Abstract
The LKB1/STK11 tumor suppressor encodes a serine/threonine kinase which coordinates cell growth, polarity, motility, and metabolism. In non-small cell lung cancer, LKB1 is somatically inactivated in 25-30% of cases, often concurrently with activating KRAS mutation. Here, we employed an integrative approach to define novel therapeutic targets in KRAS-driven LKB1 mutant lung cancers. High-throughput RNAi screens in lung cancer cell lines from genetically engineered mouse models driven by activated KRAS with or without coincident Lkb1 deletion led to the identification of Dtymk, encoding deoxythymidylate kinase which catalyzes dTTP biosynthesis, as synthetically lethal with Lkb1 deficiency in mouse and human lung cancer lines. Global metabolite profiling demonstrated that Lkb1-null cells had striking decreases in multiple nucleotide metabolites as compared to the Lkb1-wt cells. Thus, LKB1 mutant lung cancers have deficits in nucleotide metabolism conferring hypersensitivity to DTYMK inhibition, suggesting that DTYMK is a potential therapeutic target in this aggressive subset of tumors.
Keywords: LKB1, KRAS, DTYMK, CHEK1, NSCLC, GEMM-derived cell line, genome wide RNAi screen, metabolic profiling
INTRODUCTION
LKB1/STK11 functions as a master regulator of cell metabolism and energy stress responses (1, 2). Its best characterized target is AMP-activated protein kinase (AMPK), which, is directly phosphorylated and activated by LKB1 in context of low cellular ATP levels (2). AMPK in turn modulates nutrient utilization to restore energy homeostasis through phosphorylation of multiple substrates controlling nutrient uptake and metabolism (1, 2). LKB1 also activates other members of AMPK-related family of kinases, which regulate diverse aspects of cell metabolism, growth, and polarity (1, 2). LKB1/STK11 deficiency results in broad defects in metabolic control, as evidenced by primary cells and cancer cell lines lacking LKB1 being sensitized to nutrient deprivation and other types metabolic stress (3-5). LKB1 is also a major tumor suppressor which is somatically inactivated in many common types of cancer (3, 4). Human tumor data and genetic studies in mice suggest LKB1 mutant cancers are biologically distinct from those with LKB1 intact (6). Notably, Lkb1 inactivation is the single most prominent biomarker for poor outcome in cervical cancer, predicting a survival of 1 year, as compared to a 10 year survival for Lkb1 wild type tumors (7). In mouse models of lung cancer and melanoma, Lkb1 loss synergizes with active KRAS to drive a highly metastatic phenotype not seen in the context of other combinations of mutations (6, 8). Unfortunately, there are currently few drugs available for clinical use that target LKB1 loss specifically, and recent human cancer cell line screens using >130 drugs under clinical and preclinical investigation failed to identify known anti-cancer agents with strong selective activity in this subset of tumors (data not shown) (9).
Here, we sought to employ an integrative program to systematically identify novel drug targets in LKB1 mutant lung cancer using synthetic lethal RNAi screen and comprehensive metabolomics analysis. For these studies, we took advantage a series of low passage lung cancer cell lines derived from genetically engineered mouse models programmed with common mutations in KRAS and p53, alone or in combination with LKB1. Whereas the heterogeneity of human cancer cell lines can obscure synthetic lethal associations, we predicted this murine cell line panel developed in the context of a well-defined model system, would effectively enable discovery of genotype-driven sensitivities.
RESULTS
Generation of lung cancer cell lines from GEMMs
To generate isogenic lung cancer cell lines, somatic KRAS activation and p53 loss with or without LKB1 inactivation were induced in the lungs of genetically engineered mice (Kras+/LSL− G12DTrp53L/L or Kras+/LSL-G12DTrp53L/LLkb1L/L) by intranasal administration of Adenovirus-Cre as previously described (6). Inactivation of p53 was included in these models as inactivation of p53 is common in human NSCLC (>50%) (10). Tumor nodules from mice of defined genotypes were dissected, minced and cultured, resulting in the derivation of the 634, 855, and 857 lines from Kras+/LSL-G12DTrp53L/L mice (Lkb1-wt) and the t2, t4, and t5 lines from Kras+/LSL− G12DTrp53L/LLkb1L/L mice (Lkb1-null) (Supplementary Fig. S1A). Genotype, LKB1 expression, and epithelial origin of the lines were confirmed by PCR, Western blot, and pan-cytokeratin immunostaining (Supplementary Fig. S1B-S1D). These 6 lines showed similar growth rates (Supplementary Fig. S1E), and further the Lkb1-null lines exhibited lower cellular ATP levels compared to Lkb1-wt cells (Supplementary Fig. S1F).
