Abstract
Due to its high specificity, trypsin is the enzyme of choice in shotgun proteomics. Nonetheless, several publications do report the identification of semi-tryptic and non-tryptic peptides. Many of these peptides are conjectured to be signaling peptides or to have formed during sample preparation. It is known that only a small fraction of tandem mass spectra from a trypsin-digested protein mixture can be confidently matched to tryptic peptides. Leaving aside other possibilities such as post-translational modifications and single amino acid polymorphisms, this suggests that many unidentified spectra originate from semi-tryptic and non-tryptic peptides. To include them in database searches, however, may not improve overall peptide identification due to possible sensitivity reduction from search space expansion. To circumvent this issue for E-value based search methods, we have designed a scheme that categorizes qualified peptides ( i.e., peptides whose molecular weight differences from the parent ion are within a specified error tolerance) into three tiers: tryptic, semi-tryptic and non-tryptic. This classification allows peptides belonging to different tiers to have different Bonferroni correction factors. Our results show that this scheme can significantly improve retrieval performance when compared to search strategies that assign equal Bonferroni correction factors to all qualified peptides.
Introduction
Enzymatic digestion of proteins is an essential step in many protocols used in proteomics. Trypsin is often the enzyme of choice due to its high specificity for cleaving at the C-terminal of lysine and arginine, producing positive-charge-retaining tryptic peptides that are suitable for tandem mass spec-trometry.1 The specificity of trypsin has been validated by Olsen et al.2, and recently the “Keil rules” of trypsin were revised in a study by Rodriguez et al.3 Although trypsin is very specific, several publications have reported identifications of semi-tryptic peptides (having one incorrectly cleaved terminal) and non-tryptic peptides (having two incorrectly cleaved terminals) in shotgun proteomics analysis.4–10 The majority of semi-tryptic and non-tryptic peptides detected are postulated to be in-solution post-digestion cleavage products of tryptic peptides.7 This is further supported by the fact that routine analyses yield confident peptide identifications for only about 10% to 30% of all MS/MS spectra analyzed.10–14 To increase the number of explainable MS/MS spectra, it might be fruitful to include into consideration semi-tryptic and non-tryptic peptides in data analyses. Carrying out such an investigation will indeed be of interest to the proteomics community for the following additional reasons. First, many detected semi-tryptic and non-tryptic peptides are potentially signaling peptides.15,16 Second, identified semi-tryptic and non-tryptic peptides can assist with genomic anno-tation17,18 and protein identification.19 Third, searching for semi-tryptic and non-tryptic peptides can potentially increase the number of correctly explained MS/MS spectra along with more confidently identified peptides,7 hence improving the overall retrieval.
Most database search strategies (DSSs) employed for identification of semi-tryptic and non-tryptic peptides consist of multi-pass database searches of various sorts.7–9,20 For example, one may query the database with the enzyme strictness option first set to include tryptic peptides only, then set to include tryptic and semi-tryptic peptides, and lastly set to include tryptic, semi-tryptic and non-tryptic peptides. To speed up the computation required to perform repeated database searches, one can reduce the total number of searches by excluding from next-level-searches MS/MS spectra whose top-ranking peptides have scores above a predetermined cut-off. Another approach to improve speed is to reduce the size of the search space for subsequent searches by removing from the database proteins that do not contain any significantly identified tryptic peptide. One can argue that these DSSs described above are biased towards tryptic peptides.21 In the former case, a spectrum with best tryptic score barely above the threshold might get much more significant semi-tryptic or non-tryptic hits. And this strategy is surely to miss them. For the latter case, the only identifiable semi-tryptic and non-tryptic peptides are limited to the those belonging to proteins containing significant tryptic hits. To avoid this tryptic bias, one may simultaneously search for tryptic, semi-tryptic and non-tryptic candidate peptides. The problem with this strategy, when no additional features are considered, is that tryptic, semi-tryptic and non-tryptic peptides are now viewed as equally likely to appear in the database, causing a decrease in the number of significantly identified peptides. This decrease occurs because here tryptic peptides are competing against all the qualified peptides present in the database, i.e., all peptides are assigned identical Bonferroni’s correction factor (BCF). By qualified peptides, we mean peptides whose molecular weight differences from the parent ion are within the allowed parent ion molecular weight error tolerance (MWET). Evidently, the number of qualified peptides changes with the database searched as well as the parent ion molecular weight queried.
For E-value based database search methods, using an identical BCF can be problematic, especially when the size of search space is allowed to vary.22 The number of qualified peptides in a database, i.e., the size of the search space, in addition to its dependence on the parent ion molecular weight, is controlled by the database search tool’s input parameters, e.g., the enzyme specificity, the number of miscleavage sites allowed, the number of post-translational modifications requested, parent ion mass accuracy, etc. For a typical protein database and for most molecular weights considered, the number of tryptic peptides is smaller than that of semi-tryptic peptides, which is smaller than that of non-tryptic peptides. Therefore, when investigating the contribution of semi-tryptic and non-tryptic peptides to the overall number of identified peptides, it is important to have a database search strategy that is robust against changes in the size of search space. Although in shotgun proteomics different studies have reported detection of semi-tryptic and non-tryptic peptides,4,5,7 by including them in searches the gain in confident identifications has not been investigated when the proportion of false discoveries (PFDs) are computed using E-values. To initiate such an investigation and to evaluate the contribution of semi-tryptic and non-tryptic peptides to peptide identifications, we have employed a variation of an earlier described strategy22,23 for the reasons we describe below.
Several publications22–26 reported search tools with accurate statistical significance. The reasons to use RAId_aPS24 are multifold. First, it provides accurate E-values. Second, being an in-house tool, RAId_aPS can be easily customized so as not to limit the number of miscleavage sites within candidate peptides, see the “Analysis of MS/MS Spectra” subsection of the “Materials and Methods” section for why we choose to do so. Third, for each MS/MS spectrum queried, using RAId_aPS allows us to count separately the numbers of tryptic, semi-tryptic, and non-tryptic peptides by searching the database once. When using other search tools, the same effect might be achieved, but for each spectrum one needs to search the database multiple times and to post-process the results, see the “Search Strategies to be Evaluated” subsection of the “Materials and Methods” section for more details. It is important to note that one should not regard the investigation in this paper as a performance or E-value accuracy evaluation of a database search tool. The primary goal is to investigate the poten- tial gain in confident identifications by including semi-tryptic and non-tryptic peptides in the database searches when different search strategies are used. Therefore, we have also chosen to leave out of consideration identification gains from including other possible sources, such as post-translational modifications (PTMs) and single amino acid polymorphisms (SAPs).
Results from our study suggest that the major factor affecting the contribution from semi-tryptic and non-tryptic peptides to the overall number of identified peptides has to do with how different DSSs estimate the BCF. This implies that DSSs that do not discriminate among tryptic, semi-tryptic and non-tryptic peptides can cause a non-negligible decrease in the overall number of significantly identified peptides at small PFD. Our study also shows that parent ion accuracy, sample load and fractionation scheme have an effect on the contribution from semi-tryptic and non-tryptic peptides to the overall number of significantly identified peptides. However, the effect is less notable when using a DSS that is stable against changes in the size of search space. In conclusion, our study indicates that searching for semi-tryptic or non-tryptic peptides using a DSS similar to the one employed in this study has the potential to increase the overall number of significantly identified peptides.
Materials and Methods
To save the readers’ efforts in going back and forth among the acronyms and their definitions, we provide in Table 1 a complete list of acronyms used in this paper and their definitions.
Table 1. List of Acronyms used.
