Abstract
High-throughput sequencing for microRNA (miRNA) profiling has revealed a vast complexity of miRNA processing variants, but these are difficult to discern for those without bioinformatics expertise and large computing capability. In this article, we present miRNA Sequence Profiling (miRspring) (http://mirspring.victorchang.edu.au), a software solution that creates a small portable research document that visualizes, calculates and reports on the complexities of miRNA processing. We designed an index-compression algorithm that allows the miRspring document to reproduce a complete miRNA sequence data set while retaining a small file size (typically <3 MB). Through analysis of 73 public data sets, we demonstrate miRspring’s features in assessing quality parameters, miRNA cluster expression levels and miRNA processing. Additionally, we report on a new class of miRNA variants, which we term seed-isomiRs, identified through the novel visualization tools of the miRspring document. Further investigation identified that ∼30% of human miRBase entries are likely to have a seed-isomiR. We believe that miRspring will be a highly useful research tool that will enhance the analysis of miRNA data sets and thus increase our understanding of miRNA biology.
INTRODUCTION
High-throughput sequence (HTS) profiling of microRNAs (miRNA) is becoming increasingly affordable and the technology of choice for many researchers, as it not only informs on transcript abundance but also reveals the complexity of miRNA processing. The size and complexity of HTS mapped data sets imposes great challenges in efficiently processing, visualizing and sharing all the biological information. Typically, at the completion of a study, the most obvious differences identified in HTS data sets are published, but the bulk of the data is deposited in sequence data archives; the wealth of information here is accessible only to those with time and bioinformatics capabilities, and even then there is a distinct lack of published bioinformatic tools that will aid in calculating and visualizing the data. Although there are a growing list of web tools and databases that provide visualization of sequence reads to individual miRNA within data sets (1–8), their implementation is often cumbersome and not amenable to sophisticated interrogation of large data sets. Additionally, these resources do not provide a method to deeply analyze new data sets. With the challenges encountered in preparing a mapped HTS data set, it is likely that a significant majority of data sets remain underutilized.
To address these challenges, we created a pipeline that will produce a stand-alone interactive miRNA Sequence Profiling (miRspring) document. This document is small in size, yet completely reproduces the mapped data set; it provides visualization of the data as well as tools that will calculate miRNA processing statistics which were based on our previous detailed analysis of miRNA datasets (9). The document itself is a JavaScript encoded HTML file that is compatible with all major browser types across all computing platforms. We envisage that the miRspring document could increase accessibility and transparency of data sets by being used as a supplement for all HTS miRNA profiling publications.
MATERIALS AND METHODS
miRspring pipeline
The aim of the miRspring software document is to provide portability and a universal method to extract miRNA processing information from high-throughput sequencing data sets. The pipeline that generates the miRspring document is dependent on three resources: SAMTOOLS (10), miRBase database sequence files (4) and the mapped sequencing data in BAM format (Figure 1A). Although the default pipeline is designed to extract miRNA sequences present in miRBase, it is also possible to incorporate other small RNAs by adding sequence and genomic coordinates into the miRBase files (refer to web site or manual in Supplementary Data Set 1). Similarly novel miRNA precursor sequences, which cannot be identified directly in miRspring, can also be incorporated into the miRspring document.
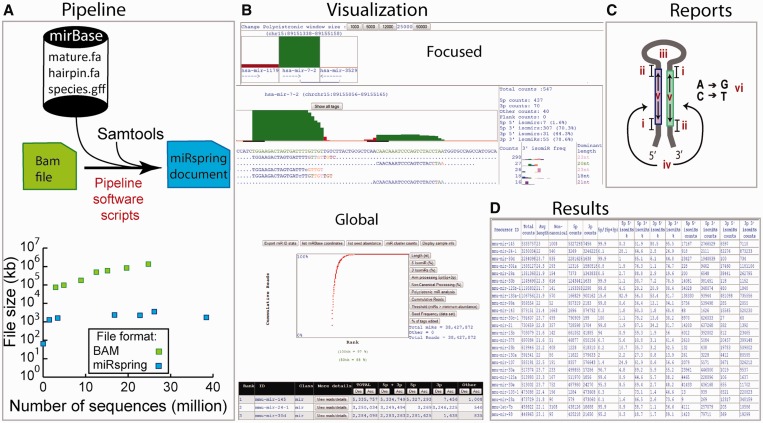
Figure 1.
