Abstract
Recent papers have provided insight into the cytoplasmic assembly of RNA polymerase II (RNA pol II) and its transport to the nucleus. However, little is known about the mechanisms governing its nuclear assembly, stability, degradation, and recycling. We demonstrate that the foot of RNA pol II is crucial for the assembly and stability of the complex, by ensuring the correct association of Rpb1 with Rpb6 and of the dimer Rpb4-Rpb7 (Rpb4/7). Mutations at the foot affect the assembly and stability of the enzyme, a defect that is offset by RPB6 overexpression, in coordination with Rpb1 degradation by an Asr1-independent mechanism. Correct assembly is a prerequisite for the proper maintenance of several transcription steps. In fact, assembly defects alter transcriptional activity and the amount of enzyme associated with the genes, affect C-terminal domain (CTD) phosphorylation, interfere with the mRNA-capping machinery, and possibly increase the amount of stalled RNA pol II. In addition, our data show that TATA-binding protein (TBP) occupancy does not correlate with RNA pol II occupancy or transcriptional activity, suggesting a functional relationship between assembly, Mediator, and preinitiation complex (PIC) stability. Finally, our data help clarify the mechanisms governing the assembly and stability of RNA pol II.
INTRODUCTION
RNA polymerase II (RNA pol II) produces all mRNAs and many noncoding RNAs but contributes less than 10% of the total RNA present in growing cells (1). It consists of 12 protein subunits with a heterodimeric subcomplex of subunits Rpb4 and Rpb7 (Rpb4/7). The catalytic core of the bacterial and eukaryotic enzymes is highly conserved through evolution. However, only five subunits have bacterial homologs (Rpb1, Rpb2, Rpb3, Rpb6, and Rpb11); the others are common to archaea but have no eubacterial homologs (2, 3). The RNA pol II transcription machinery is the most complex of those associated with the three RNA polymerases, with a total of nearly 60 polypeptides, including general transcription factors, coregulators, and specific transcription activators as well as repressors (1).
Many studies have contributed to the knowledge of physical interactions between RNA pol II and transcriptional regulators and have enabled the identification of regions that are important for transcription, from initiation to mRNA export (2, 4–12). In addition, we have recently reported the existence of five “conserved domains,” located at the surface of the structure of the complex, with poor or no conservation in their paralogs in RNA polymerases I (Rpa190 and Rpa135) and III (Rpc160 and Rpc128) and in their homologs in archaea and bacteria and demonstrate that all of them make contact with transcriptional regulators (10).
One of these regions corresponds to the foot domain (2, 10), which, in cooperation with the “lower jaw,” the “assembly” domain, and the “cleft” regions, constitutes the “shelf” module of RNA pol II, which might contribute to the rotation of the DNA as it advances toward the active center (2, 8). This domain, conserved among RNA pol II enzymes from different species, is one of the two regions in the largest subunit that differ the most in sequence among RNA polymerases I, II, and III (10, 13, 14). It should be noted that we and others have identified interactions between the foot and proteins involved in transcription initiation and/or early elongation, such as the regulators Mvp1 and Spo14 and the RNA-capping enzyme (CE) in Saccharomyces cerevisiae (13, 14). Moreover, evidence from manipulating the foot indicates that this domain contributes to specificity in the interaction of the CE with RNA pol II, as opposed to RNA polymerases I and III (14). In addition, some authors have proposed that the rpo21-4 mutation in the foot of RNA pol II of S. cerevisiae affects the assembly or the integrity of RNA pol II (15–17). However, whether this effect on the assembly of the complex is specific to this mutation or it is determined by the domain of the foot of RNA pol II remains to be elucidated.
To determine the consequences of mutating the foot of RNA pol II, we used two different S. cerevisiae mutants of this region, rpb1-84 and rpo21-4, and demonstrated that the foot of RNA pol II is essential for maintaining the stability and the assembly of the complex, in coordination with Rpb6 and the dimer Rpb4/7. In fact, both mutants affect the assembly of RNA pol II, leading to the formation of different RNA pol II intermediary subcomplexes and to a decrease in the total amount of RNA pol II, and also of Rpb1, probably as a result of Rpb1 degradation. We also demonstrate that proper assembly is crucial for maintaining correct transcriptional activity, since altering the assembly/integrity of RNA pol II affects the amount of enzyme associated to the genes, probably increases the amount of stalled RNA pol II, affects C-terminal domain (CTD) phosphorylation, and also interferes with the mRNA-capping machinery. In addition, our data show that TATA-binding protein (TBP) occupancy does not correlate with RNA pol II occupancy or transcriptional activity, suggesting a functional relationship between assembly, Mediator, and preinitiation complex (PIC) stability. Finally, our data help clarify the mechanisms governing the assembly and stability of RNA pol II.
MATERIALS AND METHODS
Yeast strains, plasmids, genetic manipulations, media, and genetic analysis.
Common yeast media, growth conditions, and genetic techniques were used as described elsewhere (13).
Strain YFN234, containing an rpb1Δ allele, strain YFN358, containing an asr1Δ allele, and strain YFN342, containing an srb10Δ allele, were obtained by chromosomal integration of a PCR product amplified using strains with these genes deleted (Euroscarf) and oligonucleotides Rpb1-507 and Rpb1-310, Asr1-501 and Asr1-301, and Srb10-501 and Srb10-301, respectively.
The rpb1::HIS3 allele of YFN191 was mutagenized into rpb1::his3::TRP1 by integrative transformation with the his3::TRP1 disrupting cassette from plasmid pHT6 (18), yielding strain YFN428.
Random mutagenesis of the conserved domain of the foot was done from plasmid pYEB220 by PCR amplification with iProof DNA polymerase (Bio-Rad), using primers Rpb1-505 and Rpb1-309, in the presence of 0.1 mM MnCl2. The PCR product was introduced into strain GR21-2d by cotransformation with a linear fragment of plasmid pYEB200 (lacking the BsmI/BspEI fragment between nucleotides [nt] 2640 and 3132) to reconstitute mutant forms of pYEB220 by recombination in vivo. The host strain used in this experiment, GR21-2d, bears the RPB1 gene of S. cerevisiae in the multicopy plasmid pFL44L (19). One viable clone that was temperature sensitive (the rpb1-84 clone) was obtained from about 500 transformants isolated on a medium lacking leucine and cured of the host plasmid by being passed onto fluoroorotic acid (FOA) medium. Strains and plasmids are listed in Tables 1 and 2.
Table 1.
S. cerevisiae strains
| Strain | Genotype | Origin or reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| D439-4d | MATα ura3-52 leu2Δ1 his3Δ200 trp1Δ63 rpb1-1 | Gift from P. Thuriaux |
| W303 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 75 |
| abd1-5 strain | MATa ade2-1 can1-100 his3-11,15 leu2-3 ura3-1 trp1-1 abd1::hisG abd1-5-TRP1 | 76 |
| abd1-8 strain | MATa ade2-1 can1-100 his3-11,15 leu2-3 ura3-1 trp1-1 abd1::hisG abd1-8-TRP1 | 76 |
| GR21-2d | MATα ura3-52 his3Δ200 leu2-3,112 trp1Δ63 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | 19 |
| LMY3.1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 trp1(YDR007w)::kanMX4 RPB4::Myc18-TRP1 | Gift from S. Chávez |
| RPB2::TAP strain | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RPB2::TAP::HIS3Mx6 | Open Biosystems |
| RPB3::TAP strain | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RPB3::TAP::HIS3Mx6 | Open Biosystems |
| SNY103 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 rpb6::LEU2 pSN266 (TRP1 CEN6 ARS1 rpb6-31) | 27 |
| Y04279 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YDR443c(srb9Δ)::kanMX4 | Euroscarf |
| Y02786 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YPL042c(srb10Δ)::kanMX4 | Euroscarf |
| Y04411 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YGL043w(dst1Δ)::kanMX4 | Euroscarf |
| Y15510 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YPR093c(asr1Δ)::kanMX4 | Euroscarf |
| Y17005 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 rpb4Δ::kanMX4 | Euroscarf |
| Y23838 | MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0/ura3Δ0 YDL140c(rpb1Δ)::kanMX4/YDL140c | Euroscarf |
| YLK25 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 3×HA TBP Mot1-Myc9-KLTRP1 | 77 |
| YSB517 | MATα his3-11,15 leu2,3-112 ura3-1 trp1-1 ceg1-250 can1-100 ade3::hisG ade2-1 | 49 |
| YVV50-4c | MATa ade2-101 ura3-52 lys2-801 trp1Δ63 his3Δ200 leu2Δ1 Rpb3-HA::KanMX4 | 78 |
| YFN116 | MATα ura3-52 his3Δ200 leu2-3,112 trp1Δ63 rpb1-Δ187::HIS3 + pYEB220 (2μm LEU2 RPB1) | This work (plasmid shuffling in GR21-2d) |
| YFN117 | MATα ura3-52 his3Δ200 leu2-3,112 trp1Δ63 rpb1-Δ187::HIS3 + pYEB220-rpo21-4 (2μm LEU2 RPB1) | This work (plasmid shuffling in GR21-2d) |
| YFN104 | MATα ura3-52 his3Δ200 leu2-3,112 trp1Δ63 rpb1-Δ187::HIS3 + pYEB220-84 (2μm LEU2 RPB1) | This work (plasmid shuffling in GR21-2d) |
| YFN167 | MATa his3Δ200 leu2Δ0 trp1Δ63 ura3Δ0 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (GR21-2d × BY4741) |
| YFN183 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 met15Δ0 rpb6Δ::kanMX4 + pFL44L-Rpb6 (2μm URA3) | From Y25602 diploid (Euroscarf) |
| YFN184 | MATα his3Δ200 leu2-3,112 met15Δ0 trp1Δ63 ura3-52 srb9Δ::kanMX4 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (GR21-2d × Y04279) |
| YFN191 | MATα his3Δ1 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 rpb4ΔkanMX4 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (YFN167 × Y17005) |
| YFN218 | MATa ade2-1 his3Δ1 leu2Δ0 trp1-1 ura3Δ0 rpb6Δ::KanMX4 + pSN266 (TRP1 CEN6 ARS1 rpb6-31) | This work (YFN183 × W303; transformed with plasmid pSN266 from strain SNY103) |
| YFN220 | MATa his3Δ1 leu2Δ0 trp1Δ63 ura3-52 RPB4::Myc18-TRP1 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (GR21-2d × LMY3.1) |
| YFN221 | MATα ade2-1 his3Δ200 leu2Δ0 trp1Δ63 ura3Δ0 rpb6Δ::KanMX4 + pSN266 (TRP1 CEN6 ARS1 rpb6-31) rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (GR21-2d × YFN218) |
| YFN234 | MATα ura3-52 his3Δ200 leu2-3,112 trp1Δ63 rpb1-Δ187::KanMX4 + pFL44-RPB1 (2μm URA3 RPB1) | This work |
| YFN236 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) ceg1-250 | This work (GR21-2d × YSB517) |
| YFN237 | MATa his3Δ1 leu2Δ0 trp1Δ63 ura3Δ0 RPB2::TAP::HIS3Mx6 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (GR21-2d × RPB2::TAP strain) |
| YFN250 | MATa ade2-1 his3Δ200 leu2-3,112 trp1Δ63 ura3-52 abd1::hisG abd1-8-TRP1 rpb1-Δ187::KanMX4 + pFL44-RPB1 (2μm URA3 RPB1) | This work (YFN234 × abd1-8 strain) |
| YFN251 | MATa his3-11,15 leu2-3,112 trp1Δ63 ura3-52 abd1::hisG abd1-5-TRP1 rpb1-Δ187::KanMX4 + pFL44-RPB1 (2μm URA3 RPB1) | This work (YFN234 × abd1-5 strain) |
| YFN271 | MATa his3Δ1 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 RPB3::TAP::HIS3Mx6 rpb1-Δ187::KanMX4 + PFL44L-RPB1 (2μm URA3 RPB1) | This work (YFN234 × RPB3::TAP strain) |
| YFN307 | MATa ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 Rpb3-HA::KanMX4 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (GR21-2d × YVV50-4c) |
| YFN339 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-52 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) 3×HATBP MOT1-Myc9-KLTRP1 | This work (GR21-2d × YLK25) |
| YFN342 | MATα ura3-52 his3Δ200 leu2-3,112 trp1Δ63 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) srb10::KanMX4 | This work |
| YFN358 | MATα ura3-52 his3Δ200 leu2-3,112 trp1Δ63 asr1Δ::kanMX4 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work |
| YFN428 | MATα his3Δ1 leu2Δ0 met15Δ0 trp1-Δ63 ura3Δ0 rpb4ΔkanMX4 rpb1-Δ187::his3::TRP1 + pFL44-RPB1 (2μm URA3 RPB1) | This work |
| YFN438 | MATa his3Δ1 leu2Δ0 ura3-52 met15Δ0 trp1Δ63 dst1Δ::KanMX4 rpb1-Δ187::HIS3 + pFL44-RPB1 (2μm URA3 RPB1) | This work (GR21-2d × Y04411) |
| Z102 | MATα ura3-52 his3Δ200 leu2-3,112 rpb2Δ297::HIS3/CEN LEU2 rpb2-6 (rpb2-R857K) | 79 |
| Z318 | MATα ura3-52 his3Δ200 leu2-3,112 rpb1Δ187::HIS3 GAL+ (AMP LEU2 CEN rpb1-19) | 79 |
Table 2.