Identification of selective essential genes in Kras/p53/Lkb1 GEMM-derived cell lines
To identify genes that induce cell death selectively in Lkb1-null lung cancers, a synthetic lethal screen was performed using a pooled 40K murine shRNA lentiviral library with each of the Lkb1-wt and Lkb1-null cell lines. The relative abundance of shRNAs in each cell line sample was determined by deep-sequencing, and for every shRNA, a log2 fold change (log2FC) value was calculated from the difference in relative abundance at a late time point after infection versus the initial shRNA-infected sample. Unsupervised hierarchical clustering analysis of the ranked hairpins from the triplicate pooled shRNA library screens revealed clear clustering of the Lkb1-wt and Lkb1-null cells into distinct groups, and the blue-color in the top-right corner represents genes for which the abundance of shRNAs is significantly reduced in all Lkb1-null cultures, suggesting a specific effect in the inhibition of Lkb1-null cell growth (Fig. 1A). We collapsed the ranked hairpins using two methods, a RIGER analysis (KS t-test based statistics) and a weighted second-best analysis to rank genes that selectively impaired proliferation/viability in Lkb1-null cells. We nominated a union of 344 genes, identified by the top 100 individual hairpins for 88 genes (Supplementary Table S1.1) and the top 200 genes from both the KS (Supplementary Table S1.2) and weighted-second best analysis (Supplementary Table S1.3), as our initial prioritized list (Fig. 1B). 340 shRNAs, targeting 70 candidate genes from this prioritized list, were chosen for validation (Supplementary Table S1.4). These 70 genes consisted of the top 10 candidates from the KS analysis, as well as 60 others involved in a range of biological processes in an attempt to represent all biological categories in the validation process. Validation was performed in an array format and identified 13 genes that displayed ≥ 2 hairpins with a significant growth disadvantage in the Lkb1-null cells (Supplementary Table S1.5). Dtymk, Chek1, Pdhb, and Cmpk1 were the top 4 candidates, each with ≥2 hairpins that scored in the validation assay (Fig. 1C and Supplementary Table S1.5).
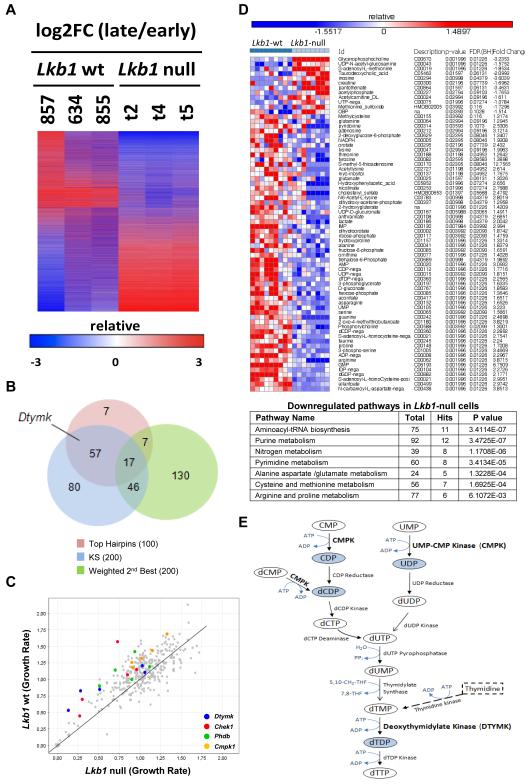
Figure 1. Identifying Dtymk.
A, Unsupervised hierarchical clustering analysis of results from triplicate pooled shRNA library screens of Lkb1-wt and Lkb1-null mouse cancer cell lines based upon log2 fold change (log2FC). Negative numbers (blue) reflect relative depletion of shRNAs at late time points.
B, Two class comparison of Lkb1-null versus Lkb1-wt cell lines was used to generate a ranked hairpin list of selectively essential hairpins in an Lkb1-null background. Hairpins were collapsed to gene values using either the weighted second best or the KS statistic in GENE-E. Venn diagram depicts the overlap of most essential genes in the Lkb1-null background nominated by the top 100 independent hairpins, and the top 200 genes from both weighted second best and KS.
C, Validation study. Relative viability of Lkb1-wt and Lkb1-null cells infected with 340 individual hairpins for 5 days. Genes of interest are highlighted by the colors indicated.
D, Metabolic signature of Lkb1-null lung cancer cells. Unsupervised clustering analysis of metabolomic data from Lkb1-wt and Lkb1-null cells. The heatmap displays those metabolites with the greatest difference between Lkb1-wt and Lkb1-null cell lines, along with compound name (ID), Description (KEGG identification number), and p-value, etc. for the comparison between the two sets of lines. The lower panel shows significantly enriched metabolic pathways in down-regulated components of the Lkb1-null metabolic signature using Pathway Analysis module from MetaboAnalist tool (http://www.metaboanalyst.ca).
E, A comprehensive metabolic map of de novo (solid line) and the salvage (dashed line) pyrimidine deoxyribonucleotide biosynthetic pathway. This map was created with CellDesigner version 4.2 using a template from Panther Classification System Database (www.pantherdb.org). DTYMK is highlighted in Bold. Metabolites depicted in light blue were significantly down-regulated in Lkb1-null cells, respectively.
Metabolomics analysis implicates Dtymk as critical gene in Lkb1-null cells
LKB1 is reported to be involved in metabolic reprogramming (4, 11), we therefore assessed the metabolic profile of Lkb1-wt and Lkb1-null cells and discovered a set of 58 metabolites, including nucleotide metabolites IMP, AMP, ADP, GMP, dGMP, UMP, UDP, CDP, dCDP, and dTDP, that were present at consistently lower levels in Lkb1-null cells (Fig. 1D). Pathway enrichment analysis demonstrated that metabolites in both purine and pyrimidine metabolism were significantly reduced in Lkb1-null compared to Lkb1-wt cells (Fig. 1D, P = 3.5 × 10−7 and 3.4 × 10−5, respectively), including multiple metabolites involved in dTTP synthesis such as dTDP (the product of DTYMK) and UDP/CDP/dCDP (products of CMPK) (Fig. 1E) (12, 13). Collectively, these two independent sets of data suggest that Lkb1 mutant lung cancer cells exhibit alterations in dTTP metabolism and are particularly sensitized to disruption of intracellular dTTP synthesis, and therefore have potential as important targets in Lkb1-null lung cancer.