Overview of the workflow for miRspring software. (A) The pipeline that creates the miRspring document uses custom scripts which access local miRBase data files, the mapped sequence BAM file and Samtools. Multiple BAM files can be used to create the final miRspring document, which has a considerably smaller file size. (B) Many parameters of the data set can be visualized through the ‘global’ or ‘focused’ visualization modes of miRspring. (C) The miRspring document reports on all aspects of miRNA-processing including (i) 5′ isomiRs, (ii) 3′ isomiRs, (iii) non-canonical processing, (iv) arm bias, (v) miRNA length and (vi) RNA editing. (D) Finally, the tabulated data sets can be copied to spreadsheet or statistical software for further in-depth analysis.
The sequencing data within a miRspring document is compressed by storing the precursor sequence once and then indexing all sequence reads with the corresponding starting position, length and number of times present in the data set. This provides a high level of data compression and makes the final miRspring document significantly smaller than an equivalent BAM file (Figure 1A). Additionally, the final document does not require any external data source or internet connectivity, thus making it completely independent and portable; it can even operate on a smart phone or tablet device. Technical documents that describe how to install the miRspring pipeline and how to use the miRspring document are located on the miRspring web site. The miRspring web site contains additional information preparing the input BAM files.
Visualization modes
The miRspring document provides navigation between ‘global’ and ‘focused’ visualization modes of miRNA data (Figure 1B), and a number of features within these views can be customized. The ‘global’ visualization mode is the first page that is loaded on opening the document and displays an XY scatter plot and a tabulated list of miRNAs and associated counts. Users can navigate from the ‘global’ to the ‘focused’ visualization mode by either selecting the ‘detail’ button located on each row of the table or selecting a data point on the XY scatter plot. The ‘focused' visualization mode displays sequence, bar graphs and isomiR processing statistics relating to sequence data of the selected miRNA. Sequencing data are ranked by abundance and displayed in either a novel 5′ condensed or traditional verbose format. The novel 5′ condensed format merges all sequences starting at a common 5′ position onto one line (Supplementary Figure S1). Variations at the 3′ end are highlighted by a colored nucleotide, and the frequency of each isomiR is graphed in the adjacent column with the corresponding color. The length of the most abundant 3′ isomiR is displayed in the rightmost column.
miRNA processing calculations
During the creation of a miRspring document, the coordinates of all sequences from a BAM file are compared to the 5′ starting position of miRBase-defined processed miRNAs. Sequences that start within 3 nt (referred to as the 5′ miRBase window) of the 5′ end of a miRBase defined processed miRNA entry are included in the total count for that miRNA. This classification system has been used by others (11), and the total counts are displayed in the main table of the global visualization page. The miRspring document uses the total counts to calculate the fraction of reads that relate to various processing features of miRNAs (Figure 1C). miRspring can be instructed to analyze the whole data set to generate a tabulated report that calculates all miRNA-processing events (Figure 1D), which can then be easily copied for downstream analysis in programs such as Excel or R. User can modify the size of the 5′ miRBase window value either within a browsing session (through the options menu) or modifying the global parameters when creating miRspring documents.
Seed analysis
Seed visualization and reporting tools transforms all transcripts encoding identical seeds into single data points. Data can be displayed as a XY scatter plot or tabulated form that be easily copied into excel or R for downstream analysis.