This table contains all the acronyms used in the manuscript.
| Acronym | Definition |
|---|---|
| BCF | Bonferroni correction factor |
| DG1 | first data group |
| DG2 | second data group |
| DSS | database search strategy |
| FP | false positives |
| MWET | molecular weight error tolerance |
| NTH | number of target hits |
| PCNTH | percentage change in the number of target hits |
| PFD | proportion of false discovery |
| PTM | post-translational modification |
| SAP | single amino acid polymorphism |
| UPS1 | Universal Proteomics Standard One |
| nmw | total number of qualified peptides in the database |
| nσ | total number of MS/MS spectra from a given experiment |
MS/MS data
The first data group (DG1) used in our study is a collection of 15 datasets adding to a total of 40,297 MS/MS spectra. The protein mixture in DG1 is the Universal Proteomics Standard (UPS1) purchased from Sigma Aldrich (St. Louis, Missouri). The UPS1 is composed of 49 known human proteins with molecular weight ranging from 6,000 to 83,000 Daltons (Da). In the samples made, cysteines were reduced with iodoacetamide and the UPS1 proteins were trypsin-digested. The spectra were acquired from an LTQ-orbitrap system (Thermo Electron) with MS acquired from orbitrap and MS/MS from ion trap. Readers interested in further experimental details can look into the Metadata_txt file, which can be downloaded together with the datasets PSM1027, PSM1028 and PSM1029 from the Pep-tidome data repository (ftp://ftp.ncbi.nih.gov/pub/peptidome/samples/PSM1nnn). Table 2 provides a summary of the number of MS/MS spectra collected for each independent analysis under each sample load.
Table 2. Breakdown of the Number of MS/MS Spectra from DG1.
This table gives the total number of MS/MS spectra for each of the three independent analyses performed under five different sample loads.
| Sample Load | Number of MS/MS Spectra | ||
|---|---|---|---|
| Ana.1 | Ana.2 | Ana.3 | |
| 5 fmol | 1,531 | 1,902 | 2,014 |
| 10 fmol | 2,026 | 2,125 | 2,253 |
| 25 fmol | 2,772 | 2,669 | 2,504 |
| 50 fmol | 3,259 | 3,406 | 2,993 |
| 100 fmol | 3,629 | 3,622 | 3,592 |
The second data group (DG2) used in our study is composed of 60 datasets adding to a total of 900,377 MS/MS spectra27 collected in a linear ion trap mass spectrometer (LTQ, Thermo Electron). The sample, prepared according to the published protocol of Whiteaker, et al.27, was composed of a complex protein mixture of human serum that was digested with trypsin. Cys-teine residues were reduced with iodoacetamide except for size fractionation. Table 3 provides a summary of the number of MS/MS spectra collected for each independent analysis under six different fractionation schemes. DG2 was downloaded from the Peptide Atlas data repository at http://www.peptideatlas.org/repository/repository_public_Hs_Plasma2.php.
Table 3. Breakdown of the Number of MS/MS Spectra from DG2.
This table gives the total number of MS/MS spectra for each of the 10 independent analyses performed under six fractionation schemes (FS): magnetic bead separation (C3, C8, WCX), size fractionation (SIZE), protein depletion (A/G) and cysteinyl-peptide enrichment (Cys).
| Number of MS/MS Spectra | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FS | Ana.1 | Ana.2 | Ana.3 | Ana.4 | Ana.5 | Ana.6 | Ana.7 | Ana.8 | Ana.9 | Ana.10 |
| C3 | 7,314 | 19,017 | 18,742 | 18,308 | 15,679 | 18,884 | 18,601 | 7,335 | 7,280 | 7,270 |
| C8 | 7,405 | 19,714 | 19,589 | 19,555 | 19,581 | 11,058 | 12,614 | 11,339 | 14,128 | 7,355 |
| WCX | 7,775 | 19,716 | 19,781 | 19,858 | 19,783 | 18,566 | 19,032 | 7,545 | 7,368 | 7,835 |
| SIZE | 7,279 | 19,738 | 20,105 | 19,571 | 19,532 | 15,837 | 18,315 | 19,743 | 19,371 | 7,305 |
| A/G | 7,395 | 19,334 | 19,387 | 18,350 | 19,021 | 13,109 | 15,141 | 17,622 | 18,635 | 7,408 |
| Cys | 7,740 | 20,251 | 20,365 | 20,015 | 20,019 | 19,409 | 19,448 | 7,610 | 7,560 | 7,735 |
PFD estimation
We wish to use the MS/MS spectra from DG1 and DG2 to evaluate the contribution of semi-tryptic and non-tryptic peptides to the overall number of identified peptides at various fixed PFD values28. Specifically, we wish to obtain NTH(PFD), the number of target hits as a function of PFD, when semi-tryptic and non-tryptic peptides are included in data analysis. For database search tools able to compute accurate E-values, which is the case for RAId_aPS24, the PFD can be routinely estimated using target database alone with the formula below29
| (1) |
In the equation above, E0 is the E-value specified for computing the PFD value, FP(E ≤ E0) is the total number of false positives with E ≤ E0, nσ is the total number of MS/MS spectra from a given experiment, and NTH(E ≤ E0) is the total number of target hits out of nσ MS/MS spectra with E ≤ E0 identified in the target database. Note that while NTH(E ≤ E0) can be obtained by directly counting the number of identified peptides, one has to estimate FP(E ≤ E0) by its expectation value E0nσ, which further emphasizes the need of accurate E-value. The E-value used above in equation (1) is a spectrum-specific measure that already includes the BCF, denoted here by nmw. That is, the reported E-value is given by
| (2) |
where nmw is the total number of qualified peptides in the database. The second BCF, nσ, multiplying the E-value in equation (1) corrects for the total number of MS/MS spectra searched in the target database.
In principle, PFD(E) might be a non-monotonic function of E-value. For example, when one has a small sample that makes FP(E ≤ E0) ≈ nσE0 a poor approximation or when the E-values reported are inaccurate. If this ever happens, one may use a strategy similar to the outlined computation30 of the q-value to make PFD a monotonically increasing function of the E-value. This will make NTH(PFD) an increasing function of PFD, since NTH(E) is an increasing function of the E-value. In all cases we studied, all the PFD(E) seem to be a monotonic function of the E-value.
Analysis of MS/MS spectra
The database searches for the MS/MS spectra were performed using a modified version of RAId_aPS24. For each MS/MS spectrum queried, RAId_aPS counts every qualified database peptide towards either tryptic (having two correctly cleaved terminals), semi-tryptic (having one incorrectly cleaved terminal), or non-tryptic (having two incorrectly cleaved terminals) counters. Thus, within one pass of the target database search, the three counters would record respectively the total numbers of tryptic, semi-tryptic, and non-tryptic peptides. These numbers were then used by RAId_aPS to form the BCFs for the DSSs that would be investigated, see Table 4 for details. As a contrast, other search tools do not further distinguish candidate peptides within a specified search space, or a specified tier. Thus, when semi-tryptic or non-tryptic searches are conducted, they only obtain the total number of candidate peptides within a search space, with the explicit counts of tryptic, semi-tryptic, and non-tryptic pep-tides unspecified. It should be a simple matter for other tool developers to set up additional counters so that the total numbers of tryptic, semi-tryptic, and non-tryptic peptides can also be enumerated with one pass of database search.
Table 4. Bonferroni Correction Factors for different search strategies.
