Abstract
Over the last two decades, several attempts to generate packaging cells for lentiviral vectors (LV) have been made. Despite different technologies, no packaging clone is currently employed in clinical trials. We developed a new strategy for LV stable production based on the HEK-293T progenitor cells; the sequential insertion of the viral genes by integrating vectors; the constitutive expression of the viral components; and the RD114-TR envelope pseudotyping. We generated the intermediate clone PK-7 expressing constitutively gag/pol and rev genes and, by adding tat and rd114-tr genes, the stable packaging cell line RD2-MolPack, which can produce LV carrying any transfer vector (TV). Finally, we obtained the RD2-MolPack-Chim3 producer clone by transducing RD2-MolPack cells with the TV expressing the anti-HIV transgene Chim3. Remarkably, RD114-TR pseudovirions have much higher potency when produced by stable compared with transient technology. Most importantly, comparable transduction efficiency in hematopoietic stem cells (HSC) is obtained with 2-logs less physical particles respect to VSV-G pseudovirions produced by transient transfection. Altogether, RD2-MolPack technology should be considered a valid option for large-scale production of LV to be used in gene therapy protocols employing HSC, resulting in the possibility of downsizing the manufacturing scale by about 10-fold in respect to transient technology.
Stornaiuolo and colleagues describe a packaging cell line for the stable production of clinical-grade lentiviral vectors for anti-HIV gene therapy. Pseudovirions generated using this cell line are able to transduce a higher proportion of progenitor CD34+ stem cells as compared with vesicular stomatitis virus glycoprotein (VSV-G) pseudovirions.
Introduction
Since the first-ever lentiviral vector (LV) phase I clinical trial against AIDS in 2001, 53 phase I/II and two phase II/III trials using HIV-based LV for gene transfer protocols have undergone authorities' scrutiny. So far, LV have been used to genetically modify, mostly, hematopoietic stem cells (HSC) and lymphocytes to correct inherited and acquired disorders. Furthermore, significant progress has been recently made using stem-cell-based gene and cell therapy approaches to treat HIV-1 infection (Kitchen et al., 2011). In this context, we have previously demonstrated that the therapeutic gene Chim3, an HIV-1 Vif-dominant negative factor, when delivered to target cells by means of Tat-conditional LV, inhibits HIV-1 replication and confers selective growth advantage to transduced cells (Porcellini et al., 2009, 2010). Chim3-based anti-HIV gene therapy is therefore a promising therapeutic intervention, which requires an adequate clinical-grade manufacturing to be further tested in preclinical animal models or in patients. Unlike gamma retroviral vector (γRV), a limitation in LV field is the lack of clinical-grade stable packaging clones. Transient transfection is instead the current technology for pilot production of LV, which is impractical for very large-scale applications under a safety, cost, and reproducibility standpoint. In fact, this technology is expensive, is difficult to standardize and scale-up, and suffers from batch-to-batch variability and low reverse transcriptase fidelity (Laakso and Sutton, 2006).
Notably, the establishment of a γRV-equivalent packaging cell line for LV is more difficult than expected because of the expression of some viral proteins, which are toxic for producer cells, such as the HIV protease (Kaplan and Swanstrom, 1991; Krausslich, 1992; Krausslich et al., 1993; Snasel et al., 2000) and the VSV-G attachment protein (Burns et al., 1993). To overcome this problem, in previous approaches, toxic genes were transcriptionally regulated by different systems, such as Tet (Kafri et al., 1999; Klages et al., 2000; Farson et al., 2001; Xu et al., 2001; Throm et al., 2009), ecdysone (Pacchia et al., 2001; Sparacio et al., 2001), Rev (Klages et al., 2000; Ni et al., 2005), and the Tet–cumate combination (Broussau et al., 2008), the removal of which has to be considered during downstream processing. Another important issue is the choice of the delivery vehicles used to integrate the genes encoding the vector components; most commonly, gag-pol, rev, and env genes are integrated by transient transfection of plasmid DNA (Carroll et al., 1994; Corbeau et al., 1996; Poeschla et al., 1996; Yu et al., 1996; Srinivasakumar et al., 1997; Kaul et al., 1998; Kafri et al., 1999; Klages et al., 2000; Farson et al., 2001; Pacchia et al., 2001; Sparacio et al., 2001; Xu et al., 2001; Ni et al., 2005; Cockrell et al., 2006; Broussau et al., 2008). This method is known to suffer eventually from gene silencing and gene loss (Bestor, 2000), which can both jeopardize the long-term stability of the producer clone. Alternative methods have been applied to STAR and GPRG-TL-20 packaging cells, in which the gag-pol and rev genes were transduced by LTR-γRV and SIN-γRV, respectively (Ikeda et al., 2003; Throm et al., 2009), with the exception, in STAR cells, of the env gene introduced by plasmid transfection (Ikeda et al., 2003).
On this premise, we conceived a new strategy to obtain RD2-MolPack-Chim3, a packaging clone for the stable production of LV carrying the Chim3 transfer vector (TV). Notably, an equal number of concentrated RD114-TR pseudovirions transduces a higher proportion of progenitor CD34+ stem cells compared with VSV-G pseudovirions.
Materials and Methods
Plasmids
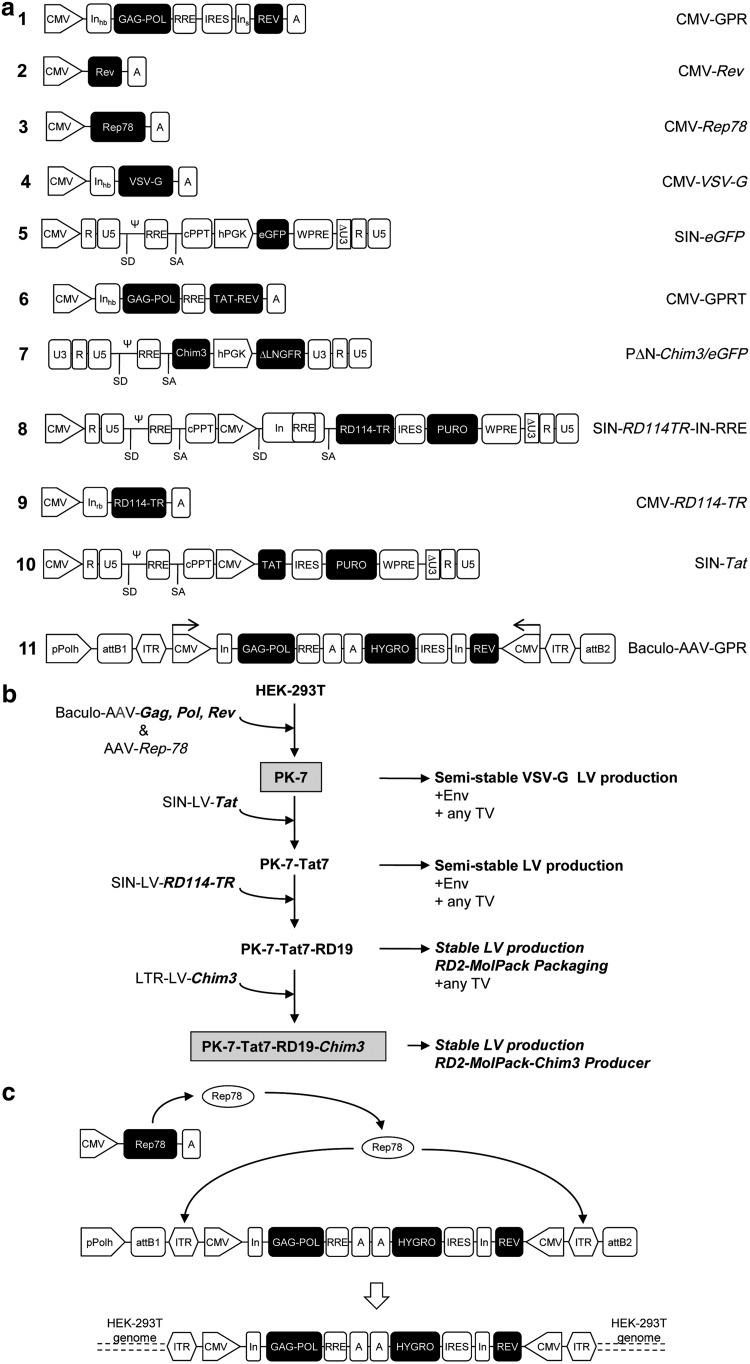
Wild-type HIV-1 gag, pol, and rev genes were derived from the pCG719-pKLgagpol (hereafter named CMV-GPR) and pCG720-pKrev (CMV-Rev) plasmids, respectively (Cell Genesys, Inc.) (Fig. 1a, schemes 1 and 2). The viral genes were inserted into the Gateway pENTR4 shuttle vector (Invitrogen, Co.) in two distinct expression cassettes tail-to-tail oriented. The first cassette expresses the gag and pol genes, whereas the second one the rev and the selection marker hygromycin resistance (hygro) gene, which was cloned downstream an internal ribosome entry site (IRES) to allow bi-cistronic translation. The two expression units of the packaging construct were introduced into the XbaI site of the recombinant pSUB201 plasmid carrying an infectious adeno-associated viral genome (Samulski et al., 1987). The resulting 5′ITR-CMV-GagPol-polyA-polyA-hygro-IRES-Rev-CMV-ITR3′ fragment was excised from the pSUB201 plasmid and inserted into the Gateway pENTR4 shuttle vector. The final recombinant hybrid baculo-AAV packaging vector (Baculo-AAV-GPR) (Fig. 1a, scheme 11) was obtained by the bacteriophage lambda site-specific recombination system between the pENTR4 shuttle entry vector, containing the two cassettes, and the BaculoDirect Linear DNA (BaculoDirect Baculovirus Expression Systems; Invitrogen, Co.). The polyhedrin gene of the baculo DNA was thereby replaced with the GPR double cassette during homologous recombination.
