Abstract
By some estimates, a eukaryotic cell must repair up to 10,000 DNA lesions per cell cycle to counteract endogenous sources of DNA damage. Exposure to environmental toxins, UV sources, or other radiations only increases this enormous number. Failure to repair such lesions can lead to a deleterious mutation rate, genomic instability, or cell death. The timely and efficient repair of eukaryotic DNA damage is further complicated by the realization that DNA lesions must be detected and repaired in the context of chromatin with its complex organization within the nucleus. Numerous studies have shown that chromatin packaging can inhibit nearly all repair pathways, and recent work has defined specific mechanisms that facilitate DNA repair within the chromatin context. In this review, we provide a broad overview of chromatin regulatory mechanisms, mainly at the nucleosomal level, and then focus on recent work that elucidates the role of chromatin structure in regulating the timely and efficient repair of DNA double-strand breaks (DSBs).
A eukaryotic cell must repair thousands of DNA lesions per cell cycle in the context of chromatin. Chromatin can prime DNA repair (e.g., repair of double-strand breaks) and coordinate it with other cellular events.
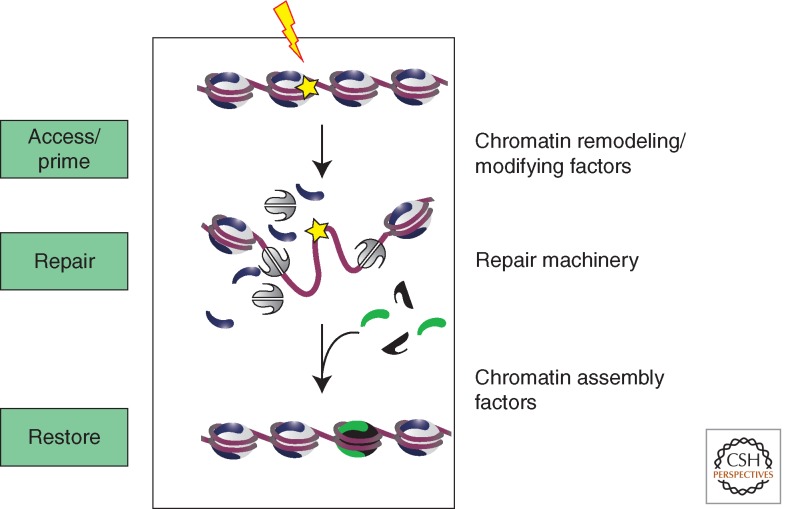
Although we tend to worry the most about environmental sources of DNA damage (e.g., chemical agents, UV radiation, ionizing radiation), it seems likely that much of the DNA repair machinery has evolved to contend with DNA lesions generated by the by-products of cellular metabolism—reactive oxygen species, endogenous alkylating agents, and DNA single- and double-strand breaks resulting from collapsed DNA replication forks or from oxidative destruction of deoxyribose residues (Lindahl and Wood 1999; Lindahl 2000). To combat the diversity of DNA lesions, cells have evolved a complex DNA damage response (DDR) that can engage many different DNA repair pathways, including nucleotide excision repair (NER), base excision repair (BER), DNA mismatch repair (MMR), single-strand annealing (SSA), nonhomologous end joining (NHEJ), and homologous recombination (HR). In eukaryotic cells, each of these repair pathways function in the context of a nucleoprotein structure, chromatin, which can potentially occlude DNA lesions from the repair machinery, and thus can influence the efficiency of repair. Early studies that focused on the response to UV damage, led to the access/repair/restore (ARR) model for repair of DNA lesions in chromatin (Green and Almouzni 2002). A central theme of this model is that chromatin inhibits the repair process, and thus it must be disrupted before or during the repair process, and then chromatin structures must be faithfully restored at the conclusion. What has become clear in the past few years, however, is that chromatin organization also serves a positive role in the DDR, to “prime” DNA repair events, functioning as a regulatory/integration platform that ensures that DNA repair is coordinated with other cellular events (Fig. 1). Here we focus on the repair of DNA double-strand breaks (DSBs), centering on the various mechanisms that facilitate this essential repair event within a chromatin context with a particular emphasis on the nucleosomal level. We envision that the concepts and themes discussed here will also be pertinent to other repair pathways, as discussed in several recent reviews (Adam and Polo 2012; Czaja et al. 2012; Lans et al. 2012; Odell et al. 2013).
Figure 1.
Access/prime/repair/restore model for the role of chromatin in the DDR. Chromatin remodeling and histone modification enzymes regulate both the accessibility of the lesion to repair factors as well as providing a platform for signaling repair events to other cellular processes. See text for details.
