Abstract
MicroRNAs (miRNAs) are small (20–22 nucleotides) regulatory non-coding RNAs that strongly influence gene expression. Most prior studies addressing the role of miRNAs in neurodegenerative diseases (NDs) have focused on individual diseases such as Alzheimer’s disease (AD), making disease-to-disease comparisons impossible. Using RNA deep sequencing, we sought to analyze in detail the small RNAs (including miRNAs) in the temporal neocortex gray matter from non-demented controls (n = 2), AD (n = 5), dementia with Lewy bodies (n = 4), hippocampal sclerosis of aging (n = 4), and frontotemporal lobar dementia (FTLD) (n = 5) cases, together accounting for the most prevalent ND subtypes. All cases had short postmortem intervals, relatively high-quality RNA, and state-of-the-art neuropathological diagnoses. The resulting data (over 113 million reads in total, averaging 5.6 million reads per sample) and secondary expression analyses constitute an unprecedented look into the human cerebral cortical miRNome at single nucleotide resolution. While we find no apparent changes in isomiR or miRNA editing patterns in correlation with ND pathology, our results validate and extend previous miRNA profiling studies with regard to quantitative changes in NDs. In agreement with this idea, we provide independent cohort validation for changes in miR-132 expression levels in AD (n = 8) and FTLD (n = 14) cases when compared to controls (n = 8). The identification of common and ND-specific putative novel brain miRNAs and/or short-hairpin molecules is also presented. The challenge now is to better understand the impact of these and other alterations on neuronal gene expression networks and neuropathologies.
Keywords: Alzheimer’s disease, deep sequencing, dementia with Lewy bodies, frontotemporal lobar dementia, hippocampal sclerosis, isomiR, microRNA, progressive supranuclear palsy
INTRODUCTION
Aberrant RNA processing can cause or exacerbate neurodegenerative diseases (NDs) via many mechanisms. RNA molecules are extremely versatile; less than 5% of total cellular RNA is messenger RNA (mRNA) coding for protein. The other 95% of non-coding RNAs have been shown to have a profound impact on gene expression regulation and also other neurochemical processes, and have been implicated as “complexity multipliers” in both normal and abnormal conditions of the human central nervous system [1–3].
One among many subtypes of non-coding RNAs are microRNAs (miRNAs), which are short (~20–22 nucleotides), conserved RNAs that have strong impact on gene expression regulation, and which have been implicated in ND pathogenesis. It has been estimated that over one-half (up to 90%) of genes are regulated in part by miRNAs [4, 5]. The impact of miRNA dys-regulation on ND pathogenesis has been a focus of much recent research [6–8]. Expression profiling has provided important insights into both ND and miRNA biology. Microarray studies and quantitative polymerase chain reaction (qPCR) studies have helped to describe which miRNAs are expressed during various normal and abnormal brain states, including in brain diseases. Although miRNA expression analyses can provide groundbreaking data for this novel research field, miRNA profiling benefits from the parallel use of multiple and different analytical techniques; there currently is no true “gold standard” because different methods (RNA isolation methods, miRNA profiling platforms, etc.) entail distinct strengths and weaknesses [9–13]. In comparison to other RNA profiling methods, deep sequencing is less biased by prior annotations of miRNAs and provides a quantitative and nucleotide-level resolution of small RNA species, although it is important to remember that each method, including deep sequencing, entail certain systematic biases [14].
As there are many species of RNA molecules, there are also pathogenetically distinct subtypes of ND that afflict elderly humans. Whereas AD is the most prevalent ND in most populations, other diseases such as dementia with Lewy bodies (DLB), frontotemporal lobar dementia (FTLD), and hippocampal sclerosis of aging (HS-Aging) comprise over 25% of aged dementia cohorts when state-of-the-art neuropathological methods are applied [15, 16]. Different NDs are often comorbid with each other [16, 17]. What all NDs have in common is that their pathobiological substrates are as yet imperfectly understood. Moreover, most studies of miRNAs in NDs have focused on individual diseases, including AD [8, 18–31], Parkinson’s disease (PD) [32–34], Huntington’s disease (HD) [35, 36], and other triplet-repeat disorders [37, 38]. Prior studies have tended not to include multiple NDs that would enable direct (within platform) comparisons across disease states.
In this study, we analyzed RNA derived from well-characterized brain samples from the University of Kentucky Alzheimer’s Disease Center (UK ADC) autopsy series. Our main aim was to augment prior studies of small RNAs in AD with a more systematic assessment that includes DLB, HS-Aging, and FTLD cases (none of the latter has been systematically assessed in the literature to date), and also rigorous bioinformatics. All cases had short postmortem interval (PMI), relatively high-quality RNA, and state-of-the-art neuropathological diagnoses.
METHODS
Case selection criteria and neuropathological assessment
Cases were selected on the bases of representing a spectrum of ND neuropathological features. RNA was extracted from snap-frozen brain tissue in the superior and middle temporal gyri (Brodmann areas 21/22) from the UK ADC biobank under a University of Kentucky IRB protocol.
Premortem clinical evaluations and pathological assessments were as described previously [12, 21, 22, 39, 40]. All included patients were Caucasians. The inclusion criteria that were applied: low PMI (<4 h wherever possible); no argyrophilic grains; no cancer in the brain parenchyma; and no large infarctions in the brain, or micro-infarcts found within 3 cm of the brain tissue samples. Neuropathological procedures were as described in detail elsewhere [16, 40]. Lewy bodies, neurofibrillary tangles, and neuritic plaques were counted as described [16]. No Lewy bodies were seen outside of amygdala in any of the non-DLB cases, except that cases D1 and F2 had sporadic Lewy body pathology in additional to progressive supranuclear palsy (PSP) and FTLD, respectively. Included patient characteristics, along with pathological lesion counts in the same cases, are shown in Tables 1 and 2. Samples used for RNA isolation were taken from tissue portions immediately adjacent to those used for neuropathological assessments.
Table 1.