FIG. 1.
Strategy of RD-MolPack development. (a) Schemes of plasmids and vectors used in this study. pPolh, polyhedrin promoter; attB1, attachment B1; ITR, inverted terminal repeat; CMV, cytomegalovirus promoter; In, intron; RRE, rev responsive element; A, polyA sequence; IRES, internal ribosome entry site; SD, splice donor; SA, splice acceptor; Ψ, packaging signal; WPRE, woodchuck hepatitis post-transcriptional regulatory element; cPPT, central polypurine tract; hPGK, human phosphoglycerate kinase promoter. (b) Flow chart of the development strategy of PK-7 and RD-MolPack-Chim3 packaging cells. (c) Cartoon of the Rep78-mediated integration of the AAV-GPR vector into HEK-293T host genome. AAV Rep78 promotes the excision of the ITR-flanked AAV-GPR cassette and facilitates the integration into human chromosomes in either single or concatameric configuration.
The pABCMV-Rep78 plasmid (CMV-AAV-Rep78) was obtained by cloning the AAV-Rep78 ORF under the CMV IE promoter of the pABS.43 vector (Recchia et al., 2004) (Fig. 1a, scheme 3). The pMD.G plasmid (CMV-VSV-G) encodes the vesicular stomatitis envelope glycoprotein (VSV-G) (Fig. 1a, scheme 4). The pCCLsin.PPT.hPGK.eGFP.WPRE.Amp (SIN-eGFP) TV and the second-generation packaging pCMV-ΔR8.74 (CMV-GPRT) construct encoding the HIV gag, pol, rev, and tat genes were kindly provided by L. Naldini (Tiget; HSR) (Fig. 1a, schemes 5 and 6). The second-generation PΔN-Chim3 TV was previously described (Porcellini et al., 2009, 2010) (Fig. 1a, scheme 7). The PΔN-eGFP was obtained by substituting the Chim3 ORF with the eGFP transgene. The SIN-RD114-TR-IN-RRE vector (Fig. 1a, scheme 8) was constructed by using the RD114-TR ORF excised from the pCMV-RD114-TR plasmid (CMV-RD114-TR) (Sandrin et al., 2002) (Fig. 1a, scheme 9). The CMV-RD114-TR-IN-RRE cassette of the SIN-RD114-TR-IN-RRE vector contains the rabbit β-globin intron derived from the CMV-RD114-TR vector in which the 230 bp RRE element, polymerase chain reaction (PCR) amplified as described in the Analytical PCR analysis section, was integrated into the ScaI site of the β-globin intron. This cassette was cloned into the MluI site of the SIN-polyMluI vector, a modified version of the SIN-eGFP vector in which the hPGK-eGFP cassette was removed and substituted with the EcoRV-MluI-SmaI-MluI-NotI-SacI-BglII-BamHI-SalI poly-linker. The SIN-Tat vector was constructed by inserting the tat gene, derived from the CMV-GPRT plasmid, into the pIRESpuro3 (Clontech Laboratories, Inc.), and then by cloning the bi-cistronic CMV-Tat-IRES-puro cassette into the MluI site of the SIN-polyMluI vector (Fig. 1a, scheme 10).
Cells
Spodoptera frugiperda (Sf9) insect cells (Invitrogen, Co.) were grown in TC-100 medium (Invitrogen, Co.) supplemented with 10% fetal calf serum (FCS; EuroClone Ltd.) and a combination of penicillin–streptomycin and glutamine (PSG) at 27°C in a non-CO2 incubator. Human embryo kidney 293T (HEK-293T) cells and its derivatives were propagated in Dulbecco's modified Eagle's medium supplemented with 10% FCS and PSG. SupT1 and CEM A3.01 T cells were grown in RPMI 1640 supplemented with 10% FCS and PSG. Human CD34+ HSC and neonatal leukocytes were purified from UCB centrifugation on a Ficoll-Hypaque gradient (Lymphoprep; Nycomed Pharma AS) after approval of the local institutional review board and written informed consent. CD34+ HSC were isolated from the UCB mononucleated cells by positive selection using CD34 MicroBeads Kit and MiniMACS Separator Columns (Miltenyi Biotec). CD34+ cells purity was >92%, as established by FACS analysis (FACSCalibur BD Bioscience) using the anti-CD34-PE antibody (BD Pharmingen) and the FlowJo software (Tree Star, Inc.). CD34+ cells and neonatal leukocytes were prestimulated for 48 hr in a cytokine cocktail and maintained in the same medium during transduction and cultivation as previously described (Porcellini et al., 2009, 2010).
Baculovirus production and baculo-GPR infection of HEK-293T cells
Baculovirus carrying the recombinant hybrid Baculo-AAV-GPR DNA genome (Fig. 1a, scheme 11) was produced following the BaculoDirect method using the Gateway-adapted Baculovirus DNA system (Invitrogen, Co.). After three cycles (p3) of recombinant baculovirus amplification in Sf9 cells, we checked the titer and the potential recombination events of the hybrid baculo-AAV DNA by plaque assay and Southern blot of viral genomic DNA (gDNA), respectively. The titer corresponded to 6×1010 pfu/ml, and a single sharp band was revealed by Southern blot, demonstrating no recombination events during virus amplification (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/hgtb). To obtain the PK-7 clone, the following parameters were individually optimized: (1) dose and time of AAV-Rep78 plasmid DNA transfection; (2) multiplicity of infection (MOI) of rh-baculo-AAV; (3) cloning strategy for infected HEK-293T cells. The best overall output was obtained by transfecting 1.5×106 cells with 4 μg of AAV-Rep78 expression plasmid and 24 hr afterward infecting the cells with the recombinant Baculo-AAV-GPR (MOI=1,000). After 4 days, a total of 5×105 cells were seeded in twenty-two 10 cm dishes in the presence of hygromycin (100 μg/ml) at serially diluted concentrations. The 22 dishes were screened by p24Gag enzyme-linked immunosorbent assay (ELISA). Only the dishes seeded with 3.7×104 cells/dish contained 40 colonies and released detectable p24Gag. Three of the 40 colonies were further characterized.
LV production, concentration, and titration
LV were obtained by transient co-transfection of HEK-293T cells with the following plasmids: the packaging constructs CMV-GPR (third-generation) [or CMV-GPRT (second-generation)], the VSV-G or RD114-TR envelope constructs, and the third-generation SIN-eGFP or the second-generation PΔN-Chim3 or PΔN-eGFP TV. LV produced from PK-7 clone were generated by co-transfecting the Env plasmid and the TV, whereas LV produced from the PK-7-Tat7-RD clones, only the TV. Supernatants were harvested 24 hr after transfection and filtered through 0.45 μm filters, and, when indicated, concentrated 100-fold by low-speed centrifugation (4,000 rpm at 4°C for 16 hr in a Multifuge 32-R). The viral pellets were resuspended in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum and frozen at −70°C (Strang et al., 2004). Titer was calculated on different cell lines by one or two cycles of spinoculation at 1,240 g for 45 min in the presence of polybrene 8 μg/ml (Sigma-Aldrich); CD34+ HSCs were transduced for 24 hr on retronectin-coated plates (Takara Bio) as previously described (Porcellini et al., 2009, 2010). Transduction efficiency was monitored by flow cytometry analysis (FACS Calibur BD Bioscience) of eGFP (SIN-eGFP) or ΔLNFGR expression (PΔN-Chim3), as previously described (Porcellini et al., 2009, 2010).
Titer-based protocol for screening of LV producer clones
All PK-7 derivative subclones were screened by calculating the LV titer of their supernatants on SupT1 cells by a small-scale transduction protocol. Briefly, 6×104 cells/well were seeded in 48-well plate and 24 hr later co-transfected with the plasmids needed to obtain LV. Forty-eight hours after transfection, 200 μl of culture supernatants was used to transduce 3×104 SupT1 cells/well seeded at 7.5×104 cells/ml. The titer threshold score was imposed ≥1×102 TU/ml.
RCL test
RCL test was carried out exactly as described by Cornetta et al. (2011). Briefly, RD2-MolPack-Chim3 supernatant (1.75 liters) was obtained by seven independent harvests from 10 T162 flasks/harvest (total 70 T162 flasks). The supernatants of each harvest were concentrated, and 1 μg p24Gag LV/harvest was used as test article in the amplification phase. The supernatants of the seven independent amplification phases were then used in seven independent indicator phases. The endpoints of the test were evaluated by the immunological assay p24Gag ELISA and the molecular psi-gag recombination PCR assay as described in the specific sections.