CHROMATIN STRUCTURE: A PRIMER
The basic unit of chromatin is the nucleosome core particle, which consists of 147 bp of DNA wrapped in left-handed superhelical turns ∼1.7 times around an octamer of histone proteins (Luger et al. 1997). The histone octamer is composed of a tetramer of histones H3 and H4 that is flanked by two heterodimers of H2A and H2B. Each histone harbors a globular, three-helix bundle called a histone fold motif, which mediates histone–histone and histone–DNA interactions. These structured histone fold domains are flanked by short flexible amino-terminal and carboxy-terminal domains or “tails,” which protrude from the nucleosome core particle. Although the histone tails are not necessarily required to form either the histone octamer or a nucleosome, they are essential for regulation of many biological processes. Numerous posttranslational modifications occur at different amino acid residues of the tails (see below), regulating key biological processes. The modifications can potentially directly affect chromatin organization. Indeed, the tails are important for both intramolecular and intermolecular folding of nucleosomal arrays to mediate different levels of compaction (Dorigo et al. 2003; Gordon et al. 2005). They can also serve as a platform to recruit factors that in turn can mediate changes (Gardner et al. 2011).
In addition to the replicative histones H2A, H2B, H3, and H4, whose expression peaks during S phase—often referred to as canonical—and are incorporated into chromatin mainly at the replication fork, eukaryotes also use a variety of histone variants that provide specialized structures and functions to chromatin (see Talbert et al. 2012) for nomenclature and evolution). In contrast to the canonical histones, the histone variants do not show a peak of expression in S phase and can be expressed at other times throughout the cell cycle. All histones are escorted by specific chaperones. In the case of the variants, particular chaperones that function in concert with ATP-dependent chromatin remodeling enzymes, such as SWR-C (see below) can mobilize the histone variants in and out of chromatin independent of DNA replication. Histone H2A has the largest number of variants, including H2A.X, H2A.Z, macroH2A, and H2ABbd. Among these variants, H2A.X and H2A.Z have been well characterized and linked to repair of DNA DSBs and regulation of transcription. H2A.X is the major form of H2A in budding and fission yeast, and it appears to be a constitutive component of mammalian chromatin, harbored by ∼10% of nucleosomes. Nucleosomal arrays that contain H2A.Z have an increased propensity to form condensed fibers compared to arrays that contain canonical H2A, suggesting that it may control fiber dynamics (Fan et al. 2002). Interestingly, nucleosomes that harbor H2A.Z flank most RNA polymerase II promoters, and these nucleosomes tend to be inherently dynamic and easily evicted during transcriptional induction (Guillemette et al. 2005; Raisner et al. 2005; Dion et al. 2007). Thus, H2A.Z may contribute to both active and inactive chromatin states. Other well-studied histone variants are related to histone H3. For instance, the centromeric variant CenH3, called Cse4 in budding yeast and CENP-A in mammals, is essential for centromere function. In both cases, this variant marks yeast and mammalian centromeres, respectively. Another H3 variant, called H3.3 is present only in metazoans, and ensures the maintenance of chromatin organization outside S phase. It is particularly critical for the replacement of protamines on sperm nuclei after fertilization and highly enriched at transcribed loci.
The primary structure of chromatin is a linear array of nucleosomes that resembles an extended “beads on a string” structure in its most decompacted state. Interactions between nucleosomes drive the folding of nucleosomal arrays into a three-dimensional secondary structure, called a 30-nm fiber, which is stabilized by the association of linker histones, such as histone H1 or H5. One key histone–histone interaction that plays a key role in chromatin condensation is an interaction between an H4 amino-terminal tail on one nucleosome with an acidic patch of residues on the H2A/H2B dimer of a neighboring nucleosome (Luger et al. 1997; Dorigo et al. 2004). Notably, this interaction can be controlled by the acetylation of histone H4 lysine 16 (H4 K16ac), a histone mark that disrupts chromatin folding and is often associated with both genomic repair and transcription (Shogren-Knaak et al. 2006). Chromatin fibers are also organized into further tertiary structures owing to self-association of the 30-nm fibers into 100- to 400-nm-thick filaments, called chromonema filaments, which are visualized by electron microscopy in interphase cells and detected by biophysical methods (Belmont and Bruce 1994; Gordon et al. 2005).
Eukaryotic chromosomes are organized into specialized domains that can regulate nuclear functions. Early cytological studies defined regions of the genome that underwent decondensation as the cells progressed from metaphase to interphase, as euchromatin. On the other hand, the regions that remained visibly condensed and deeply stained throughout the cell cycle were defined as heterochromatin. Structural features that characterize heterochromatin include the presence of repetitive DNA sequences, low or absent gene density, late S-phase replication timing, regular nucleosome spacing, decreased accessibility to nucleases, loss of nuclease hypersensitive sites, and hypoacetylation of histones. In addition, except for budding yeast, methylation of histone H3 at position K9 and its associated chromodomain-containing protein HP1 are also hallmarks of heterochromatin. Moreover, in vertebrates and plants, heterochromatin is supplemented with cytosine hypermethylation and associated proteins. For a recent, in-depth review on the role of heterochromatin proteins in the DDR, see Soria et al. (2012).
