after 4 days. (B) Increasing concentrations (>40 μM) of 1 induces death of SS05 cells over time. (C) Protein blotting time-course analysis of cell cycle regulatory genes in SS05 cells treated with 1 compared to vehicle control. (D) Cdk4 and Tuj1 protein levels in SS05 cells treated with vehicle or 1 for 4 days, then cultured for 3 additional days after washout of drug, in the presence of 1% FBS. Gapdh served as a normalization control. (Data in graphs represent ± s.d. from three independent experiments).
Ink4a/Arf plays a key role to maintain the differentiated astrocyte state
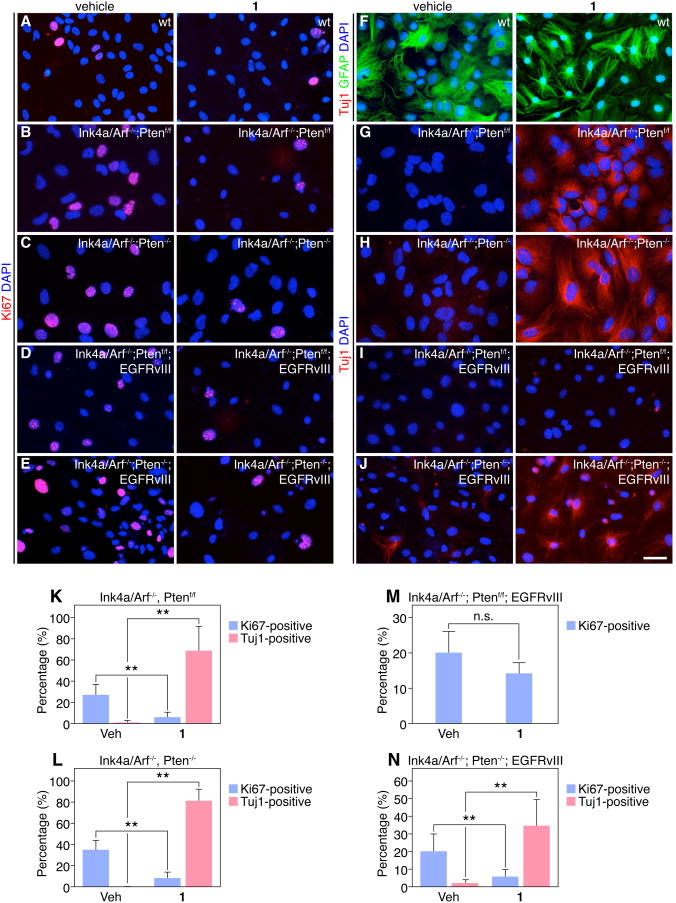
Since SS05 cells have been cultured extensively and likely harbor multiple perturbations in growth control pathways (loss of Ink4a/Arf and Pten, activation of EGFRvIII), we next assessed the effect of 1 on primary (<5 passages) Ink4a/Arf-/-; Ptenf/f and Ink4a/Arf-/-; Pten-/- astrocytes and tested their response to 1 in serum-free conditions. Compound 1 inhibited proliferation in both Ink4a/Arf-/-; Ptenf/f and Ink4a/Arf-/-; Pten-/- astrocytes (Figure 2B-C, K-L), and concomitantly increased hallmarks of neuronal differentiation, as indicated by Tuj1+ staining (Figure 2G-H, K-L) and up-regulation of pro-neuronal genes NeuroD1 and NMDA receptor 1 (NR1) consistent with activation of the neuronal lineage in 1-treated astrocytes (Figure S1). By contrast, 1 had no significant effect on the proliferation or differentiation of primary astrocytes from Ink4a/Arft+/+; Ptenf/f wild-type (wt) mice, also cultured in serum-free media (Figures 2A and 2F). These results suggest that Ink4a/Arf is required for the maintenance of the differentiated astrocytic state, either alone or in conjunction with Pten, and blocks the ability of 1 to induce the neuronal phenotype.
Figure 2.
Comparison of 1's effects in primary mouse astrocytes carrying glioma-relevant mutations. (A-E) Representative images of vehicle or 1-treated astrocytes with indicated Ink4a/Arf; Pten; EGFRvIII genotypes or wild-type (wt) astrocytes and immunostaining for Ki67 in 4-day cultures. (F-J) Representative images of vehicle or 1-treated astrocytes with indicated Ink4a/Arf; Pten; EGFRvIII genotypes or wild-type (wt) astrocytes and immunostaining for Tuj1 or GFAP in 4-day cultures. Scale bar: 25 μm. (K-N) Quantification of Ki67+ or Tuj1+ primary astrocytes after 4 days of vehicle or 1 treatment is shown. (**p < 0.001, t-test, data in graphs represent ± s.d. from three independent experiments).