Analytical PCR analysis
The 868 bp AAV-Rep78 amplicon was obtained by using 1 pg of control plasmid CMV-Rep78 DNA and 300 ng of gDNA of PK-7 cells; the following set of primers: AAV-Rep78 forward, 5′-CGG GCT GCT GGC CCA CCA GG-3′, and AAV-Rep78 reverse, 5′-ATG CCG GGG TTT TAC GAG ATT GTG-3′; and the following PCR conditions: 98°C for 7 min, 30 cycles of 94°C for 30 sec, 66°C for 30 sec, and 72°C for 1.5 min.
The 230 bp RRE amplicon was obtained by using 1 ng of SIN-eGFP vector as DNA template; the following set of primers: RRE-forward, 5′-AGT ACT GGA GCT TTG TTC CTT GGG-3′, and RRE-reverse, 5′-AGT ACT AAA TCC CCA GGA GCT G-3′; and the following PCR conditions: 98°C for 7 min, 30 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec.
To establish orientation of the two integrated GPR cassettes, PCR amplification was performed on 300 ng of gDNA of PK-7 cells with the set of primers: Rev forward, 5′- CTT GAG GAG GTC TTC GTC GC-3′; β-glob. reverse, 5′-CCC TGT TAC TTC TCC CCT TCC-3′; Rev forward nested, 5′-TGT CTC CGC TTC TTC CTG CC-3′; and β-glob. reverse nested, 5′-TTA ACC ATA GAA AAG AAG GGG-3′; and the following PCR conditions: 94°C for 2 min, 35 cycles of 94°C for 30 sec, 52°C for 30 sec, and 72°C for 1.5 min.
PCR conditions for the detection of putative psi-gag recombination events in the RCL test were carried out exactly as described in Cornetta et al. (2011).
Vector copy number quantification by quantitative PCR
The vector copy number (VCN) of the integrated vector was established by Q-TaqMan PCR using an ABI Prism 7900 FAST instrument (Applied Biosystems) and analyzed by SDS 2.3 software (Applied Biosystems). PCR conditions were the following: 2 min at 50°C and 15 min at 95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. gDNA was amplified by using the primers and probes of Supplementary Table S1.
HIV p24Gag ELISA
Physical LV production estimate was obtained by measuring p24Gag with the Alliance HIV-1 p24 Antigen ELISA kit (Perkin Elmer Life and Analytical Sciences, Inc.) following manufacturer's instructions in culture supernatants, with the assumption that 1 ng p24Gag corresponds to 1×107 physical particles (pp).
Ligation-mediated PCR
gDNA was extracted from PK-7 cells by QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's instructions and digested with BglII and BamHI at 37°C overnight. Ligation of an adaptor 76-bp oligonucleotide linker compatible with the 5′-GATC-3′ ends was performed under standard conditions. Ligation-mediated (LM)-PCR was carried out using the nested primers of Supplementary Table S2. Two rounds of LM-PCR were carried out using AmpliTaq Gold DNA Polymerase (Applied Biosystems), each comprising 30 cycles (95°C for 30 sec, 52°C for 30 sec, 72°C for 2 min). PCR amplicons were cloned using the TOPO cloning kit (Invitrogen, Co.) and plasmid colonies carrying inserts of approximately 100–200 bp were selected for sequencing. Sequence homologies were identified by BLAST search, NCBI, as previously described (Maruggi et al., 2009).
Southern and Northern blot assays
gDNA was isolated by the QIAamp Mini kit (Qiagen GmbH) according to manufacturer's instructions. Baculovirus DNA was extracted from viral particles by the QIAamp DNA micro kit (Qiagen). After overnight digestion with the indicated restriction enzymes, 10 μg/sample was run on 0.8% agarose gel, blotted by Southern capillary transfer onto nylon membranes (Duralon), and then hybridized to 1×106 dpm/ml of 32P-random primed labeled of either 600 bp CMV, 11 kb GPR, 250 bp Tat, 600 bp Chim3, or 500 bp RD114-TR-specific probe, in PerfectHyb PLUS hybridization buffer.
Total RNA was isolated by the TRIzol reagent (Ambion; Life Technologies Ltd.) according to manufacturer's instructions. About 5 μg/sample was run on denaturing formaldehyde/MOPS 1.0% agarose gel, blotted by capillary transfer onto nylon membranes (Duralon), and then hybridized to 2×106 dpm/ml of 32P-random primed labeled of either 250 bp Tat, 500 bp RD114-TR, or 270 bp ψ-specific probe, in PerfectHyb PLUS hybridization buffer.
After extensive washes, the membranes were exposed to X-ray films at −80°C or to PhosphorImager (Molecular Dynamics).
Western blot analysis
Cellular and viral proteins, the latter derived from isolated cell-free viral-like particles (VLPs) or LV, were prepared and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis as previously described (Porcellini et al., 2009, 2010). The anti-HIV serum, obtained from an AIDS patient, was used at 1:1,000 dilution; the anti-transmembrane (TM) RD114-TR rabbit serum, kindly provided by F.L. Cosset (Inserm) (Sandrin et al., 2004) was used at 1:1,000 dilution; the anti-surface RD114-TR MoAb, generated by Areta International, was used at 1:50 dilution; the HIV-1 Rev MoAb (Rev-6, sc-69730), the affinity-purified rabbit polyclonal anti-YY1 antibody (C-20, sc-281) (Santa Cruz Biotechnology, Inc.), and the mouse anti-p24Gag (Acris Antibodies) were used at 1:200, 1:1,000, and 1:500 dilution, respectively. HIV-1 Tat MoAb (1D9) was obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH, Bethesda, MD) from Dr. Dag E. Helland and used at 1:1,000 dilution. The antihuman and antimouse secondary antibodies were diluted 1:10,000, whereas the antirabbit 1:20,000.
Fluorescence in situ hybridization
Metaphase chromosomes were obtained by treating PK-7 cells with 10 μg/ml colchicine (Sigma-Aldrich) for 2 hr at 37°C. After standard phosphate buffer saline (PBS) washing, cells were kept in 75 mM KCl hypotonic solution for 6 min at room temperature, fixed with four washes of methanol/acetic acid (3:1), and then spread on a glass slide. Samples were denatured in 70% formamide for 2 min at 72°C, dehydrated by cold 70%, 85%, and 95% ethanol washes, and then air-dried. The 13 kb plasmid DNA containing the GPR-cassette-specific probe was labeled using the Random Primed DNA Labelling Kit (Roche Applied Science) with SpectrumOrange-dUTP (Vysis, Inc.), whereas the control 30 kb cosmid DNA containing the human hox4 gene was labeled using the fluorescence in situ hybridization (FISH) Bright Nucleic Acid Labelling kit (Kreatech Biotechnology). Hybridization was performed by incubating 1.25 μg of each probe in 250 μl of 50% formamide, 2×SSC, 10% dextran sulfate, and 12.5 μg of human C0T-1 DNA hybridization buffer (Invitrogen, Co.). Samples were coated with denatured probes for 10 min at 75°C, covered with Parafilm M, and incubated overnight at 37°C in a moist chamber. Samples were washed once in 0.4×SSC, pH 7, at 72°C for 2 min; once in 4×SSC, pH 7, containing 0.0025% Tween-20 for 30 sec; and twice in PBS 1× for 1 min at room temperature. Slides were counterstained with 0.02 μg/μl of 49,6-diamidino-2-phenylindole (DAPI; Sigma). Visualization and photographic images were taken with a Nikon 80i upright microscope (Nikon Instruments S.p.A.) using the FITC (green) and spectrum orange (orange) filter illumination. Images were processed with Genikon software (Nikon).