CHROMATIN DYNAMICS AND THE DDR
The recognition and orchestrated repair of DSBs requires that chromatin structure be dynamic in nature—capable of rapid unfolding, disassembly, assembly, and refolding. Currently, we know of four major mechanisms that control the dynamics of chromatin structure: (1) ATP-dependent chromatin remodeling enzymes, (2) histone variants, (3) histone modifications, and (4) histone chaperones. In the following sections, we detail how each of these mechanisms regulate chromatin structure and how they function in the context of DSB repair. Although we present them in separate parts, for the sake of simplicity, they likely work in combination.
ATP-Dependent Chromatin Remodeling Enzymes
ATP-dependent chromatin remodeling enzymes are often multisubunit complexes that can use the energy from ATP hydrolysis to actively disrupt histone–DNA interactions (for an in-depth review, see Clapier and Cairns 2009). Indeed, of the four mechanisms that regulate chromatin dynamics, these remodeling enzymes perform the “heavy lifting” of chromatin regulation, as they can disrupt heterochromatin, unfold chromatin fibers, mobilize nucleosomes, evict histones, or catalyze the incorporation or removal of histone variants. These enzymes each harbor a catalytic ATPase subunit that is related to the ancient SF2 superfamily of DNA helicases, and although these enzymes lack the ability to separate DNA strands, many enzymes proved capable to catalyze ATP-dependent DNA translocation. This translocase activity can be used to “pump” DNA over the histone octamer surface, leading to changes in nucleosome positions. This activity is also likely to be key for other activities of remodeling enzymes, such as the ATP-dependent deposition of histone variants (see below).
Biochemical studies have defined four well-characterized families of ATP-dependent remodeling enzymes—SWI/SNF, ISWI, Chd1/Mi2, and SWR-C—named after their founding members. Members of each family are able to catalyze the ATP-dependent sliding of nucleosomes in cis, whereas SWI/SNF family members (e.g., yeast or human SWI/SNF and yeast RSC) are also able to use ATP hydrolysis to evict H2A-H2B dimers or even entire histone octamers. Several members of the SWR-C family are specialized for deposition and removal of histone H2A variants. For instance, yeast SWR-C and mammalian Tip60/p400/SRCAP complexes catalyze the incorporation of the H2A.Z histone variant by removing H2A/H2B dimers from a nucleosomal substrate and exchanging with an H2A.Z/H2B dimer (Mizuguchi et al. 2004; Ruhl et al. 2006). In contrast, the yeast INO80 complex catalyzes the reverse reaction, evicting H2A.Z from nucleosomes and replacing with H2A (Papamichos-Chronakis et al. 2011).
Although ATP-dependent remodeling enzymes were discovered through studies of transcriptional regulation, it is now clear that these enzymes are rapidly recruited to DSBs and that members of each enzyme family play a unique role within the DDR. Immediately following formation of a DSB, a small number of nucleosomes (one or two) are destabilized or removed from adjacent chromatin, and this chromatin remodeling event facilitates the recruitment and function of early DNA damage responders, such as the yeast Mre11/Rad50/Xrs2 complex (Xrs2 is replaced with Nbs1 in mammals) (Shim et al. 2005, 2007; Berkovich et al. 2007; Kent et al. 2007). In budding yeast, this nucleosome loss event requires the RSC remodeling enzyme, whereas in mammals, the p400 member of the SWR-C family appears to play a key role (Xu et al. 2010). Because p400 catalyzes the incorporation of the H2A.Z variant, it seems likely that these proximal nucleosomes are destabilized by H2A.Z incorporation. In addition, work in both mammals and yeast indicates that efficient NHEJ also requires incorporation of H2A.Z at DSB chromatin (Shim et al. 2005; Xu et al. 2012). Following these early events, recent work has shown that the yeast Fun30 (mammalian Etl1) chromatin remodeling enzyme facilitates the processing of DSB ends for HR (Chen et al. 2012; Costelloe et al. 2012; Eapen et al. 2012). Fun30 is proficient in the ATP-dependent displacement of H2A-H2B dimers from nucleosome substrates (Awad et al. 2010), and it seems likely that this activity contributes to the stimulation of DSB processing.