Patient information: University of Kentucky Brain Bank
| Label | Age | Gender | Brain weight (g) |
PMI (h) | Braak stage |
CERAD plaque density |
Diagnosis | APOE alleles |
RIN |
|---|---|---|---|---|---|---|---|---|---|
| G1 | 75 | F | 1330 | 3.50 | 1 | None | Normal (no amyloid) | 3/3 | 7.1 |
| G2 | 84 | F | 1100 | 2.50 | 1 | None | Normal (no amyloid) | 3/3 | 8.1 |
| E1 | 37 | F | 760 | 3.50 | 0 | None | FTLD (TDP43 negative) | 3/3 | 5.5 |
| F1 | 53 | F | 940 | 4.75 | 2 | Mod | FTLD (TDP43 positive) | – | 7.9 |
| E2 | 48 | M | 900 | 3.75 | 0 | None | FTLD (TDP43 positive) | – | 5.3 |
| F2 | 87 | F | 970 | 4.50 | 0 | Low | FTLD (TDP43 positive) | – | 7.8 |
| D1 | 70 | M | 1240 | 3.00 | 1 | None | Progressive supranuclear palsy | 3/3 | 8.3 |
| C4 | 84 | F | 1090 | 3.00 | 2 | None | Bilateral hippocampal sclerosis | – | 8.9 |
| C3 | 84 | F | 870 | 1.75 | 1 | None | Bilateral hippocampal sclerosis | 2/3 | 8.8 |
| C2 | 93 | M | 1170 | 2.75 | 3 | Mod | Bilateral hippocampal sclerosis + Alzheimer changes | 3/4 | 8.6 |
| C1 | 101 | F | 1070 | 2.25 | 5 | None | Bilateral hippocampal sclerosis | 3/3 | 9 |
| B4 | 78 | M | 1270 | 3.75 | 1 | Low | Dementia with Lewy bodies | 3/4 | 8.3 |
| B3 | 78 | M | 1240 | 2.50 | 1 | None | Dementia with Lewy bodies | 3/3 | 8.1 |
| B2 | 85 | F | 1080 | 2.00 | 2 | High | Dementia with Lewy bodies | 2/3 | 8.9 |
| B1 | 68 | M | 1380 | 3.75 | 2 | Mod | Dementia with Lewy bodies | 3/4 | 5 |
| A5 | 83 | F | 1160 | 2.25 | 5 | High | Braak V Alzheimer | 3/4 | 8.4 |
| A1 | 86 | F | 1010 | 2.00 | 6 | High | Braak VI Alzheimer | 3/3 | 5.3 |
| A2 | 67 | F | 790 | 2.25 | 6 | High | Braak VI Alzheimer | 3/4 | 6.5 |
| A3 | 75 | F | 1050 | 2.50 | 6 | High | Braak VI Alzheimer | 3/4 | 7.9 |
| A4 | 86 | F | 1030 | 2.75 | 6 | High | Braak VI Alzheimer | 3/4 | 7.4 |
Table 2.
Patient information: Douglas Montreal Brain Bank
| Label | Age | Gender | Brain weight (g) | PMI (h) | Diagnosis | RIN |
|---|---|---|---|---|---|---|
| Ctr1 | 74 | F | 1365 | 11.00 | Normal | 7.4 |
| Ctr2 | 64 | M | 1530 | 20.75 | Normal | 6.3 |
| Ctr3 | 67 | M | 1300 | 24.75 | Normal | 6.6 |
| Ctr4 | 61 | M | 1500 | 6.25 | Normal | 6.8 |
| Ctr5 | 57 | F | 1250 | 35.75 | Normal | 6.2 |
| Ctr6 | 89 | M | 935 | 22.75 | Normal | 5.7 |
| Ctr7 | 72 | M | 1170 | 24.50 | Normal (brain infarct) | 5.4 |
| Ctr8 | 87 | M | 1190 | 29.00 | Normal (brain hypoperfusion) | 5.7 |
| FTLD1 | 74 | F | 890 | 19.00 | FTLD | 5.2 |
| FTLD2 | 62 | F | 625 | 31.50 | Pick’s disease | 6.5 |
| FTLD3 | 55 | M | 1000 | 18.75 | FTLD-U | 3.5 |
| FTLD4 | 86 | M | 1000 | 23.50 | FTLD-MAPT | 4.3 |
| FTLD5 | 67 | M | 750 | 30.00 | Pick’s disease | – |
| PSP1 | 88 | F | 1000 | 22.50 | Progressive supranuclear palsy + AD | 3.4 |
| PSP2 | 81 | F | 1045 | 40.50 | Progressive supranuclear palsy + AD | 4.7 |
| PSP3 | 65 | M | 1160 | 23.15 | Progressive supranuclear palsy | 4.5 |
| PSP4 | 60 | M | 1150 | 20.00 | Progressive supranuclear palsy | 3.7 |
| PSP5 | 61 | M | 1310 | 20.00 | Progressive supranuclear palsy | 3.8 |
| PSP6 | 72 | F | 1170 | 27.25 | Progressive supranuclear palsy | 4.6 |
| PSP7 | 69 | F | 1030 | 11.50 | Progressive supranuclear palsy | 7.4 |
| PSP8 | 63 | M | 1080 | 44.75 | Progressive supranuclear palsy | 5.6 |
| PSP9 | 92 | F | 865 | 13.50 | PSP + AD | 4.7 |
| AD1 | 79 | F | 900 | 10.75 | AD | 7.1 |
| AD2 | 85 | F | 1025 | 34.50 | AD | 5.8 |
| AD3 | 75 | F | 925 | 4.50 | AD | 4.9 |
| AD4 | 83 | M | 1137 | 28.00 | AD | 4.9 |
| AD5 | 71 | M | 1290 | 12.00 | AD + Hydrocephalus | 4.8 |
| AD6 | 79 | M | 1210 | 24.75 | AD senile | 4.9 |
| AD7 | 66 | M | 1075 | 8.50 | AD pre-senile | 7.1 |
| AD8 | 88 | M | 1240 | 24.75 | AD | 4.7 |
RNA isolation from a human cerebral cortex
Biochemical analyses were performed blind with respect to patient information. RNA was isolated as described previously in detail [12, 21, 39, 40]. Briefly, prior to RNA extraction, gray matter was dissected away from white matter and only gray matter was used for these studies. Tissue (1–3 g) that had been snap-frozen in liquid nitrogen and then transferred to a −80°C freezer was thawed in isotonic lysis buffer with RNAsin ® (Promega, Madison, WI; 250 U/ml) and Complete protease inhibitor pills (Roche, Basel Switzerland). Trizol LS (Invitrogen, Carlsbad, CA) was used according to manufacturer’s instructions, except for an added overnight −20°C precipitation step during isopropanol precipitation. The quality of total RNA was analyzed on an Agilent 2100 Bioanalyzer system...that measured RNA Integrity Number (RIN) RNA purity was confirmed using A260/A280 readings (not shown).
RNA deep sequencing and data analysis
All RNA samples were processed and analyzed by LC Sciences (Houston, TX, USA). Each received RNA sample was processed to generate a cDNA library that was then used for deep sequencing. Briefly, a small RNA library was generated from our sample using the Illumina Truseq™ Small RNA Preparation kit according to Illumina’s Truseq™ Small RNA Sample Preparation Guide (see Supplementary data; available online: http://www.j-alz.com/issues/35/vol35-2.html#supplementarydata02). The purified cDNA library was used for cluster generation on Illumina’s Cluster Station and then sequenced on Illumina GAIIx following the vendor’s instruction for running the instrument. Raw sequencing reads (40 nts) were obtained using Illumina’s Sequencing Control Studio software version 2.8 (SCS v2.8) following real-time sequencing image analysis and base-calling by Illumina’s Real-Time Analysis version 1.8.70 (RTA v1.8.70). The extracted sequencing reads were stored and then a proprietary pipeline script, ACGT101-miR v4.2 (LC Sciences),was used for sequencing data analysis as described in the Supplementary data.