Results
Development of the intermediate PK-7 clone
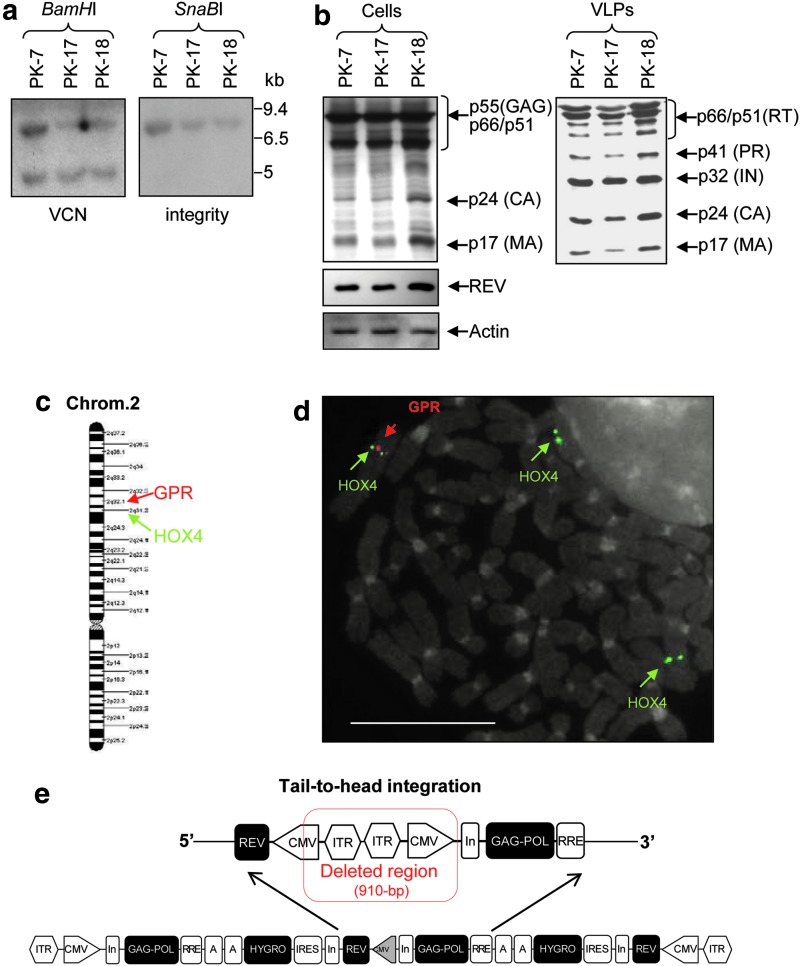
To obtain RD2-MolPack packaging cells, we developed several HEK-293T-derived intermediate clones (Fig. 1b). The first one, PK-7, was created by stable integration of HIV-1 gag-pol and rev genes by means of the recombinant hybrid baculo-AAV vector (rhBaculo-AAV-GPR) (Fig. 1a, scheme 11) in the transient presence of AAV-Rep78 protein (Fig. 1c). Only three of the 360 grown-up, PK-7, −17, and −18 clones, expressed p24Gag above the 0.1 ng/ml arbitrary threshold, indicating a low level of tolerance for the constitutive expression of Gag, Pol, and Rev proteins. Southern blot and quantitative PCR (Q-PCR) analyses revealed that each clone contains two copies of the correct-in-size vector (Fig. 2a), which produces HIV-1 viral proteins rightly processed and in the right relative proportion in both cell and VLP extracts as visualized by Western blot analysis (Fig. 2b). For further genetic manipulation, we selected the PK-7 clone based on cell morphology, growth capabilities, viability, p24Gag production, and titer of the LV produced after transfecting the VSV-G plasmid and the SIN-eGFP TV, which correspondes at the best value to 1.1×107 TU/ml (mean 3.6×106±0.6 SEM TU/ml, n=12) (Supplementary Table S3) when calculated on SupT1 cells. Next, we spotlighted the break point of the ITR-GPR cassette integration on the chromosome 2q32.1 locus by LM-PCR technology (Fig. 2c), confirming this result by FISH using the specific GPR probe and, as internal control, the human HOX4 gene probe, which is known to map on the chromosome 2q31.2 locus (Fig. 2d). We demonstrated, by using an appropriate primer set design in nested PCR, that the two copies of packaging construct are integrated in tail-to-head orientation (Fig. 2e). Sequence analysis of the amplicon encompassing the tail-to-head junction showed that a 910 bp fragment got lost likely during integration (Fig. 2e). To exclude integration of residual AAV-Rep78 plasmid DNA, we carried out AAV-Rep78-specific PCR on PK-7 gDNA detecting no amplification (Supplementary Fig. S1b). To demonstrate genetic and functional stability of PK-7 clone over time, we cultivated the cells in the presence or absence of hygromycin for 350 days and measured p24Gag production on a per cell basis (Table 1). Next, we established that PK-7 cells maintain long-term functionality by cotransfecting passage 60 (p60) and p102 cells with the VSV-G plasmid and SIN-eGFP TV either in the presence or in the absence of antibiotic (Table 1). As in both passages and conditions titer values were similar (1.3–3.2×106 TU/ml), we avoided the use of hygromycin in further experiments. Furthermore, to determine whether PK-7 clone is suitable for semi-stable LV production, we transfected in parallel PK-7 and HEK-293T cells with the plasmids necessary, in each cell type, for LV production. We measured on SupT1 and CD34+ cells the titer of second-generation LV (PΔN-Chim3) and third-generation LV (SIN-eGFP) produced from the two cell types. The titer of PK-7-derived PΔN-Chim3 LV is comparable to the HEK-293T-derived LV, whereas the titer of PK-7-derived SIN-eGFP LV is 1-log lower than the HEK-293T-derived LV (Table 2).
FIG. 2.
Characterization of PK-7 clone. (a) Southern blot analysis of the AAV-GPR vector integration. About 10 μg/sample of gDNA derived from PK clones was digested with BamHI and SnaBI restriction enzymes and hybridized with the specific probe to establish the VCN and the integrity of the cassette (integrity). (b) Western blot analysis of the viral proteins. Left panel (cells): 50 μg/sample of cell extract obtained from the PK clones was hybridized to an anti-HIV human serum-recognizing HIV-1 proteins. The membrane was sequentially hybridized with anti-Rev and anti-actin antibodies. Right panel (VLPs): 30 ng p24Gag-equivalent/sample of VLP produced by PK clones was processed similarly to the cellular extracts. (c) Schematic mapping of the GPR cassette integration by LM-PCR technique indicating the break point in the long arm of chromosome 2 at the 2q32.1 location and the known location of the HOX4 gene. (d) Co-localization of the AAV-GPR cassette and the HOX4 gene into chromosome 2. FISH analysis of PK-7 metaphase chromosomes was carried out by using Gag-specific (red) and HOX4-specific (green) probes. As HEK-293T cells are triploid, HOX4 signal is detected on the three chromosomes 2. (e) Schematic representation of the rearrangement of the two GPR-integrated cassettes in the PK-7 clone and their tail-to-head orientation. FISH, fluorescent in situ hybridization; LM-PCR, ligation-mediated polymerase chain reaction; VCN, vector copy number; VLP, viral-like particles. gDNA, genomic DNA.
Table 1.
Stability of PK-7 Clone
| |
p24Gag ng/106 cells/day |
|
|---|---|---|
| Passage | Hygro | No Hygro |
| p2 | 10.0 | 8.10 |
| p6 | 7.00 | 4.80 |
| p10 | 11.4 | 4.00 |
| p16 | 5.00 | 4.80 |
| p20 | 9.30 | 5.20 |
| p24 | 7.20 | 6.50 |
| p28 | 9.20 | 4.40 |
| p32 | 6.20 | 8.80 |
| p36 | 11.0 | 9.60 |
| p40 | 18.0 | 10.0 |
| p44 | 4.30 | 8.40 |
| p48 | 37.5 | 3.80 |
| p52 | 7.00 | 3.20 |
| p56 | 11.0 | 6.70 |
| p60 | 19.0 | 4.40 |
| p64 | 22.0 | 18.7 |
| p68 | 15.2 | 8.00 |
| p72 | 16.5 | 4.90 |
| p76 | 17.8 | 7.60 |
| p80 | 30.0 | 11.0 |
| p84 | 27.0 | 8.40 |
| p88 | 23.2 | 3.50 |
| p92 | 23.0 | 7.50 |
| p98 | 16.7 | 1.30 |
| p102 | 19.0 | 3.80 |
| Mean±SD | 15.3±8.4 | 6.70±3.5 |
| Titer (TU/ml)a | ||
| p60 | 3.2×106 | 2.0×106 |
| p102 | 2.7×106 | 1.3×106 |
| p24Gag (ng/ml)a | ||
| p60 | 86 | 38 |
| p102 | 80 | 13 |
| Infectivity (TU/ng p24Gag)a | ||
| p60 | 4.2×104 | 5.2×104 |
| p102 | 3.3×104 | 1.0×105 |
Potency values of VSV-G-pseudotyped lentiviral vectors (LV) produced after transfection of PK-7 cells with SIN-eGFP and VSV-G plasmids and tested on SupT1 cells 3 days after transduction.
Table 2.
Potency of VSV-G-Pseudotyped Second- and Third-Generation Lentiviral Vectors Produced from PK-7 Clone
| |
|
SupT1 |
CD34+ |
|---|---|---|---|
| Vector | Producer | Titer (TU/ml) | |
| PΔN-Chim3 | PK-7a | 5.8×106 | 3.5×104 |
| HEK-293Tb | 1.0×107 | 3.9×104 | |
| p24Gag (ng/ml) | |||
| PK-7 | 166 | 166 | |
| HEK-293T | 237 | 237 | |
| Infectivity (TU/ng p24Gag) | |||
| PK-7 | 3.5×104 | 2.0×102 | |
| HEK-293T | 4.2×104 | 1.6×102 | |
| SIN-eGFP | Titer (TU/ml) | ||
| PK-7a | 5.7×106 | 2.4×104 | |
| HEK-293Tb | 5.6×107 | 3.0×105 | |
| p24Gag (ng/ml) | |||
| PK-7 | 45.0 | 45.0 | |
| HEK-293T | 350 | 350 | |
| Infectivity (TU/ng p24Gag) | |||
| PK-7 | 1.2×105 | 5.3×102 | |
| HEK-293T | 1.6×105 | 8.6×102 | |
VSV-G-pseudotyped LV were produced after transfection of PK-7 cells with either the SIN-eGFP or the PΔN-Chim3 and VSV-G envelope plasmids.
