Abstract
As the gatekeepers of the eukaryotic cell nucleus, nuclear pore complexes (NPCs) mediate all molecular trafficking between the nucleoplasm and the cytoplasm. In recent years, transport-independent functions of NPC components, nucleoporins, have been identified including roles in chromatin organization and gene regulation. Here, we summarize our current view of the NPC as a dynamic hub for the integration of chromatin regulation and nuclear trafficking and discuss the functional interplay between nucleoporins and the nuclear genome.
Introduction
Nuclear pore complexes (NPCs) are large multiprotein channels embedded in the nuclear envelope (NE) that mediate the transport of macromolecules between the nucleus and the cytoplasm of eukaryotic cells [1]. Initial proteomic studies in yeast and vertebrates revealed that NPCs are composed of ~30 different nucleoporins (Nups) [2]. The recent identification of a novel transmembrane Nup in Saccharomyces cerevisiae [3•] suggests that the current list of NPC components may not be complete, and that additional Nups remain to be discovered. In general, Nups can be subdivided into at least two classes. The first class represents the stable scaffold components of the NPC, such as the Nup107/160 complex [4], which stabilize the highly curved pore membrane and are critical for nuclear pore biogenesis [5–7] and maintenance [8]. The second class includes peripheral Nups that are attached to the scaffold and often contain Phenylalanine-Glycine (FG) repeats that function directly in nucleo-cytoplasmic transport [9]. Interestingly, many nucleoporins of this group are mobile and shuttle off and on the NPC [4, 10], but it is unclear if this dynamic behavior is linked to their transport functions.
By controlling the access of transcription factors and the exit of RNA molecules, the NPC plays a critical transport-based role in gene expression [9]. In addition to the well-established roles of Nups in the transport process, it is becoming evident that the NPC components have additional functions in mitosis [11] and gene regulation, the latter being the focus of this review. Here, we will examine recent insights into our understanding of how NPC components participate in gene regulation and discuss the functional significance of these interactions for developmental processes of higher eukaryotes.
NPC components bind to genomic loci
Electron microscopy images of multiple cell types have revealed that the NE is underlined with electron-dense heterochromatin, interrupted by NPC-associated light-staining euchromatin [12]. One influential interpretation of these findings was summarized in the ‘gene gating’ hypothesis, which proposed that nuclear pores specifically interact with active genes to promote co-regulation of transcription with mRNA export [13]. Several studies performed in yeast support a potential anchoring of active genes at NPCs. For instance, a subset of inducible genes such as the GAL genes and HXK1 relocalize to the NPCs from interior sites under certain activation conditions [14, 15]. Furthermore, a genome-wide analysis in S. cerevisiae demonstrated that multiple Nups occupied regions of highly transcribed genes [17••] (Figure 1). Interestingly, conserved DNA sequences were identified that are required to target NPC-regulated genes to the nuclear periphery [16•], suggesting that NPC components play an active role in gene regulation in yeast.
Figure 1.