Validation cohort patient information and miRNA qRT-PCR
The non-dementia controls (n = 8), AD (n = 8), FTLD (n = 14, including PSP n = 9) patient brains were obtained from the Douglas Hospital Research Centre brain bank in Montreal, Canada as described previously [41, 42] (Table 2), in accordance with the national ethical committee protocols and in agreement with the local CRCHUQ ethical committee. Blocks from the temporal lobe (Brodmann area 20) were dissected and snap frozen in liquid nitrogen until use. Total RNA from was extracted on ice using Trizol® according to the manufacturer’s instructions. The mean age average of patients was: controls 71.4±11.6 y, PSP 73.5±12.2 y, FTLD 67.8±10.9 y, and AD 78.3±7.3 y. The mean PMI values were: controls 21.8±9.4 h, PSP 22.3±9.0 h, FTD 27.9±9.8 h, and AD 18.5±10.8 h. The quality of total RNA was analyzed on an Agilent 2100 Bioanalyzer system. No correlations were observed between PMI and miRNA quantifications, as documented before [19, 41]. It is noteworthy that qRT-PCR provides a robust method to measure miRNAs in samples with relatively low (≤3–4) RNA integrity (RIN) values [43, 44]. For miRNA quantifications, probe-specific Taq-Man miRNA assays (Life technologies) were used according to the manufacturer’s instructions. Relative expression was calculated by using the comparative CT method. In all experiments, hsa-miR-16 (Life Technologies) was used as a normalization control, as described previously [19, 41, 45]. Similar results were observed using let-7a as normalizing control (data not shown).
Northern blot analysis
All procedures performed as described previously [22, 46, 47]. For these experiments, we used total RNA (10 µg) extracted from the frontal cortex, cerebellum, hippocampus, substantia nigra, and superior and middle temporal gyri from non-demented controls. The gel used for these analyses was 15% urea-PAGE.
RESULTS
Characterization of small RNA species in the adult human brain
Total RNA was isolated from the gray matter of snap-frozen samples of superior and mid-temporal neocortex derived from the autopsies of 20 aged individuals, including non-demented controls (n = 2), AD Braak stage V (n = 1), AD Braak stage VI (n = 4), DLB (n = 4), HS-Aging (n = 4), and FTLD (n=5) patients. Note that FTLD patients were further subdivided into three groups: FTLD with TARDNA-binding protein 43 (TDP-43) positive inclusions (FTLD-TDP, n = 3), FTLD without TDP-43 positive inclusions (FTLD, n = 1], and PSP (n = 1). Detailed pathological, biochemical, and clinical characterization of these samples are presented in Tables 1 and 2. PMIs were low (2.95 h±0.85 [see inclusion criteria]), and RIN values ranged between 5 and 8, indicating overall high RNA quality when considering working with postmortem human brain tissue following careful gray matter dissection.
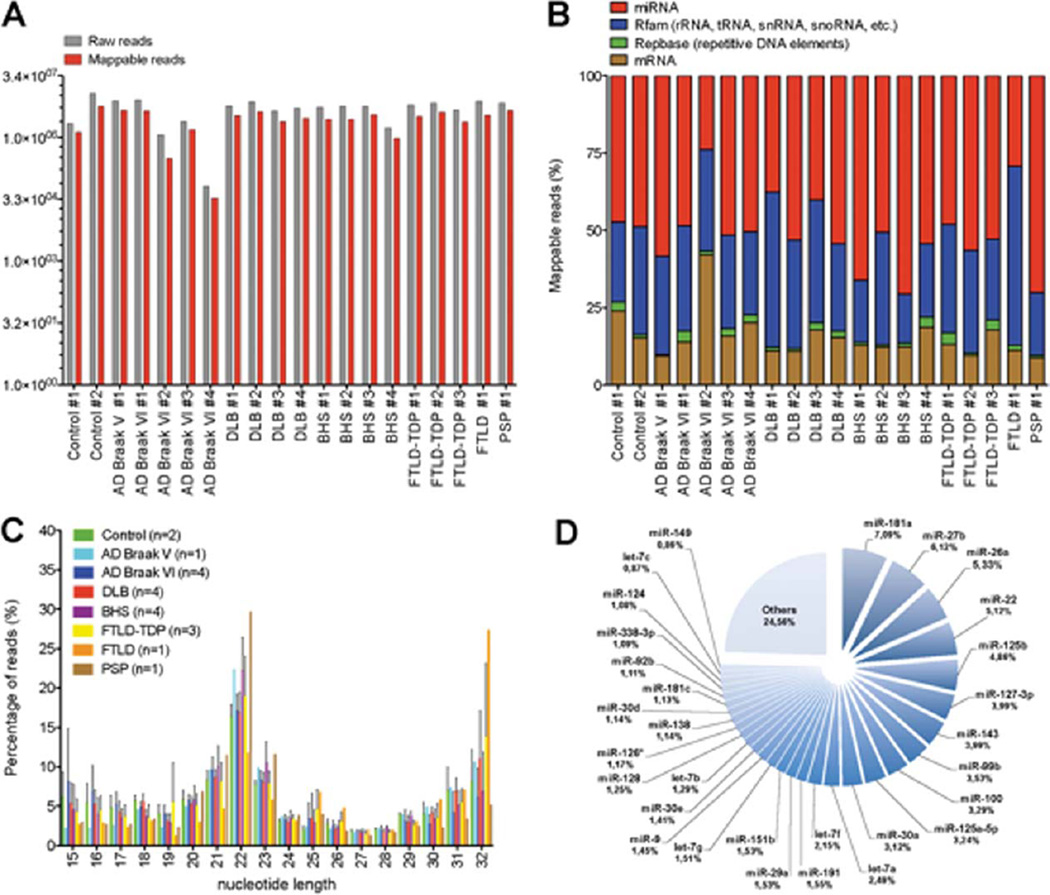
Deep sequencing of small (18–35 nt) RNA sequences was performed on an Illumina platform. An RNA library preparation kit was used in order to capture all potential small RNAs in a relatively unbiased way. A total of 113, 905, 268 raw reads (number of molecules) were obtained, with an average of 5, 695, 263 raw reads per sample. Approximately half (54±8%) of the raw reads could be mapped to the genome (Fig. 1A and Supplementary Table 1A). No significant changes were observed between raw versus mappable reads among groups. Among mappable reads, 51±12% comprised miRNAs, whereas the remaining RNA sequences were mapped to Rfam (i.e., tRNAs, rRNAs, snRNAs, snoRNAs, etc.), mRNAs, and Repbase (i.e., repetitive elements) (Fig. 1B). The global distribution of RNA length did not consistently vary among the different groups (Fig. 1C). A 22nt peak was observed in all samples, which is consistent with miRNAs being among the most abundant small RNA species. A yet uncharacterized 32nt peak was also observed.
Fig. 1.
A) Overview of raw versus mappable reads counts from each sample used in this study. B) Distribution (in percentage) of mappable small RNA species. Note that miRNAs constitute approximately half of the small RNAs in the adult human brain. C) Nucleotide length distribution (in percentage) of individual samples (by groups) used in this study. Error bars (standard deviation) are shown. D) Pie chart of individual miRNAs (including percentages) constituting approximately 75% of all bulk miRNAs. Here, calculations were based on the average number of reads from the two non-demented controls.