The constitutively activated EGF receptor cooperates with loss of Ink4a/Arf during malignant gliomagenesis [21] and is frequently observed in orthotopic and de novo mouse glioma models as well as human GBMs [18, 22]. Notably, expression of EGFRvIII in the context of Ink4a/Arf-/-; Ptenf/f astrocytes attenuated the anti-proliferation and neuronal differentiation effects of 1 (Figures 2D, 2L, and 2M). Interestingly, additional loss of Pten in Ink4a/Arf-/-; EGFRvIII astrocytes promoted partial recovery of 1-mediated neuronal differentiation and decrease in proliferation, suggesting that deletion of Pten can mitigate, at least in part, the blocking effects of EGFRvIII on 1-treated cells (Figures 2E, 2J, and 2N). Importantly, there was no change in proliferation or differentiation with 1 treatment in Ink4a/Arf+/+ astrocytes after Pten deletion, consistent with the critical role of Ink4a/Arf in maintaining the differentiated state of astrocytes and preventing de-differentiation into glioma stem-like cells (Figure S2).
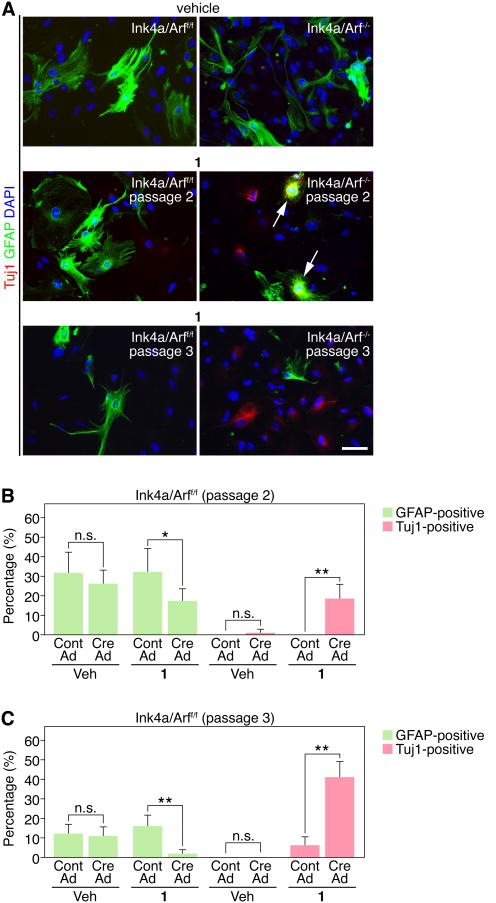
To examine whether neuronal conversion occurs at the expense of astrocytic fate, we established primary astrocytes from Ink4a/Arff/f mice and deleted Ink4a/Arf with Cre-expressing adenovirus. We treated Ink4a/Arf-deficient cells with vehicle or 1 at early passages (passage 2 and 3), when GFAP expression was still detectable. Compound 1-treated cells displayed a reduction in GFAP and increased Tuj1 expression after deletion of Ink4a/Arf, while GFAP expression was unchanged in vehicle-treated cells at passage 2, regardless of Ink4a/Arf deletion (Figure 3A-B). Interestingly, some 1-treated cells expressed both GFAP and Tuj1 (Figure3A), consistent with Ink4a/Arf-deficient astrocytes exhibiting phenotypic plasticity. The reduction in GFAP and up-regulation of Tuj1 was even more dramatic in passage 3 cells, suggesting further de-differentiation of Ink4a/Arf-deficient glioma stem-like cells over time (Figure 3A and 3C).
Figure 3.
Compound 1 induces Tuj1 at the expense of GFAP expression in primary Ink4a/Arf/-astrocytes. (A) Comparison of vehicle (passage 2) or 1 (passage 2 and 3) treatment in Ink4a/Arff/f and Ink4a/Arf-/- astrocytes. Note co-expression of Tuj1 and GFAP in a subset of 1-treated cells (arrows). Scale bar: 25 μm. (B-C) Quantification of Tuj1+ or GFAP+ in passage 2 or 3 astrocytes after 4 days of vehicle or 1 treatment is shown. (*p<0.05, **p < 0.001, t-test, data in graphs represent ± s.d. from three independent experiments).