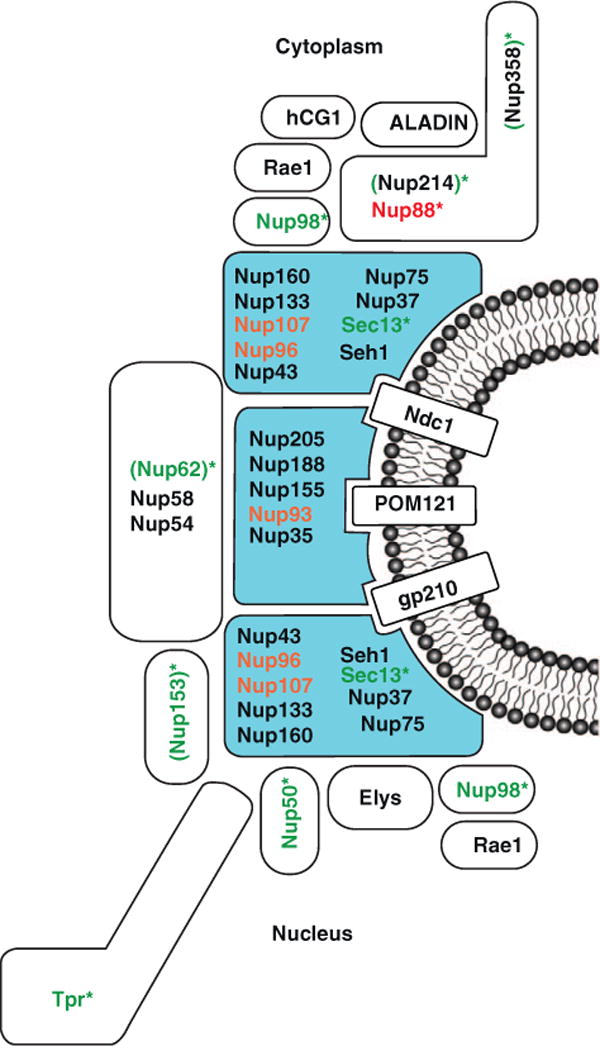
Nup-gene association. Vertebrate Nups are shown based on the functions of corresponding homologues in yeast or Drosophila. Subcomplexes of the NPC that are composed of scaffold Nups are colored in blue. Nups that show preferential association with active genes are shown in green. Nup88, the only Nup found to interact with repressed loci, is shown in red [17,18••,19••,20••]. Nups that are linked to both active and repressed genes are shown in orange [17, 47]. Nic96, the Nup93 homologue in yeast, preferentially associates with active genes. By contrast, human Nup93 targets in Hela cells are enriched for repressive histone marks. Asterisks indicate Nups with intranuclear (i.e. away from the pore) gene association. One or more of the Nups in parenthesis (Nup62, Nup153, Nup214, and Nup358), recognized by 414 antibody, interact with active genes in the nucleoplasm in Drosophila salivary glands [20••].
A link between Nups and active genes has also been discovered in multi-cellular organisms, although the situation appears to be more complicated. Analysis of chromatin binding behavior of Drosophila Nups revealed the presence of several NPC components at active genes [18••,19••,20••]. Surprisingly, the Nup-chromatin contacts were commonly found to occur in the nucleoplasm, i.e. away from the NE-embedded NPCs (Figure 1). Most of the Nups identified to participate in nucleoplasmic gene binding and regulation, including Tpr, Nup153, Nup98, Nup62 and Nup50, belong to the peripheral and dynamic NPC components [18••,19••,20••]. This is consistent with a model of dynamic Nups able to come off the nuclear pore to interact with gene loci inside the nucleus — that is, not at NE contact sites. This notion is supported by the reported transcription-dependent mobility of Nup153 and Nup98 both within the nucleus and at the nuclear pore [10].
Since the majority of active genes are located in the nuclear interior, this model could overcome a major limitation of the ‘gene gating hypothesis’, which implies that active genes have to be positioned to pores at the NE for optimal co-regulation of transcription and mRNA export. Furthermore, intranuclear gene regulation by Nups may facilitate the maintenance of the three-dimensional organization of the genome. There is emerging evidence for a regulatory role of chromatin organization in gene activities. The tethering of genomic loci to subnuclear structures such as the NPCs, the NE and various nuclear bodies may directly affect transcription and RNA processing [44]. Such interactions between Nups and genomic loci, which can change in reponse to developmental processes [19, 40], would involve global changes in chromatin organization if they occurred exclusively at the nuclear periphery. In contrast, intranuclear Nups have been observed that intranuclear Nups can achieve dynamic regulation by binding to different targets in the nuclear interior without altering the subnuclear positioning of these genes and their neighboring genetic elements (Figure 2).
Figure 2.