This table illustrates how the Bonferroni correction factors (BCF) are computed for the different database search strategies (DSSs) investigated. Denoted by T, ST and NT are respectively the numbers of qualified tryptic, semi-tryptic and non-tryptic peptides in the target database. DSS-1, DSS-2 and DSS-3 each has a single but different BCF, whereas DSS-4 and DSS-5 each contains several BCFs.
| DSS | Tier | BCF | Domain of qualified peptides |
|---|---|---|---|
| DSS-1 | T | tryptic | |
| DSS-2 | T + ST | tryptic and semi-tryptic | |
| DSS-3 | T + ST + NT | tryptic, semi-tryptic and non-tryptic | |
| DSS-4 | tryptic tier: semi-tryptic tier: |
T T + ST |
tryptic semi-tryptic |
| DSS-5 | tryptic tier: semi-tryptic tier: non-tryptic tier: |
T T + ST T + ST + NT |
tryptic semi-tryptic non-tryptic |
Each dataset analyzed shared the following search parameters in common: daughter ions’ MWET was ± 0.8 Da, cysteines were permanently modified with iodoacetamide, and the E-value cut-off was set to 250. To investigate the effect of search space changes, each of the DG1 datasets was analyzed using three different parent ion MWET: ± 0.03 Da, ± 0.15 Da, and ± 0.45 Da. The target database used for DG1 contains the 49 proteins in the UPS1 mixture. As for specific parameters used in analyzing spectra of DG2, the parent ion MWET was set to ± 3.0 Da according to the MWET specified in the original publication, while the target protein database used is RAId’s Homo sapiens database version (v.09.28.2010) downloaded from (ftp://ftp.ncbi.nlm.nih.gov/pub/qmbp/qmbp_ms/RAId/RAId_Databases/).
All database searches were performed without restricting the number of miscleavage sites within each candidate peptide. This brings in more qualified peptides per spectrum when compared with the scenario where the number of miscleavage sites are limited. Although this is not the optimal choice for realistic peptide identifications, it provides a generic case where one does not need to argue what is the appropriate limit for the number of miscleavage sites per candidate peptide. It is very possible that limiting the allowed number of miscleavages per candidate peptide can lead to better identification baseline (considering only tryptic peptides); however, the scope of this paper does not cover the investigation of the best maximum number of allowed miscleavages per candidate peptide. Since our goal is to examine the effect of including semi-tryptic and non-tryptic peptides in the database searches, not a search tool performance evaluation, the exact location of the baseline does not matter.
Search strategies to be evaluated
To investigate the possible contribution of semi-tryptic and non-tryptic peptides to the overall number of identified peptides along the PFD curve, we used five different database search strategies. We label the five DSSs as follows: DSS-1, DSS-2, DSS-3, DSS-4, and DSS-5. DSS-1 only searches the database for tryptic peptides, DSS-2 and DSS-4 search the database for tryptic and semi-tryptic peptides simultaneously, while DSS-3 and DSS-5 search the database for tryptic, semi-tryptic and non-tryptic peptides simultaneously. The DSSs described above will have database search spaces containing different numbers of peptides, which means that the BCF, nmw, will vary in magnitude, affecting the estimated PFD values computed using equation (1).
DSS-4 and DSS-5 compute the BCF, nmw, differently from DSS-1, DSS-2 and DSS-3. In DSS-1, the BCF equals the total number of qualified tryptic peptides; in DSS-2, the BCF equals the total number of qualified tryptic and semi-tryptic peptides; in DSS-3, the BCF equals the total number of qualified tryptic, semi-tryptic and non-tryptic peptides; whereas in DSS-4 and DSS-5 the BCF is computed in a more elaborate manner using a simplified version of the proposed database search strategy22,23. In essence, DSS-4 and DSS-5 categorize the database peptides (search space) into tiers, each containing a different number of peptides22,23, see Table 4 for details.
This stratification of search space can be and is best adapted by other search tools by their developers. As a user, however, one will need to run database searches for each spectrum multiple times and combine the results: assign to each candidate peptide the best E-value among all of its E-values resulting from various search space sizes. This already makes the analysis process time-consuming and impractical to implement by a non-developer. Even if it is implemented, there are other conditions that may undermine the effectiveness of this post-hoc strategy. In order for the database stratification strategy to be useful, the search engine employed must (1) report accurate statistical significances, and (2) not omit candidate peptides. The former not only is the responsibility of the tool developers but also is beyond the control of a user. The latter is especially important because if the correct candidate peptide that can be brought forward by the stratification strategy is truncated from the report list of a search tool, then even with post-processing that mimics the stratification strategy one still cannot bring out the correct identification. Unfortunately, the majority of search tools employ heuristics to limit the number of peptides reported/scored per spectrum, undermining the feasibility of the naive post-processing procedure. Therefore, we believe the same analyses using other tools are best done by their respective developers who can easily adapt DSS-4 and DSS-5 into their codes. Since our goal is to evaluate different DSSs, not to evaluate the performances of different search tools, we omit the same analyses for other tools. Let us now allude to a possible interpretation of search space stratification.
Stratifying the search space into tiers is effectively assigning to peptides in different tiers different prior probabilities of being false positives. For any given database, our design renders the search space of tryptic peptides to be smaller than that of semi-tryptic peptides, which is smaller than that of non-tryptic peptides. For a trypsin-digested sample, the majority of peptides identified with high confidence are tryptic peptides. Thus, it is logical to assign to tryptic peptides lower prior probabilities of being false positives than that to semi-tryptic and non-tryptic peptides.
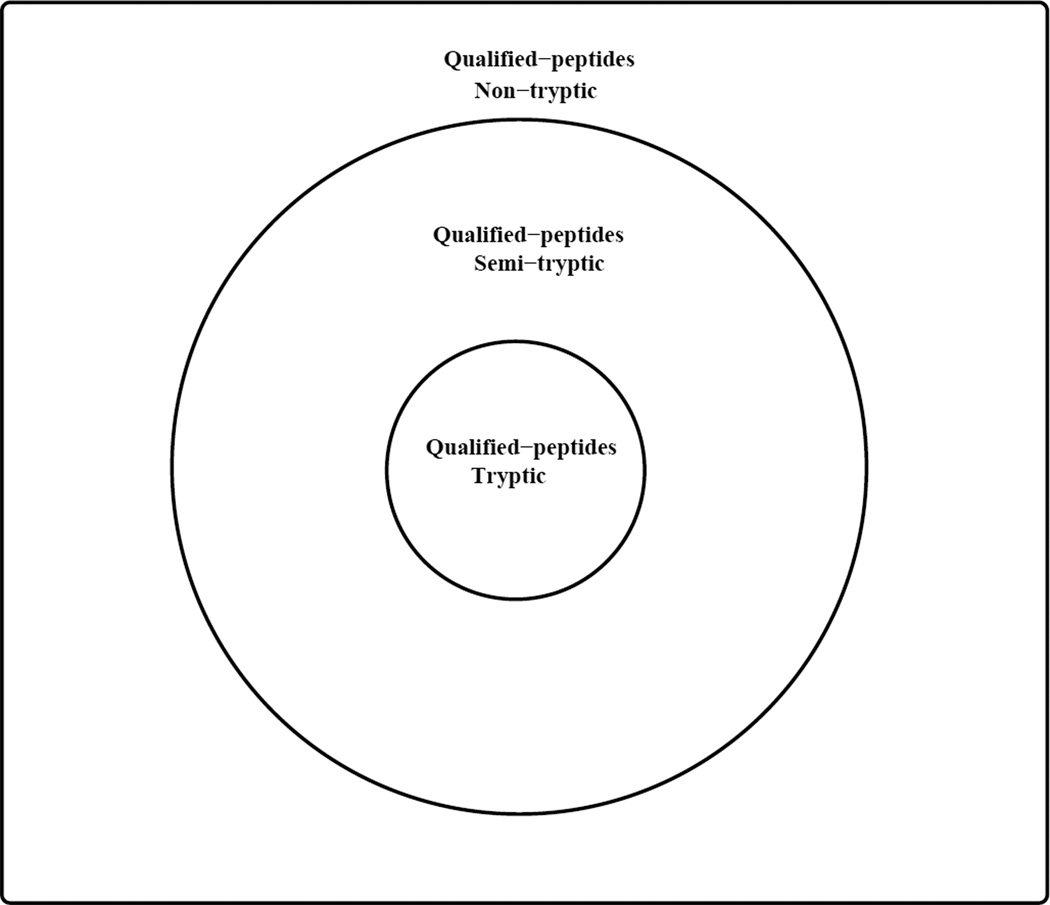
One advantage of DSS-4 and DSS-5 is that peptides belonging to different tiers are assigned different BCFs, nmws, which are used to adjust the P-values for multiple hypothesis (see eq. 2). For example, using DSS-4 and DSS-5, a qualified tryptic peptide (in the tryptic tier) will have a BCF equal to the number of qualified tryptic peptides, and likewise a qualified semi-tryptic peptide (in the semi-tryptic tier) will have a BCF equal to the number of qualified tryptic peptides plus the number of qualified semi-tryptic peptides. Figures 1 and 2 illustrate how the BCFs for DSS-4 and DSS-5 are computed from the numbers of the qualified tryptic, semi-tryptic and non-tryptic peptides.