Plasmids used
| Name | Yeast markers and ORIa | Origin |
|---|---|---|
| pFL44L | ORI (2μm) URA3 | 80 |
| pFL44-RPB1 | ORI (2μm) URA3 | 19 |
| pYEB220 | ORI (2μm) LEU2 | Gift from Pierre Thuriaux |
| pYEB220-rpo21-4 | ORI (2μm) LEU2 | This work |
| pYEB220-84 | ORI (2μm) LEU2 | This work |
| pCM185 | ORI (CEN) TRP1 | 26 |
| pCM185-RPB6 | ORI (CEN) TRP1 | This work |
| pFL44L-RPB6 | ORI (2μm) URA3 | This work |
| pGEN | ORI (2μm) TRP1 | 81 |
| pGEN-RPB7 | ORI (2μm) TRP1 | 82 |
| pCM190 | ORI (2μm) URA3 | 26 |
| pCM190-RPB4 | ORI (2μm) URA3 | This work |
| pCM190-CEG1 | ORI (2μm) URA3 | This work |
| pSL15-Sc (RPB4) | ORI (2μm) TRP1 | Gift from Pierre Thuriaux |
| pRS313-GFP-RPB4 | ORI (CEN) HIS3 | 24 |
| pHT6 | 18 | |
| pGENU-SUA7 | ORI (2μm) URA3 | Gift from Pierre Thuriaux |
CEN, centromeric vector; 2μm, multicopy vector.
Protein immunoprecipitation and TAP.
First, 100 ml of cells growing exponentially (A600, ∼0.6 to 0.8) in yeast extract-peptone-dextrose (YPD) or synthetic minimal (SD) medium was washed twice with ultrapure water and lysis buffer {50 mM HEPES [pH 7.5], 120 mM NaCl, 1 mM EDTA, 0.3% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)}. Then the cells were resuspended in 0.5 ml lysis buffer supplemented with 1× protease inhibitor cocktail (Complete; Roche), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM sodium orthovanadate, and 1 mM sodium fluoride, with 0.2 ml of glass beads (425 to 600 μm; Sigma). Whole-cell extracts were prepared by vortexing for 3 cycles of 10 min each. Immunoprecipitations were carried out as described previously (20) with some modifications: 150 μl of whole-cell extract (2,000 μg) was used, and lysis buffer was used for all washes. Also, 35 μl of Dynabeads M-280 sheep anti-mouse IgG (Invitrogen) was used with anti-Rpb1 antibody 8WG16 (1.5 μg; Covance) and anti-Rpb3 antibody 1Y26 (1.5 μg; Abcam). For tandem affinity purification (TAP), the same protocol was used with Dynabeads Pan Mouse IgG (Invitrogen). The affinity-purified proteins were released from the beads by boiling for 10 min. Eluted proteins were analyzed by Western blotting with different antibodies: anti-c-Myc (9E10; Santa Cruz Biotechnology), anti-Rpb1 (8WG16 [Covance] or y-80 [Santa Cruz Biotechnology]), PAP (Sigma), anti-POLR2C (anti-Rpb3; 1Y26; Abcam), anti-CTD-Ser5 (anti-RNA polymerase II; CTD4H8l; Millipore), and anti-Rpb6 (a gift from M. Werner).
Chromatin isolation.
Chromatin was isolated as described previously (21) with some modifications. Briefly, about 5 × 108 cells growing exponentially (A600, ∼0.6 to 0.8) were resuspended in 3 ml of 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)]-KOH (pH 9.4) containing 10 mM dithiothreitol (DTT) and 0.1% sodium azide and were then incubated at room temperature for 10 min. Cells were spun down, resuspended in 2 ml of 50 mM phosphate buffer (pH 7.5) containing 0.6 M sorbitol, 10 mM DTT, and 4 μl of 20-mg/ml Zymolyase, and incubated 10 min at 37°C in a water bath until spheroplast formation. Spheroplasts were then pelleted at 4°C, washed with 50 mM HEPES-KOH buffer (pH 7.5) containing 100 mM KCl, 2.5 mM MgCl2, and 0.4 M sorbitol, resuspended in an equal volume (∼80 μl) of EBX buffer (50 mM HEPES-KOH [pH 7.5], 100 mM KCl, 2.5 mM MgCl2, 0.25% Triton X-100, 0.5 mM PMSF, 0.5 mM DTT, and 1× protease inhibitor cocktail [Complete; Roche]), and incubated for 3 min on ice. This whole-cell extract was laid onto 400 μl of EBX-S buffer (EBX with 30% sucrose) and was spun at 12,000 rpm for 10 min. After the sucrose gradient, a chromatin pellet became visible; it was washed with 400 μl of EBX buffer and was finally resuspended in 100 μl of the same solution. A 1/10 dilution of the chromatin pellet was used for SDS-PAGE and Western blotting with antibodies against Rpb1 (8WG16; Santa Cruz), CTD-Ser5 (anti-RNA polymerase II; CTD4H8; Millipore), alpha-tubulin (T5168; Sigma-Aldrich), and Nop1 (28F2; Abcam).
Chromatin immunoprecipitation.
The following antibodies were used for chromatin immunoprecipitation (ChIP): for Rpb1, 8WG16 or y-80 (Santa Cruz) against the C-terminal or N-terminal region, respectively; for CTD-Ser5P, anti-RNA polymerase II (CTD4H8; Millipore); and for TBP, antihemagglutinin (anti-HA; 12CA5; Roche). Chromatin immunoprecipitations were performed as described previously (13). For quantitative real-time PCR (qRT-PCR), a 1:100 dilution was used for input and a 1:4 dilution was used for immunoprecipitated samples.
Genes were analyzed by qRT-PCR in triplicate with at least three independent biological replicates using SYBR Premix Ex Taq (TaKaRa).
The values found for the immunoprecipitated PCR products were compared to those of the total input, and the ratio of the value of each PCR product of a transcribed gene to the value of a nontranscribed region of chromosome V was calculated. The oligonucleotides used are listed in Table 3.
Table 3.
Oligonucleotides used
| Gene or region and primer designation | Primer sequence (5′–3′) |
|---|---|
| 18S rRNA gene | |
| 501 | CATGGCCGTTCTTAGTTGGT |
| 301 | ATTGCCTCAAACTTCCATCG |
| ACT1 | |
| 501 | GCCTTCTACGTTTCCATCCA |
| 301 | GGCCAAATCGATTCTCAAAA |
| ASR1 | |
| 501 | CTTAGTAAGTGAGAAATATGGCAAGTAAT |
| 301 | CTATATTTGAGTTTGTTCAACTGTTTTCTAT |
| CIT2 (promoter region) | |
| 502 | AAAAGGGCCCATTATTCTCG |
| 303 | CTTAATAATGAGGAACGAACACCA |
| CIT2 (ORF) | |
| 501 | AGATGGCAAAATGGGTGAAG |
| 301 | GATTAGCACGCCCATGAAGT |
| Intergenic region | |
| IntergChrV-F | TGTTCCTTTAAGAGGTGATGGTGAT |
| IntergChrV-R | TGTTCCTTTAAGAGGTGATGGTGAT |
| MFα2 | |
| 501 | CTACCGCCAGTGGGCTATT |
| 301 | CTACCGCCAGTGGGCTATT |
| PMA1 (promoter region) | |
| P-503 | AAAGGCCAAATATTGTATTATTTTCAA |
| P-302 | TTGGTGTTATAGGAAAGAAAGAGAAAA |
| PMA1 (ORF) | |
| 6(forw) | ATATTGTTACTGTCGTCCGTGTCTGGAT |
| 6(rev) | ATTAGGTTTCCTTTTCGTGTTGAGTAGA |
| PYK1 (promoter region) | |
| P-502 | ACAAGACACCAATCAAAACAAA |
| P-301 | AGTCAGAACCAGCAACAACG |
| PYK1 (ORF) | |
| 4(forw) | CTATGGCTGAAACCGCTGTCATTG |
| 4(rev) | CAGCTCTTGGGCATCTGGTAAC |
| RPB1 | |
| 507 | GATGCTGACAACGACACTGAAG |
| 310 | CGCAAGCCATGATTACTGG |
| RPL3 | |
| 501 | AAGAAGGCTCATTTGGCTGA |
| 301 | AACACCTTCGAAACCGTGAC |
| RPS2 | |
| 501 | TGGTAAGTGTGGTTCCGTCA |
| 301 | AATGGAGAAACTGGCAATGG |
| SPP2 | |
| 501 | TGCTTTACTTCGGGGTATGG |
| 301 | TGAAGCCTCCTCCACGTTTA |
| SRB10 | |
| 501 | AAGGGTATACTGAAGTTAGTAATTTTGCTTC |
| 301 | AAAAAGTTTAATGAATATAATAGTGACAGTGCT |
Extraction of mRNA and reverse transcription.