Since Ink4a/Arf are upstream regulators of Rb and p53, we further tested 1's effects on neuronal differentiation in p53-/-; Ptenf/f and p53-/-; Pten-/- astrocytes. We confirmed increased number of Tuj1+cells in p53-/-; Ptenf/f and p53-/-; Pten-/- astrocytes, although to a much lesser extent compared to Ink4a/Arf-/-; Ptenf/f or Ink4a/Arf-/-; Pten-/- astrocytes, respectively, suggesting the contribution of additional pathways (Figure S3). In addition, we used SS05 cells to confirm a second structurally distinct isoxazole analog (2) on the ability to upregulate pro-neuronal reporter genes (Figure S4). A summary of effects of 1 on proliferation and differentiation of various genotypes is shown in Table S1. Taken together, these data demonstrate that isoxazole SCMs induce the neuronal phenotype in both primary astrocytes and an established cell line, so long as they harbor the glioma-relevant Ink4a/Arf mutation.
Treatment with isoxazole SCMs leads to a delay in G1 and entry into M-phase
A critical issue is whether 1 retains its potentially therapeutic effects by upregulating neuronal gene expression and inhibiting cell growth under conditions of sustained mitogenic signaling, as present in tumors. To explore this, we assessed the impact of 1 on SS05 cells under serum conditions. Compound 1 induced a concentration-dependent decrease in SS05 cell number even in the presence of 10% FBS (Figure 4A). To determine whether this reduced cell number is a consequence of increased cell death, we performed propidium iodide (PI) staining, which stains dead/dying cells in live cultures, and observed a concentration-dependent increase in the percentage of PI-positive cells over time, but only with concentrations greater than 40 μM (Figure 4B). Thus, the negative effect of 1 on cell number is not apparently due to effects on cell viability, but on a decrease in growth rate, possibly due to effects on cell cycle progression. Next, we set out to evaluate 1's effects on cell cycle regulation.
By modulating Rb and p53 activity, Ink4a/Arf regulates the G1-S transition during the cell cycle [11, 23]. We therefore examined the expression of the G1 cyclin and Cdk proteins, cyclin D1 and Cdk4. We observed diminished expression of cyclin D1 and Cdk4 with 1 treatment, compared with vehicle-treated cultures at 4 days in serum-free conditions (Figure 4C). Interestingly, there was already increased Cdk inhibitor p21WAF1 expression at 2 days, concomitantly with increased Tuj1 after 1 treatment (Figure 4C). Compound 1's effects on cell cycle regulation appears to be restricted to the early part of G1, since cyclin E1 levels (a late G1 cyclin) remained unchanged (Figure 4C). To examine whether compound 1's effects on cell cycle gene expression was reversible, we treated SS05 cells with 1 for 4 days, then cultured cells without 1 for 3 additional days (washout). The absence of serum after 1 withdrawal led to poor cell survival (data not shown), thus we treated cells with vehicle or 1 in the presence of low serum (1% FBS). In the presence of serum, there was only a modest reduction in Cdk4 levels in 1-treated cells, compared to vehicle treatment, which did not significantly change after the 3-day washout (Figure 4D), suggesting that isoxazole's effects on the expression of cell cycle regulators are reversible, at least in a subset of cells.
To confirm the effects of 1 in the cell cycle progression of SS05 cells, we conducted flow cytometric analysis to examine DNA content of unsynchronized cells. At 1 and 2 days, SS05 cells treated with 1 showed a substantial increase in the G1 population compared to vehicle-treated cells or cells grown in 10% FBS (Figure S5). As expected, serum-starved SS05 cells arrested in G1, with both vehicle and 1-treatment, after extended time in culture. Taken together, these results suggested that the 1-mediated block in proliferation and reduction in cyclin D1 and Cdk4 is associated with a delay in G1 and entry into M-phase.