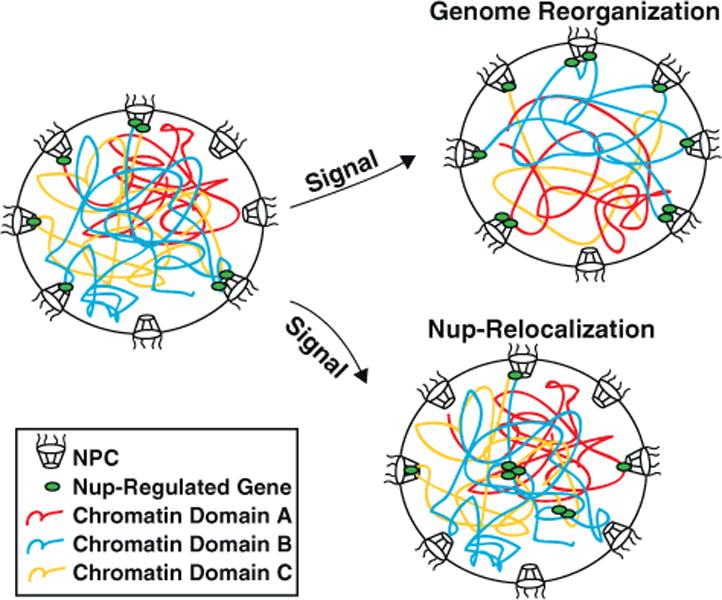
Signal-induced alterations in Nup regulation of gene activities may be achieved by genome reorganization that results in different sets of genes being positioned at the NPCs, Nup relocalization to new intranuclear loci, or both. One example of Nup relocalizing to intranuclear genes in response to signaling is shown.
As an important note, in Drosophila embryonic cells, nucleoplasmic Nup98, accounting for the majority of total detected Nup98-gene interaction, selectively interacts with highly transcribed genes [19••,20••]. By contrast, NPC-bound Nup98 targets show no enrichment for active genes [20••], which implies that the role of peripheral nuclear pores in gene regulation is more complicated than originally thought. A conclusion that should be drawn from these findings is that Nup-chromatin interactions that are identified by genome wide approaches in higher eukaryotes do not necessarily indicate peripheral association with NPCs.
Causative relationship between Nup association and transcription stimulation
The selective physical interaction of multiple Nups with active genes suggests that Nup association may directly enhance transcription. As mentioned above, NPC-localization is correlated with higher transcription activity for various yeast genes. Furthermore, fusion of multiple scaffold Nups with DNA binding domain of lexA stimulated transcription of the reporter gene, suggesting that NPC core components are capable of recruiting factors that promote transcription [25]. Moreover, Drosophila Nup153 and Megator associate with the dosage compensation complex, which stimulates transcriptional elongation, and both Nups are required for the NPC-anchoring of the male X chromosome and its gene upregulation, supporting a causative link between NPC association and gene activation [21]. It should be noted, however, that one study did not find a connection between Nups and dosage compensation, a matter that requires further clarification [22]. Additionally, there are cases in which failure of gene-NPC tethering did not cause strong transcription defects, suggesting that under some circumstances, transcription can occur without positioning at the NPC [12].
What might be the role of Nups in transcription in the nuclear interior? In salivary glands of Drosophila larvae, intranuclear Nup98 and Sec13, which are recruited to developmentally induced genes, were found to be required for transcription of their targets [19••]. Moreover, in Drosophila embryonic cells, elevating the levels of nucleoplasmic, NPC-uncoupled Nup98 increased the expression of its target genes genome-wide, whereas reducing specifically the levels of intranuclear Nup98 decreased the expression of its targets [19••,20••]. These findings suggest that intranuclear Nup98 is not only necessary but also sufficient to stimulate transcription of its targets. The mechanism of transcriptional stimulation by Nup98 and potentially other FG-Nups may involve the recruitment of the histone acetyltransferase CBP/p300 through their FG-repeats [23]. If and how Nups interact with chromatin remodeling complexes remains a subject of future research.
Transcription regulation of developmental genes by Nups
In Drosophila embryonic cells Nup98, Nup62 and Nup50 targets are enriched for genes that regulate development and cell cycle [21]. Importantly, Nup98, Sec13 and FG-Nups are recruited to developmentally induced genes in Drosophila salivary glands, and this association is required for the expression of these genes [19••]. Together, they suggest a role for Nup regulation in developmental programs in multi-cellular organisms.