Availability
The miRspring pipeline and example documents are freely available online at http://mirspring.victorchang.edu.au. Technical documents and training screen casts are also provided through the web site.
Analysis of public data sets
A number of human publically accessible data sets were used to demonstrate the versatility of the miRspring document. From hereon, we refer to these data sets as follows:
‘Tissue Altas’. This refers to data sets generated from a number of tissues using two different versions of a SOLiD library prepation kit (12). Multiple raw sequence files exist for each library, and each one was mapped using the Lifescope™ small RNA pipeline to generate BAM files. Each BAM file was converted to a separate miRspring ‘intermediate file’ (refer software manual in Supplementary Data Set 1). Intermediate files for the same library were then concatenated and converted into a final miRspring document.
‘AGO IP’. This refers to data sets generated from Argonaute immunoprecipitations (13). Together, this data set comprises three raw sequence files, one each for AGO1, AGO2 and AGO3 immunoprecipitations, which were mapped using the Lifescope™ small RNA pipeline to generate BAM files. Each BAM file was then converted into a miRspring document.
‘ENCODE’. This refers to data sets from release 1 and 2 of the CSHL encyclopedia of DNA elements (ENCODE) project (14,15). Mapped BAM data sets were downloaded directly from the ENCODE repository on the UCSC web browser and converted into miRspring documents.
The accession numbers or download paths for all the aforementioned data sets are provided as additional information (Supplementary Table S1).
Quality control analysis
Custom scripts were used to extract miRNA length, non-canonical processing and mismatches from miRspring documents. For each of these features, the average value was calculated for each rank centile. For the ‘Tissue Atlas’, we only calculated values from the Whole Transcriptome Analysis Kit (WTAK) data, which corresponds to the most recent manufacturers kit release.
Identifying processed miRNAs from miRBase-defined precursors
Each human tissue data set was mapped and converted to a miRspring document and used the ‘global view’ table sorting feature to identify abundant non-canonical processed miRNAs. We only considered those miRNAs having non-canonical counts >5000 and with no miRBase-defined entry for either the 5p or 3p arm (i.e. having a value of ‘undefined’). We identified the most abundant sequence by navigating to the ‘focused’ view for those short listed miRNAs.
Cluster analysis
Using the miRspring document for each human tissue data set, we downloaded the miRNA cluster report (via the ‘miR cluster counts’ button) for those miRNAs located <25 kb from each other. We primarily wanted to identify pre-miRs within clusters that were differentially expressed across different tissues. To do this, we used custom scripts to identify ‘active’ clusters having at least one pre-miR sequence with >100 sequences aligning to it. The expression levels of miRNAs within these miRNA-clusters were ranked for each tissue data set. We then only considered miRNA clusters that had the same rank order in data sets generated from two related library prep kits. The rank orders of each short listed miRNA cluster were then compared with the corresponding clusters other data sets. Candidate clusters were identified where a pre-miRNA rank increased or decreased by at least one rank in any tissue data set.
IsomiR and seed distribution analysis
We downloaded the seed distribution report from the miRspring document created from human Tissue Atlas data sets. Custom scripts were then used to count seed sequences identified in the data set and cross reference to miRbase entries.
RESULTS
To demonstrate the research versatility of the miRspring document, we remapped or directly obtained BAM files from previously published data sets (12–15) and converted them into miRspring documents (Supplementary Data Set 2). In total, ∼895 million sequence tags aligned to miRBase v19 precursors, which were distributed across 73 miRspring documents needing <55 megabytes of disk space (Supplementary Table S1). Our analysis focused on three areas: (i) quality control parameters, (ii) miRNA genomic clusters and (iii) miRNA seed analysis.