The RNA data was analyzed primarily to assess the number and identity of miRNAs that were expressed in the human neocortex, including comparisons across the different ND conditions. Only a small fraction (1.5%) of annotated miRNAs (based on miRBase [47] Release 18) was expressed at very high (>10, 000 raw reads) levels (Supplementary Table 1). The remaining miRNAs were expressed at relatively high (15%, 1000–9999 raw reads), moderate (30%, 100–999 raw reads), and low (55.5%, 1–99 raw reads) levels. Notably, 11 miRNAs (miR-181a > miR-27b > miR-26a > miR-22 > miR-125b > miR-127-3p > miR-143 > miR-99b > miR-100 > miR-125a-5p > miR-30a) comprised roughly 50% of all miRNA reads (Fig. 1D). An additional 19 miRNAs (let-7a > let-7f > miR-191 > miR-29a > miR-151b > let-7g > miR-9 > miR-30e > let-7b > miR-128 > miR-126* > miR-138 > miR-30d > miR-181c > miR-92b > miR-338-3p > miR-124 > let-7c > miR-149) constituted 25% of total miRNA reads across all samples. Thus, ¾ of the mature miRNA fraction detected from these samples correlated with only ~30 individual annotated miRNAs.
Northern blot analysis of selected “annotated” miRNAs
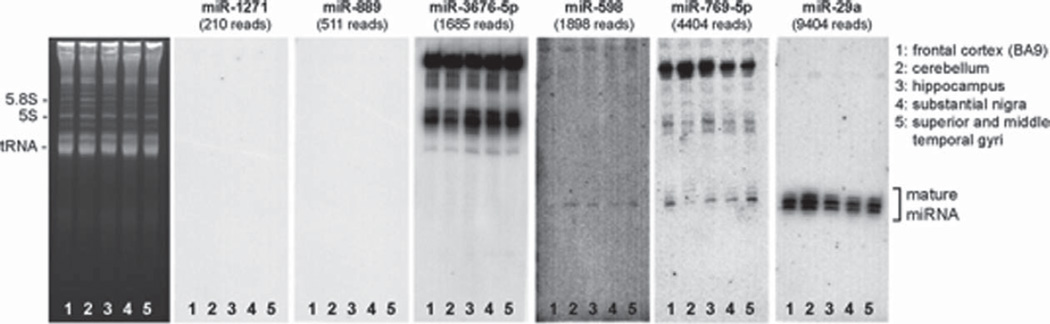
Previously, it has been shown that some “annotated” miRNAs actually represent non-specific or non-canonical RNA species, as determined by northern blot from brain tissue-derived RNA; anomalous northern blotting characteristics were observed in more recently-annotated “high number” miRNAs [12, 48]. We therefore selected a subset of miRNAs identified in the deep sequencing data, namely miR-598, miR-769-5p, miR-889, miR-1271, and miR-3676-5p for further validation. In these experiments, miR-29a was included as positive control. The northern blots demonstrated that only two miRNAs, miR-598 (1898 reads) and miR-769-5p (4404 reads), gave a positive miRNA signal (Fig. 2). No signal was observed for miR-1271 and miR-889. As with previous observations, the “miR-3676-5p” gave no miRNA-like signal. These data confirm that deep sequencing data of high-read RNA species (as with other miRNA profiling platforms) need to be corroborated with northern blotting.
Fig. 2.
Northern blot analysis of selected “annotated” miRNAs. Candidate miRNAs were chosen based on their low-to-moderate expression, to see whether they were detectable via northern blot, and if they had the ‘classic’ miRNA pattern with regard to precursor and mature miRNAs, as described in a prior study [48]. For this analysis, we used total RNA from different brain regions for comparative reasons. As shown here, only two miRNAs, miR-598 and miR-769-5p, gave positive signals. No signal was observed for miR-1271 and miR-889, which may be below detection levels. The putative “miR-3676-5p’ gave no miRNA-like signal. A representative ethidium-bromide-stained gel was photographed to convey RNA quality and loading parity among the samples; indicated on the figure are well-characterized RNA bands: tRNA (73–93nts); 5S rRNA (120nts); and 5.8S rRNA (160nts).
Analysis of miRNA and isomiR expression profiles
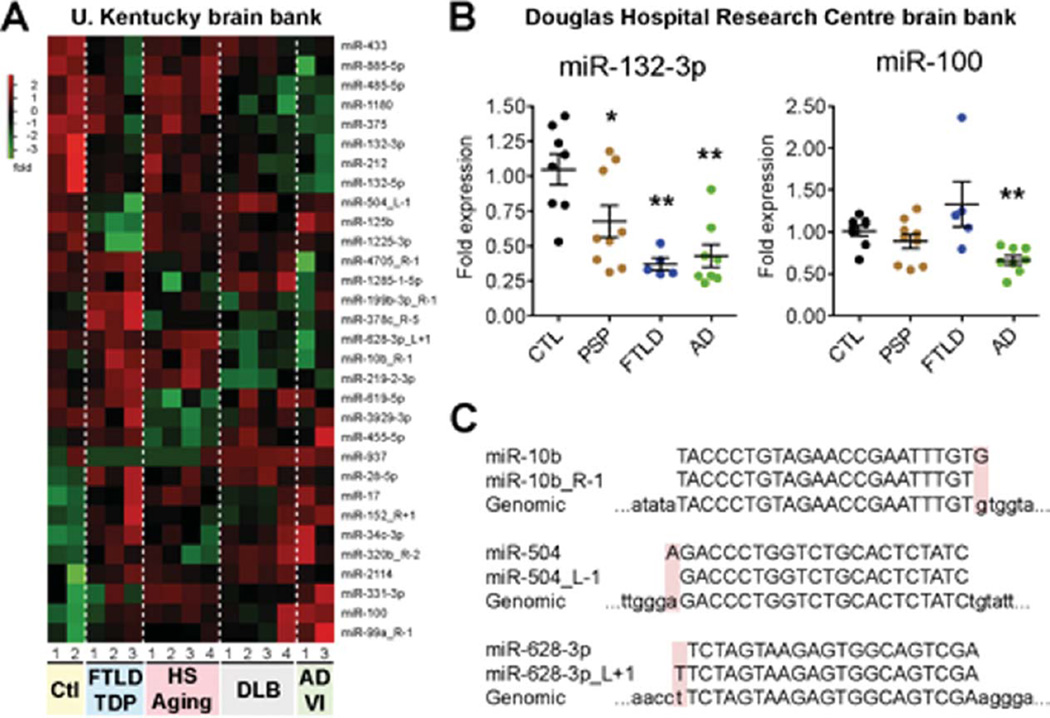
Previous studies have identified alterations in miRNA expression profiles in AD brain [8, 18–30]. We next asked whether such changes exist in our samples, and, importantly, whether certain miRNAs were robustly affected in different NDs (sample sizes were inadequate to detect highly-variable or more subtle changes across NDs). To this end, we focused on samples with comparable amounts of mappable reads, including AD Braak VI (cases #1 and 3), FTLD-TDP (cases #1–3), DLB (cases #1–4), and HS-Aging (cases #1–4) pathologies. From a total of 795 miRNAs expressed in the human brain (and 721 isomiRs, see below), we identified 31 human miRNAs to be expressed differently (p < 0.05, ANOVA, without Bonferroni correction for multiple comparisons) in disease conditions when compared to non-demented controls (cases #1–2) (Fig. 3A and Supplementary Table 2). A number of these miRNAs have previously been associated with AD and other NDs, including miR-132/212 family members, but also miR-34c, miR-125b, and miR-106b. Other miRNAs hitherto associated with AD (e.g., miR-29a, miR-29b, miR-103/107, miR-181c, miR-9) were not statistically different in these analyses. We note that the sample size of the current study was limited, and both miR-29 and miR-107 paralogues, for instance, were lower in AD samples than controls, but the variability was too high to achieve statistical significance. Finally, we also performed disease versus disease comparisons where a number of miRNAs had different expression between samples, but none of these survived a Bonferroni correction for multiple comparisons (Supplementary Table 2).