Once the DSB ends have been processed into long single-strand DNA (ssDNA) tails, formation of a Rad51-ssDNA filament initiates a search for an homologous DNA duplex that can be used for DSB repair by HR (for review, see Symington and Gautier 2011). This homology search step appears to require several ATP-dependent remodeling enzymes, including yeast INO80 and SWI/SNF (Sinha et al. 2009; Neumann et al. 2012). Interestingly, the SWI/SNF enzyme only plays a key role in this step if repair must use a condensed, heterochromatic duplex. In this case, SWI/SNF catalyzes the eviction of the yeast Sir3 heterochromatin protein, facilitating strand invasion and subsequent repair (Chai et al. 2005; Sinha et al. 2009). Finally, the human INO80, hSNF2H, and CHD4 remodeling enzymes have also been shown to be recruited to DSBs and to play key roles in their ultimate repair, although the steps and roles that they play remain unclear (for a recent review, see Papamichos-Chronakis and Peterson 2012). Thus, a plethora of ATP-dependent remodeling enzymes are targeted to DSBs where they can mobilize, evict, or disrupt both nucleosomes and nonhistone proteins, facilitating nearly every step of DSB repair.
Histone Variants
As discussed above, the dynamic incorporation of the H2A.Z histone variant within DSB chromatin impacts early steps of NHEJ and HR. Recent work also implicates this histone variant with later checkpoint events. Key initial components of the checkpoint-signaling pathway in humans are the two members of the PIKK family of kinases, ATM and ATR (ScTel1 and ScMec1, respectively, in yeast) (Abraham 2001). ATM binds to unprocessed or minimally processed ends of broken DNA, whereas recruitment of ATR at damaged DNA requires extensive ssDNA formation by DNA end-processing factors and binding of the single-stranded binding protein, RPA. The activation of the checkpoint kinases triggers a phosphorylation cascade that impacts, among other factors, the mediator proteins scRad9/Crb2/53BP1 and MDC1, which ultimately activate the key transducer kinases Rad53/Chk2 and Chk1, essential to disperse the signal to a multitude of downstream targets and arrest the cell cycle.
Early during the DDR in budding yeast, the H2A.Z variant histone is incorporated by the SWR-C chromatin remodeling complex, replacing either γH2AX or H2A (Papamichos-Chronakis et al. 2006; Kalocsay et al. 2009). Although the SWR-C enzyme remains associated with DSB chromatin, the presence of H2A.Z at the DSB is only transient—the subsequent recruitment of the yeast INO80 complex is believed to reverse SWR-C action by exchanging nucleosomal H2A.Z/H2B dimers with free H2A/H2B (Papamichos-Chronakis et al. 2006). These H2A.Z dynamics appear to be an important control switch during the DDR—if a DSB is not repaired in a timely fashion, a SUMOylated form of H2A.Z has been proposed to mediate the anchoring of the DSB to the nuclear periphery (Kalocsay et al. 2009), whereas the removal of H2A.Z by INO80 promotes proper checkpoint signaling by an unknown mechanism (Papamichos-Chronakis et al. 2006).
Histone Posttranslational Modifications
Each of the core histones is subject to a vast array of posttranslational modifications, including lysine acetylation, methylation, SUMOylation and ubiquitylation, serine/threonine phosphorylation, proline isomerization, and arginine methylation (for a review, see Gardner et al. 2011). Most of the well-characterized histone modifications occur within the extended amino-terminal or carboxy-terminal histone tail regions, but there are also a growing number of posttranslational modifications within the nucleosome core. Histone modifications control chromatin dynamics by two broad mechanisms. First, histone modifications can either create or eliminate binding sites for nonhistone proteins that influence the structure and function of the chromatin fiber. For instance, methylation of histone H3 at lysine 9 promotes the anchoring of the HP1 protein that facilitates formation of condensed heterochromatin structures. Second, histone modifications may directly impact either the stability of individual nucleosomes or influence the ability of chromatin fibers to fold into higher order structures. For instance, one histone modification, acetylation of histone H4 at lysine 16, has been shown to directly impact chromatin structure. In this case, acetylation of this one lysine residue is sufficient to block formation of 30-nm fibers (Shogren-Knaak et al. 2006).