Quality control
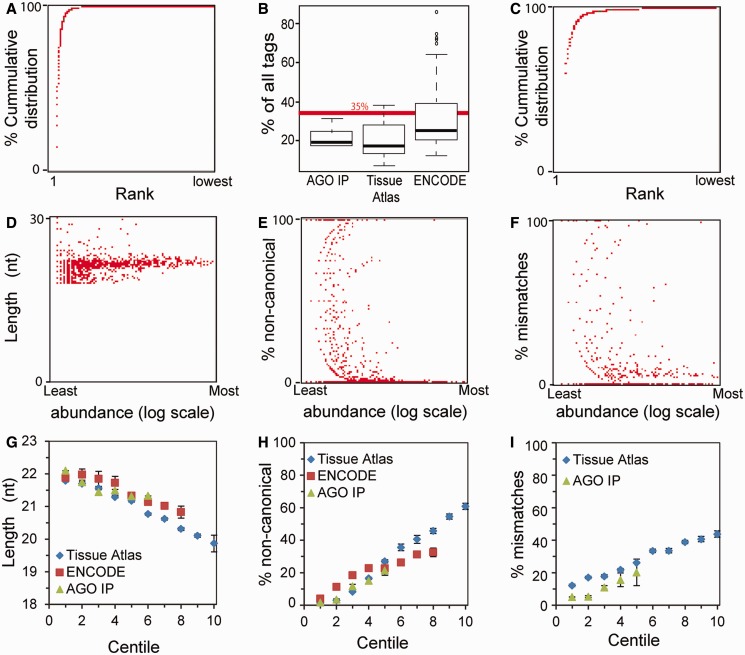
A number of reports have highlighted systematic transcript selection biases introduced into library preparations (16–18), highlighting the importance of using the same preparative method when comparing different data sets. However, to our knowledge, there are no quality control measures that reflect the efficiency of individual library preparations. In examining numerous miRspring documents generated from different library preparations and sequenced on different sequencing platforms, we identified a number of parameters that reflect the efficiency, variation and quality of those preparations. From the miRspring cumulative distribution XY-scatter plot, we noticed that in most data sets, a small number (<50) of miRNAs contribute to a large portion of miRBase mapped tags (Figure 2A). In the majority of data sets analyzed, the most abundant miRNA represented <35% of all miRBase mapped tags (Figure 2B). This was an informative parameter, as in a small number of data sets, the most abundant miRNA was >50%, which suggests that low abundant miRNAs may have been poorly sampled (Figure 2C).
Figure 2.
Quality control visualization parameters provided by the miRspring document. (A) Cumulative distribution of miRNAs for a typical data set. (B) Examination of numerous data sets identified that the most abundant miRNA represented <35% of all reads. Data sets where the majority of sequence tags are taken by a single miRNA as in (C) have to be treated with caution, as any low abundant miRNAs are poorly represented. (D) In individual data set, we noticed that the less abundant miRNAs have a large distribution in length, whereas abundant miRNAs have a more uniform length. Additionally, we also noticed low abundant miRNAs are not processed as defined in miRBase and therefore considered non-canonically processed (E). Furthermore, more of the low abundant miRNAs tend to have 1 nt mismatches (F). Average, (G) length, (H) non-cannonical processing and (I) mismatches for each rank centile were calculated from the analyzed data sets.
The XY-scatter plots of miRNA length, non-canonical processing and mismatches are also useful in assessing the quality of small RNA libraries. In many data sets, we noticed that low abundant miRNAs have a broad range of lengths, whereas the more abundant miRNAs are more uniform in length (Figure 2D). Furthermore, low abundant miRNAs have a greater proportion of reads that are not defined in miRBase, and therefore classified as non-canonically processed (Figure 2E). Some of these data points are likely to represent novel processed miRNAs that are derived from the arms of miRNA stem-loop sequences that have no annotated entry in miRBase. We therefore used the tabulated list of miRNA on the ‘global’ visualization page to determine whether any abundant non-annotated miRBase entries existed in a data set. From the human tissue data sets (12), we identified 34 miRNAs that were derived from miRBase defined hairpins, but had no annotation (Supplementary Table S2). Finally, we also identified that many of low abundant miRNAs had a 1 nt mismatch to the reference (Figure 2F), and these were not primarily due to RNA editing events. We next calculated the average length (Figure 2G), non-canonical (Figure 2H) and mismatches (Figure 2I) in data sets for each centile rank. This analysis confirmed the observations from individual miRspring documents. In most data sets, variation in miRNA length, non-canonical processing and mismatches increased in miRNAs with a centile rank greater than 3. Using these data sets as a guide, we conclude that the top 100 ranked miRNA of high-quality miRNA library preps should have an average length of 22 nt and have an average of <10% non-canonical processing. Furthermore, miRNAs with a rank >300 are most likely to have an average length <21 nt and >20% non-canonical processing. Together, these findings highlight the value of the miRspring document in defining parameters for assessing the quality of small RNA data sets.