Figure 1. A schematic illustration of the search space.
The Venn diagram shows how different tiers of qualified peptides are related. When computing the Bonferroni’s correction factor (BCF) using DSS-4 and DSS-5, qualified peptides in the tryptic tier are assigned the same BCF equal to the number of qualified tryptic peptides. However, for qualified peptides in the semi-tryptic tier, their BCF equals the tryptic tier BCF plus the number of qualified semi-tryptic peptides. And for qualified peptides in the non-tryptic tier, their BCF equals the semi-tryptic BCF plus the number of qualified non-tryptic peptides.
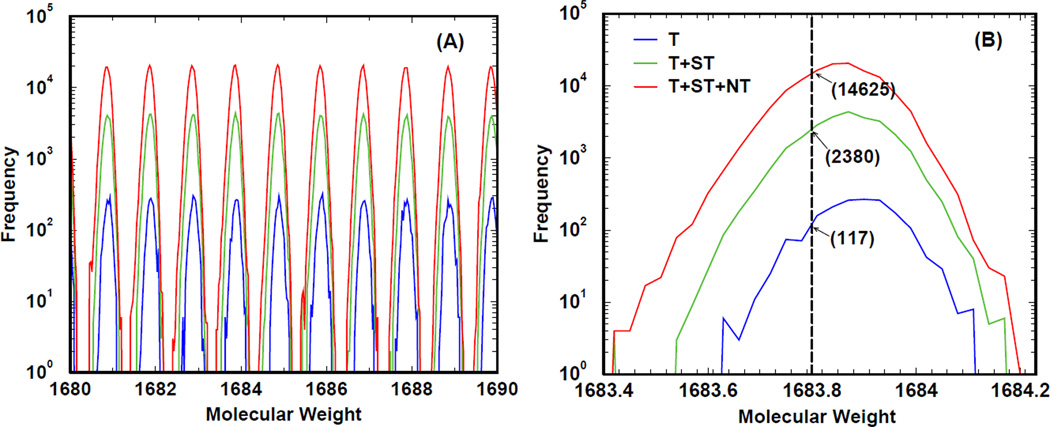
Figure 2. An explicit illustration of the search space sizes in human protein database.
As described in the caption of Table 4, we respectively denote by T, ST and NT the numbers of qualified tryptic, semi-tryptic and non-tryptic peptides in the database. In panel A, we plot as a function of parent ion molecular weight the BCFs of different tiers: tryptic in blue, semi-tryptic in green, and non-tryptic in red. The counts associated with molecular weight w are obtained by summing up the occurrences of qualified peptides whose molecular weights are in the range [w, w + 0.03Da]. Tryptic peptides contribute to all three tiers; semi-tryptic peptides contribute to the semi-tryptic tier and the non-tryptic tier; non-tryptic peptides contribute only to the non-tryptic tier. Note that due to atomic compositions of amino acids, there is a quasi-periodic pattern in terms of number of peptides as a function of the molecular weight. Panel B provides an explicit example of BCFs at a given parent ion 1683.8 Da.
To quantify the contribution of semi-tryptic and non-tryptic peptides to the overall number of identified peptides at different PFD values, we use the percentage change in the number of target hits (PCNTH)
| (3) |
where represents the average of NTHi(PFD) over either three (DG1) or ten (DG2) independent MS/MS experiments. In equation (3), the subscript i in stands for one of the five aforementioned DSSs. For example, i = 1 refers to DSS-1 considering only tryptic peptides, and i = 2 refers to DSS-2 considering both tryptic and semi-tryptic peptides . Also, all the PCNTH values were computed relative to . is the baseline for the current investigation that considers only tryptic peptides from the database.
Analyses and Results
Our two data groups, DG1 and DG2, are composed of multiple independent MS/MS replicas. DG1 consists of five different sample loads of a known protein mixture, and for each sample load analysis via a high-resolution instrument LTQ-Orbitrap (Thermo Electron) were repeated three times. The 15 datasets from DG1 were used to investigate the effects that sample concentration and parent ion mass resolution have on the contribution of semi-tryptic and non-tryptic peptides to the overall number of identified peptides along the PFD curve. DG2 represents a sample of a real, complex biological mixture investigated under different fractionation schemes. For each fractionation scheme, the sample was independently analyzed 10 times. Having 10 independent analyses for each fractionation scheme allowed us to compute expectation values for the contributions of semi-tryptic and non-tryptic pep-tides to the overall number of identified peptides along the estimated PFD curves of each fractionation scheme.