Isoxazole's effects on proliferation and differentiation is irreversible in a subset of glioma stem cells
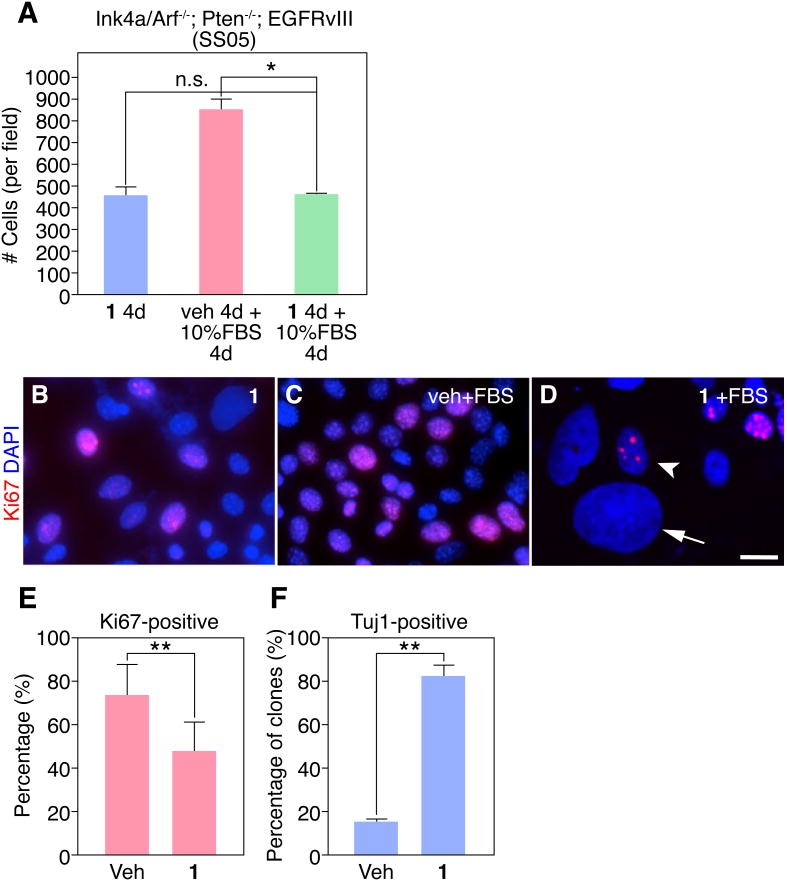
To evaluate the potential utility of 1 as an anti-glioma agent, a key question that comes from these above experiments is whether its anti-proliferation and differentiation effects are reversible? To investigate this, we pre-treated SS05 cells with vehicle or 1 for 4 days in serum-free conditions, then switched the media to 10% FBS for an additional 4 days. In a parallel experiment, we treated SS05 cells with 1 for 4 days, also in the absence of serum. As expected, we observed an increase in cell number when cells pre-treated with vehicle are switched to 10% FBS, compared to 1 treatment alone (Figure 5A). Strikingly, when cells were pre-treated with 1 and then stimulated with serum we saw no increase in cell number, suggesting 1 pre-treatment can irreversibly block serum-induced proliferation.
Figure 5.
Ink4a/Arf-/-; PTEN-/-; EGFRvIII astrocytes (SS05 cells) display an irreversible block in proliferation with 1 treatment. (A) Quantification of the total number of cells/field after vehicle or 1 pre-treatment is shown. (B-D) Shown are representative images of SS05 cells treated with 1 for 4 days, pre-treated with vehicle for 4 days and post-treated with 10% FBS for 4 days, or pre-treated with 1 for 4 days and post-treated with 10% FBS for 4 days. Note in (D): 1 pre-treated cultures and post-treated with FBS contain a mixture of Ki67+ cells with small round nuclei (arrowhead) and Ki67- cells with large flat nuclei (arrow). Scale bar: 10 μm. (E) Quantification of the percentage of Ki67+ cells is shown. (F) Quantification of the percentage of clones expressing Tuj1 is shown. (*p < 0.05 or **p < 0.001, t-test, data in graphs represent ± s.d. from three independent experiments).
These conclusions were confirmed by Ki67 staining of vehicle- or 1 pre-treated cells and post-treated with FBS when we observed the distribution of cells with two types of nuclear morphologies in 1 pre-treated cultures: (1) cells with small, round Ki67+ nuclei and (2) cells with large, flat Ki67-nuclei, suggesting a subset of 1 pre-treated cells still retained the ability to proliferate after serum challenge (Figure 5B-D). The mixed responsiveness of cells to 1 treatment could be a consequence of heterogeneity of the starting population. To address this, we performed clonal analysis of vehicle or 1-treated SS05 cells (n>100 clones per treatment). SS05 cells were dissociated and single cells were plated and treated with vehicle or 1 for 4 days. By scoring individual clones derived from a single cell, we observed ∼70% Ki67+ cells within vehicle-treated clones, compared with only ∼45% Ki67+ 1-treated clones (Figure 5E). Since we failed to observe clones that were entirely positive or negative for Ki67 (data not shown), these results suggest that the response to vehicle or 1 treatment is uniform across SS05 cells. Upon staining for Tuj1, we observed clones either positive or negative for Tuj1, consistent with 1 treatment of the bulk population. We therefore scored individual vehicle-or 1-treated clones and determined there were ∼18% Tuj1+ clones with vehicle treatment compared to ∼80% Tuj1+ clones with 1 treatment (Figure 5F). Based on these data from bulk cultures and single cells, we conclude that the changes in neuronal gene expression and reduction of proliferation SS05 cells in response to 1 is irreversible, at least in a subpopulation of cells.