Characterizing expression levels within miRNA clusters
Recent reports have identified sequence modifications and interactions that regulate miR-1 and let-7 pre-miRNA processing (19–22). Both miR-1 and let-7 are produced from polycistronic primary miRNAs (pri-miRs); as the regulatory proteins target the pre-miR loop, a structural component common to all miRNAs, many other pre-miRs within polycistronic pri-miRs could be similarly regulated. We propose miRNA HTS profiling can identify other candidates by comparing the relative expression among pri-miRs. To our knowledge, miRspring is the first software tool that calculates the relative expression level within a genomic cluster of miRNAs, which is suitable for this analysis. We examined public data sets to annotate expression levels within miRNA clusters and potentially identify clusters that may be differentially processed between tissues. For this examination, we looked for reproducibility from small RNA library replicates made from two different releases of the manufacturer’s library preparation kit (12).
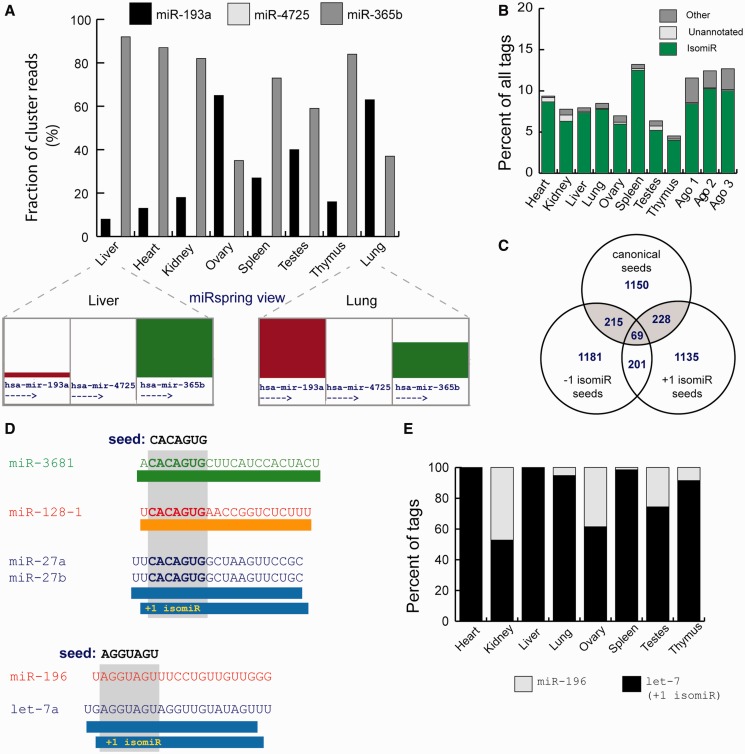
Suggested genomic distances that define a miRNA cluster range from 6 to 50 Kb (23,24), and owing to the lack of a strict definition, miRspring offers the user a number of distances that can be used to define the boundaries of a genomically clustered miRNAs. Using the output generated from the miRspring ‘miRNA cluster counts’ and custom scripts, we identified that the expression pattern within many miRNA clusters was conserved across tissues, and that this was independent of transcription direction (Supplementary Table S3). For one potential pri-miR polycistronic cluster, we identified a ranked expression difference of a individual miRNA in different tissues (Figure 3A). With the exception of lung and ovary, miR-365b was the predominant miRNA expressed from this cluster and suggests that this precursors of this cluster may be differentially processed.