The effect of sample load on the detection of semi-tryptic and non-tryptic pep-tides
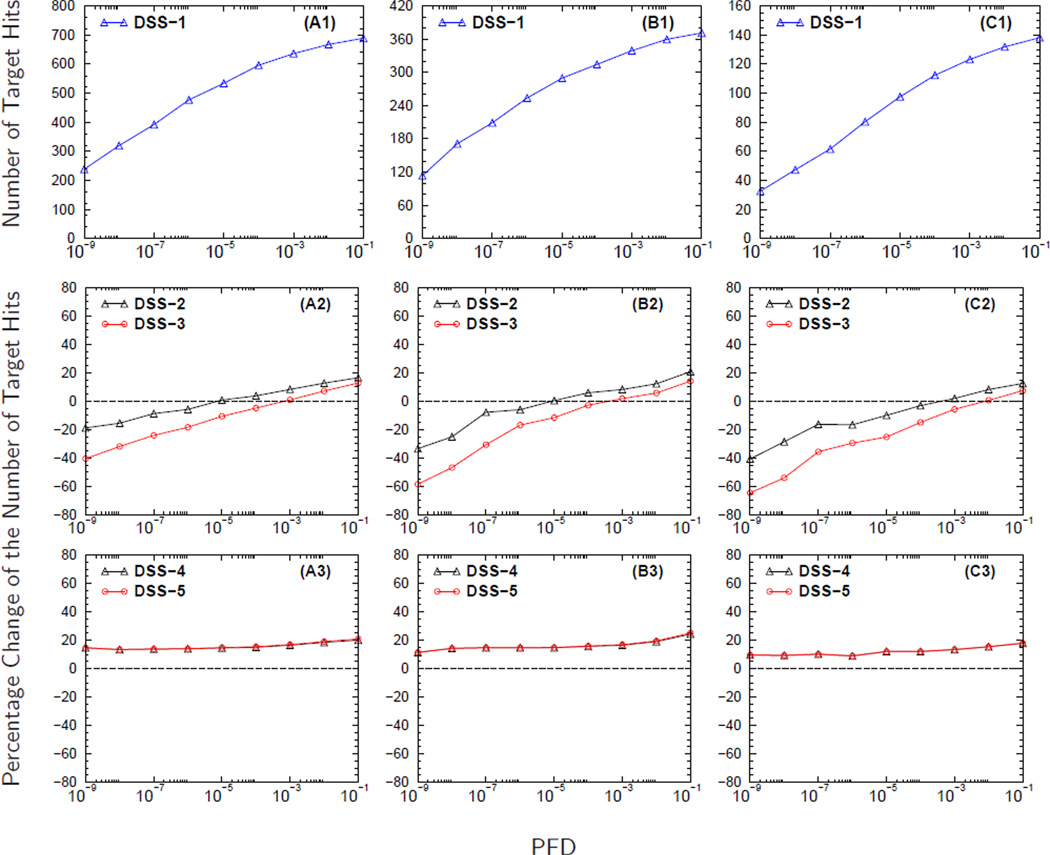
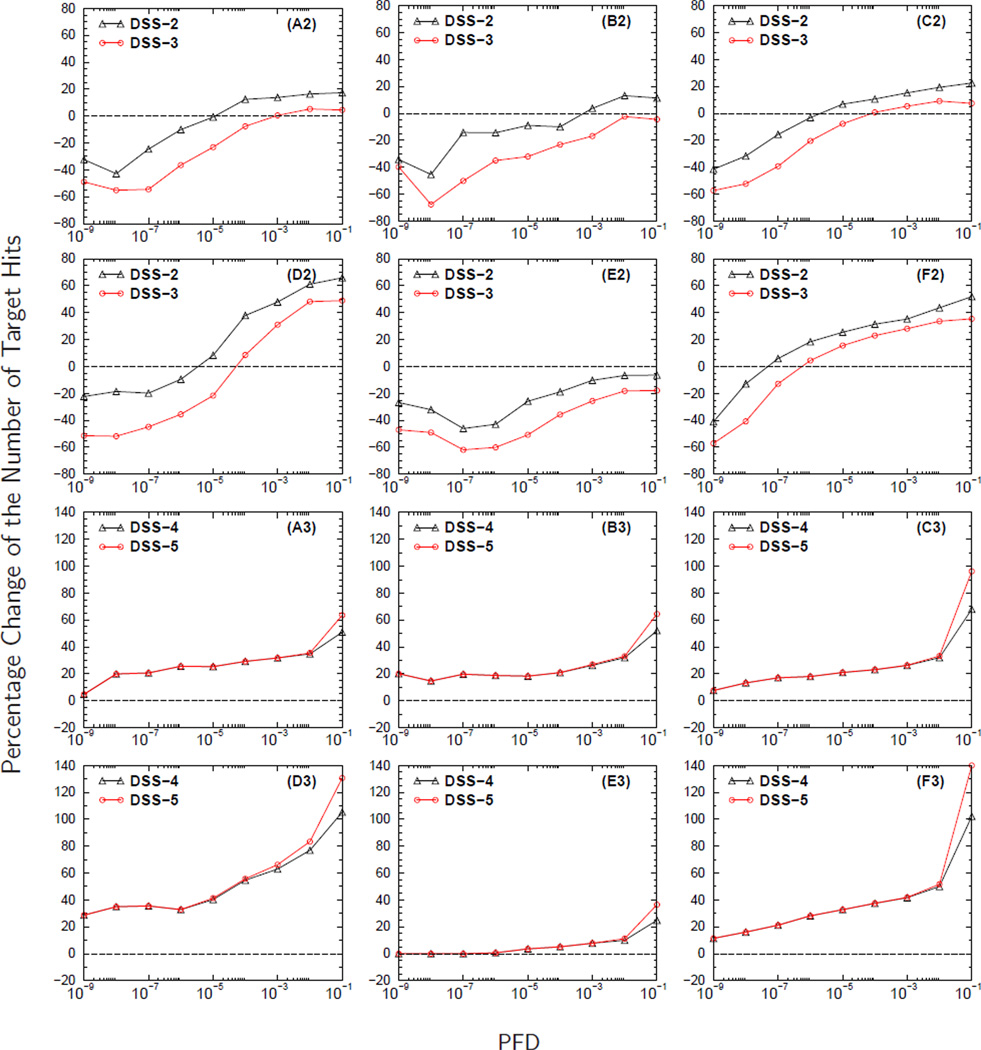
Figure 3 shows the results from analyzing the MS/MS spectra from DG1. The number of MS/MS spectra collected for each independent MS/MS experiment for a fixed sample load is provided in Table 2. The curves in Figures 3A2–3C2 demonstrate the effect that sample load has on the contribution of semi-tryptic and non-tryptic peptides to the PCNTH using DSS-2 and DSS-3. The trend of the curves in Figures 3A2–3C2 shows that as the sample load decreases, from 100 fmol to 5 fmol, it becomes more challenging to identify non-tryptic peptides. The curves in Figure 3A2 have higher starting PCNTH values at small PFD than the curves in Figures 3B2–3C2, indicating that the contribution of significantly identified semi-tryptic and non-tryptic peptides to the overall number of identified peptides decreases as the sample load decreases. A similar trend is observed when a comparison is made among the five sample loads (figure not shown).
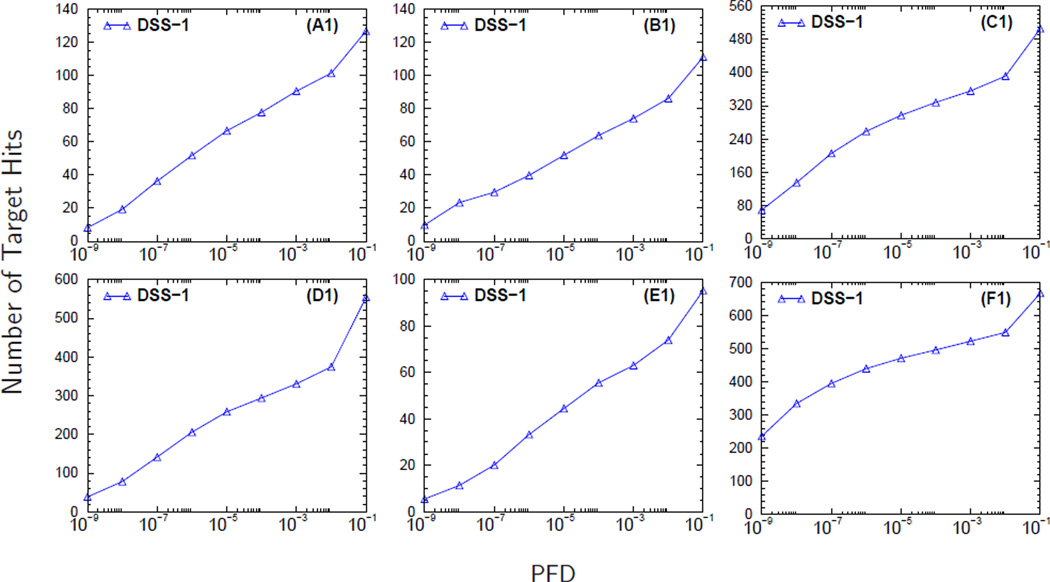
Figure 3. The effect of sample load on the identification of semi-tryptic and non-tryptic peptides.