Neuronal gene expression in malignant astrocytes is triggered by HDAC inhibition
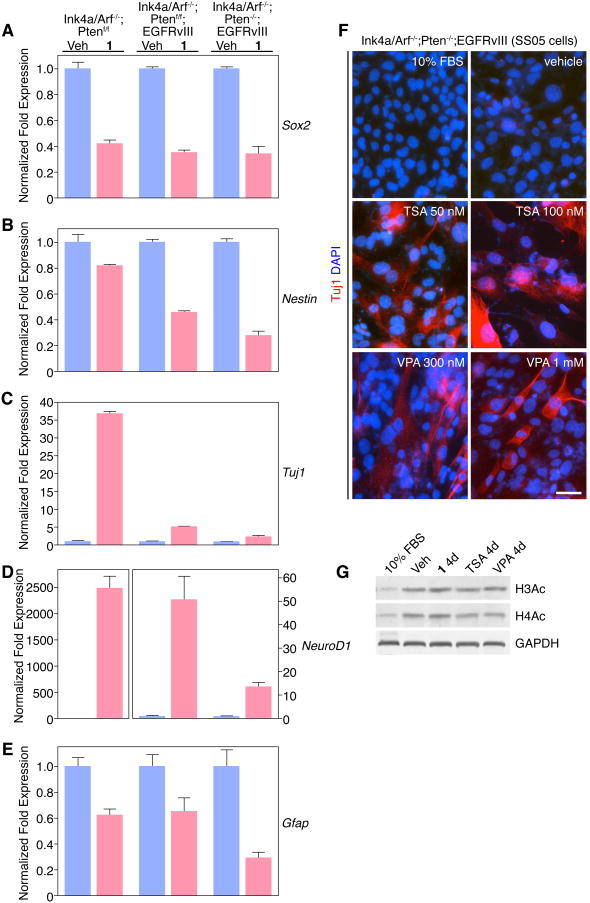
To explore the molecular basis of 1-mediated anti-proliferation and up-regulation of neuronal gene expression in malignant astrocytes, we considered two non-exclusive hypotheses. Namely, that 1 regulates the expression of stem cell reprogramming factors or that 1 acts via an epigenetic mechanism. To address the first possibility, we performed quantitative real time PCR (Q-PCR) to evaluate the expression changes of candidate stem/progenitor, neuronal, and astrocyte lineage mRNAs in both genotypes of EGFRvIII-expressing Ink4a/Arf-/- (with and without Pten) astrocytes compared to Ink4a/Arf-/-; Ptenf/f astrocytes between vehicle- and 1-treated cells. We observed 1-mediated down-regulation of NSC/progenitor genes (e.g., Sox2 and Nestin) and the astrocyte gene (e.g., GFAP) (Figures 6A-B and 6E). Besides Sox2, we interrogated the expression of the remaining stem cell reprogramming/pluripotency factors [24, 25] (e.g., Oct4, Klf4, c-Myc and Nanog) but did not observe a significant change after 1 treatment (data not shown). Consistent with marker staining, 1 induced robust up-regulation of the neuronal lineage gene Tuj1 in Ink4a/Arf-/-; Ptenf/f astrocytes (Figure 6C). By contrast, Tuj1 is only modestly increased in both Ink4a/Arf-/-; Ptenf/f; EGFRvIII and Ink4a/Arf-/-; Pten-/-; EGFRvIII astrocytes, 5- and 2.5-fold, respectively, in vehicle-treated cells compared with 1 treatment (Figure 6C). The reason for this modest Tuj1 fold-change, particularly in Ink4a/Arf-/-; Pten-/-; EGFRvIII astrocytes is due to higher baseline Tuj1 levels upon serum-withdrawal, consistent with marker staining (Figure 2J and Figure S6). Finally, we observed similar results with a second neuron-specific gene NeuroD1, confirming isoxazole SCMs as neuronal inducers of stem-like malignant astrocytes (Figure 6D).
Figure 6.