Fig. 3.
A) Heatmap of significantly (p < 0.05) misregulated human miRNAs in the disease groups when compared to non-demented controls. For these calculations, we focused on samples with relatively high and comparable reads numbers: controls #1 and 2, AD Braak VI #1 and 3, FTLD-TDP #1–3, DLB #1–4, and HS-Aging #1–4. Normalization details are found in Supplementary data. B) Real-time quantitative RT-PCR of mature miR-132-3p or miR-100 in control (n = 8), FTLD (n = 5), and PSP (n = 9) patients from the Canadian cohort. Here, total RNA extracted from temporal cortex was used. The miRNA miR-16 was used as normalization control (using the average of non-demented controls as 1 fold). Statistics were calculated using a Mann-Whitney t test, where *p < 0.05 and **p < 0.01. C) Representative examples of significantly changed isomiRs that are actually expressed at higher levels than their miRNA reference counterpart (http://mirbase.org/). In all cases, the added or deleted nucleotide corresponds to the genomic sequence (see shaded boxes).
In an attempt to provide validation for some of these changes, we measured miR-132-3p (the most abundant miR-132/212 family member) and miR-100 in a completely different cohort of patients (Table 2). For these studies, we increased the number of cases and disease subtypes, which included non-demented controls (n = 8), AD (n = 8), and FTLD (n = 14). The FTLD group was further divided into FTLD (n = 5) and PSP (n = 9) patients. By miRNA quantitative RT-PCR, we observed a significant downregulation of miR-132-3p in AD (p < 0.01, Mann-Whitney test), FTLD (p < 0.01, Mann-Whitney test), and PSP (p < 0.05, Mann-Whitney test) cases when compared to non-demented controls (Fig. 3B). MiR-100 was statistically lower in AD versus non-demented controls (p < 0.05, Mann-Whitney test), reflecting some disease-specific changes as suggested by the ANOVA analysis.
MiRNAs with subtle sequence changes in relation to annotated miRNAs, also known as isomiRs, can significantly contribute to miRNA target specificity and function [35, 49–51]. A number of isomiRs were present in the list of misregulated miRNAs (names ending with R [right] or L [left]) (Fig. 3A). However, further analysis demonstrated that all affected isomiRs constituted equal or increased expression with regard to the consensus (reference) miRNA (Fig. 3C and Supplementary Table 3), an observation consistent with previous studies (see e.g., [35]). Thus, no significant changes in human isomiR expression per se were observed in our disease groups.
Evidence for mature miRNA editing
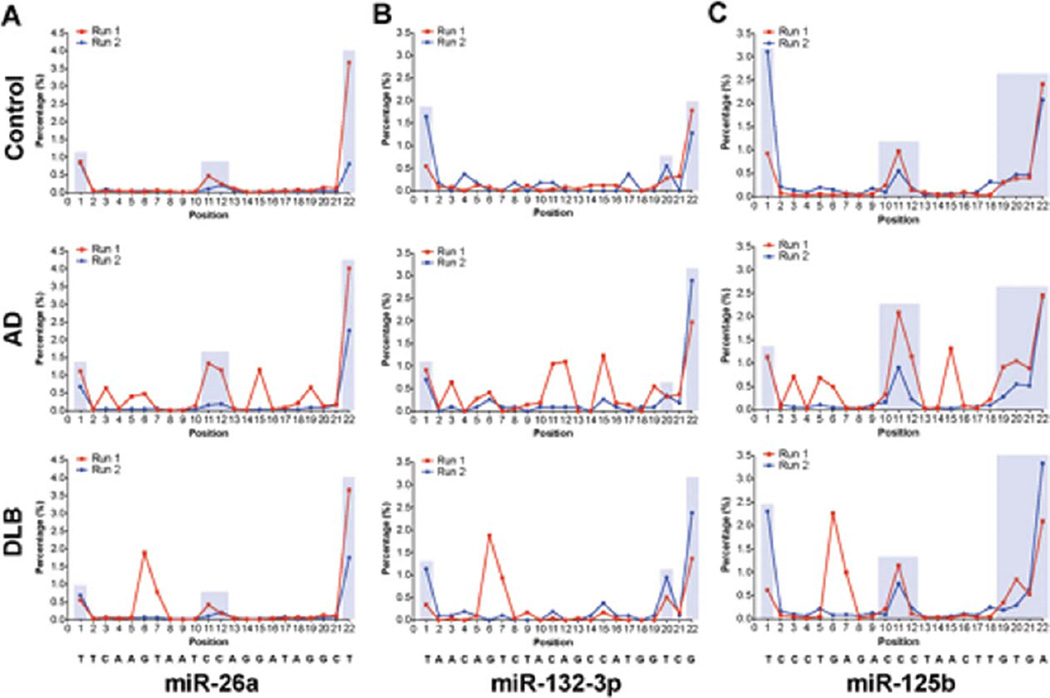
RNA editing is a mechanism that allows for posttranscriptional modification of RNA sequences. MiRNA transcripts are recognized targets for RNA editing enzymes, and single-nucleotide changes through editing can impact miRNA target specificity and therefore function [52–54]. Previously, Martì et al. [35] identified a number of edited nucleotides within mature miRNA sequences in the human brain. We asked whether such modifications could be reproduced in our sample sets, and whether differential RNA editing could be correlated with pathological brain conditions. We found evidence of limited edited nucleotides located in the mature miRNA (Fig. 4). Representative results are shown for miR-26a, miR-125b, and miR-132-3p. The highest editing rate occurred at 5′ and 3′ ends (positions 1–2 and 19–22) of the mature miRNA, which is consistent with prior results [35], and which may also be linked to the technical process of RNA sequencing itself (e.g., adapter ligation, PCR, and informatics). Additional editing “hot spots” were also present at position 11 and to a lesser degree at position 12.
Fig. 4.
Distribution of edited nucleotides for (A) miR-26a, (B) miR-132-3p, and (C) miR-125b. A representative example was taken from non-demented controls, AD Braak VI, and DLB. Note that most “disease-specific” editing patterns (in shaded boxes) could not be reproduced when sequencing the same RNA samples independently (run 1 versus run 2). However, some “hot-spot” editing patterns were consistently observed, for instance at positions 1, 11, 12, 20, 21, and 22. These likely represent physiological editing patterns, although representing only ~1–4% of total reads. The mature miRNA sequences are presented below. Notably, editing patterns were not significantly different between control and disease groups, as determined using a two-way ANOVA with Bonferroni post-hoc test (not shown). Y-Axis; nucleotide position of the mature miRNA sequence. X-Axis, percentage of edited miRNAs. “Physiological” (consistent) editing patterns are highlighted in light blue boxes.