Panels A1-C1 display the number of target hits (NTH) versus the proportion of false discoveries (PFD) of the baseline search strategy DSS-1. In panels A2-C2 and A3-C3 are the curves for the percentage change in the NTH (PCNTH) versus the PFD for different database search strategies, DSS2-DSS5, computed using equation (3). The curves in panels headed by A, B, and C were computed from analyzing the MS/MS spectra collected from sample loads (DG1) of 100 fmol, 25 fmol, and 5 fmol respectively. The molecular weight error tolerance of the parent ion was set to ± 0.03 Da. during database searches for these three sample loads.
One of the factors that influences the signal to noise ratio of MS/MS spectra is the amount of peptide present in the sample. Therefore, as the sample load decreases, it becomes more difficult to identify peptides regardless of the peptide type. However, the troublesome aspect of the curves in Figures 3A2–3C2 is that the contribution of semi-tryptic and non-tryptic petides to the overall number of identified peptides causes PCNTH to be negative for almost the entire PFD range plotted. The negative value for the PCNTH indicates a problem associated with the DSS employed. A desirable DSS should remain stable against changes in the size of search space, producing always non-negative PCNTH values.
Although the search space of DSS-2 and DSS-3 are equal to that of DSS-4 and DSS-5 respectively, the PCNTH curves obtained from using DSS-4 and DSS-5 have the correct trend of an acceptable DSS showing an increase in the total number of identified peptides. Even though the search space of DSS-5 is larger than that of DSS-4, DSS-5 produces PCNTH curves that are comparable to the PCNTH curves obtained using DSS-4. The curves in Figures 3A3–3C3 also show that as the sample load decreases, the PCNTH curves also decrease, going from an average gain of approximately 20% when the sample load is 100 fmol (Figure 3A3) to an average gain of approximately 15% when the sample load is 5 fmol (Figure 3C3).
The effect of MWET on the detection of semi-tryptic and non-tryptic peptides
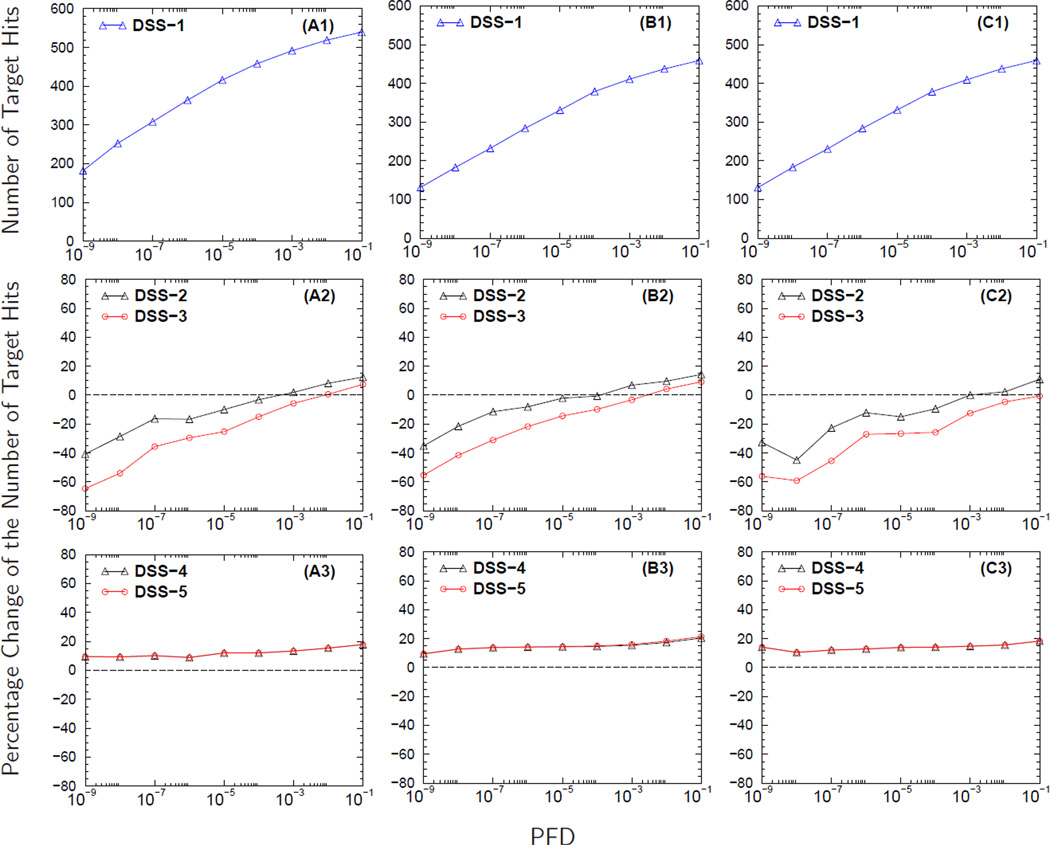
Figure 4 shows how the accuracy of parent ions’ molecular weights affects the detection of semi-tryptic and non-tryptic peptides using the MS/MS spectra from DG1. Figures 4A2–4C2 show that as the parent ions’ MWET increases from±0.03 Da to ±0.45 Da, there is also a decrease in the PCNTH values because relaxing the MWET of the parent ion induces an increase in the search space. This increase in search space causes the BCF to be larger for database searches performed at large MWET. As the BCF increases, the total number of identified peptides with E-value less than or equal to a preset cutoff decreases. The decrease in the number of statistically significant identified peptides causes the estimated value of PFDi to increase regardless of the DSS used.
Figure 4. The effect of molecular weight accuracy of the parent ion on the identification of semi-tryptic and non-tryptic peptides.
Panels A1-C1 display the number of target hits (NTH) versus the proportion of false discoveries (PFD) of the baseline search strategy DSS-1. In panels A2-C2 and A3-C3 are the curves for the percentage change in the NTH (PCNTH) versus the PFD for different database search strategies, DSS2-DSS5, computed using equation (3). Computed from analyzing the MS/MS spectra collected from a sample load of 50 fmol (DG1), curves in panels headed by A, B, and C result from setting the MWET of the parent ion to ± 0.03 Da, ± 0.15 Da and ± 0.45 Da respectively during database searches.
Database search strategies DSS-2 and DSS-3, as shown in Figures 4A2–4C2, produce undesirable PCNTH curves, showing a decrease in the number of statistically significant peptide identifications even when database searches are conducted with small MWET (i.e., ±0.03 Da). Since the MS/MS spectra were acquired from orbitrap instruments that are believed to have good molecular weight accuracy, searching a database with small MWET is expected to perform the best. Nevertheless, DSS-4 and DSS-5 do produce PCNTH curves that have the correct trend, showing an increase in the number of significantly identified peptides. For the three MWET used in our study, the average gain by also searching for semi-tryptic and non-tryptic peptides is about 15% with a possible gain of 20% at 0.1 PFD, as shown in Figures 4A3–4C3. Consistent results for the above mentioned data were obtained for the five sample loads from DG1.