Stem cell and lineage-specific gene expression in primary Ink4a/Arf-/-; PTENf/f, Ink4a/Arf-/-; PTENf/f; EGFRvIII and Ink4a/Arf-/-; PTEN-/-; EGFRvIII astrocytes after 1 treatment. (A-E) Q-PCR analysis of stem cell, pro-neuronal, and pro-astrocytic gene expression in glioma stem cells treated with vehicle or 1 for 24 hours. Gapdh served as a normalization control. (F) Histone deacetylase inhibitors TSA or VPA induce Tuj1 in SS05 cells. Scale bar: 25 μm. (G) Histone H3 and H4 acetylation levels in SS05 cells after 4 days of treatment with various compounds. (Data in graphs represent ± s.e.m. from three independent experiments).
With no support for the reprogramming hypothesis, we next asked whether epigenetic modulation, at least in part, correlates with 1-mediated neuronal gene expression. Histone acetylation is a critical epigenetic regulatory mechanism controlled by the balance of histone acetyltransferases (HATs) and deacetylases (HDACs) [26, 27]. To investigate the possibility that modulating histone acetylation was involved in the neuronal reprogramming of malignant astrocytes, we therefore treated SS05 cells with specific HDAC inhibitors – either trichostatin A (TSA) or valproic acid (VPA). We observed strong induction of Tuj1+ cells, as well as an inhibition of cell proliferation, after either TSA or VPA treatment, similar to 1 and consistent with an epigenetic reprogramming model (Figure 6F and data not shown). Moreover, confirming HDAC inhibition, treatment of cells with 1, TSA, VPA, or serum withdrawal alone (vehicle) resulted in increased histone H3 and H4 acetylation (Figure 6G). Our data demonstrate that HDAC inhibition recapitulates the effect of 1 in malignant astrocytes, and is sufficient to induce gene expression and morphologic changes consistent with neuronal differentiation.
Discussion
One of the most surprising and exciting discoveries in neural developmental biology this past decade is the finding that NSCs possess characteristics of glial identity [28, 29]. Radial glia give rise to neurons in the developing telecenphalon across vertebrate species and radial-like stem cells in the postnatal hippocampus and lateral ventricle subependymal zone differentiate into neurons throughout life [30]. Why do some glial cells possess neurogenic potential, while other glia apparently lack it? This is a critical question, especially in the context of brain tumors of glial origin, such as glioblatoma multiforme (GBM). The question remains whether GBMs arise from de-differentiated astrocytes, committed astrocyte progenitors, or undifferentiated NSCs? The identification of NSCs in adult brain has led to the model that poorly differentiated GBM may result from transformation of a NSC or early glial progenitor, rather than progressive de-differentiation of a mature astrocyte. Distinction among these possibilities may be important in predicting the probable genetic targets involved in malignant transformation and tumor progression. Indeed, the state of glial cell differentiation may be an important factor that governs whether a given genetic mutation exerts its full oncogenic potential. Thus, identifying and understanding specific tumor suppressor genes and/or oncogenes, in conjunction with novel pharmacological tools, that alter the phenotypic plasticity of mature astrocytes, may shed light on the etiology of malignant glioma.
Here, we provide compelling evidence that even a subset of highly malignant Ink4a/Arf-/-; Pten-/-; EGFRvIII-expressing astrocytes can halt their proliferation and upregulate neuronal gene expression mediated by isoxazole SCMs. We confirmed a role of Ink4a/Arf alone, or together with Pten, in the maintenance of the mature astrocyte state, consistent with the emerging concept that tumor suppressor genes represent a barrier for reprogramming to the pluripotent state in iPS cells [8, 9, 31, 32]. In addition, our study is consistent with work from a number of laboratories that have demonstrated that Ink4a is important for maintaining NSC renewal and that Ink4a suppression is associated with increased neuronal differentiation [33-35]. In our studies, although we observed expression of the four pluripotency-inducing transcription factors, Oct4, Sox2, Klf4, and c-Myc in the context of Ink4a/Arf and Pten deletion, only Sox2 was significantly downregulated after 1 treatment, suggesting that both transcriptional and epigenetic mechanisms may be involved in the reprogramming of mature astrocytes. Our work also establishes that potent oncogenes, such as EGRvIII, can inhibit the ability of isoxazole SCMs to induce the neuronal phenotype in de-differentiated astrocytes and that additional tumor suppressor gene mutations, such as loss of Pten, is important to overcome the EGFRvIII-mediated block of 1's effects on growth and differentiation. These studies underscores the need to identify downstream genes that are regulated by the combinatorial actions of Ink4a/Arf, Pten, and EGFRvIII, alone and in conjunction with 1, to fully understand the mechanism of astrocyte de-differentiation and subsequent anti-proliferation and induction of neuronal gene expression. Our model is shown in Figure 7.