Interestingly, our initial deep sequencing run provided evidence for atypical miRNA editing patterns in disease conditions (Fig. 4). In order to validate these findings, we performed a second, independent sequencing run on the same RNA samples. In the second sequencing run (performed several months hence), most of the original “disease-specific” editing patterns could not be replicated (run 1 versus run 2), implying some sequencing errors associated with this technology. By contrast, most of the “physiological” editing profiles (see above) could be reproduced (e.g., nucleotides 1, 11, 19, 20, 21, and 22), although these modifications were not statistically different between disease conditions or when compared to non-demented controls (not shown).
Analyses of putative novel miRNAs
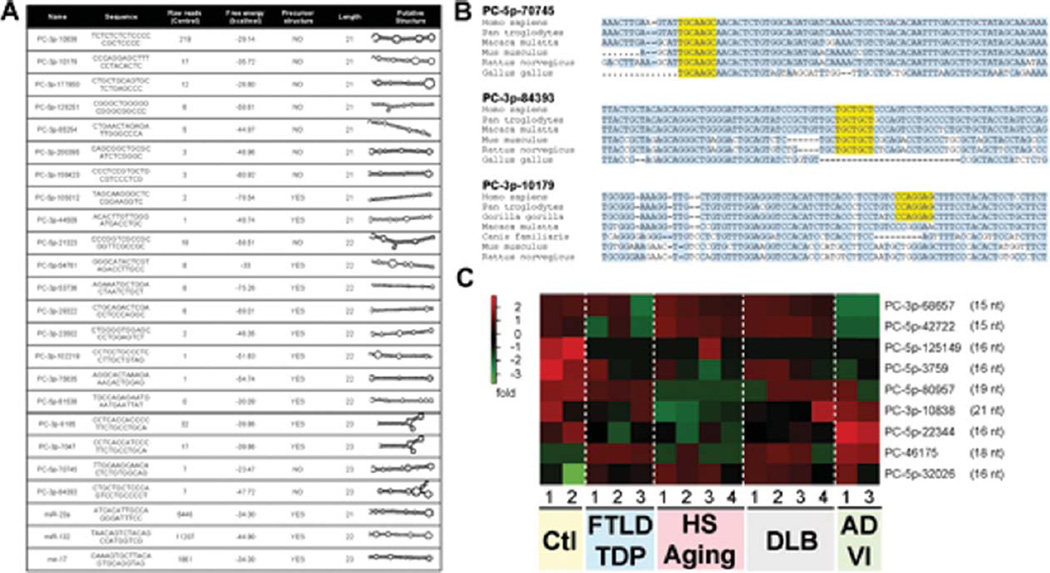
Bioinformatics analysis identified a subset of putative novel miRNAs derived from predicted RNA hairpins (Supplementary Table 1), from which only 1.18% (87 out of 7381) were expressed at ≥10 raw reads in one or more samples. Twenty-three percent (20 out of 87) of putative novel miRNA hairpin RNAs had between 21–23nt in length (Fig. 5A). Secondary structure analysis of corresponding putative precursor miRNAs (pre-miRs) indicated free energies between −23 and −78 kcal/mol. For comparative purposes, we included three recognized miRNAs, miR-23a, miR-132-3p, and miR-17, chosen arbitrarily. Conservation alignments demonstrated that most putative novel miRNAs (with focus on the seed sequences) were human or primate-specific, with the exception of PC-5p-70745 and PC-3p-84393 (Fig. 5B and Supplementary Fig. 2). Interestingly, a number of putative novel miRNAs were apparently misregulated in disease conditions (Fig. 5C). Only PC-3p-10838 was exactly 21nt in length, while the remaining candidates were 15–19nt in length. Given technical issues with regard to the detection of mature miRNAs in the brain (as shown above), it will be challenging to validate conclusively whether these candidate hairpin RNAs represent genuine miRNAs with important biological functions. Nonetheless, the potential diagnostic value of these molecules remains an interesting possibility.
Fig. 5.
A) Table demonstrating putative novel miRNAs with 21–23 nucleotides. The candidate miRNA names (“PC-”), sequence, number of reads (average n = 2 of controls), precursor miRNA (pre-miR) free energy (calculated using RNAfold: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), miRNA precursor properties (calculated using MirEval: http://tagc.univ-mrs.fr/mireval), and putative pre-miR structure (generated using RNAfold). Range of read numbers in these samples was from 0 (PC-3p-81538) to 219 (PC-3p-10838). A “0” reads count indicates that no putative miRNA molecule was detected in this sample. For comparative reasons, we included miR-23a (21nt), miR-132-3p (22nt), and miR-17 (23nt) which all had >1000 reads. B) Conservation alignment of three putative novel miRNAs. The seed sequence (nucleotides 2–7 of the mature sequence) is shown in yellow. Conserved nucleotides are shown in light blue. C) Heatmap of significantly misregulated putative novel miRNAs in the different disease groups when compared to non-demented controls. Note that only one candidate miRNA has 21 nucleotides.
DISCUSSION
The present study involves the analysis small RNAs isolated from the superior/mid-temporal gyri (gray matter) of 20 individuals including pathologically verified examples of AD, DLB, FTLD, HS-Aging, and non-demented control subjects. The method of RNA sequencing is powerful because it is unbiased by prior annotated miRNA reads, and has previously been applied to analyze the human brain, albeit in different contexts [55, 56]. Here, although we found no obvious sequence (e.g., isomiR) defining small RNA changes in these samples to distinguish a particular ND or control subjects, bioinformatics and independent qRT-PCR analyses did help refine our understanding of the specificity of quantitative miRNA changes in AD. We also found solid evidence of physiologic miRNA editing in human brain, as documented before [35]. Together, these data constitute important primary information about the human brain miRNome with new insights into the complex world of small non-coding RNA neurochemistry, in both normal and pathogenic conditions.
Caveats inherent to the study design include the small number of samples in each case category, which severely limited statistical power in terms of categorical comparisons. This consideration needs to be kept in mind in interpreting the results, as stated below. Another cautionary technical point is that the RNA preparation stages of the distinct miRNA profiling platforms entail key differences that affect results [13]. For example, deep sequencing of small RNAs involves multiple potential sources of technical biases, including those referent to the application of T4 RNA ligase, which is required for small RNA ligations and has base-specific bias, and PCR, which also introduces biases in the course of multi-step amplifications [11, 14, 57, 58]. These factors may help explain some of the differences when comparing RNA deep sequencing results and profiling data derived from other platforms as we previously described (for example, the unexpectedly low relative abundance of miR-124 in comparison to other brain miRNAs; see [9]). There is some evidence that RNA degrades postmortem and that particular miRNAs decay at different rates in human brain [26], although it is also notable that brain miRNAs have been described to be very robust under other circumstances [59]. Because of these technical considerations and limitations, the present study cannot be seen as a definitive quantitative profiling experiment comparing the miRNA repertoire across ND conditions. Instead, the goals of the study were to assess the complexity of small RNA from this area of the brain, and to use those data to help define broader aspects of similarity and difference between the different ND, and to evaluate the differential influence of RNA modifications and novel RNAs.
Another note of caution pertains to the current lack of insights into what induces miRNA expression variation in the adult human brain. In addition to disease states, there are as-yet uncharacterized potential contributions to miRNA expression variability from the heterogeneous influence of gender, anatomical region, age, medication, diet, concomitant diseases (e.g., diabetes), agonal events, genetic polymorphisms, and many other factors. Some of these factors have been hinted at in prior studies [60–63]. An assumption underlying the current study is that we need to “start somewhere”, and with these limitations in mind, we can now begin to characterize the small RNA repertoire from human temporal neocortex in various NDs.