The effect of fractionation scheme on the detection of semi-tryptic and non-tryptic peptides
In Figures 5 and 6 we display the results of analyzing the DG2 MS/MS spectra obtained using the six fractionation schemes shown in Table 3. The curves in Figures 6A2–6F2 show that for DSS-2 and DSS-3 at the low PFD range there is a deterioration in performance when semi-tryptic and non-tryptic peptides are included in database searches. The curves in panels A2-C2 of Figures 3 and 4, also using DSS-2 and DSS-3, show the same problem as well. This deterioration occurs because either DSS-2 or DSS-3 assign equal BCF (or prior probabilities of being false positives) to all candidate peptides. Since the search space of DSS-3 is larger than that of DSS-2, the BCFs of DSS-3 are greater than those of DSS-2, causing the PCNTH for DSS-3 to be worse than that for DSS-2. The PCNTH curves obtained from using DSS-2 or DSS-3 are undesirable. Ideally, a DSS like DSS-3 that covers a larger portion of the peptide space should perform better or comparably to a DSS like DSS-2 that covers a smaller portion of the peptide space.
Figure 5. The retrieval curves of baseline search strategy DSS-1 for different fractionation schemes.
Panels headed by A, B, C, D, E, and F correspond to fractionation schemes C3, C8, WCX, SIZE, A/G, and Cys respectively.
Figure 6. The effect of fractionation scheme on the identification of semi-tryptic and non-tryptic peptides.
Panels headed by A, B, C, D, E, and F correspond to fractionation schemes C3, C8, WCX, SIZE, A/G, and Cys respectively. In panels A2-F2 and A3-F3 are the curves for the percentage change in the number of target hits (PCNTH) versus the proportion of false discoveries (PFD) for the different database search strategies, DSS2-DSS5, computed using equation (3). The curves in panels A2-F2 (A3-F3) were computed from analyzing the MS/MS spectra collected from a complex biological mixture (DG2) with molecular weight error tolerance (MWET) of the parent ion set to ± 3 Da during database searches.
Figures 6A3–6F3 display the PCNTH curves for DSS-4 and DSS-5. The curves obtained from using DSS-4 and DSS-5 have the trend of a desirable DSS. As shown in panels A3-C3 of Figure 3 and Figure 4, adding semi-tryptic and non-tryptic peptides to search space has a positive contribution to the PCNTH curves. Another important result shown by the curves in Figures 6A3–6F3 is that the contribution from semi-tryptic and non-tryptic peptides to the PCNTH curves depends on the fractionation scheme. We must point out that the abrupt increases in the PCNTH of DSS-4 and DSS-5 at large PFD values should not be taken literally. If we carefully examine equation (1), we note that the denominator NTH(E ≤ E0) is dominated by the TP counts at small E-values. At that region, the PCNTH is essentially given by TP/TPbaseline −1, where TP and TPbaseline refer respectively to the number of TP hits of DSS-(4,5) and DSS-1 at the same PFD value. As E-value increases, however, NTH(E ≤ E0) may have comparable contributions from the TP counts and FP counts and the PCNTH is given by (TP+nσE)/(TPbaseline +nσEbaseline)−1, where E and Ebaseline refer respectively to the E-values of DSS-(4,5) and DSS-1 at the same PFD value. At this region, a small discrepancy between the correct E-value and the reported E-value can cause a much larger deviation of the reported PCNTH from its correct value.
The curves in Figure 6D3 show that the MS/MS spectra obtained from using size fractiona-tion scheme have the highest contribution from semi-tryptic and non-tryptic peptides to the PCNTH curves. When using size fractionation scheme, the contribution from semi-tryptic and non-tryptic pep-tides to the PCNTH curves starts with PCNTH around 20% when PFD is equal to 10−9 and reaching a PCNTH greater than75% when PFD reaches10−2 (see Figure 6D3). Figure 6E3 shows that for A/G depletion fractionation scheme, the PCNTH curves benefit the least from the contribution of semi-tryptic and non-tryptic peptides. When using A/G depletion fractionation scheme, the contribution of semi-tryptic and non-tryptic peptides to the PCNTH curves, as shown by the curves in Figure 6E3, starts to become significant only when PFD is greater than 10−4 where the PCNTH reaches 5%. The observed differences in the PCNTH curves from different fractionation schemes are due to different protocols used27. Therefore, the fractionation scheme employed in shotgun proteomics should be taken into consideration while making decisions on whether or not to include semi-tryptic and non-tryptic peptides in database searches.
Comments on search strategies
We should note that for each spectrum, every candidate peptide’s P-value was assigned by RAId_aPS, and upon multiplying by the corresponding BCF (see eq. (2)), the candidate peptide’s E-value was determined. That is, for a given spectrum, as long as the BCFs assigned to a candidate peptide were the same, that candidate peptide would have received the same E-value regardless the search strategies used. As an example, since a semi-tryptic candidate peptide has the same BCF from both DSS-2 and DSS-4, its E-values assigned by DSS-2 and DSS-4 are the same.
When viewing Figures 3–6 as a whole, one observes an interesting trend among search strategies DSS-2, DSS-3, DSS-4, and DSS-5. In general, at low PFD values, DSS-2 yields more target hits than DSS-3. This may be attributed to an enlarged search space (in DSS-3) that reduces the sensitivity. On the other hand, DSS-5 holds up about the same number of target hits when compared to DSS-4. This is because the search space stratification can retain retrieval sensitivity by assigning different BCFs to tryptic, semi-tryptic, and non-tryptic peptides. In the Figures, we have always shown the PCNTH of various search strategies with respect to the baseline, DSS-1. Even though we did not show comparison between DSS-2 (DSS-3) and DSS-4 (DSS-5), it can be argued that under the approximation of eq. (1) all target hits from DSS-2 (DSS-3) are already included in DSS-4 (DSS-5). Consequently, along with the PFD curve of DSS-1, the difference between PCNTHs of DSS-2 (DSS-3) and DSS-4 (DSS-5) with respect to DSS-1 renders directly the number of target hits that are identified via DSS-4 (DSS-5) but not DSS-2 (DSS-3) for the given PFD threshold. We present the arguments below.
Using the approximation, FP(E ≤ E0) ≈ nσE0, in eq. (1), let us consider a fixed E-value threshold Et and compare the target hits from DSS-2 and DSS-4. From the example in the first paragraph of this section, one can infer that the semi-tryptic target hits with E ≤ Et are the same for DSS-2 and DSS-4. For DSS-2, however, its tryptic target hits with E ≤ Et can only be a subset of that for DSS-4. This is because the BCF for tryptic peptides is larger under DSS-2 than under DSS-4. This implies that target hits with E ≤ Et for DSS-2 is always a subset of that for DSS-4. For a given threshold Et , since the numerator in the approximation in eq. (1) remains the same for both DSS-2 and DSS-4 while the denominator is larger for DSS-4, the PFD of DSS-4 is smaller than the PFD of DSS-2. To reach the same PFD value for DSS-2 and DSS-4, one would need to choose a larger E-value cutoff for DSS-4. This would further expand the set of target hits for DSS-4. Therefore, at the same PFD value, every target hit of DSS-2 is also a target hit of DSS-4, but not vice versa. Similar arguments can be used to show that every target hit of DSS-3 is also a target hit of DSS-5 and every target hit of DSS-4 is also a target hit of DSS-5.
Concluding Summary and Outlook
Our study has shown that for database searches, assigning a fixed BCF to all peptides of similar molecular weights decreases the contribution of semi-tryptic and non-tryptic peptides to the overall number of identified peptides. Thus, we have proposed a database search strategy that assigns different BCFs to peptides depending on where they reside in the stratified search space. The proposed database search strategy was shown to be robust when evaluated using different sample loads, parent ion MWET and fractionation schemes. The results from our investigation suggest that existing E-value reporting search tools can benefit from adopting a database search strategy that assigns an adjusted BCF to a qualified peptide based on the tier it belongs to in the stratified peptide space.