Dtymk is synthetic lethal gene selectively required for Lkb1-null cell proliferation
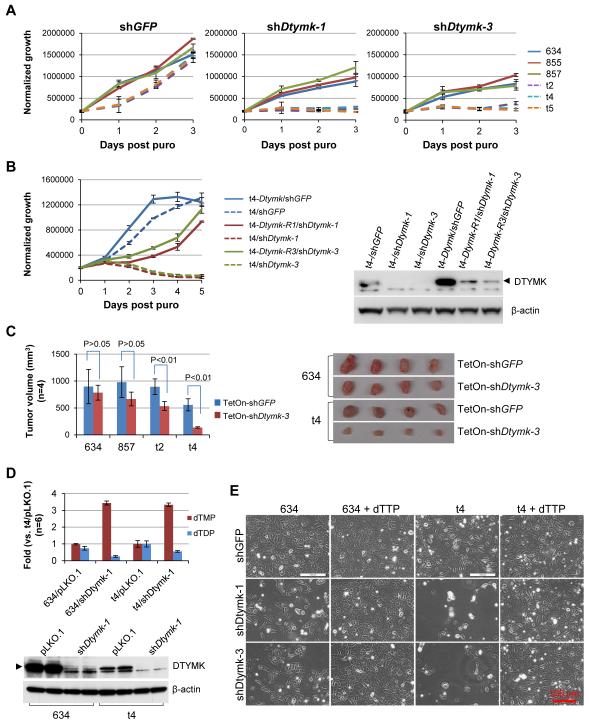
To further examine the role of DTYMK in lung tumorigenesis, we screened 5 shDtymks and identified shDtymk-1 and shDtymk-3 knocking down DTYMK to nearly undetectable levels (Supplementary Fig. S2A and Table S2). Compared to shGFP, both shDtymk-1 and shDtymk-3 strongly inhibited the growth of the Lkb1-null (t2, t4, and t5), while producing a weaker effect in the Lkb1-wt (634, 855, and 857) cell lines (Fig. 2A and Supplementary Fig. S2B). To see if overexpression of shRNA-resistant Dtymk can rescue shDtymk effect, Dtymk-R1 and Dtymk-R3 were cloned into pLenti6 vector and then transduced into Lkb1-null t4 cells. Blasticidin-resistant cells were pooled and further transduced with shGFP, shDtymk-1, or shDtymk-3, respectively. Consistently, shDtymk-1 and shDtymk-3 killed Lkb1-null t4 cells within 3 days, whereas Dtymk-R1 and Dtymk-R3 expression largely restored the growth of shDtymk-1 and shDtymk-3 transduced t4 cells (Fig. 2B). Western blot analysis revealed lower DTYMK signals in t4-Dtymk-R1/shDtymk-1 and t4-Dtymk-R3/shDtymk-3 cells, suggesting that some of the blasticidin-resistant cells were not DTYMK-R positive and thus were killed by shDtymk (Fig. 2B), which likely accounted for the significant but incomplete rescue by Dtymk-R1 or Dtymk-R3. To extend these findings to tumorigenesis in vivo, Lkb1-wt (634 and 857) and Lkb1-null (t2 and t4) cells were transduced with doxycycline-inducible (TetOn) shGFP or shDtymk-3 and then implanted into athymic nude mice. Consistent with the in vitro proliferation assay, doxycycline-induced expression of shDtymk-3 for 3 weeks resulted in a marked impairment in the growth of Lkb1-null tumors while producing more modest effects in the Lkb1-wt tumors (Fig. 2C).
Figure 2. Dtymk is the synthetic lethal target of Lkb1 loss.
A, Lkb1-wt (634, 855, and 857) and Lkb1-null (t2, t4, and t5) cells were transduced with the indicated shRNA for 2 days and then plated into 96-well plates at 2000 cells/well in 150 μl medium with 3 μg/ml puromycin (puro). Viable cells were measured daily using Promega’s CellTiter-Glo Assay. The data represent mean ± SD for 3 replicates.
B, Lkb1-null t4 cells were first transduced with pLenti6-Dtymk, pLenti6-Dtymk-R1, or pLenti6-Dtymk-R3, and selected with blasticidin. The blasticidin-resistant cells were pooled and further transduced with the indicated shRNA for 2 days and then plated for proliferation assay as described in A. The data represent mean ± SD for 3 replicates.
C, 1×106 Lkb1-wt (634 and 857) and Lkb1-null (t2 and t4) cells transduced with the indicated shRNA were implanted into athymic nude mice for 3 weeks. Tumor volume (mm3) was calculated as (length × width2)/2. The data represent mean ± SD for 4 mice. Lkb1-wt 634 and Lkb1-null t4 tumors with the indicated shRNAs were shown.
D, Graph of dTMP and dTDP levels in Lkb1-wt 634 and Lkb1-null t4 cells transduced with the indicated shRNA for 3 days. The data represent mean ± SD for 6 replicates. Expression of DTYMK in these cells at the time of metabolite extraction was determined by Western blotting.
E, Morphology of Lkb1-wt 634 and Lkb1-null t4 cells transduced with the indicated shRNA and then cultured in medium with or without additional 100 μM dTTP for 4 days.