Total RNA from yeast cells was prepared as described previously (22). First-strand cDNA was synthesized using 1 μg of RNA with an iScript cDNA synthesis kit (Bio-Rad) by following the manufacturer's protocol. As a negative control for genomic DNA contamination, each sample was subjected to the same reaction without reverse transcriptase.
Quantitative real-time PCR.
Real-time PCR was performed in a CFX-96 real-time PCR instrument (Bio-Rad) with an EvaGreen detection system (SsoFast EvaGreen Supermix; Bio-Rad). Reactions were performed in 96- or 384-well plates with optical sealing tape (Bio-Rad) in a 10-μl total volume containing cDNA corresponding to 0.1 ng of total RNA. Each PCR was performed at least three times with three independent biological replicates in order to make a representative average. The 18S rRNA gene was used as a normalizer. The oligonucleotides used are listed in Table 3.
Immunolocalization and fluorescence microscopy.
Immunolocalization was performed as described previously (23) with some modifications. Cells were grown at 30°C in SD medium (A600, ∼0.5 to 0.7), fixed with 37% (wt/vol) formaldehyde at room temperature for 2 h with slow shaking, and then centrifuged and washed twice with phosphate-buffered saline (PBS). Cells were resuspended in spheroplasting buffer (1.2 M sorbitol, 0.1 M K-phosphate buffer [pH 6.5]), and the cell wall was digested with 125 μg/ml Zymolyase 20T (US Biological) and 22.7 mM 2-mercaptoethanol (Sigma) by incubation for 1 h at 37°C without shaking. The spheroplasts were washed twice with PBST (PBS with 0.05% Tween 20) and were then resuspended in the same solution. The cell suspension was added to a 3-aminopropyltriethoxysilane (AAS; Sigma) slide, incubated at room temperature until the slide was dry, and washed twice with PBST. Then 50 μl of PBS-bovine serum albumin (BSA; 1 mg/ml) was added. After incubation for 30 min in a humid chamber, slides were washed three times with PBS. Next, 50 μl of a 1:100 dilution of the primary antibody (8WG16; Santa Cruz) or 12CA5 anti-HA antibody (Roche) in PBS-BSA was added, and the slides were incubated for 2 h at room temperature in a humid chamber.
The slides were then washed three times with PBS and were incubated for 1 h in the dark at room temperature in a humid chamber with 50 μl of a 1:100 dilution of the secondary antibody (Alexa Fluor 488-conjugated goat anti-mouse IgG [H+L]). The slides were washed three times with PBS and were incubated for 5 min with 50 μl of 1-μg/ml 4′,6-diamidino-2-phenylindole (DAPI) (in PBS). After three washes with PBS, the slides were finally covered with Vectashield (Vector Laboratories) mounting solution.
For Rpb4 localization, strains were transformed with the centromeric vector expressing a Gfp-Rpb4 fusion protein (24). Cells were grown at 30°C in SD medium lacking histidine (A600, ∼0.5 to 0.7). The slides were finally covered with a Vectashield (Vector Laboratories) mounting solution containing DAPI.
Fluorescence intensity was scored with a fluorescence microscope (Olympus BX51).
Structure modeling.
Figure 1 was produced using the PyMOL program (DeLano Scientific LLC).
Fig 1.
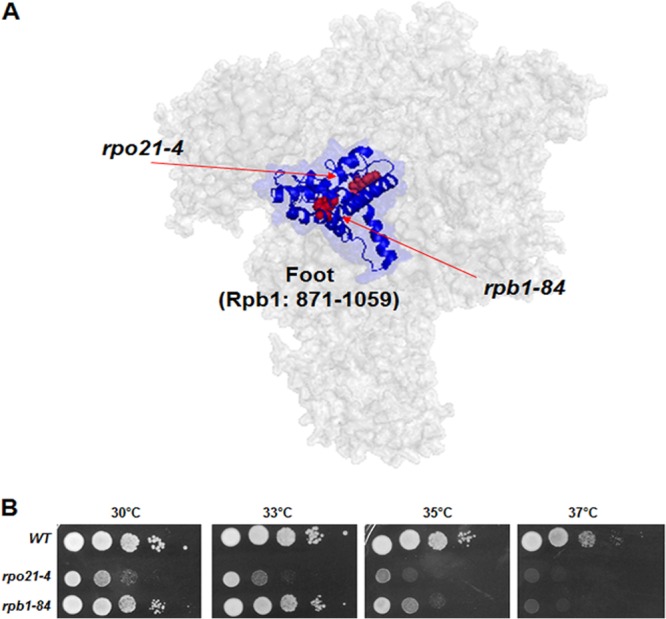
Localization and phenotypes of the rpo21-4 and rpb1-84 mutations. (A) Schematic view of the conserved region of the foot (dark blue ribbon) and the foot domain (light blue) of S. cerevisiae RNA pol II in the structure of the complex and the positions of foot mutations (red ribbons and arrows). (B) Growth of wild-type (WT) S. cerevisiae and of the rpo21-4 and rpb1-84 mutants.
RESULTS
Generation and characterization of RNA pol II foot mutants.
To decipher the role of the foot domain in transcription and to determine whether it mediates the assembly and/or stability of RNA pol II, we used two different mutants with alterations in the “conserved domain of the foot.” This domain, which was defined by us previously, contains residues 881 to 1044 of the largest subunit of S. cerevisiae RNA pol II, Rpb1 (10, 13), and corresponds to the majority of the RNA pol II “foot” (2).
The rpb1-84 mutant allele was generated by random mutagenesis (see Materials and Methods). This mutant contains three nucleotide replacements in the RPB1 gene (A2956G, T3077C, and A3096G), resulting in the amino acid substitutions I986V and L1026S. The second mutant allele, rpo21-4, has been reported previously to contain a linker-insertion (LELE) between positions W954 and P955 of Rpb1 (16). As shown in Fig. 1A, neither the I986 and L1026 residues nor the W954 and P955 residues of Rpb1 are exposed on the surface of the structure of the RNA pol II complex. Furthermore, I986 and L1026 correspond to helices α31 and α33, respectively, while W954 and P955 are in the region between α29 and α30.
The effects of these mutations were analyzed by growing the rpb1-84 and rpo21-4 mutants at different temperatures. As shown in Fig. 1B, mutant cells were temperature sensitive, although the rpo21-4 mutant grew more slowly. In agreement with this finding, pleiotropic phenotypes for the rpo21-4 mutant have been described previously (16).
Overexpression of RPB6, RPB4, and RPB7 suppresses the temperature sensitivity of RNA pol II foot mutants.
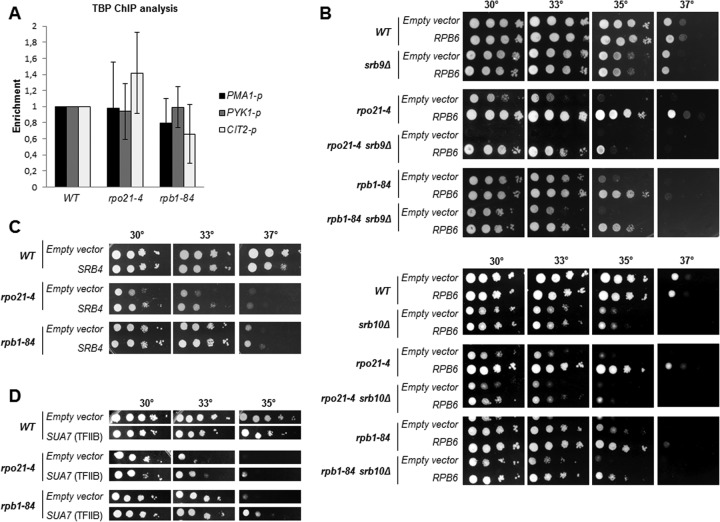
In an attempt to uncover novel genes that can suppress the temperature sensitivity of rpb1-84 and rpo21-4, we undertook a high-copy-number suppressor screening. Cells were transformed with a 2μm-based (pFL44L) multicopy genomic DNA library (25) and were selected for growth at 37°C. From about 8,000 independent transformants for each screening, plasmids containing the wild-type RPB1 gene were isolated, as expected. A second plasmid, containing the RPB6 gene, suppressed the temperature sensitivity of rpb1-84. Then we cloned the RPB6 gene into the centromeric plasmid pCM185 (26) and demonstrated that its overexpression corrects the growth defects of both the rpb1-84 and rpo21-4 mutant strains (Fig. 2A).
Fig 2.

Genetic interaction of rpo21-4 and rpb1-84 mutants with RPB6, RPB4, and RPB7. (A) Growth of cells transformed with an empty vector or with a vector overexpressing RPB6, RPB4, or RPB7. (B) Growth of single and double mutants in YPD medium (left) or in SD medium containing FOA (right).
RPB6 overexpression also suppresses the inositol auxotrophy of the rpo21-4 mutant (17, 27). In addition, it has been proposed that Rpb6 is essential for maintaining the assembly of RNA pol II and the stability of its largest subunit, Rpb1 (15, 27). RPB6 mutation lowers the steady-state level of Rpb1 in vivo, pointing to a relationship between Rpb6 and Rpb1 in maintaining the stability of RNA pol II (27). However, the mechanisms governing this disassembly and its effect on transcription are poorly understood.
We explored in more detail the possibility that RPB6 suppression in foot mutants are related to a defect in the assembly and/or integrity of RNA pol II, and we analyzed the effect of overexpressing RPB4, RPB7, or both genes together, since an rpb6 mutant lacked the dimer Rpb4/7 (28). As shown in Fig. 2A, both RPB4 and RPB7 suppressed the temperature sensitivity of mutant strains, although to different extents. It is worth noting that RPB6 is the strongest suppressor. Furthermore, suppression by the overexpression of RPB6 is specific to foot mutants, since RPB6 overexpression does not suppress the temperature sensitivity of other RNA pol II mutants, such as the rpb1-1, rpb1-19, and rpb2-6 mutants (Fig. 2A). In addition, we also analyzed the growth of both foot mutants in combination with the rpb6-31 mutation, which results in failure to assemble correctly into RNA pol II (27), or in combination with RPB4 deletion. The rpb6-31 mutation aggravates not only the growth phenotype of the rpb1-84 mutant but also that of the rpo21-4 mutant, as reported previously (27) (Fig. 2B). Similarly, the deletion of RPB4 causes synthetic lethality in combination with the rpb1-84 and rpo21-4 alleles (Fig. 2B).
These data, taken together, suggest that the effect of mutating the foot of RNA pol II on the assembly and/or integrity of the complex is not specific to the rpo21-4 allele but extends to other mutants of this domain and that assembly depends on the correct association not only of Rpb6 but also of the dimer Rpb4/7 with the complex.