Although in this investigation we have only classified qualified peptides into three tiers, the pro- cedure utilized in our study can be generalized to provide finer stratifications. For example, within the tryptic tier, one can introduce another dimension called the number of miscleavage sites. This will separate tryptic peptides into different subtiers, each of which contains qualified tryptic peptides containing different numbers of miscleavage sites. It is also possible to incorporate searching of SAPs and PTMs. These generalizations enlarge the search space, and thus peptide candidates containing SAPs or PTMs will have larger BCFs under the proposed strategy. For features that can be learned from experimental data such as peptide hydrophobicity, the proposed search strategy can also use them for refined stratifications. Many of these possible generalizations await to be further investigated by the proteomics community.
Based on the current study, the DSS-4 and DSS-5 strategies can facilitate the identification of semi-tryptic and non-tryptic peptides, and can be used as a means to improve the overall number of statistically significant identifications of peptides. We believe further investigations along this line may eventually lead to non-negligible improvement in protein identification, the discoveries of signaling peptides, genome annotations and more.
Acknowledgement
We thank the administrative group of the NIH Biowulf Clusters, where all the computational tasks were carried out. This work was supported by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health. Funding for Open Access publication charges for this article was provided by the National Institutes of Health.
References
- 1.Why b, y’s? Sodiation-induced tryptic peptide-like fragmentation of non-tryptic peptides. International Journal of Mass Spectrometry. 2007;268:181–189. Protein Mass Spectrometry: New Technologies and Biological Applications. A Special Issue Honoring Peter Roepstorff on his 65th Birthday. [Google Scholar]
- 2.Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez J, Gupta N, Smith RD, Pevzner PA. Does trypsin cut before proline? J. Proteome Res. 2008;7:300–305. doi: 10.1021/pr0705035. [DOI] [PubMed] [Google Scholar]
- 4.Tsur D, Tanner S, Zandi E, Bafna V, Pevzner PA. Identification of post-translational modifications via blind search of mass-spectra. Proc IEEE Comput Syst Bioinform Conf. 2005:157–166. doi: 10.1109/csb.2005.34. [DOI] [PubMed] [Google Scholar]
- 5.Strader MB, Tabb DL, Hervey WJ, Pan C, Hurst GB. Efficient and specific trypsin digestion of microgram to nanogram quantities of proteins in organic-aqueous solvent systems. Anal. Chem. 2006;78:125–134. doi: 10.1021/ac051348l. [DOI] [PubMed] [Google Scholar]
- 6.Bandeira N, Tsur D, Frank A, Pevzner PA. Protein identification by spectral networks analysis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6140–6145. doi: 10.1073/pnas.0701130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alves P, Arnold RJ, Clemmer DE, Li Y, Reilly JP, Sheng Q, Tang H, Xun Z, Zeng R, Radivojac P. Fast and accurate identification of semi-tryptic peptides in shotgun proteomics. Bioinformatics. 2008;24:102–109. doi: 10.1093/bioinformatics/btm545. [DOI] [PubMed] [Google Scholar]
- 8.Wilmarth PA, Riviere MA, David LL. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J Ocul Biol Dis Infor. 2009;2:223–234. doi: 10.1007/s12177-009-9042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tharakan R, Edwards N, Graham DR. Data maximization by multipass analysis of protein mass spectra. Proteomics. 2010;10:1160–1171. doi: 10.1002/pmic.200900433. [DOI] [PubMed] [Google Scholar]
- 10.Ning K, Fermin D, Nesvizhskii AI. Computational analysis of unassigned high-quality MS/MS spectra in proteomic data sets. Proteomics. 2010;10:2712–2718. doi: 10.1002/pmic.200900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcotte EM. How do shotgun proteomics algorithms identify proteins? Nat. Biotechnol. 2007;25:755–757. doi: 10.1038/nbt0707-755. [DOI] [PubMed] [Google Scholar]
- 12.Webb-Robertson BJ, Cannon WR. Current trends in computational inference from mass spectrometry-based proteomics. Brief. Bioinformatics. 2007;8:304–317. doi: 10.1093/bib/bbm023. [DOI] [PubMed] [Google Scholar]
- 13.Searle BC, Turner M, Nesvizhskii AI. Improving Sensitivity by Probabilistically Combining Results from Multiple MS/MS Search Methodologies. Journal of Proteome Research. 2008;7:245–253. doi: 10.1021/pr070540w. [DOI] [PubMed] [Google Scholar]
- 14.Cannon WR, Rawlins MM, Baxter DJ, Callister SJ, Lipton MS, Bryant DA. Large Improvements in MS/MS-Based Peptide Identification Rates using a Hybrid Analysis. Journal of Proteome Research. 2011;10:2306–2317. doi: 10.1021/pr101130b. [DOI] [PubMed] [Google Scholar]
- 15.Gupta N, Tanner S, Jaitly N, Adkins JN, Lipton M, Edwards R, Romine M, Oster-man A, Bafna V, Smith RD, Pevzner PA. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain MR, Bian S, Liu T, Hu J, Elkabes S, Li H. Altered proteolytic events in experimental autoimmune encephalomyelitis discovered by iTRAQ shotgun proteomics analysis of spinal cord. Proteome Sci. 2009;7:25. doi: 10.1186/1477-5956-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U.S.A. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanner S, Shen Z, Ng J, Florea L, Guigo R, Briggs SP, Bafna V. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansson M, Warell K, Levander F, James P. Membrane protein identification: N-terminal labeling of nontryptic membrane protein peptides facilitates database searching. J. Proteome Res. 2008;7:659–665. doi: 10.1021/pr070545t. [DOI] [PubMed] [Google Scholar]
- 20.Sheng Q, Dai J, Wu Y, Tang H, Zeng R. BuildSummary: Using a Group-Based Approach To Improve the Sensitivity of Peptide/Protein Identification in Shotgun Proteomics. Journal of Proteome Research. 2012;11:1494–1502. doi: 10.1021/pr200194p. [DOI] [PubMed] [Google Scholar]
- 21.Bern M, Kil YJ. Comment on “Unbiased statistical analysis for multi-stage proteomic search strategies”. J. Proteome Res. 2011;10:2123–2127. doi: 10.1021/pr101143m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alves G, Ogurtsov AY, Yu YK. Assigning statistical significance to proteotypic peptides via database searches. J Proteomics. 2011;74:199–211. doi: 10.1016/j.jprot.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alves G, Ogurtsov AY, Yu YK. RAId_DbS: peptide identification using database searches with realistic statistics. Biol. Direct. 2007;2:25. doi: 10.1186/1745-6150-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alves G, Ogurtsov AY, Yu YK. RAId_aPS: MS/MS analysis with multiple scoring functions and spectrum-specific statistics. PLoS ONE. 2010;5:e15438. doi: 10.1371/journal.pone.0015438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klammer AA, Park CY, Noble WS. Statistical Calibration of the SEQUEST XCorr Function. J. Proteome Res. 2009;8:2106–2113. doi: 10.1021/pr8011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiteaker JR, et al. Head-to-head comparison of serum fractionation techniques. J. Proteome Res. 2007;6:828–836. doi: 10.1021/pr0604920. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 29.Sorić B. Statistical “Discoveries” and Effect-Size Estimation. Journal of the American Statistical Association. 1989;84:608–610. [Google Scholar]
- 30.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]