Figure 7.
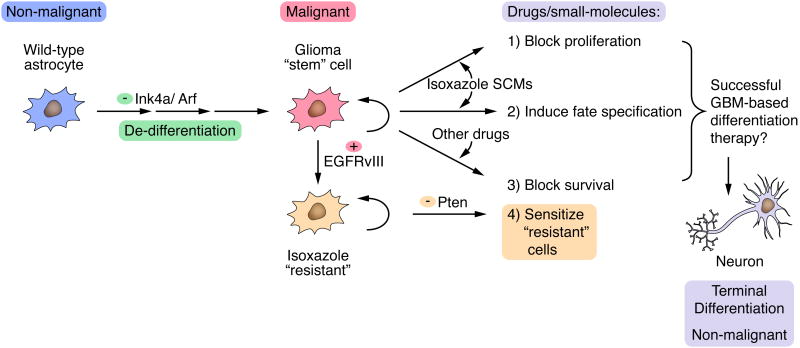
Model showing isoxazole SCM-mediated growth arrest and neuronal differentiation in malignant astrocytes. Upon loss of Ink4a/Arf tumor suppressor genes, wild-type astrocytes undergo de-differentiation into glioma “stem” cells. The inclusion of SCMs (e.g., 1 or 2) can be used to block proliferation and induce terminal neuronal differentiation, alone or in conjunction with other small-molecules/drugs that induce cell death, will likely facilitate successful GBM differentiation-based therapy. The subsequent activation of potent oncogenes in Ink4a/Arf-/- astrocytes (e.g., EGFRvIII) may promote the emergence of 1-resistant cells that may require additional loss of tumor suppressor genes (e.g., Pten) to sensitize 1-resistant cells and enhance the growth inhibitory/pro-differentiation effects of 1.
What could be the mechanism mediating changes in astrocyte plasticity induced by 1? Recent evidence point to a handle of neuronal-specific transcription factors, when ectopically expressed, can convert non-neuronal cell types directly to express neuronal lineage genes [36, 37]. By altering transcriptional/epigenetic regulatory networks, 1 may be sufficient to activate salient features of the neuronal transcriptional program, at least in part. Indeed, our prior mechanistic studies suggested that 1 is a neurotransmitter-like excitation signal in NSCs that regulates the neuronal genome through a transcriptional/epigenetic network that involves histone deacetylase 5 (HDAC5) and myocyte enhancer factor-2 to control early neuronal cell fate events, including activation of the neurogenic transcription factor NeuroD1 [17]. Although at present, the precise molecular targets of isoxazoles remain unclear, 1 treatment of wt mouse NSCs is associated with increased global and gene-specific histone acetylation, but does not directly appear to block HDAC activity (data not shown). However, isoxazole SCMs may functionally belong to a growing class of “epi-drugs” such as DNA methyltransferase inhibitors, HDAC inhibitors, HAT activators, and inhibitors of methyltransferases or demethylases. Indeed, the HDAC inhibitor sodium butyrate was used as a positive control in our high-throughput screen based on its strong and reliable activation of the Nkx2.5-luc transgene [16]. Thus, it is not entirely surprising that isoxazoles are capable of epigenetic modulation of gene expression in the context of normal and cancer stem cells, which provide rationale, in retrospect, for the discovery of small molecules with epi-drug-like effects from the original screen. It will be intriguing to identify the isoxazole target, especially in the context of cancer stem cells, which may shed light on the underlying biology of malignant glioma.
Interestingly, in recent years there has been some progress with epi-drugs to treat cancer, such as the demethylation agents azacitidine (Vidaza) and decitabine (Dacogen) in the treatment of myelodysplastic syndrome and AML, and the HDAC inhibitor SAHA (Zolinza) for therapy of cutaneous T-cell lymphoma [38-40]. However, the biology of these drugs and how they target cancer cells, or cancer stem cells, is not clear. Moreover, from the clinical perspective, epi-drugs have not yet met the expectations placed on them, due to nonspecific cytotoxic effects and/or unanticipated effects of enhanced proliferation of cancer stem/progenitor cells [41, 42]. In the context of GBM, epigenetic alterations can affect the expression of cancer genes alone, or in combination with genetic mechanisms [43]. Combination therapy with both epi-drugs and traditional chemotherapeutic agents is likely to be a powerful strategy against GBM.
In conclusion, our findings indicate that isoxazole SCMs can be delivered to malignant astrocytes, to induce neuronal gene expression in addition to suppressing tumor cell proliferation. Future structure-activity relationship studies and improvements on 1 pharmacokinetics will pave the way for target identification and in vivo testing. These results demonstrate the feasibility of chemical biology approaches to provide novel pharmacological tools for reprogramming pathway discovery that can also serve as entry points for differentiation agents to treat GBM.