The RNA pol II stability defects of foot mutants are offset by RPB6 overexpression.
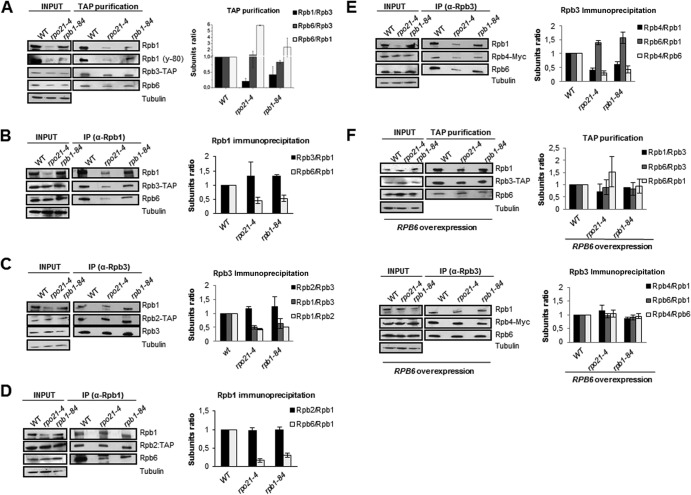
To further characterize the effect of mutations of the foot of RNA pol II on the stability of the complex, we purified RNA pol II containing tagged forms of different RNA pol II subunits from wild-type and rpb1-84 and rpo21-4 mutant strains by immunoprecipitation or TAP, and we analyzed the composition of RNA pol II and the amounts of the subunits by Western blotting. Because tagging more than one subunit altered the growth of the mutant strains, we used different strains for the different analyses. As shown in Fig. 3A, the amounts of Rpb1 in whole-cell crude extracts were lower in the mutants than in the wild-type strain, in contrast to Rpb3. In addition, the amount of Rpb1 was also lower in the rpo21-4 mutant than in the rpb1-84 mutant, a difference consistent with the differences in their growth. These data correlated with the smaller amount of Rpb1 associated with Rpb3 in foot mutants containing Rpb3-TAP (Fig. 3A). These results were corroborated by using the anti-Rpb1 antibody y-80, recognizing the amino-terminal region (amino acids 1 to 80) of Rpb1 (Fig. 3A). Furthermore, the decrease in the amount of Rpb1 did not result from a difference in the RPB1 mRNA level (not shown). In contrast, the Rpb1/Rpb3 ratios were not significantly altered in the mutants when Rpb1 immunoprecipitation was performed (Fig. 3B). The Rpb1/Rpb2 ratios were also lower for the mutants when Rpb3 immunoprecipitation was performed (Fig. 3C). In contrast, these ratios were similar for mutant and wild-type strains under Rpb1 immunoprecipitation (Fig. 3D). In addition, the amounts of Rpb2 in whole-cell crude extracts showed no significant differences (Fig. 3C and D).
Fig 3.
RNA pol II foot mutations affect the assembly of RNA pol II. (A to E) RNA pol II was purified by TAP or immunoprecipitated (IP) from the wild type or foot mutants containing Rpb2-TAP, Rpb3-TAP, or Rpb4-Myc and was analyzed by Western blotting. (F) The wild type or foot mutants containing Rpb3-TAP or Rpb4-Myc were transformed with vectors overexpressing RPB6 and were then treated as for panels A to E.
Also, we analyzed Rpb6 by using specific anti-Rpb6 antibodies. Rpb3 TAP resulted in similar Rpb6/Rpb3 ratios for the mutant and wild-type strains but a higher Rpb6/Rpb1 ratio for the mutants (Fig. 3A). Furthermore, under Rpb1 immunoprecipitation, the Rpb6/Rpb1 ratios decreased in the foot mutants (Fig. 3B and D). Similar results were found for the Rpb6/Rpb1 ratios when RNA pol II was immunoprecipitated with Rpb3-specific antibodies (Fig. 3E).
Finally, we analyzed Rpb4 from strains containing functional Myc-tagged Rpb4. Rpb3 immunoprecipitation showed that not only the ratio of Rpb4 to Rpb1 but also the ratio of Rpb4 to Rpb6 clearly decreased in the mutants from that in the wild-type strain (Fig. 3E). As with Rpb2, Rpb3, and Rpb6, the amounts of Rpb4 in whole-cell crude extracts showed no significant differences between mutant and wild-type strains.
All these data, taken together, demonstrate that mutations in the foot of RNA pol II affect the assembly and stability of RNA pol II, decreasing the amount of Rpb1 and rendering different types of subcomplexes. Our data suggest that all subcomplexes contain Rpb2 and Rpb3. In addition, most, if not all, Rpb1 subunits in the mutants are in complexes with Rpb2 and Rpb3, and the data point to the existence of Rpb2-Rpb3 subcomplexes lacking Rpb1. Furthermore, our data suggest that these mutations affect the association of Rpb6 with the rest of the complex, as well as the association of Rpb6 and the dimer Rpb4/Rpb7 with the rest of the complex, pointing to a lower yield of Rpb4 than of the rest of the RNA pol II complex. These data indicate that most, if not all, Rpb6 subunits in the mutants are in complexes with Rpb2-Rpb3 and that some complexes containing Rpb1-Rpb2-Rpb3 lack Rpb6. Finally, our data suggest that mutations in the foot of RNA pol II lead to the formation of subcomplexes containing at least Rpb1-Rpb2-Rpb3-Rpb6 but lacking Rpb4.
To ascertain whether RPB6 overexpression suffices to correct the assembly and stability defects caused by mutations in the foot of RNA pol II, we overexpressed RPB6 in some of the strains mentioned above and analyzed the composition and amount of RNA pol II as before. Notably, an increasing dosage of Rpb6 was sufficient to overcome the observed defect (Fig. 3F).
Rpb1 degradation, but not an Rpb1 assembly defect, modifies RNA pol II composition, and disassembled RNA pol II subunits shuttle to the cytoplasm.
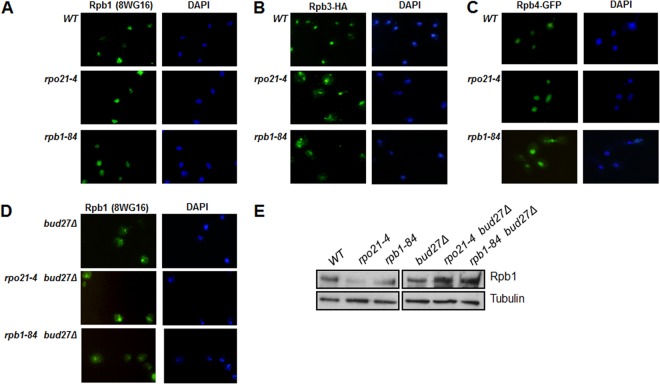
Our data show that the rpo21-4 and rpb1-84 phenotypes were associated with reduced amounts of Rpb1. Furthermore, a defect in the steady-state level of Rpb1 has been reported previously for rpo21-4 (15). To establish whether the lower accumulation of Rpb1 in mutant cells was the consequence of degradation of the subunit incorrectly assembled with the rest of the complex, we first performed immunocytochemistry experiments with the wild-type and mutant strains. As expected, fluorescence in the wild-type strain was restricted to the nucleus (Fig. 4A). Similar Rpb1 localization was observed for the rpo21-4 and rpb1-84 mutants. In contrast, Rpb3 (by use of anti-HA antibodies) clearly displayed cytoplasmic accumulation in the mutant strains (Fig. 4B). These data support the idea that Rpb3 subcomplexes lacking Rpb1 are ejected from the nucleus. Similar results were found for Rpb2 (not shown).
Fig 4.
Rpb1 degradation, but not Rpb1 assembly, modifies the composition of RNA pol II. (A and B) Immunocytochemical experiments for Rpb1 (A) and Rpb3-HA (B) in wild-type and foot mutant cells at 30°C in YPD medium. (C) Live-cell imaging of Gfp-Rpb4 in the wild type and foot mutants at 30°C in SD medium. (D) Rpb1 immunolocalization in the wild type and foot mutants with BUD27 deleted at 30°C in YPD medium. (E) Analysis of Rpb1 in whole-cell crude extracts by Western blotting with 8WG16.
In the foot mutants, Rpb4 dissociates from RNA pol II, although the total amounts of Rpb4 do not change, suggesting that Rpb4 could shuttle between the nucleus and the cytoplasm, as reported previously (24, 29). Analysis of Rpb4 localization in vivo by using a Gfp-Rpb4 construction (24) demonstrated that, at least for the rpb1-84 mutant, Rpb4 accumulates in the cytoplasm (Fig. 4C). All these data, taken together, show that nonassociated Rpb4 shuttles to the cytoplasm, although we cannot rule out the possibility that a fraction remains in the nucleus. In addition, a similar mechanism probably accounts for Rpb7.
Recent observations in yeast and humans are consistent with RNA pol assembly in the cytoplasm as a prerequisite for its nuclear import (23, 30–32). Thus, our results do not eliminate the possibility that RNA pol II foot mutations affect the correct assembly of Rpb1 with the rest of the complex and that, thus, nonassociated Rpb1 could be degraded in the cytoplasm. To help elucidate this possibility, we tested the localization and accumulation of Rpb1 upon deletion of BUD27, a prefoldin whose inactivation alters the assembly of RNA pol II, leading to its cytoplasmic accumulation (23). In immunocytochemistry experiments, clear Rpb1 cytoplasmic accumulation was found in a bud27Δ mutant strain, although nuclear staining was also detected, indicating that RNA pol II was partially mislocalized (Fig. 4D). Notably, deletion of BUD27 in rpo21-4 and rpb1-84 cells showed similar Rpb1 cytoplasmic accumulation (Fig. 4D), correlating with increased amounts of Rpb1 (Fig. 4E). These data indicate that the Rpb1 subunit is not degraded in the cytoplasm but rather that degradation may occur in the nucleus.
Nuclear Rpb1 degradation suggests an Asr1-independent mechanism.
Rpb1 ubiquitylation and proteasome-mediated degradation of RNA polymerase II stalling at a transcribed gene have been reported both in humans and in yeast (33, 34). In yeast, two major proteins have been found to be involved in these processes: Rsp5 and Asr1. Asr1, a RING finger ubiquitin-ligase, associates with the CTD in the context of chromatin, in a Ser5 phosphorylation-dependent manner (35). In addition, Asr1-mediated ubiquitylation alters the composition of RNA pol II, favoring the ejection of Rpb4/7 from the enzyme and thus leading to inactivation of the polymerase function (35).