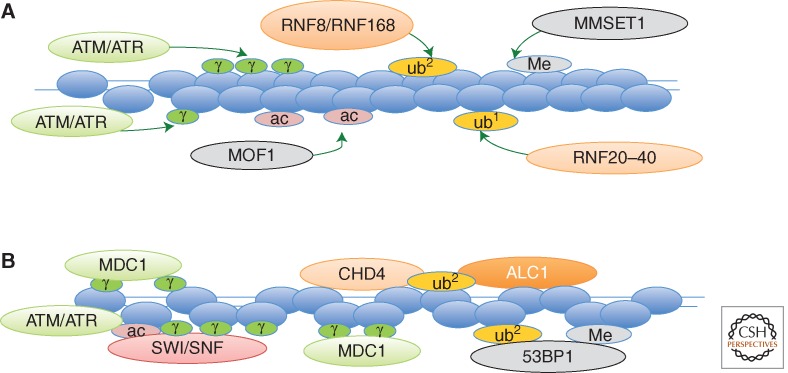
During recent years, it has been clear that formation of a DSB induces a host of histone posttranslational modifications (Table 1), and functional studies are consistent with the establishment of a complex regulatory chromatin platform. One of the most intensively studied DSB-induced histone modifications is the phosphorylation of the histone variant H2A.X. The phosphorylation of H2AX at S139 in mammalian histones (termed γH2AX; S129 in budding yeast) by the DDR kinases, ATM (yeast Tel1), ATR (yeast Mec1), and DNA-dependent protein kinase (DNA-PK), is one of the earliest events at a DSB, and this mark spreads over at least a megabase of chromatin adjacent to each DSB in mammalian cells and up to 50 kb on each side of a DSB in yeast (Rogakou et al. 1999; Shroff et al. 2004). This large domain of γH2AX promotes a robust DNA damage checkpoint pathway that halts the cell cycle and coordinates repair of damage with cell-cycle transitions. In mammalian cells, γH2AX provides a host of binding sites for MDC1, which enhances retention of this key checkpoint mediator within DSB chromatin, facilitating efficient cell-cycle arrest (Fig. 2) (Stucki et al. 2005).
Table 1.
Histone posttranslational modifications associated with the DDR
| Histone-modifying enzymes | Name (organism) | Proposed molecular function—targets |
|---|---|---|
| K-kinases | ATM/ATR (Sc-h) | H2A.X S139 (h), S129 (Sc) |
| WSTF (h) | H2A.X Y142 | |
| Cdc7 (Sc) | H3 T45 | |
| Aurora B | H3 S10, S28 | |
| Bub1 | H2A S121 | |
| Haspin | H3 T3 | |
| K-acetyltransferases | Hat1 (Sc) | H4 K5, K12 |
| NuA4/Tip60 (Sc, Dm, h) | H4 K5, K8, K12, K16, | |
| H2A.Z K3, K8, K10, K14 | ||
| Gcn5 (Sc, h) | H3 K9, K14, K18 | |
| CBP/p300 | H3 K18, H4 K5, K8, K12, K16 | |
| RTT109 (Sc) | H3 K56ac | |
| K-deacetylases | Rpd3 (Sc, h) | H3, H4 |
| Hda1 (Sc) | H2A.Z, H2B, H3 | |
| Sir2/SIRT1, 6 (Sc, h, m) | H4 K16, | |
| HDAC1, 2, 4 (h) | H2A, H2B, H3, H4 | |
| K-methyltransferases | SET1 (Sc) | H3 K4 |
| PR-Set7/Set8 (h) | H3 K20me | |
| MMSET1 (h) | H4 K20me2, 3 | |
| EZH2 (h) | H3 K27 | |
| Dot1 (Sc) | H3 K79 | |
| Ubiquitilases | RNF8 (h) | H2A |
| RNF168 (h) | H2A | |
| Rad6/RNF20-40 (Sc, h) | H2B K120ub (h), K123 (Sc) |
Figure 2.
Histone modifications orchestrate binding of key DDR factors to DSB chromatin. (A) Histone modifications and their associated enzymes at DSB chromatin. (B) Histone modifications and their binding partners or regulators. See text for details. Note that the figure illustrates the ubiquitination-dependent recruitment of 53BP1 to DSBs, but it is not thought to directly bind to ubiquitinylated proteins.
Other histone modifications and histone-modifying enzymes play an elaborate role in promotion of the MDC1-mediated checkpoint pathway. For instance, acetylation of H4K16 by the human MOF acetyltransferase has been suggested to regulate binding of MDC1 to γH2AX domains, presumably owing to the chromatin unfolding function of this histone mark (Fig. 2) (Li et al. 2010; Sharma et al. 2010). Furthermore, following its recruitment to DSB chromatin, MDC1 binds and recruits the E3 ubiquitin ligase RING finger protein 8 (RNF8) to DSB chromatin and RNF8 functions in concert with a second ubiquitin ligase, RNF168, to promote assembly of checkpoint regulators 53BP1 and BRCA1 at damaged chromatin (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007). These enzymes are likely to target histones for ubiquitinylation, although nonhistone substrates should also be considered. A combination of RNF8 action and polyADP ribosylation is also believed to facilitate the recruitment of the CHD4 and ALC1 chromatin remodeling enzymes (Fig. 2). And finally, dimethylation of histone H4 (H4K20me2), a constitutive histone mark that is increased at laser irradiation-induced foci and induced at single DSBs, also facilitates recruitment of the 53BP1 checkpoint mediator to DSB chromatin (Pei et al. 2011). Thus, multiple histone marks play key roles within DSB chromatin to recruit various mediators of the cell-cycle checkpoint response, orchestrating their functions with the repair process (Fig. 2).