Dtymk knockdown alters pyrimidine metabolism
DTYMK catalyzes the phosphorylation of dTMP to form dTDP, and it is the first merged step of both the de novo and salvage pathways in the production of dTTP (Fig. 1E). We expected that knockdown of Dtymk would inhibit this pathway and lead to accumulation of the substrate dTMP and decrease of the product dTDP. Corresponding metabolite analysis of Lkb1-wt 634 and Lkb1-null t4 cells transduced with shDtymk-1 revealed the expected significant increase in dTMP and moderate decrease in dTDP levels in both cell lines (Fig. 2D), indicating that DTYMK is a major source of dTDP in the cells and underscores the importance of this gene in cancer cell proliferation, as dTDP is required for production of dTTP for DNA synthesis.
dTTP rescues shDtymk growth phenotype
To investigate whether adding dTTP to the medium can rescue shDtymk-induced cell death, Lkb1-wt 634 and Lkb1-null t4 cell lines were transduced with shGFP, shDtymk-1, or shDtymk-3 and cultured in the presence or absence of 100 mM dTTP for 4 days (14). Consistently, shDtymk-1 and shDtymk-3 killed more Lkb1-null t4 than Lkb1-wt 634 cells; but not the cells cultured in medium containing exogenous dTTP (Fig. 2E; confirmation of Dtymk knockdown and incorporation of the exogenous dTTP into DNA are shown in Supplementary Fig. S3A and S3B). Collectively, our data indicate that dNTP metabolism is impaired in Lkb1-deficient lung cancer cells, and that targeting Dtymk is synthetically lethal in this setting.
Lkb1-null cells are more prone to DNA damage than Lkb1-wt cells
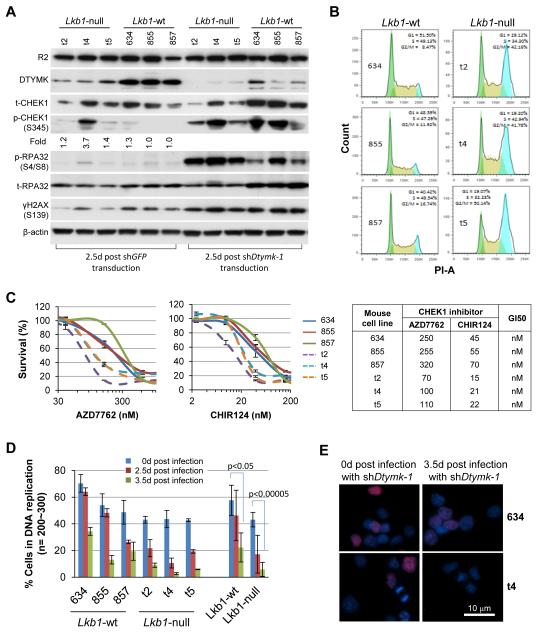
Knockdown of Dtymk will consequently reduce dTTP but also increase dUTP levels. Such changes have been previously linked to dUTP misincorporation and DNA damage, when high expression levels of ribonucleotide reductase R2 subunit, activating nucleotide excision repair (15-18). We noted that while, Lkb1-null and Lkb1-wt cells have similar R2 expression, Lkb1-null cells have much lower DTYMK expression (Fig. 3A), potentially creating a cellular state favorable for dUTP misincorporation. Supportively, Lkb1-null cells have a large 4N peak in DNA content (Fig. 3B) and more sensitive to CHEK1 inhibition (Fig. 3C and Supplementary Fig. S4A), and slightly increased basal phospho-CHEK1 (Fig. 3A), consistent with the activation of a G2 DNA damage checkpoint during replication in Lkb1-null cells (19-22). This pathway appears relevant in vivo. Lkb1-null tumors exhibited increased γH2AX and phospho-CHEK1 signals as compared to Lkb1-wt tumors (Supplementary Fig. S5A-S5C), suggesting there is more DNA damage in vivo than under in vitro culture conditions. In line with this evidence for more DNA damage in Lkb1-null cells, it is notable that Chek1 ranked the second (Fig. 1C and Supplementary Table S1.5), suggesting a dependence of Lkb1-null survival on CHEK1 function. Knockdown of Dtymk shortly (i.e. 2.5 days post shDtymk-transduction) resulted in comparable increases in the phosphorylation of CHEK1 and H2AX in both cell types, whereas the phosphorylation of RPA32 was much more pronounced in Lkb1-null cells (Fig. 3A), suggesting more DNA damage and elevation in nucleotide excision repair in Lkb1-null cells (23). Interestingly, the expression of total RPA32 was increased in Lkb1-wt cells (Fig. 3A), suggesting that LKB1 may positively regulate RPA32 expression following Dtymk knockdown and more DNA damage. Collectively, these data suggest that Lkb1 loss sensitizes cells to DTYMK-depletion induced DNA damage and replication stress, as equivalent knockdown of Dtymk in Lkb1-null and Lkb1-wt cells leads to more robust DNA damage in the Lkb1-null cell lines.
Figure 3. Characterizations of Lkb1-wt and Lkb1-null cell lines.
A, Western blot analyses of the indicated protein expression in Lkb1-wt (634, 855, and 857) and Lkb1-null (t2, t4, and t5) cell liens after shDtymk-1 knockdown. Phospho-CHEK1 Western blot bands were quantified by ImageJ.
B, Lkb1-wt (634, 855, and 857) and Lkb1-null (t2, t4, and t5) cell lines in log-phase growth were fixed with cold 70% ethanol, stained with PI, and then analyzed with flow cytometry. 20,000 cells per line were analyzed.