A fundamental observation is that a small number of miRNAs represents the large bulk of neocortical tissue miRNA in these samples. More than ¾ of the miRNAs corresponded to only 30 different individual mature miRNA species. Moreover, the top 50 expressed miRNAs constituted 91% and the top 100 expressed miRNAs 97% of total miRNAs. These findings are broadly compatible with prior studies [35, 64–67]. It is notable that these samples represent multiple populations of cells (neurons, oligodendrocytes, astrocytes, and endothelial cells), yet, the number of miRNAs expressed in high amounts within a given sample is not large. Only a small number (1.5% of total) miRNAs were present in large (>10,000 raw reads) amounts. This consideration is directly relevant to miRNA gene-function analyses. For example, it seems likely that much of the functional impact of miRNAs will be correlated with this small list of gene products, and that relatively small “fold-changes” in these robustly-transcribed genes may be more important than larger-percentage changes in expression among miRNAs that are present in far lower levels (three or more orders of magnitude lower expression). Even among fairly highly-represented miRNAs, our results also underscore the importance of using northern blots to verify outcomes of deep sequencing, qPCR, and microarray profiling experiments, since even annotated miRNAs can have non-canonical (or even non-existent!) banding patterns.
A key strength of the current study was the use of RNA deep sequencing technology that enables the study of RNA editing. Collectively, our observations and analyses strengthen the hypothesis that miRNA editing is indeed a physiological event. However, the overall percentage of edited miRNAs was low (less than 4% of total miRNA reads). The functional relevance of these edited miRNAs remains unknown. Furthermore, the presence of isomiRs needs to be critically assessed using other platforms and RNA samples. Whether miRNA editing occurs at a higher rate in other biological contexts (e.g., during brain development) or other areas of the brain also remains to be determined. Based on these observations, we did not find strong evidence of ND-linked RNA editing of miRNAs.
The analyses of multiple NDs enable us to contextualize some of the prior work in the field. These new data can be compared with prior studies of miRNA perturbation linked with individual NDs, with the abovementioned caveat that the sample size of this study was problematic in terms of categorical comparisons. For the most part, our data are compatible with prior work. However, the small sample sizes precluded new evaluation of the prior studies that show significant but not “qualitative” changes in AD brain miRNAs such as miR-29 paralogues, miR-15/107 genes, miR-153, and miR-146a [19, 41, 68, 69]. However, we note that the general trends were as previously published accounting for the inadequate statistical power. Whether these are affected in only a subset of patients, or in specific brain regions, remains to be explored. The observation that miR-29a/b-1 is downregulated in AD patients displaying increased BACE1 protein levels is in line with this hypothesis [17]. Another source of discrepancy could reside in the technology used for miRNA profiling (e.g., deep-sequencing versus hybridization). The advantage of the current study’s design is to enable detection of miRNAs that are altered across a range of diseases, such as miR-132, that previously has been shown to regulate splicing of the microtubule associated protein tau gene [41, 70]. MiR-132 has also been shown to regulate cognitive function in other experimental models including neuronal/synaptic integrity and the brain’s response to stressors [71–75]. Interestingly, miR-132 downregulation was recently observed in another cohort of FTLD-TDP brains [76], which is consistent with our results (and which helps illustrate the importance of having disease controls as well as cognitively intact controls). This provides us with insights that some miRNAs may have more general ‘neuroprotective’ functions whereas others have direct roles in specific subtypes of pathologies.
There are many future challenges and unanswered questions with regard to small noncoding RNA research in the human brain; it is probable that new paradigms and small RNA species remain to be discovered. These efforts will profit from resources that help define the repertoire of human brain RNAs across a gamut of conditions and anatomical areas. We hope that data from the current study, comprising over 113 million different reads from human neocortex (averaging almost 6 million read per sample), will be a resource that will be useful to our colleagues in this exciting field, as more hypotheses are generated and tested with regard to small non-coding RNA in the human brain in health and disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Alzheimer Society of Canada and the Fonds de recherche du Québec – Santé (FRQS) and by grants from the National Institutes of Health (NS061933, AG036875, and AG028383). We gratefully acknowledge the research volunteers and the clinical collaborators whose contributions enabled this work.
Footnotes
Supplementary data available online: http://www.j-alz.com/issues/35/vol35-2.html#supplementarydata02
Authors’ disclosures available online (http://www.j-alz.com/disclosures/view.php?id=1651).
REFERENCES
- 1.Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: Insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport SI, Nelson PT. Biomarkers and evolution in Alzheimer disease. Prog Neurobiol. 2011;95:510–513. doi: 10.1016/j.pneurobio.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson PT, Keller JN. RNA in brain disease: No longer just “the messenger in the middle”. J Neuropathol Exp Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- 4.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebert SS, De Strooper B. Molecular biology. miRNAs in neurodegeneration. Science. 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- 8.Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Hebert SS, Nelson PT. Studying microRNAs in the brain: Technical lessons learned from the first ten years. Exp Neurol. 2012;235:397–401. doi: 10.1016/j.expneurol.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podolska A, Kaczkowski B, Litman T, Fredholm M, Cirera S. How the RNA isolation method can affect microRNA microarray results. Acta Biochim Pol. 2011;58:535–540. [PubMed] [Google Scholar]
- 11.Sorefan K, Pais H, Hall AE, Kozomara A, Griffiths-Jones S, Moulton V, Dalmay T. Reducing sequencing bias of small RNAs. Silence. 2012;3:4. doi: 10.1186/1758-907X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson PT, Wang WX, Wilfred BR, Tang G. Technical variables in high-throughput miRNA expression profiling: Much work remains to be done. Biochim Biophys Acta. 2008;1779:758–765. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang F, Fuchs RT, Sun Z, Zheng Y, Robb GB. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012;40:e54. doi: 10.1093/nar/gkr1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker WW, Luis CA, Kashuba A, Luis M, Harwood DG, Loewenstein D, Waters C, Jimison P, Shepherd E, Sevush S, Graff-Radford N, Newland D, Todd M, Miller B, Gold M, Heilman K, Doty L, Goodman I, Robinson B, Pearl G, Dickson D, Duara R. Relative frequencies of Alzheimer disease, Lewy body, vascular and frontotemporal dementia, and hippocampal sclerosis in the State of Florida Brain Bank. Alzheimer Dis Assoc Disord. 2002;16:203–212. doi: 10.1097/00002093-200210000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Nelson PT, Jicha GA, Schmitt FA, Liu H, Davis DG, Mendiondo MS, Abner EL, Markesbery WR. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: Neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–1146. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider JA, Aggarwal NT, Barnes L, Boyle P, Bennett DA. The neuropathology of older persons with and without dementia from community versus clinic cohorts. J Alzheimers Dis. 2009;18:691–701. doi: 10.3233/JAD-2009-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer’s amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PLoS ONE. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer’s disease brain neocortex: Validation study. J Alzheimers Dis. 2010;21:75–79. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 24.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogue AI, Li YY, Cui JG, Zhao Y, Kruck TP, Percy ME, Tarr MA, Lukiw WJ. Characterization of an NF-kappaB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J Inorg Biochem. 