The proposed role of Nups in developmental gene regulation is further supported by the finding that deletions or mutations in various Nups caused tissue-specific developmental defects or diseases [9]. The tissue-specific nature of defects with Nup mutations might seem surprising given the indispensable role of the NPC in nucleocytoplasmic transport, but could be explained by their regulation of development-specific or disease-specific genes.
Coordination of transcription and export at the NPC
In addition to transcription stimulation, gene association with NPC components may function to promote mRNA export. It has been shown that diffusion of mRNAs to NPC is not rate limiting for export and NPC localization may not enhance mRNA export efficiency by reducing gene-pore distances [24]. Instead, gene tethering at the NPC may promote mRNA export by the coordination of transcription and export. For instance, cotranscriptional recruitment of mRNA processing and packaging factors as well as NPC targeting may facilitate the formation of messenger ribonucleoprotein particles (mRNPs) with a conformation ready for transport. If mRNP formation was complete before its movement to the NPC for translocation to the cytoplasm, energy-dependent structural alterations such as unfolding and refolding of certain ribonucleoprotein domains might have to occur at the NPC for optimal interaction with transport factors and for adaptation to the shape of the NPC channel. By contrast, if mRNPs are formed while it is in contact with transport factors and NPC components, the transport-ready conformation of the particle can possibly be achieved in one step and no subsequent protein unfolding and refolding is necessary. Moreover, if transcribed genes are localized to NPCs, transcriptional feedback can be more efficiently relayed in response to mRNP malformation and export defects [25].
On chromatin, there are two complexes implicated in cotranscriptional NPC tethering and subsequent coordination of transcription and export; the SAGA and THO complexes (Figure 3). Discovered as a histone acetyltransferase complex at promoters, SAGA is linked through Sus1 to the NPC-anchored complex TREX-2 (Sac3/Thp1/Sus1/Cdc31), which is required for mRNA export [26, 27]. The interplay between NPC tethering, transcriptional activation and mRNA export is supported by a series of mutational studies. SAGA components (Ada2, Sgf73, and Sus1), TREX-2 components (Sac3 and Thp1) as well as the Sac3-associated mRNA exporter Mex67 are all required for stimuli-induced repositioning of GAL genes [28–30]. As the physical linker, Sus1 is required for both the expression of ~9% of yeast genes and GAL1 induction and mRNA export [26, 28, 29]. Moreover, Thp1 and Mex67 disruption caused transcriptional defects under some circumstances [31]. Recently, other than transcription activation at the promoter, SAGA has also been implicated in transcriptional elongation, pointing to a potential chromatin-NPC link by Sus1 at gene coding regions [32].
Figure 3.

NPCs function in gene looping, organization of chromatin domains, and coordination of transcription and export. Vertebrate Nups are shown based on the functions of corresponding homologues in yeast or Drosophila. Nup2 (vertebrate homologue Nup153) and Megator (vertebrate homologue Tpr) can protect active regions from heterochromatin spreading, and the SAGA complex may mediate the barrier activity of NPCs [39–43]. Mlp1 (vertebrate homologue Tpr) is required for the maintenance of memory gene loops and possibly sequestration of transcription factors during repression, allowing the associated gene to be reactivated faster [45]. In addition, NPCs may be cotranscriptionally tethered to genes by THO/TREX and/or SAGA-TREX-2 complexes [33••,20••]. SAGA complex is also present at coding regions apart from promoters [32], which is omitted here for simplicity. Mlp1 (vertebrate homologue Tpr) can be copurified with SAGA complex, and they associate with the same region of GAL promoters [45]. Sac3 (TREX-2 component) copurified with Nup1 (vertebrate homologue Nup153), and requires Nup1 for NPC anchoring [27]. SAGA-TREX-2 and THO/TREX complexes may physically interact [27, 46]. THO mutation resulted in a stalled mRNA intermediate associated with Nup60 (vertebrate homologue Nup153) and Nup116 (related to vertebrate Nup98), suggesting that THO coordination of mRNP formation and export may involve these two Nups [34••]. Abbreviations: 98, Nup98; 153, Nup153; polII: RNA polymerase II.