C, Lkb1-wt (634, 855, and 857) and Lkb1-null (t2, t4, and t5) cell lines were plated into 96-well plates at 2000 cells/well in 150 ml medium containing the indicated concentrations of AZD7762 or CHIR124 for 3 days. Viable cells were then counted with Dojindo’s Cell Counting Kit-8 assay. The data represent mean ± SD for 3 repeats. GI50 was calculated with GraphPad.
D, Lkb1-wt and Lkb1-null cells in 6-well plates were transduced with shDtymk-1. Two sets of the cells were plated into multiple chamber slides: one was 2 days and the other was 3 days post transduction. After overnight culturing, the cells were labeled with 100 μM IdU for 20 min then fixed for indirect immunofluorescence staining with anti-BrdU. The data represent mean ± SD for 200~300 cells.
E, Representative merged images from the cells stained with IdU (red) and DAPI (blue) as described in D are shown.
DNA replication is more sensitive to Dtymk knockdown in Lkb1-null than in Lkb1-wt cells
To examine how the knockdown of Dtymk affects DNA synthesis, Lkb1-wt and Lkb1-null cells were pulse-labeled with IdU for 20 min at 0, 2.5, and 3.5 days post-transduction with shDtymk-1. As shown in Fig. 3D, the proportion of IdU-labeled cells dropped upon Dtymk knockdown regardless of Lkb1 status, although the decrease was much greater in the Lkb1-null cells (dropping from 43.1% to 5.8% in 3.5 days, a decrease of 86.5%) as compared to those with Lkb1-wt (decreasing from 57.7% to 22.3%, a decrease of 61.2%). The lower degree of labeling of Lkb1-null compared to Lkb1-wt cells observed under basal conditions (43.1% vs. 57.7%) may be related to the broad reductions in dNTP metabolism in Lkb1-null cells. After shDtymk transduction, Lkb1-null cells appeared normal within 3 days; whereas by day 4 there was massive cell death leaving virtually no surviving cells, although there was no evidence of apoptosis (data not shown). Here Lkb1-null t4 cells after 3.5 day knockdown of Dtymk, the remaining cells showed deformed and fragmented nuclei, indicative of thymineless death (Fig. 3E) (24-27).
LKB1 mutant human NSCLC cell lines are hypersensitive to DTYMK knockdown
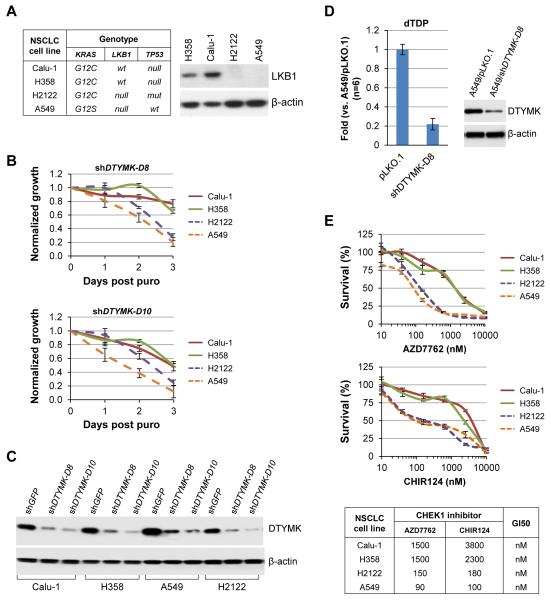
We further sought to determine whether our observations in mouse lung cancer cells could be recapitulated in human LKB1-deficient NSCLC cell lines. We first screened 5 shRNAs targeting DTYMK and identified two, shDTYMK-D8 and shDTYMK-D10, gave efficient knockdown (Supplementary Fig. S6A and Table S2). Next we screened LKB1-wt and LKB1-deficient NSCLC cell lines (Fig. 4A) for proliferation in response to DTYMK knockdown and found that LKB1-deficient H2122 and A549 cell lines had heightened sensitivity as compared to LKB1-wt H358 and Calu-1 cell lines (Fig. 4B). Knockdown of DTYMK was confirmed by Western blot (Fig. 4C). shDTYMK-D10 transduction demonstrated an increased lethality in LKB1-wt H358 and Calu-1 cells, trending towards that of the LKB1-deficient cell lines (Fig. 4B). One possible explanation could involve a differential threshold of DTYMK knockdown, as the remaining DTYMK protein levels after shDTYMK-D10 transduction were lower than that of shDTYMK-D8 (Fig. 4C). This suggests that there may be a differential sensitivity to absolute DTYMK reduction between LKB1-deficient and LKB1-wt cells. DTYMK was reported to be synthetic lethal with doxorubicin in colon cancer cells independent to p53 (28). Consistently, the synthetic lethal interaction of LKB1-deficient and DTYMK knockdown may be independence to p53 status, as LKB1-deficient cell lines with (A549) or without functional p53 (H2122) behave similarly (Fig. 4B).
Figure 4. Knockdown of DTYMK in LKB1-wt and LKB1-mutant NSCLC cell lines.
A, Western blot analyses of LKB1 expression in LKB1-wt (H358 and Calu-1) and LKB1-deficient (H2122 and A549) NSCLC cell lines.