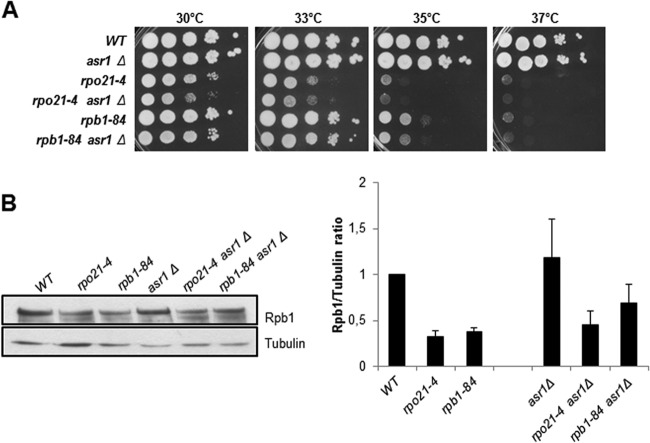
Based on the effect of Asr1 on the composition of RNA pol II, we investigated whether an Asr1-dependent mechanism would cause Rpb1 degradation in the RNA pol II foot mutants, considering the abnormal CTD Ser5 phosphorylation observed (see below). If CTD Ser5 phosphorylation were indeed the source of Asr1-mediated Rpb1 degradation, then deletion of ASR1 would at least partially restore the growth defect of the rpo21-4 and rpb1-84 mutants. However, as shown in Fig. 5A, ASR1 deletion even slightly aggravated the growth defect of foot mutants. These results agree with data from Western blotting showing that Asr1 inactivation did not restore the amount of Rpb1 in the mutants (Fig. 5B). In addition, it should be noted that Asr1 inactivation did not increase CTD Ser5 phosphorylation (data not shown). These data suggest that the degradation of Rpb1 in RNA pol II foot mutants is mediated by an Asr1-independent mechanism.
Fig 5.
Rpb1 degradation is independent of Asr1. (A) Growth of wild-type and mutant strains in YPD medium. (B) Analysis of Rpb1 amounts by Western blotting (with 8WG16) of whole-cell crude extracts at 30°C in YPD medium. Tubulin was used as an internal control.
Transcriptional defects caused by RNA pol II foot mutations correlate with decreases in the amounts of RNA pol II associated with the genes.
We and others have shown that the foot of RNA pol II associates with proteins involved in transcription initiation or early elongation (13, 14). However, the role of this region in transcription remains to be elucidated.
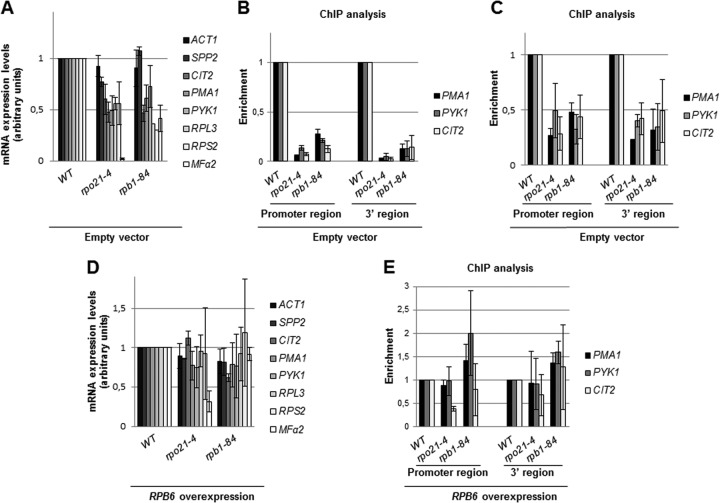
Therefore, to gain insight into the consequences of mutating the foot of RNA pol II, we used quantitative RT-PCR (qRT-PCR) to test the expression levels of different genes in the mutant and wild-type strains containing an empty vector. As shown in Fig. 6A, except for the constitutively expressed ACT1 gene and SPP2, the mRNA levels of most of the other genes analyzed fell to about 50% of the wild-type level in the rpo21-4 and rpb1-84 mutants. It is worth noting that the accumulation of MFα2 mRNA was dramatically diminished in the rpo21-4 mutant.
Fig 6.
Foot mutations affect gene expression and the amount of DNA associated with RNA pol II. (A and D) Quantitative RT-PCR analysis of mRNA levels for different genes in the wild type and foot mutants transformed with an empty vector (A) or a vector overexpressing RPB6 (D). (B, C, and E) ChIP analysis of different genes in the wild type and foot mutants transformed with an empty vector (B and C) or a vector overexpressing RPB6 (E). ChIP was performed with antibody 8WG16 (B and E) or y-80 (C). Cells were grown in SD medium.
Because RNA pol II foot mutations lead to reductions in the amount of RNA pol II, we investigated whether this could account for the decrease in the accumulation of mRNA observed. Thus, we performed ChIP on the wild type and foot mutants with antibodies 8WG16 and y-80 against the C-terminal and amino-terminal regions of Rpb1, respectively, and analyzed RNA pol II occupancy on the PMA1, PYK1, and CIT2 genes. As shown in Fig. 6B (for 8WG16) and C (for y-80), Rpb1 cross-linking decreased drastically both at promoters and at open reading frames (ORFs) in both mutants, although to different extents, suggesting that mRNA accumulation correlates with the amount of chromatin-associated RNA pol II. To test this assumption, we analyzed mRNA accumulation and RNA pol II association under conditions in which RNA pol II assembly defects are corrected, i.e., RPB6 overexpression. As shown in Fig. 6D, a higher Rpb6 dosage in both mutants restored the amount of mRNA to wild-type levels for most of the genes tested. Again, MFα2 mRNA accumulated differently in the rpo21-4 mutant than in the rpb1-84 mutant, although its level was also augmented more than 13-fold. In agreement with this, the cross-linking of Rpb1 to the promoters and ORFs of the genes tested (by using antibody 8WG16) revealed no significant differences from the wild-type strain (Fig. 6E).
Taken together, these data support the view that mutations in the foot of RNA pol II alter the assembly of the complex, lowering the amount of the enzyme associated with the genes and thus causing a transcription defect. However, this effect is not a general consequence of mutating the foot of RNA pol II; thus, neither the inducible expression of the GAL1 gene (expressed as the fold change [not shown]) nor the constitutive expression of the ACT1 gene (see above) was significantly altered.
TBP occupancy does not correlate with RNA pol II occupancy and transcriptional activity.
It has been established previously that TBP and RNA pol II recruitment to and occupancy at the promoters correlate strongly with transcriptional output and that this association occurs in concert (36, 37). To test whether the decrease in RNA pol II occupancy at the promoter region correlated with a loss of TBP occupancy, we performed chromatin immunoprecipitation assays for TBP and RNA pol II occupancy in wild-type, rpo21-4, and rpb1-84 cells containing HA-tagged TBP. Surprisingly, TBP occupancy at the genes tested in mutant strains is not significantly different from that in the wild-type strain (Fig. 7A). In agreement with this finding, overexpression of the SPT15 gene, coding for TBP, had no effect on the growth of mutant strains (not shown). Deletion of SRB10 (the gene coding for Mediator subunit Srb10) (38), as well as mutations in TBP (39), has a greater impact on promoter occupancy by Pol II than on promoter occupancy by TBP. In addition, the Mediator head module interacts with the RNA pol II subunits Rpb4/7 and with the clamp, triggering structural rearrangements of the enzyme that affect promoter engagement and transcriptional activity (40). Thus, we tested for conditional synthetic interactions between rpo21-4 or rpb1-84 and mutations of the Mediator machinery. As shown in Fig. 7B, the deletion of SRB9 or SRB10 aggravates the growth defect of the rpo21-4 and rpb1-84 mutants. Notably, RPB6 overexpression partially overcomes the slow-growth phenotype of double mutants (Fig. 7B). In accordance, SRB4 overexpression (Fig. 7C), but not SRB10 overexpression (not shown), partly suppresses the growth defect of foot mutants. To help clarify the relationship between TBP, RNA pol II, and Mediator, we analyzed the effect of overexpressing SUA7 (coding for TFIIB) in foot mutants, since Mediator plays a critical role in preinitiation complex (PIC) assembly, and Mediator and TFIID stabilize each other in the PIC and stimulate TFIIB assembly (41). Notably, as shown in Fig. 7D, SUA7 overexpression partially reverses the slow-growth phenotype of the rpo21-4 and rpb1-84 mutants.
Fig 7.
TBP occupancy does not correlate with RNA pol II occupancy and transcriptional activity. (A) TBP occupancy at the promoters of different genes in the wild-type strain and foot mutants transformed with an empty vector. (B) Growth of wild-type and mutant strains, transformed with an empty vector or a vector overexpressing RBP6, in SD medium. (C) SRB4 overexpression in SD medium. (D) SUA7 overexpression in SD medium.
All these data, taken together, suggest that impairing the correct assembly of RNA pol II affects the concerted association of TBP and RNA pol II to regulate transcription, probably altering PIC stabilization.
Abnormal CTD Ser5 phosphorylation may indicate an increase in the amount of stalled RNA pol II.
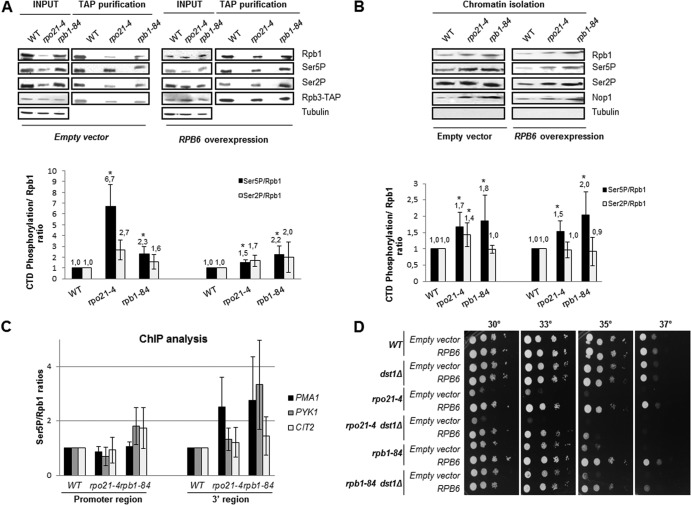
We have shown previously that defects in transcription initiation or early elongation associated with the deletion of proteins interacting with the foot of RNA pol II are accompanied by increased CTD Ser5 phosphorylation (13). Thus, we investigated whether mutations in the foot of RNA pol II would also lead to this phenotype. We used TAP to purify RNA pol II containing a functional TAP-tagged form of Rpb3 from wild-type, rpo21-4, and rpb1-84 cells, and we analyzed the amount of Rpb1 and the level of CTD Ser5 phosphorylation (with antibody CTD4H8). Notably, the relative CTD Ser5P/Rpb1 ratios were higher in the mutants than in the wild-type strain (Fig. 8A). To rule out the possibility that a decrease in the amount of the free hypophosphorylated pool of RNA pol II led to these increases in the mutant strains, we also analyzed the CTD Ser5P/Rpb1 ratios in chromatin fractions. As shown in Fig. 8B, the amounts of CTD Ser5P were also higher in DNA-associated RNA pol II in both mutants, although these ratios were lower than those in the purified enzymes. In contrast, CTD Ser2 phosphorylation analysis showed no significant differences between wild-type and mutant strains (Fig. 8A and B).
Fig 8.