Histone Chaperones
Histone chaperones, by definition, are proteins that transfer histones without necessarily being part of the final product (Loyola and Almouzni 2004; De Koning et al. 2007). They can be general without specificity for variants or specific of particular variants. Considering the different aspects of chromatin dynamics within the general framework of the access/repair/restore model, one can immediately envisage that they will play critical roles throughout the process to assist all aspects from the destabilization, eviction, to help present histone for their modification, and for the restoration to deposit new histones or recycle older ones. In the above discussion related to H2AZ and H2AX, we actually already referred to histone chaperones as part of larger complexes as shown with the INO80 complex that contains several histone chaperones. This example illustrates the tight interaction in the function of remodeling complexes and histone chaperones. In addition, in the context of DDR, the histone chaperone FACT (facilitates chromatin transcription) should also be considered. Indeed, initially identified for its ability to mobilize H2A-H2B during transcription (Belotserkovskaya et al. 2003), it can also mediate H2A.X exchange. Following genotoxic stress, FACT is polyADP-ribosylated, which disrupts its ability to interact with nucleosomes (Heo et al. 2008); as a consequence, H2A.X/H2A exchange is inhibited.
We now consider chaperones associated with H3-H4 and their possible roles during the DDR. Concerning the dynamics of H3 variants in the DDR, the most documented role for a histone chaperone relates to CAF-1 (chromatin assembly factor-1). Indeed, CAF-1, initially identified as a factor stimulating histone deposition during replication (Stillman 1986), was also shown to restore chromatin organization on repaired DNA in vitro (Gaillard et al. 1996), and later defined as a dedicated chaperone for the replicative H3.1 deposition (Tagami et al. 2004; Ray-Gallet et al. 2011). The transient expression of epitope-tagged H3.1 variants in human cells further revealed a CAF-1--dependent new histone deposition at both UVC- and laser-induced damage sites (Polo et al. 2006). Thus, H3.1 deposition is not restricted to replication but also takes place at sites of DNA repair synthesis in vivo. Most importantly, new histone deposition at DNA damage sites implies that there is not a simple recycling of preexisting histones, and this potentially helps to replace old histones. Importantly, new soluble histones differ from preexisting nucleosomal histones in terms of their posttranslational modifications (Loyola et al. 2006), and as a consequence their incorporation into chromatin will dilute parental marking. This alteration of local chromatin marks may subsequently affect the expression of genes in the damaged chromatin region. This possibility is supported by recent findings in the context of DNA replication in chicken cells deficient for the specialized polymerase REV1 (reversionless 1) involved in the bypass of roadblocks such as G4-DNA sequences. In these cells, the uncoupling of DNA replication and histone recycling at the time of replication leads to a local increase of new histone incorporation and thereby a loss of parental marks, which alters the transcriptional status of the loci (Sarkies et al. 2010, 2012). In light of these findings, a tight control of histone recycling along with new histone deposition is likely critical. In this context, the histone chaperone ASF1 (antisilencing function 1) involved in both recycling parental histones and providing new histones at the replication fork (Groth et al. 2007; Jasencakova et al. 2010) represents an attractive histone chaperone for such a role at repair sites.
Interestingly, there is also an interesting connection between chromatin restoration and termination of DNA damage signaling, as shown in budding yeast where CAF-1 and ASF1 orthologs function redundantly to promote the recovery from checkpoint arrest (Kim and Haber 2009). Therefore, new H3.1 histone incorporation coupled to DNA repair participates in the restoration of nucleosomal organization after DNA damage and possibly modulates checkpoint termination.
The recently identified CenH3 histone chaperone HJURP (Holliday junction recognition protein (Dunleavy et al. 2009; Foltz et al. 2009; Shuaib et al. 2010) has also been connected to the DDR. This protein was initially characterized on the basis of its ability to bind Holliday junctions (recombination intermediates) in vitro and showed an increase of expression after DNA damage in human cells in a manner that depends on the DSB sensor kinase ATM (Kato et al. 2007). Furthermore, HJURP and CenH3 expression levels correlate with cell sensitivity to radiation in vitro and in vivo, high levels of HJURP being predictive for increased sensitivity to radiotherapy in breast cancer patients (Hu et al. 2010). Further characterization of HJURP and CenH3 properties will be necessary to determine whether distinct or similar features as those required for their centromeric function could be at work in the DDR.
Interestingly, a potential role for other H3 chaperones deserved to be explored. Indeed, in fission yeast, the ortholog of the HIRA (histone regulator A) complex, a critical H3.3 chaperone in mammals, is required for protection against genotoxic agents, as shown by mutating Hip1, Slm9, and Hip3 subunits (Anderson et al. 2009). The possible link between H3.3 and genome stability is further supported by the recent identification in high-grade pediatric brain tumors and in pancreatic tumors of mutations in H3.3 itself as well as in DAXX-ATRX (death domain-associated protein-α-thalassemia/mental retardation syndrome X-linked), another H3.3-specific chaperone (Jiao et al. 2011; Schwartzentruber et al. 2012). Given that nucleosome rearrangements in damaged chromatin can expose stretches of naked DNA independently of DNA repair synthesis, H3.1 deposition mechanism may not suffice. It will be particularly interesting to investigate whether the nucleosome gap-filling function recently proposed for human H3.3 as a salvage pathway for chromatin integrity (Ray-Gallet et al. 2011) applies to sites of DNA damage.