B, LKB1-wt (H358 and Calu-1) and LKB1-deficient (H2122 and A549) NSCLC cell lines were transduced with the indicated shRNAs for 1 day and then selected with 5 μg/ml puromycin (puro) for 2 days in 6-well plates. The cells were collected by trypsin and re-plated into 96-well plates at 2000 cells/well in 150 μl medium containing 5 μg/ml puromycin (puro) and measured daily using Promega’s CellTiter-Glo Assay. The data represent mean ± SD for 3 replicates. The cells left from the re-plating were lysed for Western blot analysis of DTYMK expression (C).
D, Graph of dTDP levels in A549 cells transduced with the indicated shRNA for 4 days. The data represent mean ± SD for 6 replicates. Expression of DTYMK in these cells at the time of metabolite extraction was determined by Western blotting.
E, LKB1-wt (H358 and Calu-1) and LKB1-deficient (H2122 and A549) NSCLC cell lines were plated into 96-well plates at 2000 cells/well in 150 ml medium containing the indicated concentrations of AZD7762 or CHIR124 for 3 days. Viable cells were counted daily using Dojindo’s Cell Counting Kit-8 Assay. The data represent mean ± SD for 3 repeats. GI50 was calculated with GraphPad.
Next we showed that knockdown of DTYMK in A549 cells reduced dTDP levels (Fig. 4D), suggesting DTYMK is a major source of dTDP in human lung cancer cells. We further showed that LKB1-deficient H2122 and A549 were more sensitive than LKB1-wt H358 and Calu-1 cell lines to the treatment with the selected CHEK1 inhibitors (Fig. 4E and Supplementary Fig. S4B), suggesting more DNA damage in LKB1-deficient than in LKB1-wt cell lines. This pathway appears relevant in vivo since LKB1 loss was associated with elevated CHEK1 expression in KRAS-mutant NSCLCs (Supplementary Fig. S6B).
DISCUSSION
In the current study, we create cell lines using Lkb1-null lung tumor nodules and performed multiple screens that identified Dtymk as putative synthetic lethal candidate with Lkb1 loss. Furthermore, we demonstrated that depletion of DTYMK in mouse and human NSCLC cells diminished the dTDP pool and led to greater growth inhibition in Lkb1/LKB1-deficient cells; and that LKB1 loss in mouse and human linked to more DNA damage. These results suggest that DTYMK is a potential therapeutic target in LKB1-mutant human cancer. Additionally, the parallel results observed in both mouse and human cell lines suggest that GEMM-derived tumor cell lines can be used successfully for in vitro synthetic lethal screening.
One possible explanation for the synthetic lethality of Lkb1 loss and Dtymk knockdown is in part because of the lower expression of DTYMK in Lkb1-null cell lines, leading them to be more dependent on the dTTP synthesis pathway. ShDtymk depletes the absolute amount of DTYMK protein below a critical threshold, resulting in thymine-less death in Lkb1-null cells but not in Lkb1-wt cells (Supplementary Fig. S7). Additionally, differing from Lkb1-wt cell lines, Lkb1-null cell lines lack feedback upregulation of RPA32 expression upon Dtymk knockdown (Supplementary Fig. S7). Because RPA32 is involved in binding and stabilizing single-stranded DNA during repair and replication, the lack of feedback upregulation of RPA32 expression may hinder DNA repair. A preliminary study revealed lower Dtymk and Chek1 transcripts in Lkb1-null cell lines (Supplementary Fig. S8A and S8B), suggesting that transcriptional regulation contributes to the lower DTYMK and CHEK1 protein levels in Lkb1-null cell lines. More work will be needed to decipher the roles of LKB1 in regulation of the DTYMK, CHEK1, and RPA32 expression. Although LKB1-deficient NSCLC cell lines did not apparently show less DTYMK expression, the shDTYMK data still suggested a differential sensitivity to absolute DTYMK reduction between LKB1-deficient and LKB1-wt cells. As an essential gene governing dTTP biosynthesis and DNA replication, DTYMK is necessary to all dividing cells and over depletion of DTYMK below the threshold is lethal to all dividing cells, especially to the tumor cells carrying low levels of deoxynucleotide pools yet maintaining fast growth rate. This may explain the eventual death of Lkb1/LKB1-wt cells after shDtymk/DTYMK.
Human DTYMK was cloned by functional complementation of a Saccharomyces cerevisiae cell cycle mutant cdc8, an essential gene for DNA synthesis (29). DTYMK is the first enzymatic step following the convergence of the de novo and salvage pathways in dTTP biosynthesis. In the de novo pathway, the DTYMK substrate dTMP is synthesized from methylation of dUMP by thymidylate synthase (TS). In the salvage pathway, dTMP is produced from phosphorylation of thymidine by thymidine kinase (TK). The next step in both pathways is the DTYMK-mediated phosphorylation of dTMP to form dTDP (30, 31). The production of dTDP is in contrast to that of the other deoxyribonucleotides used in DNA synthesis—dADP, dGDP, dCDP, and dUDP, which are synthesized from ADP, GDP, CDP, and UDP by ribonucleotide reductase (12, 32). Therefore, the unique dTTP biosynthesis pathway made itself good targets for drugs. There are multiple precedents of inhibition of the key enzymes in the de novo dTTP synthesis pathway, including thymidylate synthase by 5-fluorouracil or pemetrexed (15) and ribonucleotide reductase by hydroxyurea (33). We have targeted TS and ribonucleotide reductase in both the mouse and human NSCLC Lkb1/LKB1 cell lines using the same drugs and have not seen any selective effect on Lkb1/LKB1-deficient cell growth, likely because of an escape mechanism from the salvage pathway (Supplementary Fig. S9). In summary, the lack of redundant pathways for dTTP biosynthesis and the vital role of DTYMK in this process together make DTYMK a new anti-cancer target. In this regard, expression of DTYMK is increased in the majority of lung adenocarcinomas in comparison to normal lung (Supplementary Fig. S10A), and elevated DTYMK levels are correlated with poorer survival (Supplementary Fig. S10B). Unfortunately, LKB1 mutation status was not determined in these data sets.