Figure 3.
Analysis of published data sets using the miRspring document. (A) Differential expression or processing of miRNA precursors within a miRNA cluster across different human tissues. (B) Proportion of non-miRBase defined seeds identified in human tissues. Majority are derived from isomiRs of defined-processed miRNAs, and the remainder is derived from miRNAs that are processed from miRBase precursor arms that in miRBase v19 have no defined mature sequence (unannotated) or other processed sequences. (C) Vennn diagram showing the number of miRNAs and their isomiRs that have identical seeds. Seed-isomiRs are highlighted in gray. (D) Examples of interesting seed-isomiRs. The +1 isomiR of miR-27b has the same seed as miR-128 and miR-3681. Similarly, the +1 isomiR derived from the let-7 family has the same seed as miR-196 family. (E) The proportion of miRNAs and their isomiRs that have the canonical miR-196 seed (AGGUAGU) in different human tissues.
Seed-isomiR
One of the first detailed miRNA profiling studies identified that a small number miRNAs produced 5′ isomiRs, and owing to the altered seed-sequence, they are proposed to have a different spectrum of targets (25).
The importance of the miRNA seed sequence in identifying targets inspired the incorporation of seed analysis tools in the miRspring document. We used the miRspring ‘list seed abundance’ reporting feature to obtain a list of all miRNA seeds and their abundance from each of the human tissue data sets. From this, we determined that in each tissue between 4 and 14% of all miRNAs had seeds that were not defined in miRBase (Figure 3B). The majority (>99%) of non-miRBase seeds were generated by isomiRs of miRNAs (defined in miRBase), and <1% were derived from precursor arms not defined in miRBase, whereas the remainder were processed from non-canonically processed RNA. Similar analysis on small RNA data sets from Argonaute immunoprecipitations (13) confirms that a similar proportion is incorporated in the RNA induced silencing complex (RISC) complex (Figure 3B). It has not previously been appreciated that the small number of miRNAs that produce 5′ isomiRs represents a large proportion of miRNA seeds within human tissues. We believe this highlights a need to have existing public target identification software tools that primarily focus on canonically processed miRNAs (26) to expand their repertoire to include 5′ isomiRs.
We visualized the seed frequency on the miRspring XY scatter plot and observed that the seed sequences of an isomiR of one miRNA family could be identical to a seed sequence of a canonically processed miRNA of another family. These occurrences were rare, but most prevalent among low abundance miRNAs, with a few exceptions noted later in the text. We predict that miRNAs having identical seeds could be important in enhancing target selection or in providing some level of target redundancy. We term the relationship of miRNAs that have this trait ‘seed-isomiRs’. When comparing all human canonical miRNAs and the +/− 1 nt isomiRs (miRBase v19), 30.8% of canonical miRNA seed sequences had possible seed-isomiRs (Figure 3C). In other species, the proportion of seed-isomiRs correlated with the number of miRNAs identified (Supplementary Table S4), which may suggest a smaller need of redundancy and target enhancement in lower organisms.
In the analysis of publically available data sets, nine seed-isomiRs were expressed predominantly by the isomiR rather than the canonical miRNA (Table 1). The two most notable examples (as identified from the XY scatter plots) were seed-isomiRs found to be present in many tissues and cell lines: miR-196-5p/let-7-5p +1 isomiR and the miR-128/miR-27a +1 isomiR (Figure 3D). The seed-isomiR of let-7 and miR-196 are particularly intriguing, given that both these miRNAs play important roles late in development (27,28), and that HMGA2, a well-described target of let-7, can also be targeted by miR-196a (29). In all human tissues analyzed, the let-7-5p +1 isomiR contributed a significant proportion of reads that contained the AGGUAGU seed (Figure 3E). We note that while only ∼1% of the let-7-5p sequence tags are the +1 isomiR, we predict that the combined output of all let-7 family members together could effectively target a subset of mRNAs of the canonical miR-196 family.