VSV-G-pseudotyped LV were produced after transfection of HEK-293T cells with CMV-GPR, SIN-eGFP (or PΔN-Chim3), and VSV-G envelope plasmids. LV were tested on target cells after 3–5 days from transduction.
Development of RD2-MolPack stable packaging cells
To generate RD2-MolPack packaging clone (Fig. 1b), we carried out stable integration of the tat gene into PK-7 cells by means of SIN-Tat LV delivery (Fig. 1a, scheme 10). Cells were SIN-Tat transduced, puromycin selected, and then cloned by limiting dilution. We measured Tat expression by Western blot in 5 of the 11 screened clones producing ≥5 ng p24Gag/1×106 cells. Both PK-7-Tat5 and PK-7-Tat7 clones showed elevated expression of Tat, released 110 and 130 ng p24Gag/1×106 cells, and contained 12 and 6 copies of SIN-Tat, respectively (Supplementary Fig. S2). After small-scale transfection of the TV and vsv-g plasmids, we selected PK-7-Tat7 to integrate the RD114-TR envelope because, in view of a similar titer (Supplementary Table S4), the level of p24Gag of PK-7-Tat7 is lower and the number of copies of SIN-Tat lower than those of PK-7-Tat5. Next, we stably integrated the rd114-tr gene into PK-7-Tat7 cells by SIN-RD114-TR-IN-RRE LV transfer. The rationale of the complicated design of this vector is described elsewhere (Stornaiuolo et al., in preparation). After SIN-RD114-TR-IN-RRE LV transduction, PK-7-Tat7 cells were cloned by limiting dilution, and three out of nine selected clones, PK-7-Tat7-RD3, -RD12, and -RD19, were screened as done for PK-7-Tat cells. We decided on PK-7-Tat7-RD19 clone because it produces the highest titer (3×105 TU/ml) (Supplementary Table S5) and the highest amount of RD114-TR transmembrane and surface subunits, and contains the highest copies of SIN-RD114-TR vector (Supplementary Fig. S2b). We named the PK-7-Tat7-RD19 clone, RD2-MolPack, which is a stable packaging cell line for the production of any LV.
Development of RD2-MolPack-Chim3 producer clone
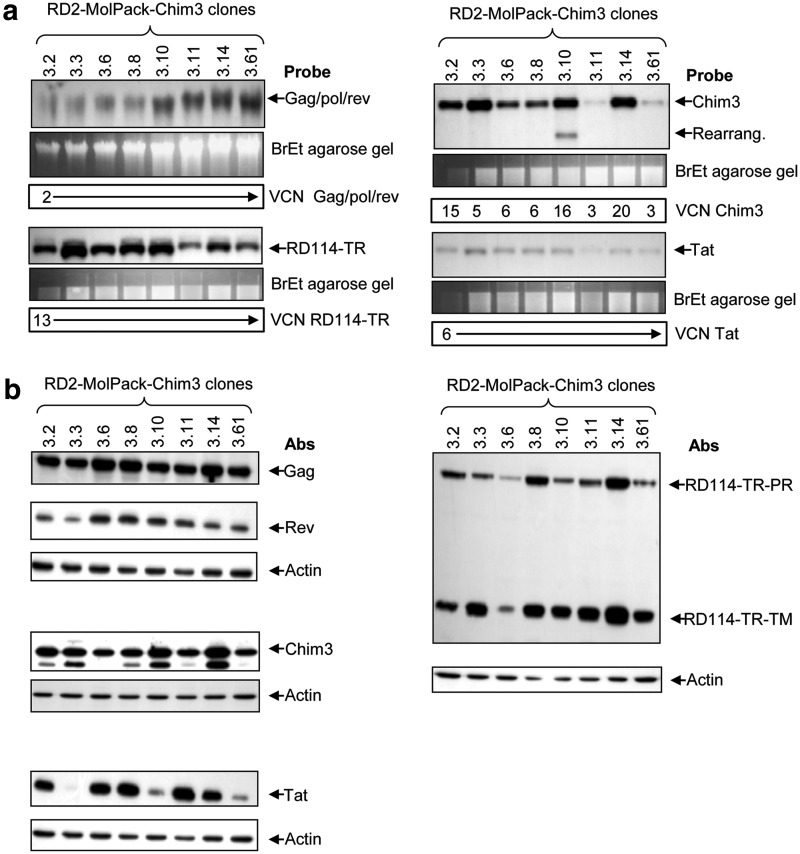
To construct the RD2-MolPack-Chim3 producer clone from the packaging cells, we stably integrated the PΔN-Chim3 TV by transduction of the VSV-G pseudotyped Chim3 LV. After the standardized screening protocol, we analyzed the potency of LV produced from four clones, RD2-MolPack-Chim3.2, -Chim3.3, -Chim3.14, and -Chim3.25, on SupT1 and CD34+ HSC (Table 3). We selected RD2-MolPack-Chim3.14 and -Chim3.25 clones for further characterization because they provided the best titer on CD34+ cells (5.1×105 and 1×106 TU/ml, respectively) (Table 3). Remarkably, the potency of the vectors produced from both clones is higher than that of the corresponding RD114-TR pseudovirions produced by transient transfection (Table 3). We then checked the integrity of the exogenous genes into eight producer cells by Southern and Western blot analyses demonstrating that each gene maintains its integrity in all, but one (Chim3 TV in the -Chim3.10 clone), clones (Fig. 3a, right panel), and that the amount and size of the corresponding proteins are correctly detected in all clones (Fig. 3b). These findings indicate that the use of SIN-LVs to integrate the viral components in an LV packaging cell itself is feasible and safe when the integration of the viral genes occurs in a sequential order. To further address the safety of RD2-MolPack-Chim3.25 cells, we carried out Northern blot analysis looking for potential mobilization of the RD114-TR and Tat expression cassettes, which could be generated by an host promoter reading through the 5′LTR of the vector and, so doing, incorporating the packaging signal. We did not detect aberrant transcripts using the psi probe, suggesting no mobilization going on (Supplementary Fig. S3). Additionally, we verified the possible mobilization of RD114-TR, Tat, E1A, and SV40 LTA in CD34+ transduced cells by Q-PCR for the corresponding potentially integrated DNA sequences, detecting no amplification (Supplementary Table S6). In particular, we carried out a deep analysis of the possible mobilization of the SIN-LV encoding the Tat and RD114-TR genes by Q-PCR. We first calculated the expected frequency of the two SIN-LV inside the Chim3-LV based on the number of integrated copies and the known mobilization frequency of SIN-LV (1:1,000–3,000) (Hanawa et al., 2005) (Supplementary Table S7). We then calculated the true mobilization frequency into viral particles by measuring the percentage of the specific Tat and RD114-TR RNA respect to that of Chim3 after in vitro retrotranscription (Supplementary Table S7). We measured a frequency of 0.47% for RD114-TR and 0.99% for Tat that takes into account the contribution of both the specific gRNA and mRNA encapsidated into the viral particles. Finally, we calculated the expected and the real number of mobilized copies present in CD34+ cells transduced >90%, detecting no signal for either SIN-LV, indicating that the amount of retrotranscribed RNA contained into the LV corresponds mostly to mRNA or small, nonfunctional gRNA sequences (Supplementary Table S7). We also verified the functional stability of RD2-MolPack-Chim3.25 clone as previously done for PK-7 cells. The RD2-MolPack-Chim3.25 clone persists in continuous culture with ≥80% viability for longer than 3 months, producing steady LV titer until p20 (87 days) that corresponds to slightly less than 3 months (Supplementary Table S8). Finally, we demonstrated the absence of RCL on a medium scale production lot equivalent to 7,146 ng p24Gag LV produced by RD2-MolPack-Chim3.25 cells by two virus detection assays: the immunological p24Gag ELISA (Supplementary Table S9) and the molecular analytical PCR for possible psi-gag recombination (Supplementary Fig. S4).
Table 3.
Potency of Lentiviral Vectors Produced from RD2-MolPack-Chim3 Clones
| |
SupT1 |
CD34+ |
|---|---|---|
| Clones | Titer (TU/ml) | |
| RD2-MolPack-Chim3.2 | 1.9×105 | 1.1×105 |
| RD2-MolPack-Chim3.3 | 7.2×105 | 2.6×105 |
| RD2-MolPack-Chim3.14 | 5.0×105 | 5.1×105 |
| RD2-MolPack-Chim3.25 | 1.2×106 | 1.0×106 |
| PK-7a | 1.5×104 | 2.5×104 |
| HEK-293Tb | 2.0×105 | 8.6×104 |
| p24Gag (ng/ml) | ||
| RD2-MolPack-Chim3.2a | 112 | 112 |
| RD2-MolPack-Chim3.3 | 120 | 120 |
| RD2-MolPack-Chim3.14 | 127 | 127 |
| RD2-MolPack-Chim3.25 | 95.0 | 95.0 |
| PK-7 | 128 | 128 |
| HEK-293T | 275 | 275 |
| Infectivity (TU/ng p24Gag) | ||
| RD2-MolPack-Chim3.2 | 1.7×103 | 9.8×102 |
| RD2-MolPack-Chim3.3 | 6.0×103 | 2.1×103 |
| RD2-MolPack-Chim3.14 | 4.0×103 | 4.0×103 |
| RD2-MolPack-Chim3.25 | 2.4×104 | 1.0×104 |
| PK-7 | 1.2×102 | 1.9×102 |
| HEK-293T | 7.2×102 | 3.1×102 |
Bold indicates the selected clones.