In addition to identifying DTYMK as a potential therapeutic target for LKB1-deficient lung cancer, we have also provided proof of principle that GEMM-derived cancer cell lines can provide a genetically homogeneous and therefore tractable substrate for high-throughput screens to identify novel therapeutic targets. To date, the weakness of genome-wide RNAi screening has been high false discovery rates and cell line-specific off-target effects, and consequently, despite the implementation of multiple very large scale cell line screening programs, few actionable genotype-associated sensitivities have been uncovered to date. Our study suggests that integrating RNAi screening with metabolite profiling is an effective strategy to leverage the strengths and mitigate the weaknesses of each approach. We propose that this approach will have broad applicability, and enable more rapid development of additional targeted therapeutics for an array of genetic abnormalities occurring in cancer.
METHODS
Detailed protocols for all sections are described in Supplementary Methods.
RNAi screening and metabolite profiling
Large-scale pooled screening and data analysis were performed at the Broad Institute’s RNAi Platform as recommended (34), and metabolite extraction and targeted mass spectrometry analysis were performed as reported (35, 36).
Cell lines and cell culture
Fresh murine lung tumor nodules were minced and cultured in 100 mm dishes with RPMI 1640/10% FBS/1% pen-strep. Calu-1, H358, H2122, and A549 (obtained from ATCC) were cultured in 1640/10% FBS/1% pen-strep; and 293ft (Invitrogen) was cultured in DMEM/10% FBS/1% pen-strep. All cells were cultured at 37°C in a humidified incubator with 5% CO2.
Plasmid constructs and mutagenesis
pLKO.1-shRNAs were purchased from Broad Institute. DTYMK (BC030178) cDNAs was purchased from Thermo Scientific. ShRNA-resistant DTYMKs were made by mutagenesis PCR and subcloned into the BamH I and XhoI sites of pLenti6 vector (Invitrogen). All mutagenized cDNAs were confirmed by sequencing.
Multiple routine in vitro studies
Lentiviral production and target cell transduction, proliferation assay, RT-qPCR, Western blot, flow cytometry, and immunofluorescence microscopy were performed as described in the Supplementary Methods.
In vivo study
Lkb1-wt and Lkb1-null cells were transduced with pTetOn-shGFP (puro) or pTetOn-shDtymk-3 (puro) and then 1 million of puro-resistant cells per transduction were implanted into athymic nude mice. When tumors grew to a diameter of 3 mm, the mice were maintained on doxycycline diet for 3 weeks to allow 634/shGFP tumors reach about 1000 mm3.
Supplementary Material
Acknowledgements
We thank Ozan Alkan for helping RNAi screen; Hin-Koon Woo, Min Yuan and Susanne Breitkopf for conducting mass spectrometry; and Jacob B. Reibel for proofreading.
Grant Support. This work is supported by the NIH (CA122794, CA140594, CA163896, CA166480, CA154303, and Lung SPORE P50CA090578), United against Lung Cancer Foundation, American Lung Association, and Susan Spooner Research Fund (KKW); and by the NIH CA142794 as well as Damon Runyon Cancer Research Foundation (WYK)
Footnotes
Disclosure of Potential Conflicts of Interest: Dr Wong owns equity in, receives compensation from G1Therapeutics, serves as a consultant to Molecular MD, and sponsored research agreement with AstraZeneca, Infinity Pharmaceuticals and Millennium Pharmaceuticals. Dr. Cantley owns equity in, receives compensation from Agios Pharmaceuticals, and serves on the Board of Directors and Scientific Advisory Board of Agios Pharmaceuticals. Agios Pharmaceuticals is identifying metabolic pathways of cancer cells and developing drugs to inhibit such enzymes in order to disrupt tumor cell growth and survival. Kevin Marks, Sung E Choe, and Edward M Driggers are employees of Agios Pharmaceuticals. Dr. Kimmelman is a consultant for Forma Therapeutics.
The Editor-in-Chief of Cancer Discovery (Lewis C. Cantley) is an author of this article. In keeping with the AACR’s Editorial Policy, the paper was peer reviewed and a member of the AACR’s Publications Committee rendered the decision concerning acceptability.
SIGNIFICANCE Using cell lines derived from the lung cancers occurring in genetically engineered mice, we conducted an integrative genome-wide shRNA and metabolite screen to identify DTYMK as a potential therapeutic target in Kras/Lkb1 mutant lung cancer. We believe that DTYMK is tractable for the development of novel therapeutics, and demonstrate an integrative approach to target identification that reduces false-positive candidates and should have broad applicability for the development of targeted therapeutics.