The THO complex (Tho2/Hpr1/Mft1/Thp2) is present at gene coding regions and traditionally considered to promote transcriptional elongation [31]. Sub2 helicase and Yra1, the adaptor for the mRNA exporter Mex67, associate with THO to form the THO/TREX complex, linking transcription and export [33••]. Deletion of each THO/TREX component impaired mRNA export, and Sub2and Yra1 are required for the transcription of at least some genes [33••]. Additionally, for a subset of yeast genes, THO/Sub2 was found to coordinate mRNA 3′-end processing with gene-NPC interaction [34••].
It is not known whether the promoter-and-coding-region-associated SAGA and the coding-region-associated THO complexes act on the same set of genes for NPC interaction. In yeast, the SAGA-associated TREX-2 component Sac3 co-purified with THO/TREX components [27]. Moreover, both SAGA and THO components are recruited to GAL or GAL: reporter genes upon transcription induction, suggesting coregulation of GAL genes by the two complexes [26,33••]. Interestingly, Drosophila SAGA and THO complexes also have a common component, E(y) 2, the Sus1 homologue. However, SAGA and THO mutations caused different phenotypic manifestations of the E(y)2 mutation, suggesting that E(y)2-SAGA and E(y)2-THO do not have identical targets and that subsets of genes may employ different machineries for cotranscriptional tethering to the NPCs [46].
In addition, the pattern and mechanism of gene–NPC interaction may be dynamically regulated by chromatin structure. Upon induction of histone acetylation, the percentage of genes with Nup93 association at promoters increased, while the percentage of genes with Nup93 association at coding regions decreased [47]. This observation raised the possibility that genes could gain promoter-NPC tethering mediated by SAGA complexes in response to chromatin opening.
Apart from SAGA and THO complexes, a screen of a yeast viable knockout library identified other factors essential for NPC-tethering of GAL-reporter during transcriptional induction and short periods of repression after removal of the induction signal. For example, ISW1 chromatin remodeling proteins were found to mediate NPC-association of transcribing genes and components of the RNA-degrading exosome were shown to be required for posttranscriptional NPC interaction [35••].
NPC, gene looping and organization of chromatin domains
One newly discovered functional significance of NPC-gene interaction is to maintain genes in a ‘ready-to-transcribe’ status through gene looping (Figure 3). In yeast, the gene HXK1 relocalizes to the NPC and is looped by association of its 5′-end and 3′-end when induced. Both NPC localization and the gene loop persist for some time after removal of the inducing signal, allowing HXK1 to be reactivated faster when re-induced. The NPC component Mlp1 was shown to interact with the HXK1 promoter and 3′-end, and be required for looping and rapid reactivation of HXK1 [36••].
In addition, looping of genetic elements by NPCs suggests a role in the establishment and/or maintenance of chromatin domains. Euchromatin regions might be looped out by association with NPCs, and thus insulated from the silencing environment of adjacent heterochro-matin. According to such a model, NPC components are expected to associate with boundaries of different chromatin domains, and Nup deletions should cause ectopic spreading of silencing factors into active genomic regions and/or aberrant activation of silent domains. In fact, some yeast Nups are in contact with both active genes and repressed regions such as telomeres, suggesting a positioning of NPCs in between active and silent chromatin domains [37–40]. Furthermore, a reporter gene placed within a heterochromatic environment was insulated from heterochromatin spreading when tethered to the NPC at both ends by Nup2 [39]. Consistent with a role of inhibiting heterochromatin spreading, Tpr knockdown caused failure of heterochromatin exclusion under NPCs along the NE’s nuclear surface [41]. Moreover, several SAGA components that potentially tether euchromatin–heterochromatin boundary elements to the NPCs have been found to exhibit activities that protect euchromatic regions from silencing [42,43]. In conclusion, NPCs may function to establish an active chromatin subdomain in the otherwise silent environment under the NE at euchromatin-heterochromatin boundaries (Figure 3).