LV were produced after transfection of PK-7 cells with the PΔN-Chim3 transfer vector and RD114-TR envelope plasmids.
LV were produced after transfection of HEK-293Tcells with CMV-GPR, PΔN-Chim3 transfer vector, and RD114-TR envelope plasmids. LV were tested on target cells 5 days after transduction.
FIG. 3.
Characterization of RD2-MolPack-Chim3 clones. (a) Southern blot analysis of the integrity of the individual genes in eight independent RD2-MolPack-Chim3 clones. About 8 μg/sample of gDNA was digested with the appropriate restriction enzyme to analyze the corresponding integrity of the different integrated cassettes and hybridized with the specific probes as indicated. (b) Western blot analysis of cellular extracts derived from the eight RD2-MolPack-Chim3 clones. About 40 μg/sample of cellular extracts was probed with the specific antibodies as indicated. After stripping, the membranes were hybridized with an antiactin antibody as control of protein loading.
RD2-MolPack-Chim3 LV transduce more efficiently CD34+ cells than VSV-G pseudotyped Chim3 LV produced transiently
To assess the transduction efficiency in CD34+ cells of RD2-MolPack-Chim3 LV compared with VSV-G pseudotyped LV produced by standard transfection, we performed a comparative study using equal amount of pp of each type of LV, carrying the Chim3 TV. LV were concentrated by low-speed centrifugation (Strang et al., 2004) and then 73, 7.3, and 0.73 ng p24Gag of each LV were delivered to CD34+ cells and CEM A3.01, as control cells. After 5 and 14 days, transduction values of PΔN-Chim3 LV were calculated (Table 4). Notably, we observed that 73 ng p24Gag of RD114-TR pseudovirions transduces 1.93-fold more CD34+ cells compared with the same amount of pp of VSV-G pseudovirions after 5 days from transduction. The difference increases with the decrement of the amount of pp used (3.2-fold more with 7.3 ng and 8.2-fold more with 0.73 ng p24Gag). In contrast, in CEM A3.01 cells the ratios of transduction efficiency of RD114-TR- versus VSV-G-pseudovirions are 0.94 (73 ng), 0.52 (7.3 ng), and 0.24 (0.73 ng). These data clearly establish the power of RD114-TR-pseudovirions in HSC, but not in CEM A3.01 cells. Again, in CD34+ cells, 2-log less RD114-TR LV (0.73 ng p24Gag) than VSV-G LV (73 ng p24Gag) pp is sufficient to obtain equivalent VCN (1.05 and 0.95, respectively) whereas, in CEM A3.01 cells, 1-log more RD114-TR LV (7.3 ng p24Gag) than VSV-G LV (0.73 ng p24Gag) pp is required to obtain similar VCN (0.44 and 0.50, respectively) (Table 4). The percentage of positivity of CD34+-derived cells transduced with RD114-TR pseudovirions is more stable over time than that of CD34+-derived cells transduced with VSV-G pseudovirions.
Table 4.
Comparison of Transduction Efficiency of RD114-TR Versus VSV-G-Pseudotyped LV on CD34+ and CEM A3.01 Cells
| |
Target cellsa: CD34+ |
|||
|---|---|---|---|---|
| LV | p24Gag (ng) | Transdb5d (%) | Transdb14d (%) | VCNc14d (n) |
| RD114-TR-pseudot.d (RD2-MolPack-Chim3) | 73.0 | 99.25 | 96.5 | 16.9 |
| 7.30 | 91.50 | 86.5 | 7.80 | |
| 0.73 | 42.65 | 40.0 | 1.05 | |
| VSV-G-pseudot.d (HEK-293T cells) | 73.0 | 51.50 | 30.0 | 0.95 |
| 7.30 | 27.90 | 20.5 | 0.51 | |
| 0.73 | 05.20 | 02.8 | 0.06 | |
| |
Target cellsa: CEM A3.01 |
|||
|---|---|---|---|---|
| LV | p24Gag (ng) | Transdb5d (%) | Transdb14d (%) | VCNc14d (n) |
| RD114-TR-pseudot.d (RD2-MolPack-Chim3) | 73.0 | 93.0 | 85.0 | 1.60 |
| 7.30 | 50.0 | 51.0 | 0.44 | |
| 0.73 | 12.0 | 13.0 | 0.07 | |
| VSV-G-pseudot.d (HEK-293T cells) | 73.0 | 99.0 | 99.0 | 15.7 |
| 7.30 | 96.0 | 97.0 | 2.96 | |
| 0.73 | 49.0 | 50.0 | 0.50 | |
One hit of transduction was performed in the presence of retronectin on 4×104 CD34+ or CEM A3.01 cells in 96-well plates. Seventy-three ng p24Gag of VSV-G LV, whose titer was calculated on CEM A3.01 cells, corresponds to MOI=100.
Transduction efficiency was calculated by fluorescence-activated cell sorting analysis of ΔLNGFR expression at the indicated time points. Values for CD34+ cells are the means of two normal donors.
Vector copy number (VCN) was calculated 14 days after transduction by quantitative polymerase chain reaction.
LV were produced from the RD2-MolPack-Chim3 cells (RD114-TR LV) in the presence of 2 mM Na-butyrate, and from transfection of HEK-293T cells of second-generation packaging Chim3 TV and VSV-G plasmids (VSV-G LV).
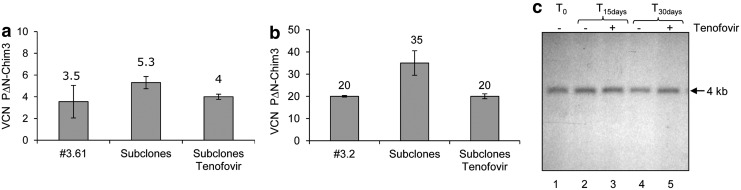
Tenofovir prevents autotransduction in RD2-MolPack-Chim3 cells
To evaluate whether RD2-MolPack-Chim3 cells can be autotransduced by the LV they themselves produce, we measured the fold of increase of PΔN-Chim3 VCN over 1 month of culture in four RD2-MolPack-Chim3 subclones by Q-PCR. After 30 days of continuous culture, we quantified 1.3-, 1.9-, 1.4-, and 1.7-fold increase of VCN relative to that calculated for each subclone at the beginning of their culture (Supplementary Table S10), indicating that RD2-MolPack-Chim3 cells endure autotransduction during long-term cultivation. Next, we demonstrated that autotransduction is blocked by the Nucleoside Analog Reverse Transcriptase inhibitors tenofovir, which is a drug authorized in clinic for the treatment of HIV infection. First, we established that tenofovir is the most effective, with no toxicity at the most efficacious dose (50 μM), among other NRTI we tested, abacavir and lamivudine, in preventing PΔN-Chim3 integration in RD2-MolPack cells (Supplementary Fig. S5). Remarkably, we determined that RD2-MolPack-Chim3 subclones raised in the presence of tenofovir conserve the VCN of the parental clone in both low-VCN (Chim3.61-derived, which carries 3.5 PΔN-Chim3 VCN) and high-VCN (Chim3.2-derived, which carries 20 PΔN-Chim3 VCN) packaging cells (Fig. 4a, b). Finally, we verified that tenofovir is not toxic for the TV integrity by carrying out Southern blot analysis with the representative subclone RD2-MolPack-Chim3.61, cultivated in the presence or absence of tenofovir and showing in all samples a correct-in-size 4 kb band (Fig. 4c). Altogether, these data indicate that tenofovir can be safely used for preventing autotransduction in stable LV production.
FIG. 4.
Tenofovir prevents autotransduction in RD2-MolPack-Chim3 clones. Quantitative PCR analysis of PΔN-Chim3 VCN in two RD2-MolPack-Chim3 clones, the RD2-MolPack-Chim3.61 clone, carrying 3.5 copies (a), and the RD2-MolPack-Chim3.2 clone, carrying 20 copies (b), and in several subclones obtained either in the presence or absence of tenofovir. (c) Southern blot analysis of gDNA of the representative RD2-MolPack-Chim3.61clone, grown either in the presence or in the absence of tenofovir, and digested with SacI to monitor vector integrity over time at three different time points as indicated.
Discussion
We report a new technological strategy to develop packaging cells for stable production of HIV-based LV. In particular, we developed the RD2-MolPack-Chim3 producer clone primarily to go forward with the preclinical anti-HIV gene therapy studies based on Chim3-LV (Porcellini et al., 2009, 2010), and secondly to pave the way for the future development of a third-generation stable packaging cell line, RD3-MolPack. The feasibility of HSC-based anti-HIV gene therapy has been demonstrated for different therapeutic genes and payloads (Kitchen et al., 2011; Mitsuyasu et al., 2011; Bovolenta et al., 2012). Nevertheless, a frequent downside for the production of LV carrying anti-HIV transgene is the transgene- or payload-mediated inhibition of LV production itself (i.e., Rev-M10) (Bahner et al., 2007). In this context, we have previously established that the dominant negative Vif protein Chim3 exerts its antiviral action only in the presence of wt HIV Vif (Porcellini et al., 2009, 2010). Here, we strengthen this result showing no impairment of Chim3-LV production from any of the RD2-MolPack-Chim3 clones, thus validating the feasibility of large-scale manufacturing of RD2-MolPack-Chim3-derived LV.