2009;103:1591–1595. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 27.Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 29.Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, Bahari-Javan S, Burkhardt S, Sananbenesi F, Fischer A. microRNA-34c is a novel target to treat dementias. EMBO J. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shioya M, Obayashi S, Tabunoki H, Arima K, Saitoh Y, Ishida T, Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neuron navigator-3. Neuropathol Appl Neurobiol. 2010;36:320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 31.Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-beta precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun. 2011;404:889–895. doi: 10.1016/j.bbrc.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Marti E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 34.Santosh PS, Arora N, Sarma P, Pal-Bhadra M, Bhadra U. Interaction map and selection of microRNA targets in Parkinson’s disease-related genes. J Biomed Biotechnol. 2009;2009:363145. doi: 10.1155/2009/363145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marti E, Pantano L, Banez-Coronel M, Llorens F, Minones-Moyano E, Porta S, Sumoy L, Ferrer I, Estivill X. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Bio-chemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 38.Gambardella S, Rinaldi F, Lepore SM, Viola A, Loro E, Angelini C, Vergani L, Novelli G, Botta A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J Transl Med. 2010;8:48. doi: 10.1186/1479-5876-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang WX, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, Nelson PT. Focus on RNA isolation: Obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008;1779:749–757. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson PT, Abner EL, Schmitt FA, Kryscio RJ, Jicha GA, Santacruz K, Smith CD, Patel E, Markesbery WR. Brains with medial temporal lobe neurofibrillary tangles but no neuritic amyloid plaques are a diagnostic dilemma but may have pathogenetic aspects distinct from Alzheimer disease. J Neuropathol Exp Neurol. 2009;68:774–784. doi: 10.1097/NEN.0b013e3181aacbe9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buee L, Hebert SS. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum Mol Genet. 2011;20:4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- 42.Smith P, Al Hashimi A, Girard J, Delay C, Hebert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011;116:240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 43.Yu SL, Chen HY, Yang PC, Chen JJ. Unique MicroRNA signature and clinical outcome of cancers. DNA Cell Biol. 2007;26:283–292. doi: 10.1089/dna.2006.0555. [DOI] [PubMed] [Google Scholar]
- 44.Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, Jung K. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 45.Davoren PA, McNeill RE, Lowery AJ, Kerin MJ, Miller N. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol Biol. 2008;9:76. doi: 10.1186/1471-2199-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson PT, Wilfred BR. In situ hybridization is a necessary experimental complement to microRNA (miRNA) expression profiling in the human brain. Neurosci Lett. 2009;466:69–72. doi: 10.1016/j.neulet.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: White matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Valverde SL, Taft RJ, Mattick JS. Dynamic isomiR regulation in Drosophila development. RNA. 2010;16:1881–1888. doi: 10.1261/rna.2379610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cloonan N, Wani S, Xu Q, Gu J, Lea K, Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, Krishnan K, Gardiner B, Wang X, Nones K, Steen JA, Matigian NA, Wood DL, Kassahn KS, Waddell N, Shepherd J, Lee C, Ichikawa J, McKernan K, Bramlett K, Kuersten S, Grimmond SM. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H, Arcila ML, Li Z, Lee EJ, Henzler C, Liu J, Rana TM, Kosik KS. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012;40:5864–5875. doi: 10.1093/nar/gks247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amariglio N, Rechavi G. A-to-I RNA editing: A new regulatory mechanism of global gene expression. Blood Cells Mol Dis. 2007;39:151–155. doi: 10.1016/j.bcmd.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 54.Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. Noncoding RNAs in long-term memory formation. Neuroscientist. 2008;14:434–445. doi: 10.1177/1073858408319187. [DOI] [PubMed] [Google Scholar]
- 55.Shao NY, Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Li N, Chen W, Khaitovich P. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics. 2010;11:409. doi: 10.1186/1471-2164-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, Hu H, Lachmann M, Zeng R, Chen W, Khaitovich P. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–1218. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, Ojo T, Luo S, Schroth G, Tuschl T. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munafo DB, Robb GB. Optimization of enzymatic reaction conditions for generating representative pools of cDNA from small RNA. RNA. 2010;16:2537–2552. doi: 10.1261/rna.2242610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lanceta J, Prough RA, Liang R, Wang E. MicroRNA group disorganization in aging. Exp Gerontol. 2010;45:269–278. doi: 10.1016/j.exger.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 61.Pandey AK, Agarwal P, Kaur K, Datta M. MicroRNAs in diabetes: Tiny players in big disease. Cell Physiol Biochem. 2009;23:221–232. doi: 10.1159/000218169. [DOI] [PubMed] [Google Scholar]
- 62.Ibanez-Ventoso C, Driscoll M. MicroRNAs in C. elegans Aging: Molecular Insurance for Robustness? Curr Genomics. 2009;10:144–153. doi: 10.2174/138920209788185243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gascon E, Gao FB. Cause or effect: Misregulation of microRNA pathways in neurodegeneration. Front Neurosci. 2012;6:48. doi: 10.3389/fnins.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babiarz JE, Hsu R, Melton C, Thomas M, Ullian EM, Blelloch R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA. 2011;17:1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cristino AS, Tanaka ED, Rubio M, Piulachs MD, Belles X. Deep sequencing of organ- and stage-specific microR-NAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae) PLoS ONE. 2011;6:e19350. doi: 10.1371/journal.pone.0019350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ling KH, Brautigan PJ, Hahn CN, Daish T, Rayner JR, Cheah PS, Raison JM, Piltz S, Mann JR, Mattiske DM, Thomas PQ, Adelson DL, Scott HS. Deep sequencing analysis of the developing mouse brain reveals a novel microRNA. BMC Genomics. 2011;12:176. doi: 10.1186/1471-2164-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shinohara Y, Yahagi K, Kawano M, Nishiyori H, Kawazu C, Suzuki N, Manabe R, Hirase H. miRNA profiling of bilateral rat hippocampal CA3 by deep sequencing. Biochem Biophys Res Commun. 2011;409:293–298. doi: 10.1016/j.bbrc.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 68.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: Evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Long JM, Ray B, Lahiri DK. MicroRNA-153 physiologically inhibits expression of amyloid-beta precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J Biol Chem. 2012;287:31298–31310. doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hebert SS, Sergeant N, Buee L. MicroRNAs and the regulation of tau metabolism. Int J Alzheimers Dis. 2012;2012:406561. doi: 10.1155/2012/406561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, Edbauer D, Sur M. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14:1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, Roth RH, Edbauer D, Kleiman RJ, Wahlestedt C. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Neill LA. Boosting the brain’s ability to block inflammation via microRNA-132. Immunity. 2009;31:854–855. doi: 10.1016/j.immuni.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 74.Shaltiel G, Hanan M, Wolf Y, Barbash S, Kovalev E, Shoham S, Soreq H. Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetyl-cholinesterase target. Brain Struct Funct. 2013;218:59–72. doi: 10.1007/s00429-011-0376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wanet A, Tacheny A, Arnould T, Renard P. miR-212/ 132 expression and functions:Within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, Busch JI, Akle S, Grossman M, Van Deerlin V, Trojanowski JQ, Lee VM. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.