CONCLUDING REMARKS
Following successful repair of the DSB, repair factors and chromatin regulators are removed from the site of repair, the checkpoint response is inactivated, and the epigenetic landscape largely returns to its original state. Several studies have shown that γH2AX is targeted for dephosphorylation following repair in both yeast and mammalian cells, and its removal is key for down-regulating the DNA damage checkpoint response (Chowdhury et al. 2005; Keogh et al. 2006; Nakada et al. 2008). It is not clear whether the entire host of histone modifications survive the repair event and thus must be actively removed, or whether the final, replication-dependent steps of HR lead to displacement of the modified parental nucleosomes. This replicative step leads to the assembly of new nucleosomes that harbor the histone mark H3-K56Ac in yeast. Histone H3-K56Ac is known to be associated with histone turnover at gene promoter regions, perhaps providing a molecular explanation for why newly repaired chromatin is more labile (Xu et al. 2010). Interestingly, the deposition of nucleosomes that harbor H3-K56Ac is required for inactivation of the DNA damage checkpoint. It has been proposed that the H3-K56ac-containing chromatin is sensed by the DNA damage checkpoint machinery (Chen et al. 2008), but an alternative possibility is that incorporation of newly synthesized histones, driven by H3-K56 acetylation, may lead to the eviction of nucleosomal histones that contain marks required for checkpoint activation or maintenance, thus switching off the checkpoint. How these dynamics apply to metazoans will have to be examined.
Although initial thoughts on the role of chromatin in DNA repair were driven by simple paradigms derived from transcriptional studies (e.g., “access, repair, restore”), it is clear that chromatin dynamics orchestrate a much more complex and intriguing set of roles in the DDR. It is now clear that histone modifications do not simply promote access of repair factors to the lesion, but in fact they play essential roles in mediating the cell-cycle checkpoint response. This role underlines the importance of histone modifications and histones themselves as signaling modules. Likewise, recent work also implicates key roles for ATP-dependent chromatin remodeling enzymes in directing large-scale movements of chromosomes that facilitate the homology search step of recombinational repair (Dion et al. 2012; Miné-Hattab and Rothstein 2012; Neumann et al. 2012). Many of the current studies that focus on chromatin and the DDR have focused on identifying enzymes that are recruited to DNA lesions, rather than understanding how these events interface with the biochemistry of DNA repair in the context of the global control of cellular responses. Like the case in transcription, it is likely that much functional redundancy exists among chromatin regulators, and thus such mechanistic studies may need to use sophisticated genetics, biochemical and cellular approaches in an integrated fashion to uncover specific roles for each enzyme or chromatin alteration within the DDR. An exciting challenge will be to figure out during development and in various cell types how these properties can be modulated, in particular in stem cells that show distinct chromatin and repair properties.
ACKNOWLEDGMENTS
We thank members of the Peterson and Almouzni laboratories for helpful discussions, and Manolis Papamichos-Chronakis (Curie Institute, Paris) for Table 1. Research in the Peterson laboratory is funded by grants from the National Institutes of Health, and work in the Almouzni laboratory is funded by Centre National de la Recherche Scientifique and Institute Curie as well as EU grants (ERC and NoE EpiGeneSys) and Labellisation Ligue.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Abraham RT 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Adam S, Polo SE 2012. Chromatin dynamics during nucleotide excision repair: Histones on the move. Int J Mol Sci 13: 11895–11911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson HE, Wardle J, Korkut SV, Murton HE, López-Maury L, Bähler J, Whitehall SK 2009. The fission yeast HIRA histone chaperone is required for promoter silencing and the suppression of cryptic antisense transcripts. Mol Cell Biol 29: 5158–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad S, Ryan D, Prochasson P, Owen-Hughes T, Hassan AH 2010. The Snf2 homolog Fun30 acts as a homodimeric ATP-dependent chromatin-remodeling enzyme. J Biol Chem 285: 9477–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Bruce K 1994. Visualization of G1 chromosomes: A folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol 127: 287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Kastan MB 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 [DOI] [PubMed] [Google Scholar]
- Chai B, Huang J, Cairns BR, Laurent BC 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev 19: 1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK 2008. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu C-D, Tang J, Chen K, Pan X, Ira G 2012. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489: 576–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Keogh M-C, Ishii H, Peterson CL, Buratowski S, Lieberman J 2005. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell 20: 801–809 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. 2012. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489: 581–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaja W, Mao P, Smerdon MJ 2012. The emerging roles of ATP-dependent chromatin remodeling enzymes in nucleotide excision repair. Int J Mol Sci 13: 11954–11973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning L, Corpet A, Haber JE, Almouzni G 2007. Histone chaperones: An escort network regulating histone traffic. Nat Struct Mol Biol 14: 997–1007 [DOI] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol 14: 502–509 [DOI] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Bystricky K, Richmond TJ 2003. Chromatin fiber folding: Requirement for the histone H4 N-terminal tail. J Mol Biol 327: 85–96 [DOI] [PubMed] [Google Scholar]
- Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ 2004. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306: 1571–1573 [DOI] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Eapen VV, Sugawara N, Tsabar M, Wu W-H, Haber JE 2012. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol 32: 4727–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW 2009. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G 1996. Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor I. Cell 86: 887–896 [DOI] [PubMed] [Google Scholar]
- Gardner KE, Allis CD, Strahl BD 2011. Operating on chromatin, a colorful language where context matters. J Mol Biol 409: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon F, Luger K, Hansen JC 2005. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J Biol Chem 280: 33701–33706 [DOI] [PubMed] [Google Scholar]
- Green CM, Almouzni G 2002. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep 3: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Corpet A, Cook AJL, Roche D, Bartek J, Lukas J, Almouzni G 2007. Regulation of replication fork progression through histone supply and demand. Science 318: 1928–1931 [DOI] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W 2008. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell 30: 86–97 [DOI] [PubMed] [Google Scholar]
- Hu Z, Huang G, Sadanandam A, Gu S, Lenburg ME, Pai M, Bayani N, Blakely EA, Gray JW, Mao J-H 2010. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res 12: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Scharf AND, Ask K, Corpet A, Imhof A, Almouzni G, Groth A 2010. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell 37: 736–743 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S 2009. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33: 335–343 [DOI] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y 2007. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res 67: 8544–8553 [DOI] [PubMed] [Google Scholar]
- Kent NA, Chambers AL, Downs JA 2007. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem 282: 27693–27701 [DOI] [PubMed] [Google Scholar]
- Keogh M-C, Kim J-A, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, et al. 2006. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439: 497–501 [DOI] [PubMed] [Google Scholar]
- Kim J-A, Haber JE 2009. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci 106: 1151–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Vermeulen W 2012. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Corsa CAS, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y 2010. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol 30: 5335–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T 2000. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res 462: 129–135 [DOI] [PubMed] [Google Scholar]
- Lindahl T, Wood RD 1999. Quality control by DNA repair. Science 286: 1897–1905 [DOI] [PubMed] [Google Scholar]
- Loyola A, Almouzni G 2004. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 1677: 3–11 [DOI] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell 24: 309–316 [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Miné-Hattab J, Rothstein R 2012. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14: 510–517 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Nakada S, Chen GI, Gingras A-C, Durocher D 2008. PP4 is a γH2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep 9: 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann FR, Dion V, Gehlen LR, Tsai-Pflugfelder M, Schmid R, Taddei A, Gasser SM 2012. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev 26: 369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell ID, Wallace SS, Pederson DS 2013. Rules of engagement for base excision repair in chromatin. J Cell Physiol 228: 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL 2012. Chromatin and the genome integrity network. Nat Rev Genet 14: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Krebs JE, Peterson CL 2006. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev 20: 2437–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL 2011. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144: 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z 2011. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470: 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127: 481–493 [DOI] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, Schultz DC, Pchelintsev NA, Adams PD, Jansen LET, et al. 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell 44: 928–941 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 146: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC 2006. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45: 5671–5677 [DOI] [PubMed] [Google Scholar]
- Sarkies P, Reams C, Simpson LJ, Sale JE 2010. Epigenetic instability due to defective replication of structured DNA. Mol Cell 40: 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE 2012. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res 40: 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang D-AK, Tönjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231 [DOI] [PubMed] [Google Scholar]
- Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al. 2010. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 30: 3582–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Ma J-L, Oum J-H, Yanez Y, Lee SE 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol 25: 3934–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Hong SJ, Oum J-H, Yanez Y, Zhang Y, Lee SE 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol 27: 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuaib M, Ouararhni K, Dimitrov S, Hamiche A 2010. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci 107: 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL 2009. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell 138: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G 2012. Prime, repair, restore: The active role of chromatin in the DNA damage response. Mol Cell 46: 722–734 [DOI] [PubMed] [Google Scholar]
- Stillman B 1986. Chromatin assembly during SV40 DNA replication in vitro. Cell 45: 555–565 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271 [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Ahmad K, Almouzni G, Ausió J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SWL, et al. 2012. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD 2010. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol 191: 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD 2012. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell 48: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]