RD2-MolPack-Chim3 has been designed to produce LV for targeting HSC since it carries the RD114-TR envelope, which has been previously (Sandrin et al., 2002; Neff et al., 2004; Di Nunzio et al., 2007) and in this study shown to have a preferential tropism for CD34+ cells compared with VSV-G envelope. As LV titer mainly depends on the type and number of envelope molecules mounted on each LV and on the number of specific envelope receptors present on the cell type used for its calculation, HSCs contain an higher number of RD114-TR receptor than that present, for example, on CEM A3.01 cells. The titer of nonconcentrated RD2-MolPack-Chim3.25-derived LV (1.0×106 TU/ml on both SupT1 and CD34+ cells) is comparable to that obtained from nonconcentrated RD114-PR LV produced by STAR packaging cell line (∼5.0×106 on HT1080 cells) (Ikeda et al., 2003) and even higher to that produced by transient protocols (5.2×104 on HT1080 cells) (Kim et al., 2010; Trobridge et al., 2010). These data confirm and extend our previous observations demonstrating an higher fraction of human hematopoietic clonogenic progenitors transduced with RD114-TR pseudovirions than those transduced with VSV-G LV (Di Nunzio et al., 2007). These results appear to be at odds with those of Kim et al. (2010), reporting an opposite behavior for VSV-G and RD114-TR envelopes in CD34+ cells. We explain this discrepancy by the different experimental conditions applied in the two studies: we used concentrated RD114-TR pseudovirions LV derived from a stable packaging clone instead of LV transiently produced; UCB rather than BM- and mPB; and thereby a different cytokine activation protocol. The lower amount of p24Gag ng/ml (4.7-fold less) produced from RD2-MolPack-Chim3 cells (mean=61.2±8.5 SEM ng/ml, n=12) that likely permits the constitutive expression of gag/pol genes compared with that produced transiently from HEK-293T cells (mean=291.8±58.4 SEM ng/ml, n=6) is balanced out by the fact that RD114-TR LV transduce CD34+ cells much more robustly than VSV-G LV do: 100-fold less pp are needed to get equivalent VCN. This implies that the manufacturing scale of RD2-MolPack-Chim3 supernatant could be ∼1/10 respect to the current scale of 25L/lot for the production of VSV-G pseudovirions. Of note, in contrast to VSV-G pseudovirion–transduced stem cells, HSC transduced with RD114-TR LV maintain unaltered expression after 14 days, which likely foresees a better engraftment in patients.
The rationale of integrating the core genes by the Rep78/baculo-AAV hybrid vector system was to hit the AAVS1 region on human chromosome 19, which is the specific and transcriptionally active target for Rep78-mediated AAV integration (Kotin et al., 1990, 1991, 1992; Lamartina et al., 2000). Although the vector landed, instead, into chromosome 2, we obtained results functionally equivalent to those expected for AAVS1 since the 2q32.1 locus is long-term (1 year) transcriptionally active as well. In contrast to other delivery vehicles, for example, plasmid DNA, an integrating vector is expected to persist indefinitely embedded on the host genome. The baculo-AAV vector is tail-to-head oriented, which is the natural configuration of the integrated wild-type AAV and most rAAV vector concatamers (Cheung et al., 1980; Nakai et al., 1999; Musatov et al., 2002). The majority of the vector–vector and vector–cellular genome recombination events occurs within the AAV ITR sequences during concatamerization and integration processes (Nakai et al., 1999; Musatov et al., 2002). Spontaneous deletion and mobilization of the rAAV cassette from the genome integration locus is mediated by rearrangements between the ITRs that define the boundaries of the individual AAV genome subunits (Musatov et al., 2002). Therefore, in PK-7 cells, the documented partial deletion of the 910-bp fragment, central in the concatamer, predicts an unlikely additional event of spontaneous mobilization of the baculo-AAV vector explaining the long-term genetic stability of the packaging cells.
Interestingly, PK-7 clone and RD2-MolPack-Chim3 constitutively express gag/pol and rev genes with no cellular toxicity. This is a rare and precious benefit among stable packaging cells because it allows the continuous production of VLP without needing inducers, that is, tetracycline or others, the absence of which must be documented after downstream purification processing.
Furthermore, despite the presence of several homology regions among the viral constructs (i.e., RRE, packaging signal, part of the gag gene, and the cPPT element, which is normally contained in the pol gene), the sequential strategy we adopted assures biosafety of the system by reducing the risk of recombination events.
Finally, we have ascertained that autotransduction does not interfere with the integrity, functionality, safety, and efficacy of the vectors. We have characterized numerous clones of RD2-MolPack-Chim3 carrying either high (20) or low (3) copies of the TV Chim3 without finding correlation between the number of original integrants and the amplitude of the autotransduction process: for both types of packaging cells, the fold of increment of VCN Chim3 integrants is equal to 1.6. To prevent autotransduction, the use of reverse transcriptase inhibitors such as AZT in previous studies (Vogt et al., 2001; Brandtner et al., 2008) and tenofovir in our study has been proven to be very efficacious.
In conclusion, RD2-MolPack-Chim3 is the proof of principle that the combination of the Chim3 transgene and the RD114-TR envelope, which does not transduce germline cells (Green et al., 2004), creates a feasible and valid manufacturing tool to be applied potentially in vivo (i.e., intrabone delivery) for preclinical studies in animals or clinical applications in humans for HSC-based therapies as anti-HIV gene therapy (Strang et al., 2004; Trobridge et al., 2010).
Supplementary Material
Acknowledgments
We wish to thank Alessandra Recchia (University of Modena, Italy) for helpful suggestions on AAV-Rep78-mediated integration, and Giuliana Vallanti (MolMed S.p.A.) for helpful suggestions on Q-PCR technology. This work was supported in part by Takara Bio (Otsu, Japan).
Author Disclosure Statement
A.S., B.M.P., S.B., E.Z., S.C., F.S., Cl.B., G.P.R., and Ch.B. are employees of MolMed S.p.A.; F.M. served as consultant to MolMed S.p.A.