Foot mutations may lead to an increase in the amount of stalled RNA pol II. (A) (Top) Rpb1 (8WG16) and Rpb1-CTD Ser5P (CTD4H8) were analyzed by Western blotting of RNA pol II purified by TAP from cells transformed with an empty vector or overexpressing RPB6 at 30°C in SD medium. Tubulin was used as an internal control. (Bottom) CTD Ser5 phosphorylation/Rpb1 ratios calculated from the Western blot data. *, P < 0.05. (B) (Top) Rpb1 and Rpb1-CTD Ser5P were analyzed by Western blotting of chromatin fractions of cells transformed with an empty vector or overexpressing RPB6 at 30°C in SD medium. Tubulin and Nop1 were used as internal controls for whole-cell crude extracts and chromatin, respectively. (Bottom) CTD Ser5 phosphorylation/Rpb1 ratios calculated from the Western blot data. *, P < 0.05. (C) Analysis by ChIP of Rpb1 (y-80) and Rpb1-CTD Ser5P (CTD4H8) in cells transformed with an empty vector or overexpressing RPB6 at 30°C in SD medium. (D) Analysis of the interaction of foot mutants, transformed with an empty vector or with a vector overexpressing RBP6, with the dst1Δ mutation in SD medium.
These data indicate that the mutation of the foot of RNA pol II led to abnormal phosphorylation, at least of CTD Ser5P. To explore whether correct assembly of RNA pol II is important for the modulation of CTD phosphorylation, we overexpressed RPB6 in the rpo21-4 and rpb1-84 mutants. TAP of RNA pol II from wild-type, rpo21-4, and rpb1-84 cells (Rpb3-TAP) overexpressing RPB6 was performed, and the amounts of Rpb1 and CTD Ser5 phosphorylation were analyzed by Western blotting. As shown in Fig. 8A, the relative CTD Ser5P/Rpb1 ratio for the rpo21-4 mutant decreased significantly from that for the mutant containing the control empty vector, although it did not decline to the level of the wild-type strain overexpressing RPB6. In contrast to the result for the rpo21-4 mutant, the CTD Ser5P/Rpb1 ratio for the rpb1-84 mutant did not decrease, maintaining a level similar to that with the empty vector (Fig. 8A). Similar experiments, performed using chromatin fractions, demonstrated similar behavior (compare Fig. 8A and B). Again, CTD Ser2 phosphorylation showed no significant differences (Fig. 8A and B). We also explored the relative CTD Ser5P/Rpb1 ratios for different genes by performing ChIP analysis and compared the levels of CTD Ser5P and Rpb1 occupancy. As shown in Fig. 8C, with antibody y-80, the CTD Ser5P/Rpb1 ratios were higher in the rpb1-84 and rpo21-4 mutants than in the wild-type strain at the ORFs of the PMA1, PYK1, and CIT2 genes (similar but increased ratios were obtained with antibody 8WG16 [not shown]). Interestingly, this finding is especially true toward the 3′ ends of these coding regions, where CTD Ser5P levels are normally very low. In addition, these data agree with the synthetic lethality caused by deleting the RTR1 gene (coding for the recently described Ser5 CTD phosphatase Rtr1 [42]) or by tagging Ssu72 (the classical Ser5 CTD phosphatase [43]) in foot mutants (not shown).
These data point to the importance of the correct assembly of RNA pol II for maintaining correct CTD phosphorylation. However, the foot domain is also important for modulating CTD phosphorylation, since the synthetic lethality caused by deleting the RTR1 gene in foot mutants is not overcome by RPB6 overexpression (not shown).
All these data, taken together, suggest an increase in the amount of stalled RNA pol II, probably as a consequence of a defect in the transition from initiation to elongation. In accordance with this idea, a strong genetic interaction between foot mutations and the deletion of DST1 was observed (Fig. 8D). DST1 codes for the TFIIS transcription factor, which stimulates RNA cleavage by RNA polymerase II by allowing backtracked enzymes to be released from stall sites and to resume transcription elongation (44, 45). Again, RPB6 overexpression suppressed the growth phenotype of the double mutants, suggesting that incorrect RNA pol II assembly affects the amount of stalled enzyme at the promoter-proximal region. Similarly, deleting the CCR4 and NOT5 genes, components of the Ccr4-Not complex that stimulate the resumption of RNA pol II elongation from an arrested state (46), aggravates the growth phenotype of foot mutants (not shown).
RNA pol II foot mutants synthetically interact with components of the mRNA-capping machinery.
In S. cerevisiae, Ser5 phosphorylation occurs first in the CTD in coordination with the recruitment of Ceg1 (47), which binds the foot of RNA pol II in S. cerevisiae (14). In agreement, we demonstrated that the foot is in contact with proteins that participate in transcription initiation and/or early elongation (13) and that interact genetically with CEG1. Furthermore, Abd1, which methylates the cap to form m7GpppRNA, binds directly to the phosphorylated CTD, and its inactivation causes Ser5 hyperphosphorylation (48).
In an attempt to clarify the functional relationship between the assembly of RNA pol II, the foot domain, and mRNA capping, we first tested for conditional synthetic interactions between foot mutants and the ceg1-250 mutant (49), and we showed a synthetic growth defect (Fig. 9A). Similarly, the abd1-5 and abd1-8 mutations (50) aggravated the growth phenotype of the rpo21-4 and rpb1-84 mutants (Fig. 9B), but to a greater extent than the ceg1-250 mutant. Notably, overexpression of RPB6 partially suppressed the slow-growth phenotype of the double mutants (Fig. 9C).
Fig 9.

Genetic interactions between foot mutants and elements of the mRNA-capping machinery. (A and B) Growth of the wild-type strain and foot mutants in combination with the ceg1-250 (A), abd1-5, or abd1-8 (B) mutation in YPD medium. (C) Growth of wild-type and mutant strains, transformed with an empty vector or overexpressing RPB6, in SD medium. (D) Growth of the wild-type strain and foot mutants, transformed with an empty vector or overexpressing CEG1, at 30°C in SD medium.
Suh et al. have reported previously that mutating the foot domain significantly reduced the amount of CE associated with RNA pol II in S. cerevisiae (14). Thus, if the impairment of the Ceg1 interaction with RNA pol II is responsible for the slow-growth phenotypes of the rpo21-4 and rpb1-84 mutants, overexpression of CEG1 might compensate for these phenotypes. However, CEG1 overexpression did not restore the growth of foot mutants (Fig. 9D) but even stunted it slightly, suggesting that the rpo21-4 and rpb1-84 foot mutations did not alter Ceg1 recruitment in vivo.
These data, taken together, suggest that correct RNA pol II assembly is important for mRNA capping.
DISCUSSION
The foot of RNA pol II is a conserved domain among RNA pol II enzymes (10, 13, 14), which, in cooperation with other regions, constitutes the “shelf” module of the enzyme. This module might contribute to the rotation of the DNA as it advances toward the active center (2, 8). In this work, we demonstrate that the foot of RNA pol II is essential for maintaining the integrity or stability of the complex, in coordination with Rpb6 and also with the dimer Rpb4/7. We also demonstrate that proper assembly is crucial to maintain correct transcriptional activity. In fact, altering the assembly/integrity of RNA pol II affects the amount of enzyme associated with the genes, suggesting an increase in the amount of stalled RNA pol II, and affects CTD phosphorylation, leading to Rpb1 degradation by an Asr1-independent mechanism, but also interferes with mRNA-capping machinery. Our data also show a lack of correlation between TBP occupancy and RNA pol II occupancy and suggest a functional relationship between assembly, Mediator, and PIC stability. Finally, our data also provide new information concerning the mechanisms governing the nuclear disassembly of RNA pol II.
The foot domain of RNA pol II, the association between Rpb1 and Rpb6, and the correct association of the dimer Rpb4/7 are important for maintaining the assembly of the complex.
Our data demonstrate that the foot of RNA pol II is important for maintaining the correct assembly of the complex. The association between Rpb1 and Rpb6 must be a prerequisite for maintaining the correct assembly of RNA pol II, as well as for ensuring the correct association of the dimer Rpb4/7. In fact, RPB6 overexpression suppresses the growth phenotype of the rpo21-4 and rpb1-84 mutants, increases the amount of Rpb1 to wild-type levels, and rescues the RNA pol II assembly defect. In agreement with this, tagging of Rpb6 at its C-terminal end is lethal in combination with the rpo21-4 mutation (not shown). It should be noted that the importance of Rpb1-Rpb6 association for the assembly of RNA pol II has been proposed previously (15–17, 27, 28). Furthermore, the foot of RNA pol II makes physical contact with Rpb6 in the structure of the enzyme (2). In addition, the importance of the Rpb6 bacterial homolog ω subunit in RNA pol assembly has been proposed (51).
Our data are also consistent with the connection of Rpb6 and the dimer Rpb4/7 with the base of the mobile clamp of RNA pol II (2, 5, 52). In addition, Rpb6 and Rpb4/7 also make contact (5, 52, 53), and some effects described for some RPB6 mutants have been suggested to be caused by the unstable Rpb4/7 dimer (28). In agreement with this, the Rpb4/7 stalk can dissociate from the pol II core in solution (54), and the pol III subcomplex Rpc25/c17 can dissociate from the core during native mass spectrometry (55). Furthermore, Rpb4 is involved in enzyme activity and stability but not in enzyme assembly (56–58). Hence, we cannot rule out the possibility that Rpb4/7 dissociation partly accounts for the smaller amount of Rpb1, the decreased activity of RNA pol II, and the reduction in the amount of RNA pol II associated with DNA. In fact, RNA pol II lacking Rpb4/7 interacts less stably with DNA (35, 59).
RNA pol II foot mutations cause enzyme dissociation and the accumulation of intermediary assembly subcomplexes.
Assembly of the RNA polymerases in both humans and yeast is proposed to occur in the cytoplasm as a prerequisite for their nuclear import with the action of several factors (23, 30, 32, 60–62). However, little is known about the disassembly, degradation, and recycling of the RNA polymerases.
Mutation of the foot of RNA pol II affects assembly, leading to the accumulation of intermediary subcomplexes at the permissive temperature, in contrast to other mutations affecting assembly (27, 63–65). The accumulation of these intermediaries in foot mutants results from the dissociation of the enzyme, accompanied by a significant reduction in the steady-state level of Rpb1. All the subcomplexes contain the Rpb2-Rpb3 dimer, in agreement with experiments that identified Rpb1/2/3, Rpb2/3, or Rpb2/3/10/11/12 subcomplexes (31, 62, 63, 66, 67). Notably, subcomplexes containing Rpb1 include those lacking Rpb4/7, as well as those lacking Rpb6-Rpb4/7. However, in contrast to Rpb6, Rpb4 seems not to be determinant for RNA pol II assembly (56). The association of Rpb6 with Rpb2/3 suggests the action of an assembly factor, such as the prefoldin Bud27, which participates in the assembly of Rpb5 and Rpb6 to the RNA polymerases (23).