Supplementary Material
Acknowledgments
We thank Yan Jiang, Yan Wu, and HK Jeong for technical assistance and Jose Cabrera for artwork. We also thank Gerard Evan, Sarah Comerford, Chun-li Zhang, and Ondine Cleaver for helpful comments on the manuscript. This work was funded by grants from the Cancer Prevention & Research Institute of Texas (RP100674), the Welch Foundation (I-1660), and National Institutes of Health (AG032383).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costa MR, Gotz M, Berninger B. What determines neurogenic competence in glia? Brain Res Rev. 2010;63:47–59. doi: 10.1016/j.brainresrev.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Buffo A, Rite I, Tripathi P, et al. Origin and progeny of reactive gliosis: A source of multipotent cells in the injured brain. Proc Natl Acad Sci U S A. 2008;105:3581–3586. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buffo A, Vosko MR, Erturk D, et al. Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. Proc Natl Acad Sci U S A. 2005;102:18183–18188. doi: 10.1073/pnas.0506535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum R, Heinrich C, Sanchez R, et al. Neuronal Network Formation from Reprogrammed Early Postnatal Rat Cortical Glial Cells. Cereb Cortex. 2010 doi: 10.1093/cercor/bhq107. [DOI] [PubMed] [Google Scholar]
- 5.Heins N, Malatesta P, Cecconi F, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 6.Berninger B, Costa MR, Koch U, et al. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Collado M, Villasante A, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Utikal J, Polo JM, Stadtfeld M, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 12.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 13.Kamb A, Gruis NA, Weaver-Feldhaus J, et al. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 14.Ueki K, Ono Y, Henson JW, et al. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–153. [PubMed] [Google Scholar]
- 15.Bachoo RM, Maher EA, Ligon KL, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 16.Sadek H, Hannack B, Choe E, et al. Cardiogenic small molecules that enhance myocardial repair by stem cells. Proc Natl Acad Sci U S A. 2008;105:6063–6068. doi: 10.1073/pnas.0711507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider JW, Gao Z, Li S, et al. Small-molecule activation of neuronal cell fate. Nat Chem Biol. :2008. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- 18.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 19.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang HS, Nagane M, Klingbeil CK, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 21.Zhu H, Acquaviva J, Ramachandran P, et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maher EA, Furnari FB, Bachoo RM, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 23.Koh J, Enders GH, Dynlacht BD, et al. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Silva J, Nichols J, Theunissen TW, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 27.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 28.Goldman S. Glia as neural progenitor cells. Trends Neurosci. 2003;26:590–596. doi: 10.1016/j.tins.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Mori T, Buffo A, Gotz M. The novel roles of glial cells revisited: the contribution of radial glia and astrocytes to neurogenesis. Curr Top Dev Biol. 2005;69:67–99. doi: 10.1016/S0070-2153(05)69004-7. [DOI] [PubMed] [Google Scholar]
- 30.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banito A, Rashid ST, Acosta JC, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawamura T, Suzuki J, Wang YV, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruggeman SW, Hulsman D, Tanger E, et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 34.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molofsky AV, He S, Bydon M, et al. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinrich C, Blum R, Gascon S, et al. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 39.Mann BS, Johnson JR, Cohen MH, et al. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 40.Nebbioso A, Clarke N, Voltz E, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 41.Mai A, Altucci L. Epi-drugs to fight cancer: from chemistry to cancer treatment, the road ahead. Int J Biochem Cell Biol. 2009;41:199–213. doi: 10.1016/j.biocel.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Kristensen LS, Nielsen HM, Hansen LL. Epigenetics and cancer treatment. Eur J Pharmacol. 2009;625:131–142. doi: 10.1016/j.ejphar.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 43.Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol. 2009;19:188–197. doi: 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Hsieh J, Aimone JB, Kaspar BK, et al. IGF-I instructs multipotent adult neural progenitor cells to become oligodendrocytes. J Cell Biol. 2004;164:111–122. doi: 10.1083/jcb.200308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer TD, Markakis EA, Willhoite AR, et al. Fibroblast growth factor-2 activates a latent neurogenic program in neural stem cells from diverse regions of the adult CNS. J Neurosci. 1999;19:8487–8497. doi: 10.1523/JNEUROSCI.19-19-08487.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh J, Nakashima K, Kuwabara T, et al. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.