Table 1.
Abundant human seed isomiRs identified from miRspring documents of public data sets
| SEED | miRBase seed | seed-isomiR | Data set | Notable examples |
|---|---|---|---|---|
| AAGUGCU | miR-302-3p miR-520-3p | miR-106-5p +1 isomiR | ENCODE | A549 and K562 |
| miR-20-5p +1 isomiR | ||||
| GCACCAU | miR-767-5p | miR-29-3p +1 isomiR | Tissue Atlas | Placenta |
| AGGUAGU | miR-196-5p | let-7-5p +1 isomiR | Tissue Atlas | Thymus (WTAK) |
| CACAGUG | miR-128-3p | miR-27-3p +1 isomiR | Tissue Atlas | Placenta |
| UAUACAA | let-7a-3p | miR-381-3p −1 isomiR | ENCODE | Ag04450 |
| CUGGCUC | miR-149-5p | miR-24-3p −2 isomiR | ENCODE | HeLaS3 |
| UUAUCAG | miR-374-3p | miR-21 +3 isomiR | ENCODE | HepG2 |
| UAGCACC | miR-5682-3p | miR-29-3p −1 isomiR | ENCODE | Prostate |
| GAGCUUA | hsa-miR-27b-5p | miR-590 −5p −1 nt isomeR | ENCODE | Sknshra |
DISCUSSION
Here, we describe miRspring a method that reproduces a miRNA-Seq data set combined with powerful research analysis tools that identify all aspects of miRNA biogenesis. The indexing algorithm programmed into the miRspring document significantly compresses the sequencing data without compromising computation speed in displaying or performing detailed sequencing analysis. The end result is a highly portable document that is independent of servers and internet connectivity. Together, these features bring a new level of transparency and accessibility to the field of miRNA research.
Although miRspring has similar visualization features to existing online databases and web servers, it does provide a number of significant improvements to these tools. To our knowledge, it is the first software that provides a simple but powerful method to succinctly visualize from a whole miRNA data set the processing features, seed distribution and relative expression levels of genomic clustered miRNAs. Most importantly, the miRspring document quantifies and generates detailed reports on all miRNA-processing parameters that can then be used for downstream statistical analysis. This detailed level of analysis can be applied to any data sets from any species and removes the dependency of web servers to derive this information.
We used the reporting feature of the miRspring software to analyze public data sets and discovered new insights into quality control parameters that are stored within sequencing data. Importantly, we also discovered new aspects of miRNA biology that had not been identified or previously described. This included discovery of miRNAs not annotated in miRBase, quantifying the total abundance of 5′ isomiRs within human tissues, relative expression levels within genomic clustered miRNA and the discovery of seed-isomiRs.
The seed-isomiR is an intruiging class of miRNA and potentially highlights the inbuilt redundancy built into miRNAs. There is a maximum of 16 384 possible 7nt seed sequences of which the 2042 human miRNAs in miRBase v19 encode 1662 combinations. The identification of family members, which are defined as those having identical seed sequences (4), highlights a level of redundancy that is built into the system. The discovery of isomiRs significantly expanded the diversity of seed sequence possibilities. However, our finding that ∼30% of 5′ isomiRs that are shifted by 1 nt encode the same seed sequence of miRBase defined miRNAs emphasizes the importance of miRNA seed redundancy. We predict that the targets of seed-isomiRs would have significant overlap to those of the miRBase defined miRNAs, and as such, the seed-isomiRs may provide a fine tuning mechanism for biological systems to enhance repression of specific targets through their up or downregulation.