In contrast to Rpb1, which is found only in the nucleus, Rpb3 and Rpb2 accumulate in the cytoplasm in foot mutants. Similar results have been found in mammalian cells treated with α-amanitin, in which Crm1-mediated export is blocked (31). Rpb4 (and probably Rpb7) dissociates from RNA pol II and translocates to the cytoplasm. However, the mechanism involved in transport once is disassembled is unknown. In addition, our data on immunolocalization tempt us to speculate that a fraction of Rpb4 (and probably Rpb7) remains nuclear upon dissociation.
Rpb1 degradation is nuclear.
We suggest that Rpb1 is degraded in the nucleus and propose that its assembly with the rest of RNA pol II does not depend on its instability. This assumption is based on the fact that altering the cytoplasmic assembly of RNA pol II by deleting BUD27 (23) causes cytoplasmic Rpb1 accumulation, which reaches wild-type levels in mutant strains. Consistent with this fact, in α-amanitin- and leptomycin B-treated cells, Rpb1 accumulated in the cytoplasm and associated with components of the R2TP complex (31). Notwithstanding, other disassembled subunits (Rpb2, Rpb3, Rpb4, and Rpb6) remain unaltered, suggesting that they are not degraded.
Rpb1 ubiquitylation and proteasome-mediated degradation have been reported in both humans and yeast (33, 34). Rpb1 degradation in both foot mutants is independent of Asr1, a RING finger ubiquitin-ligase recognizing the CTD Ser5 phosphorylated complex in the context of chromatin (35), although mutation of the foot of RNA pol II is accompanied by greater CTD Ser5 phosphorylation. Furthermore, we cannot rule out the possibility that Rsp5, an E3 ubiquitin-protein ligase, mediates this degradation (68), although some authors have proposed that Rsp5 cannot associate with pol II when Ser5 is phosphorylated (34). Considering these proposals together, we suggest that additional mechanisms may be involved in the degradation of Rpb1, as has recently been shown in human cells (69).
Correct assembly of RNA pol II is a prerequisite for proper maintenance of multiple steps of transcription.
The defect in the assembly/integrity of RNA pol II, which is accompanied by a drastic reduction in the level of Rpb1, correlates with a decrease in RNA pol II activity and with the reduction in the amount of RNA pol II associated with DNA. Notably, however, TBP occupancy does not correlate with the amount of RNA pol II in the promoter-proximal regions. These data are highly important, since it has been established that recruitment of TBP and RNA pol II to the promoters and their promoter occupancy correlate strongly with transcriptional output and that TBP and RNA pol II association occurs in concert (36, 37). Our data also agree with the results of deletion of the Mediator subunit Srb10 (38) and point to a functional relationship between correct assembly, PIC formation, and Mediator association. In accordance with a role for Mediator in PIC stabilization and TFIIB assembly (41), overexpression of SUA7 slightly recovers the slow-growth phenotype of foot mutants. We could also speculate that assembly defects influence the association between RNA pol II and Mediator, in accordance with the interaction of the Mediator head module with RNA pol II subunits Rpb4/7 and the clamp (40) and in accordance with the slight suppression occurring with Srb4 but not Srb10 overexpression (not shown). Furthermore, the decrease in RNA pol II occupancy is not observed for stress-related genes, probably reflecting a defect in PIC formation (A. I. Garrido-Godino et al., unpublished data). However, we cannot rule out the possibility that this reflects normal PIC formation with structurally altered RNA pol II.
Our data suggest an increase in the amount of stalled RNA pol II, probably as a consequence of a defect in the transition from initiation to elongation, in accordance with the increase in CTD Ser5 phosphorylation, with the fact that deleting RTR1 or tagging Ssu72 is lethal in combination with foot mutants, with the strong genetic interaction with DST1, CCR4, and NOT5 (44, 45), and with our previous data for mutants of proteins associated with the foot (13). In addition, the uninduced heat shock in Drosophila indicates the existence of a paused elongation complex phosphorylated at Ser5 (70).
Notably, the increase in CTD Ser5 phosphorylation occurs along the gene and is especially true toward the 3′ ends of these coding regions, where CTD Ser5P levels are normally very low (71). We cannot rule out the possibility that the increase in CTD Ser5 phophorylation accounts for a defect in transcription termination, inhibiting reinitiation, impeding subsequent assembly into the preinitiation complex, and then leading to decreased levels of RNA pol II in the ORF (43). In fact, Rtr1 and Ssu72 have been proposed to participate in transcription termination, and foot mutant phenotypes mimic those of the rtr1Δ mutant (42, 43).
These data also point to the importance of the correct assembly of RNA pol II in maintaining correct CTD phosphorylation and are in line with results showing contact between Rpb4, the CTD, and the CTD phosphatase Fcp1 (72). Notably, the genetic interactions with ceg1 and abd1 mutants (48, 49) also point to the correlation between correct RNA pol II assembly and mRNA capping. In agreement, the foot of RNA pol II contacts the mRNA-capping enzyme (CE) in S. cerevisiae (14), which is recruited to the CTD in coordination with Ser5 phosphorylation (47). Furthermore, Ceg1 is stabilized by its interaction with the CTD (73). It is worth noting that Abd1 inactivation causes a defect in promoter clearance and/or early elongation, accompanied by a failure to dephosphorylate Ser5 residues normally (48).
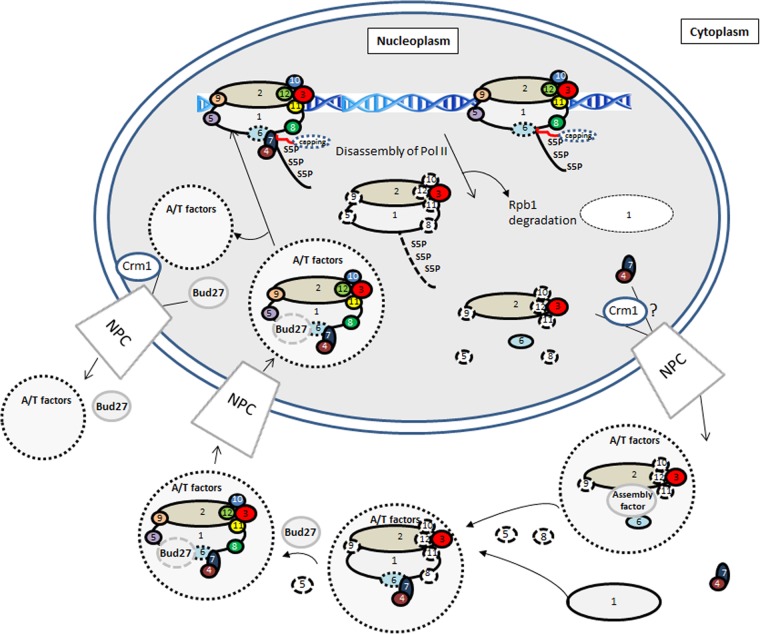
Finally, we propose the model shown in Fig. 10. RNA pol II foot mutations affect Rpb1-Rpb6 association, leading to the dissociation of Rpb4/7, which is translocated to the cytoplasm, although we cannot rule out the possibility that a fraction remains nuclear. A stalled enzyme and/or an enzyme engaged in the PIC could disassemble, leading to the formation of different intermediary subcomplexes and to Rpb1 degradation by an Asr1-independent mechanism. Disassembled subcomplexes move to the cytoplasm by an unknown mechanism that could involve Crm1. Rpb2/Rpb3 remain associated, and Rpb6 may associate with this intermediary, pointing to the action of an assembly factor, which could be the prefoldin Bud27. Reassociation of different subunits in the cytoplasm with newly synthesized Rpb1 occurs sequentially, involving different assembly and transport factors (30, 32, 60–62); it is suggested that Bud27 acts in a final step for correct assembly of Rpb5 (23). Whole RNA pol II associated with assembly and transport factors enters the nucleus, and these shuttle it again to the cytoplasm, some of them by a Crm1-dependent mechanism, Bud27 by a Crm1-independent mechanism (23). Another possibility to be considered is that the incompletely assembled enzyme is never targeted to the nucleus and just remains in the cytoplasm, allowing nonassociated Rpb4 to shuttle between the nucleus and the cytoplasm. Finally, we cannot rule out the possibility that a phosphorylated RNA pol II subcomplex could be free in the nucleus, as has been suggested previously (74).
Fig 10.
Model for the RNA pol II assembly/disassembly pathway and Rpb1 degradation. The model is based on the previous proposal by Wild and Cramer (62). S5P, phosphorylated Ser5; A/T factors, assembly and transport factors; NPC, nuclear pore complex; numbers in circles, Rpb subunits; circles with dotted borders, assembly and transport factors associated to the RNA pol II.
ACKNOWLEDGMENTS
We thank M. Werner, F. Estruch, M. Choder, S. Chávez, D. Bentley, S. Buratowski, and R. Parker for providing antibodies. strains. and constructions. We thank the Centro de Intrumentación Científico-Técnico, CICT, Universidad de Jaén for technical support. We also thank F. Estruch and Olga Calvo for critical reading of the manuscript.