Outlook
Regulation of gene expression is a crucial area of cell and developmental biology, which has the potential to transform highly diverse and important areas of biomedical research, such as differentiation, stem cell and regenerative biology, cancer mechanisms, aging processes and many epigenetic events as they relate to environmental influences, memory formation and developmental outcomes. Recent data suggest a view of the NPC as one of the central hubs at which crucial gene regulatory pathways converge [44]. Import of transcription factors, assembly of transcription machinery and distant gene loci, mRNA processing and export processes, and finally, the memory of these activating events through cell divisions may all in some ways connected to the NPC. In order to begin to understand this complex interplay, it will be necessary to dissect the molecular events and interactions that individual NPC components control. Understanding the novel role of Nups in directly regulating transcription and how it relates to transport pathways, epigenetic memory and cancer has the potential to considerably advance our knowledge on the regulation of gene expression.
Acknowledgments
We would like to thank Maya Capelson, Jesse Vargas, Brandon Toyama, Tobias Franks and Robbie Schulte for helpful suggestions. This work was supported by a grant from the American Cancer Society RSG 09-178-01-DDC.
Footnotes
Papers of particular interest, published within the period of review, have been highlighted as:
of special interest
of outstanding interest
References and recommended reading
- 1.Hetzer MW, Walther TC, Mattaj IW. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- 2.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J Cell Biol. 2010;189:795–811. doi: 10.1083/jcb.200910043. Identification of a novel trnasmembrane Nups, suggesting that more NPC components might be discovered in the future. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 5.D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 6.Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 7.Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 8.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2003;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–2230. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 13.Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci U S A. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore—promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 16•.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. Identification of evolutionary conserved DNA sequences that are required for the targeting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. Genome-wide study in yeast showing that components of the nuclear transport machinery interact with the genome. [DOI] [PubMed] [Google Scholar]
- 18••.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. The three papers [18••,19••,20••] provide first evidence that NPC components are required for gene expression in higher eukaryotes. [DOI] [PubMed] [Google Scholar]
- 21.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Grimaud C, Becker PB. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev. 2009;23:2490–2495. doi: 10.1101/gad.539509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taddei A. Active genes at the nuclear pore complex. Curr Opin Cell Biol. 2007;19:305–310. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 2005;24:813–823. doi: 10.1038/sj.emboj.7600527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 27.Fischer T, Strasser K, Racz A, Rodriguez-Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 29.Chekanova JA, Abruzzi KC, Rosbash M, Belostotsky DA. Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP. RNA. 2008;14:66–77. doi: 10.1261/rna.764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 34••.Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 35••.Vodala S, Abruzzi KC, Rosbash M. The nuclear exosome and adenylation regulate posttranscriptional tethering of yeast GAL genes to the nuclear periphery. Mol Cell. 2008;31:104–113. doi: 10.1016/j.molcel.2008.05.015. The three papers [33••,34••,35••] provide detailed insights into how transcription and mRNA export are coupled and provide molecular details about how mRNP complexes are tethered to the NPC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. This study provides compelling evidence for a role of NPC components in DNA looping. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feuerbach F, Galy V, Trelles-Sticken E, Fromont-Racine M, Jacquier A, Gilson E, Olivo-Marin JC, Scherthan H, Nehrbass U. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat Cell Biol. 2002;4:214–221. doi: 10.1038/ncb756. [DOI] [PubMed] [Google Scholar]
- 38.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 39.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 40.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–1673. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurshakova M, Maksimenko O, Golovnin A, Pulina M, Georgieva S, Georgiev P, Krasnov A. Evolutionary conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell. 2007;27:332–338. doi: 10.1016/j.molcel.2007.05.035. [DOI] [PubMed] [Google Scholar]
- 43.Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol Cell Biol. 2004;24:1956–1967. doi: 10.1128/MCB.24.5.1956-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 45.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 46.Kopytova DV, Orlova AV, Krasnov AN, Gurskiy DY, Nikolenko JV, Nabirochkina EN, Shidlovskii YV, Georgieva SG. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010;24:86–96. doi: 10.1101/gad.550010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]