HTS profiling of small RNA has proven to be a powerful tool, and as the technology becomes more affordable, its use will only increase. Analysis of mapped data sets has been limited to high-performance computers with large storage capacity, and there are only selective data sets making it onto public web databases. The miRspring document provides a revolutionary solution, as it replicates the entire mapped data set and provides user-friendly way of presenting sequencing data along with inbuilt novel analysis tools that can globally assess the whole data set. As the document can be used on personal computers and mobile devices, we anticipate that miRspring will increase the speed and depth of HTS data analysis and provide complete and compact data transparency at the time of publishing.
AVAILABILITY
For archival purposes, version 1.0 of the software is included as Supplementary Data file 1, but it is recommended to use the latest version available through the website: http://mirspring.victorchang.edu.au.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figure 1 and Supplementary Data 1–2.
FUNDING
CMS is a Future Research Fellow of the Australian Research Council (FT120100097). Supported by the Victor Chang Cardiac Research Institute. Funding for open access charge: The Victor Chang Cardiac Research Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank G.Humphreys, W.Ritchie, N.Cloonan and C.Hynes for critical discussions and technical suggestions, and R.Shuttleworth for technical assistance.
REFERENCES
- 1.Cheng WC, Chung IF, Huang TS, Chang ST, Sun HJ, Tsai CF, Liang ML, Wong TT, Wang HW. YM500: a small RNA sequencing (smRNA-seq) database for microRNA research. Nucleic Acids Res. 2013;41:D285–D294. doi: 10.1093/nar/gks1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho S, Jang I, Jun Y, Yoon S, Ko M, Kwon Y, Choi I, Chang H, Ryu D, Lee B, et al. MiRGator v3.0: a microRNA portal for deep sequencing, expression profiling and mRNA targeting. Nucleic Acids Res. 2013;41:D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2009;37:W68–W76. doi: 10.1093/nar/gkp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 7.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–D130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun Z, Wu J. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38:W392–W397. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphreys DT, Hynes CJ, Patel HR, Wei GH, Cannon L, Fatkin D, Suter CM, Clancy JL, Preiss T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012;7:e30933. doi: 10.1371/journal.pone.0030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakib MN, Tang J, Zheng WJ, Huang CT. Improving transmission efficiency of large sequence alignment/map (SAM) files. PLoS One. 2011;6:e28251. doi: 10.1371/journal.pone.0028251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief. Bioinform. 2009;10:490–497. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloonan N, Wani S, Xu Q, Gu J, Lea K, Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158–177. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium EP. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alon S, Vigneault F, Eminaga S, Christodoulou DC, Seidman JG, Church GM, Eisenberg E. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011;21:1506–1511. doi: 10.1101/gr.121715.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toedling J, Servant N, Ciaudo C, Farinelli L, Voinnet O, Heard E, Barillot E. Deep-sequencing protocols influence the results obtained in small-RNA sequencing. PLoS One. 2012;7:e32724. doi: 10.1371/journal.pone.0032724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 23.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung WS, Lin MC, Cheung DW, Yiu SM. Filtering of false positive microRNA candidates by a clustering-based approach. BMC Bioinformatics. 2008;9(Suppl. 12):S3. doi: 10.1186/1471-2105-9-S12-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 27.Mondol V, Pasquinelli AE. Let's make it happen: the role of let-7 microRNA in development. Curr. Top. Dev. Biol. 2012;99:1–30. doi: 10.1016/B978-0-12-387038-4.00001-X. [DOI] [PubMed] [Google Scholar]
- 28.Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 29.De Martino I, Visone R, Fedele M, Petrocca F, Palmieri D, Martinez Hoyos J, Forzati F, Croce CM, Fusco A. Regulation of microRNA expression by HMGA1 proteins. Oncogene. 2009;28:1432–1442. doi: 10.1038/onc.2008.495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.