REFERENCES
- 1.Wodarz A, Nathke I. Cell polarity in development and cancer. Nature cell biology. 2007;9:1016–24. doi: 10.1038/ncb433. [DOI] [PubMed] [Google Scholar]
- 2.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nature cell biology. 2011;13:1016–23. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–63. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes & development. 2009;23:537–48. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 7.Wingo SN, Gallardo TD, Akbay EA, Liang MC, Contreras CM, Boren T, et al. Somatic LKB1 mutations promote cervical cancer progression. PloS one. 2009;4:e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Monahan KB, Pfefferle AD, Shimamura T, Sorrentino J, Chan KT, et al. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer cell. 2012;21:751–64. doi: 10.1016/j.ccr.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mogi A, Kuwano H. TP53 mutations in nonsmall cell lung cancer. Journal of biomedicine & biotechnology. 2011;2011:583929. doi: 10.1155/2011/583929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen M, Ten Klooster JP, Offerhaus GJ, Clevers H. LKB1 and AMPK family signaling: the intimate link between cell polarity and energy metabolism. Physiological reviews. 2009;89:777–98. doi: 10.1152/physrev.00026.2008. [DOI] [PubMed] [Google Scholar]
- 12.Su JY, Sclafani RA. Molecular cloning and expression of the human deoxythymidylate kinase gene in yeast. Nucleic acids research. 1991;19:823–7. doi: 10.1093/nar/19.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Rompay AR, Johansson M, Karlsson A. Phosphorylation of deoxycytidine analog monophosphates by UMP-CMP kinase: molecular characterization of the human enzyme. Molecular pharmacology. 1999;56:562–9. doi: 10.1124/mol.56.3.562. [DOI] [PubMed] [Google Scholar]
- 14.Taricani L, Shanahan F, Pierce RH, Guzi TJ, Parry D. Phenotypic enhancement of thymidylate synthetase pathway inhibitors following ablation of Neil1 DNA glycosylase/lyase. Cell cycle. 2010;9:4876–83. doi: 10.4161/cc.9.24.14155. [DOI] [PubMed] [Google Scholar]
- 15.Van Triest B, Pinedo HM, Giaccone G, Peters GJ. Downstream molecular determinants of response to 5-fluorouracil and antifolate thymidylate synthase inhibitors. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2000;11:385–91. doi: 10.1023/a:1008351221345. [DOI] [PubMed] [Google Scholar]
- 16.Marenstein DR, Wilson DM, 3rd, Teebor GW. Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA repair. 2004;3:527–33. doi: 10.1016/j.dnarep.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Pogribny IP, Muskhelishvili L, Miller BJ, James SJ. Presence and consequence of uracil in preneoplastic DNA from folate/methyl-deficient rats. Carcinogenesis. 1997;18:2071–6. doi: 10.1093/carcin/18.11.2071. [DOI] [PubMed] [Google Scholar]
- 18.Hu CM, Yeh MT, Tsao N, Chen CW, Gao QZ, Chang CY, et al. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer cell. 2012;22:36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 19.Lossaint G, Besnard E, Fisher D, Piette J, Dulic V. Chk1 is dispensable for G2 arrest in response to sustained DNA damage when the ATM/p53/p21 pathway is functional. Oncogene. 2011;30:4261–74. doi: 10.1038/onc.2011.135. [DOI] [PubMed] [Google Scholar]
- 20.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20:7453–63. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 21.Tse AN, Rendahl KG, Sheikh T, Cheema H, Aardalen K, Embry M, et al. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:591–602. doi: 10.1158/1078-0432.CCR-06-1424. [DOI] [PubMed] [Google Scholar]
- 22.Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Molecular cancer therapeutics. 2008;7:2955–66. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 23.Friedberg EC. How nucleotide excision repair protects against cancer. Nature reviews Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer research. 2002;62:3659–62. [PubMed] [Google Scholar]
- 25.Knock E, Deng L, Wu Q, Lawrance AK, Wang XL, Rozen R. Strain differences in mice highlight the role of DNA damage in neoplasia induced by low dietary folate. The Journal of nutrition. 2008;138:653–8. doi: 10.1093/jn/138.4.653. [DOI] [PubMed] [Google Scholar]
- 26.Fonville NC, Vaksman Z, DeNapoli J, Hastings PJ, Rosenberg SM. Pathways of resistance to thymineless death in Escherichia coli and the function of UvrD. Genetics. 2011;189:23–36. doi: 10.1534/genetics.111.130161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuong KJ, Kuzminov A. Disintegration of nascent replication bubbles during thymine starvation triggers RecA- and RecBCD-dependent replication origin destruction. The Journal of biological chemistry. 2012;287:23958–70. doi: 10.1074/jbc.M112.359687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu CM, Chang ZF. Synthetic lethality by lentiviral short hairpin RNA silencing of thymidylate kinase and doxorubicin in colon cancer cells regardless of the p53 status. Cancer research. 2008;68:2831–40. doi: 10.1158/0008-5472.CAN-07-3069. [DOI] [PubMed] [Google Scholar]
- 29.Huang SH, Tang A, Drisco B, Zhang SQ, Seeger R, Li C, et al. Human dTMP kinase: gene expression and enzymatic activity coinciding with cell cycle progression and cell growth. DNA and cell biology. 1994;13:461–71. doi: 10.1089/dna.1994.13.461. [DOI] [PubMed] [Google Scholar]
- 30.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annual review of biochemistry. 1988;57:349–74. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 31.Arner ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacology & therapeutics. 1995;67:155–86. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 32.Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends in biochemical sciences. 1992;17:119–23. doi: 10.1016/0968-0004(92)90249-9. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu Rev Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 34.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nature protocols. 2012;7:872–81. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Analytical chemistry. 2011;83:7074–80. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.