This work was supported by grants from the Spanish Ministry of Science and Innovation and FEDER (BFU2010-21975-C03-02 Spain) and from the Junta de Andalucía (BIO258, PI10-CVI6521, and P08-CVI-03508) to F.N. A.I.G.-G. is a recipient of predoctoral fellowships from MEC.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Hahn S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11:394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramer P, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863–1876 [DOI] [PubMed] [Google Scholar]
- 3.Kwapisz M, Beckouet F, Thuriaux P. 2008. Early evolution of eukaryotic DNA-dependent RNA polymerases. Trends Genet. 24:211–215 [DOI] [PubMed] [Google Scholar]
- 4.Bushnell DA, Cramer P, Kornberg RD. 2002. Structural basis of transcription: α-amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. U. S. A. 99:1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armache KJ, Mitterweger S, Meinhart A, Cramer P. 2005. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 280:7131–7134 [DOI] [PubMed] [Google Scholar]
- 6.Meyer PA, Ye P, Suh MH, Zhang M, Fu J. 2009. Structure of the 12-subunit RNA polymerase II refined with the aid of anomalous diffraction data. J. Biol. Chem. 284:12933–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cramer P. 2006. Mechanistic studies of the mRNA transcription cycle. Biochem. Soc. Symp. 73:41–47 [DOI] [PubMed] [Google Scholar]
- 8.Zaros C, Briand JF, Boulard Y, Labarre-Mariotte S, Garcia-Lopez MC, Thuriaux P, Navarro F. 2007. Functional organization of the Rpb5 subunit shared by the three yeast RNA polymerases. Nucleic Acids Res. 35:634–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venters BJ, Pugh BF. 2009. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 44:117–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-López MC, Navarro F. 2011. RNA polymerase II conserved protein domains as platforms for protein-protein interactions. Transcription 2:193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. 2011. Direct interaction of RNA polymerase II and Mediator required for transcription in vivo. Science 331:1451–1454 [DOI] [PubMed] [Google Scholar]
- 12.Suh MH, Ye P, Zhang M, Hausmann S, Shuman S, Gnatt AL, Fu J. 2005. Fcp1 directly recognizes the C-terminal domain (CTD) and interacts with a site on RNA polymerase II distinct from the CTD. Proc. Natl. Acad. Sci. U. S. A. 102:17314–17319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-López MC, Pelechano V, Miron-Garcia MC, Garrido-Godino AI, Garcia A, Calvo O, Werner M, Perez-Ortin JE, Navarro F. 2011. The conserved foot domain of RNA pol II associates with proteins involved in transcriptional initiation and/or early elongation. Genetics 189:1235–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suh MH, Meyer PA, Gu M, Ye P, Zhang M, Kaplan CD, Lima CD, Fu J. 2010. A dual interface determines the recognition of RNA polymerase II by RNA capping enzyme. J. Biol. Chem. 285:34027–34038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nouraini S, Xu D, Nelson S, Lee M, Friesen JD. 1997. Genetic evidence for selective degradation of RNA polymerase subunits by the 20S proteasome in Saccharomyces cerevisiae. Nucleic Acids Res. 25:3570–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Archambault J, Drebot MA, Stone JC, Friesen JD. 1992. Isolation and phenotypic analysis of conditional-lethal, linker-insertion mutations in the gene encoding the largest subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Gen. Genet. 232:408–414 [DOI] [PubMed] [Google Scholar]
- 17.Archambault J, Schappert KT, Friesen JD. 1990. A suppressor of an RNA polymerase II mutation of Saccharomyces cerevisiae encodes a subunit common to RNA polymerases I, II, and III. Mol. Cell. Biol. 10:6123–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross FR. 1997. ‘Marker swap' plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647–653 [DOI] [PubMed] [Google Scholar]
- 19.García-López MC, Miron-Garcia MC, Garrido-Godino AI, Mingorance C, Navarro F. 2010. Overexpression of SNG1 causes 6-azauracil resistance in Saccharomyces cerevisiae. Curr. Genet. 56:251–263 [DOI] [PubMed] [Google Scholar]
- 20.Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, Werner M. 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol. Cell. Biol. 26:4920–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang C, Stillman B. 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11:3375–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nonet M, Scafe C, Sexton J, Young R. 1987. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7:1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirón-García MC, Garrido-Godino AI, Garcia-Molinero V, Hernandez-Torres F, Rodriguez-Navarro S, Navarro F. 2013. The prefoldin bud27 mediates the assembly of the eukaryotic RNA polymerases in an rpb5-dependent manner. PLoS Genet. 9:e1003297. 10.1371/journal.pgen.1003297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M. 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 19:3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. 1993. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 239:169–176 [DOI] [PubMed] [Google Scholar]
- 26.Garí E, Piedrafita L, Aldea M, Herrero E. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837–848 [DOI] [PubMed] [Google Scholar]
- 27.Nouraini S, Archambault J, Friesen JD. 1996. Rpo26p, a subunit common to yeast RNA polymerases, is essential for the assembly of RNA polymerases I and II and for the stability of the largest subunits of these enzymes. Mol. Cell. Biol. 16:5985–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan Q, Prysak MH, Woychik NA. 2003. Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II, and III. Mol. Cell. Biol. 23:3329–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forget A, Chartrand P. 2011. Cotranscriptional assembly of mRNP complexes that determine the cytoplasmic fate of mRNA. Transcription 2:86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czeko E, Seizl M, Augsberger C, Mielke T, Cramer P. 2011. Iwr1 directs RNA polymerase II nuclear import. Mol. Cell 42:261–266 [DOI] [PubMed] [Google Scholar]
- 31.Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, Lamond AI, Bertrand E. 2010. HSP90 and its R2TP/prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39:912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forget D, Lacombe AA, Cloutier P, Al-Khoury R, Bouchard A, Lavallee-Adam M, Faubert D, Jeronimo C, Blanchette M, Coulombe B. 2010. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteomics 9:2827–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daulny A, Tansey WP. 2009. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA Repair (Amst.) 8:444–448 [DOI] [PubMed] [Google Scholar]
- 34.Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. 2005. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121:913–923 [DOI] [PubMed] [Google Scholar]
- 35.Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP. 2008. Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc. Natl. Acad. Sci. U. S. A. 105:19649–19654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuras L, Struhl K. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609–613 [DOI] [PubMed] [Google Scholar]
- 37.Lee SK, Fletcher AG, Zhang L, Chen X, Fischbeck JA, Stargell LA. 2010. Activation of a poised RNAPII-dependent promoter requires both SAGA and Mediator. Genetics 184:659–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu H, Hu C, Yoon S, Natarajan K, Swanson MJ, Hinnebusch AG. 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24:4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choukrallah MA, Kobi D, Martianov I, Pijnappel WW, Mischerikow N, Ye T, Heck AJ, Timmers HT, Davidson I. 2012. Interconversion between active and inactive TATA-binding protein transcription complexes in the mouse genome. Nucleic Acids Res. 40:1446–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai G, Chaban YL, Imasaki T, Kovacs JA, Calero G, Penczek PA, Takagi Y, Asturias FJ. 2012. Interaction of the Mediator head module with RNA polymerase II. Structure 20:899–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31:337–346 [DOI] [PubMed] [Google Scholar]
- 42.Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. 2009. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell 34:168–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. 2012. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J. Biol. Chem. 287:8541–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez-Herreros F, de Miguel-Jimenez L, Morillo-Huesca M, Delgado-Ramos L, Munoz-Centeno MC, Chavez S. 2012. TFIIS is required for the balanced expression of the genes encoding ribosomal components under transcriptional stress. Nucleic Acids Res. 40:6508–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17:103–112 [DOI] [PubMed] [Google Scholar]
- 46.Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC. 2011. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 25:581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu M, Rajashankar KR, Lima CD. 2010. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure 18:216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley D. 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13:377–387 [DOI] [PubMed] [Google Scholar]
- 49.Cho EJ, Takagi T, Moore CR, Buratowski S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwer B, Saha N, Mao X, Chen HW, Shuman S. 2000. Structure-function analysis of yeast mRNA cap methyltransferase and high-copy suppression of conditional mutants by AdoMet synthase and the ubiquitin conjugating enzyme Cdc34p. Genetics 155:1561–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. 2001. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. U. S. A. 98:892–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armache KJ, Kettenberger H, Cramer P. 2003. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 100:6964–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bushnell DA, Kornberg RD. 2003. Complete, 12-subunit RNA polymerase II at 4.1-Å resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. U. S. A. 100:6969–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards AM, Kane CM, Young RA, Kornberg RD. 1991. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem. 266:71–75 [PubMed] [Google Scholar]
- 55.Lorenzen K, Vannini A, Cramer P, Heck AJ. 2007. Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure 15:1237–1245 [DOI] [PubMed] [Google Scholar]
- 56.Woychik NA, Young RA. 1989. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol. 9:2854–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maillet I, Buhler JM, Sentenac A, Labarre J. 1999. Rpb4p is necessary for RNA polymerase II activity at high temperature. J. Biol. Chem. 274:22586–22590 [DOI] [PubMed] [Google Scholar]
- 58.Pillai B, Verma J, Abraham A, Francis P, Kumar Y, Tatu U, Brahmachari SK, Sadhale PP. 2003. Whole genome expression profiles of yeast RNA polymerase II core subunit, Rpb4, in stress and nonstress conditions. J. Biol. Chem. 278:3339–3346 [DOI] [PubMed] [Google Scholar]
- 59.Jensen GJ, Meredith G, Bushnell DA, Kornberg RD. 1998. Structure of wild-type yeast RNA polymerase II and location of Rpb4 and Rpb7. EMBO J. 17:2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R, Svejstrup JQ. 2011. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J. Biol. Chem. 286:35553–35561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carré C, Shiekhattar R. 2011. Human GTPases associate with RNA polymerase II to mediate its nuclear import. Mol. Cell. Biol. 31:3953–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wild T, Cramer P. 2012. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 37:99–105 [DOI] [PubMed] [Google Scholar]
- 63.Kolodziej PA, Young RA. 1991. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol. Cell. Biol. 11:4669–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawakami K, Ishihama A. 1980. Defective assembly of ribonucleic acid polymerase subunits in a temperature-sensitive alpha-subunit mutant of Escherichia coli. Biochemistry 19:3491–3495 [DOI] [PubMed] [Google Scholar]
- 65.Rubbi L, Labarre-Mariotte S, Chedin S, Thuriaux P. 1999. Functional characterization of ABC10α, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J. Biol. Chem. 274:31485–31492 [DOI] [PubMed] [Google Scholar]
- 66.Corden J. 2011. Going nuclear: transcribers in transit. Mol. Cell 42:143–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kimura M, Ishiguro A, Ishihama A. 1997. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J. Biol. Chem. 272:25851–25855 [DOI] [PubMed] [Google Scholar]
- 68.Huibregtse JM, Yang JC, Beaudenon SL. 1997. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U. S. A. 94:3656–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manzo SG, Zhou ZL, Wang YQ, Marinello J, He JX, Li YC, Ding J, Capranico G, Miao ZH. 2012. Natural product triptolide mediates cancer cell death by triggering CDK7-dependent degradation of RNA polymerase II. Cancer Res. 72:5363–5373 [DOI] [PubMed] [Google Scholar]
- 70.Nechaev S, Adelman K. 2011. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Komarnitsky P, Cho EJ, Buratowski S. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kimura M, Suzuki H, Ishihama A. 2002. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 22:1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fuda NJ, Buckley MS, Wei W, Core LJ, Waters CT, Reinberg D, Lis JT. 2012. Fcp1 dephosphorylation of the RNA polymerase II C-terminal domain is required for efficient transcription of heat shock genes Mol. Cell. Biol. 32:3428–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan HY, Cheng KK, Klein HL. 1996. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1Δ of Saccharomyces cerevisiae. Genetics 142:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schroeder SC, Schwer B, Shuman S, Bentley D. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geisberg JV, Holstege FC, Young RA, Struhl K. 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 21:2736–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Mullem V, Wery M, Werner M, Vandenhaute J, Thuriaux P. 2002. The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and Elongator histone acetyltransferases. J. Biol. Chem. 277:10220–10225 [DOI] [PubMed] [Google Scholar]
- 79.Scafe C, Martin C, Nonet M, Podos S, Okamura S, Young RA. 1990. Conditional mutations occur predominantly in highly conserved residues of RNA polymerase II subunits. Mol. Cell. Biol. 10:1270–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, Lacroute F. 1991. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 7:609–615 [DOI] [PubMed] [Google Scholar]
- 81.Shpakovski GV, Acker J, Wintzerith M, Lacroix JF, Thuriaux P, Vigneron M. 1995. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol. Cell. Biol. 15:4702–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaros C, Thuriaux P. 2005. Rpc25, a conserved RNA polymerase III subunit, is critical for transcription initiation. Mol. Microbiol. 55:104–114 [DOI] [PubMed] [Google Scholar]