References
- Bahner I. Sumiyoshi T. Kagoda M., et al. Lentiviral vector transduction of a dominant-negative Rev gene into human CD34+ hematopoietic progenitor cells potently inhibits human immunodeficiency virus-1 replication. Mol. Ther. 2007;15:76–85. doi: 10.1038/sj.mt.6300025. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. Gene silencing as a threat to the success of gene therapy. J. Clin. Invest. 2000;105:409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovolenta C. Porcellini S. Alberici L. Therapeutic genes for Anti-HIV/AIDS Gene Therapy. Curr. Pharm. Biotechnol. 2012 doi: 10.2174/138920101405131111104009. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Brandtner E.M. Kodajova P. Knapp E., et al. Quantification and characterization of autotransduction in retroviral vector producer cells. Hum. Gene Ther. 2008;19:97–102. doi: 10.1089/hum.2007.071. [DOI] [PubMed] [Google Scholar]
- Broussau S. Jabbour N. Lachapelle G., et al. Inducible packaging cells for large-scale production of lentiviral vectors in serum-free suspension culture. Mol. Ther. 2008;16:500–507. doi: 10.1038/sj.mt.6300383. [DOI] [PubMed] [Google Scholar]
- Burns J.C. Friedmann T. Driever W., et al. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll R. Lin J.T. Dacquel E.J., et al. A human immunodeficiency virus type 1 (HIV-1)-based retroviral vector system utilizing stable HIV-1 packaging cell lines. J. Virol. 1994;68:6047–6051. doi: 10.1128/jvi.68.9.6047-6051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.K. Hoggan M.D. Hauswirth W.W. Berns K.I. Integration of the adeno-associated virus genome into cellular DNA in latently infected human Detroit 6 cells. J. Virol. 1980;33:739–748. doi: 10.1128/jvi.33.2.739-748.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell A.S. Ma H. Fu K., et al. A trans-lentiviral packaging cell line for high-titer conditional self-inactivating HIV-1 vectors. Mol. Ther. 2006;14:276–284. doi: 10.1016/j.ymthe.2005.12.015. [DOI] [PubMed] [Google Scholar]
- Corbeau P. Kraus G. Wong-Staal F. Efficient gene transfer by a human immunodeficiency virus type 1 (HIV-1)-derived vector utilizing a stable HIV packaging cell line. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14070–14075. doi: 10.1073/pnas.93.24.14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornetta K. Yao J. Jasti A., et al. Replication-competent lentivirus analysis of clinical grade vector products. Mol. Ther. 2011;19:557–566. doi: 10.1038/mt.2010.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nunzio F. Piovani B. Cosset F.L., et al. Transduction of human hematopoietic stem cells by lentiviral vectors pseudotyped with the RD114-TR chimeric envelope glycoprotein. Hum. Gene Ther. 2007;18:811–820. doi: 10.1089/hum.2006.138. [DOI] [PubMed] [Google Scholar]
- Farson D. Witt R. McGuinness R., et al. A new-generation stable inducible packaging cell line for lentiviral vectors. Hum. Gene Ther. 2001;12:981–997. doi: 10.1089/104303401750195935. [DOI] [PubMed] [Google Scholar]
- Green B.J. Lee C.S. Rasko J.E. Biodistribution of the RD114/mammalian type D retrovirus receptor, RDR. J. Gene Med. 2004;6:249–259. doi: 10.1002/jgm.517. [DOI] [PubMed] [Google Scholar]
- Hanawa H. Persons D.A. Nienhuis A.W. Mobilization and mechanism of transcription of integrated self-inactivating lentiviral vectors. J. Virol. 2005;79:8410–8421. doi: 10.1128/JVI.79.13.8410-8421.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y. Takeuchi Y. Martin F., et al. Continuous high-titer HIV-1 vector production. Nat. Biotechnol. 2003;21:569–572. doi: 10.1038/nbt815. [DOI] [PubMed] [Google Scholar]
- Kafri T. Van Praag H. Ouyang L., et al. A packaging cell line for lentivirus vectors. J. Virol. 1999;73:576–584. doi: 10.1128/jvi.73.1.576-584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A.H. Swanstrom R. The HIV-1 gag precursor is processed via two pathways: implications for cytotoxicity. Biomed. Biochim. Acta. 1991;50:647–653. [PubMed] [Google Scholar]
- Kaul M. Yu H. Ron Y. Dougherty J.P. Regulated lentiviral packaging cell line devoid of most viral cis-acting sequences. Virology. 1998;249:167–174. doi: 10.1006/viro.1998.9327. [DOI] [PubMed] [Google Scholar]
- Kim Y.S. Wielgosz M.M. Hargrove P., et al. Transduction of human primitive repopulating hematopoietic cells with lentiviral vectors pseudotyped with various envelope proteins. Mol. Ther. 2010;18:1310–1317. doi: 10.1038/mt.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen S.G. Shimizu S. An D.S. Stem cell-based anti-HIV gene therapy. Virology. 2011;411:260–272. doi: 10.1016/j.virol.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages N. Zufferey R. Trono D. A stable system for the high-titer production of multiply attenuated lentiviral vectors. Mol. Ther. 2000;2:170–176. doi: 10.1006/mthe.2000.0103. [DOI] [PubMed] [Google Scholar]
- Kotin R.M. Siniscalco M. Samulski R.J., et al. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotin R.M. Menninger J.C. Ward D.C. Berns K.I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- Kotin R.M. Linden R.M. Berns K.I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich H.G. Specific inhibitor of human immunodeficiency virus proteinase prevents the cytotoxic effects of a single-chain proteinase dimer and restores particle formation. J. Virol. 1992;66:567–572. doi: 10.1128/jvi.66.1.567-572.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich H.G. Ochsenbauer C. Traenckner A.M., et al. Analysis of protein expression and virus-like particle formation in mammalian cell lines stably expressing HIV-1 gag and env gene products with or without active HIV proteinase. Virology. 1993;192:605–617. doi: 10.1006/viro.1993.1077. [DOI] [PubMed] [Google Scholar]
- Laakso M.M. Sutton R.E. Replicative fidelity of lentiviral vectors produced by transient transfection. Virology. 2006;348:406–417. doi: 10.1016/j.virol.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Lamartina S. Sporeno E. Fattori E. Toniatti C. Characteristics of the adeno-associated virus preintegration site in human chromosome 19: open chromatin conformation and transcription-competent environment. J. Virol. 2000;74:7671–7677. doi: 10.1128/jvi.74.16.7671-7677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruggi G. Porcellini S. Facchini G., et al. Transcriptional enhancers induce insertional gene deregulation independently from the vector type and design. Mol. Ther. 2009;17:851–856. doi: 10.1038/mt.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuyasu R.T. Zack J.A. MacPherson J.L. Symonds G.P. Phase I/II clinical trials using gene-modified adult hematopoietic stem cells for HIV: lessons learnt. Stem Cells Int. 2011;2011:393698. doi: 10.4061/2011/393698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musatov S.A. Dudus L. Parrish C.M., et al. Spontaneous mobilization of integrated recombinant adenoassociated virus in a cell culture model of virus latency. Virology. 2002;294:151–169. doi: 10.1006/viro.2001.1267. [DOI] [PubMed] [Google Scholar]
- Nakai H. Iwaki Y. Kay M.A. Couto L.B. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J. Virol. 1999;73:5438–5447. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff T. Peterson L.J. Morris J.C., et al. Efficient gene transfer to hematopoietic repopulating cells using concentrated RD114-pseudotype vectors produced by human packaging cells. Mol. Ther. 2004;9:157–159. doi: 10.1016/j.ymthe.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Ni Y. Sun S. Oparaocha I., et al. Generation of a packaging cell line for prolonged large-scale production of high-titer HIV-1-based lentiviral vector. J. Gene Med. 2005;7:818–834. doi: 10.1002/jgm.726. [DOI] [PubMed] [Google Scholar]
- Pacchia A.L. Adelson M.E. Kaul M., et al. An inducible packaging cell system for safe, efficient lentiviral vector production in the absence of HIV-1 accessory proteins. Virology. 2001;282:77–86. doi: 10.1006/viro.2000.0787. [DOI] [PubMed] [Google Scholar]
- Poeschla E. Corbeau P. Wong-Staal F. Development of HIV vectors for anti-HIV gene therapy. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11395–11399. doi: 10.1073/pnas.93.21.11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcellini S. Alberici L. Gubinelli F., et al. The F12-Vif derivative Chim3 inhibits HIV-1 replication in CD4+ T lymphocytes and CD34+-derived macrophages by blocking HIV-1 DNA integration. Blood. 2009;113:3443–3452. doi: 10.1182/blood-2008-06-158790. [DOI] [PubMed] [Google Scholar]
- Porcellini S. Gubinelli F. Alberici L., et al. Chim3 confers survival advantage to CD4+T cells upon HIV-1 infection by preventing HIV-1 DNA integration and HIV-1-induced G2 cell-cycle delay. Blood. 2010;115:4021–4029. doi: 10.1182/blood-2009-09-243030. [DOI] [PubMed] [Google Scholar]
- Recchia A. Perani L. Sartori D., et al. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 2004;10:660–670. doi: 10.1016/j.ymthe.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Samulski R.J. Chang L.S. Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrin V. Boson B. Salmon P., et al. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood. 2002;100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- Sandrin V. Muriaux D. Darlix J.L. Cosset F.L. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J. Virol. 2004;78:7153–7164. doi: 10.1128/JVI.78.13.7153-7164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snasel J. Shoeman R. Horejsi M., et al. Cleavage of vimentin by different retroviral proteases. Arch. Biochem. Biophys. 2000;377:241–245. doi: 10.1006/abbi.2000.1776. [DOI] [PubMed] [Google Scholar]
- Sparacio S. Pfeiffer T. Schaal H. Bosch V. Generation of a flexible cell line with regulatable, high-level expression of HIV Gag/Pol particles capable of packaging HIV-derived vectors. Mol. Ther. 2001;3:602–612. doi: 10.1006/mthe.2001.0296. [DOI] [PubMed] [Google Scholar]
- Srinivasakumar N. Chazal N. Helga-Maria C., et al. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J. Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang B.L. Ikeda Y. Cosset F.L., et al. Characterization of HIV-1 vectors with gammaretrovirus envelope glycoproteins produced from stable packaging cells. Gene Ther. 2004;11:591–598. doi: 10.1038/sj.gt.3302189. [DOI] [PubMed] [Google Scholar]
- Throm R.E. Ouma A.A. Zhou S., et al. Efficient construction of producer cell lines for a SIN lentiviral vector for SCID-X1 gene therapy by concatemeric array transfection. Blood. 2009;113:5104–5110. doi: 10.1182/blood-2008-11-191049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobridge G.D. Wu R.A. Hansen M., et al. Cocal-pseudotyped lentiviral vectors resist inactivation by human serum and efficiently transduce primate hematopoietic repopulating cells. Mol. Ther. 2010;18:725–733. doi: 10.1038/mt.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt B. Roscher S. Abel B., et al. Lack of superinfection interference in retroviral vector producer cells. Hum. Gene Ther. 2001;12:359–365. doi: 10.1089/10430340150503984. [DOI] [PubMed] [Google Scholar]
- Xu K. Ma H. McCown T.J., et al. Generation of a stable cell line producing high-titer self-inactivating lentiviral vectors. Mol. Ther. 2001;3:97–104. doi: 10.1006/mthe.2000.0238. [DOI] [PubMed] [Google Scholar]
- Yu H. Rabson A.B. Kaul M., et al. Inducible human immunodeficiency virus type 1 packaging cell lines. J. Virol. 1996;70:4530–4537. doi: 10.1128/jvi.70.7.4530-4537.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.