Abstract
Huntingtin-interacting protein 1 (HIP1) binds inositol lipids, clathrin, actin, and receptor tyrosine kinases (RTKs). HIP1 is elevated in many tumors, and its expression is prognostic in prostate cancer. HIP1 overexpression increases levels of the RTK epidermal growth factor receptor (EGFR) and transforms fibroblasts. Here we report that HIP1 is tyrosine phosphorylated in the presence of EGFR and platelet-derived growth factor β receptor (PDGFβR) as well as the oncogenic derivatives EGFRvIII, HIP1/PDGFβR (H/P), and TEL/PDGFβR (T/P). We identified a four-tyrosine “HIP1 phosphorylation motif” (HPM) in the N-terminal region of HIP1 that is required for phosphorylation mediated by both EGFR and PDGFβR but not by the oncoproteins H/P and T/P. We also identified a tyrosine residue (Y152) within the HPM motif of HIP1 that inhibits HIP1 tyrosine phosphorylation. The HPM tyrosines are conserved in HIP1's only known mammalian relative, HIP1-related protein (HIP1r), and are also required for HIP1r phosphorylation. Tyrosine-to-phenylalanine point mutations in the HPM of HIP1 result in proapoptotic activity, indicating that an intact HPM may be necessary for HIP1's role in cellular survival. These data suggest that phosphorylation of HIP1 by RTKs in an N-terminal region contributes to the promotion of cellular survival.
INTRODUCTION
Huntingtin-interacting protein 1 (HIP1) was originally identified as a protein that binds Huntingtin, whose gene is mutated in Huntington's disease (1, 2). HIP1 was later found to be an endocytic protein that binds clathrin, AP2 (3–6), and actin (7). It also binds inositol lipids via an AP180 N-terminal homology (ANTH) domain (8–10). Knockout of HIP1 alone (11, 12) or with its only known mammalian relative, HIP1-related protein (HIP1r) (13, 14), results in degenerative mouse phenotypes. These phenotypes, which are more severe in double-knockout mice, include testicular degeneration, spinal defects, weight loss, and cataracts; however, the mechanisms underlying these phenotypes have yet to be delineated.
HIP1 was linked to receptor tyrosine kinase (RTK) signaling as a result of its identification as part of a chromosomal translocation involving the coding sequences for the transmembrane and tyrosine kinase domains of the platelet-derived growth factor β receptor (PDGFβR) in a leukemia patient (15). This HIP1/PDGFβR (H/P) translocation is a member of a large family of chromosomal translocations involving the PDGFβR gene (16–18). The first identified translocation in this family was the t(5;12) translocation, which encodes the TEL/PDGFβR (T/P) fusion oncoprotein (19). These translocations lead to the formation of constitutively active RTK fusion proteins that transform hematopoietic cell lines to interleukin-3 (IL-3)-independent growth (20) and result in hematopoietic neoplasms in mice (21, 22).
In addition to the transformation activity displayed by the HIP1/PDGFβR fusion protein, overexpression of HIP1, but not that of its lipid-binding domain deletion mutant (HIP1/ΔANTH), transforms fibroblasts (23) and prostate epithelial cells (24). This transforming activity remains linked to RTKs, as HIP1-transformed cells display increased levels of epidermal growth factor receptor (EGFR) (23). Indeed, HIP1 prolongs the half-lives of RTKs such as EGFR and PDGFβR (9). Additionally, treatment of the HIP1-transformed cells with an EGFR inhibitor reverses the transformed phenotype (23). High levels of the HIP1 protein have also been correlated with overexpression of EGFR and other RTKs in a series of tumors from brain and breast cancer patients (23, 25). One possible explanation for EGFR overexpression in these cancers is that HIP1-dependent stabilization of the RTKs occurs via clathrin sequestration. Low levels of available clathrin would result in diminished endocytosis-mediated receptor degradation. Another possible explanation for concomitantly increased RTK and HIP1 overexpression involves a currently unknown mechanism whereby HIP1 promotes cell survival of specific cells with increased RTK levels, further promoting their tumorigenesis (26). In this study, we investigated the interaction of HIP1 with EGFR and other RTKs and characterized the tyrosine phosphorylation of HIP1. We defined key tyrosines necessary for this phosphorylation by using site-directed mutagenesis.
MATERIALS AND METHODS
Cell culture.
Experiments were performed with HIP1-transformed NIH 3T3 cells (23) or human HeLa and embryonic kidney 293T (HEK293T) cells (ATCC). These cells were maintained at 37°C in Dulbecco's minimal essential medium (Gibco) supplemented with 10% fetal bovine serum, penicillin-streptomycin, and GlutaMAX (Gibco). For the “cold-load” experiments, 100 nM EGF was added to both the 4°C load medium and the 37°C endocytosis medium.
Kinase and phosphatase inhibitor treatment.
In experiments where cells were treated with the phosphatase inhibitor sodium orthovanadate (NaVa), cells were incubated with 2 mM NaVa in normal medium for 30 min to 2 h before collection. AG1478 (also known as tyrphostin; Calbiochem) was present in the tissue culture medium, at a final concentration of 1 μM. Imatinib (Novartis) was prepared as described previously (22) and incubated with cells for 30 min, using a final concentration of 5 μM (27).
DNA constructs.
pcDNA3.1-V5-His constructs were generated by PCR amplification of the various EGFR family members (EGFR, ErbB2 [a gift from Linda Pike, Washington University, St. Louis, MO], and EGFRvIII [a gift from Paul Mischel, UCLA]), and then the products were subcloned into the pcDNA3.1-V5-His vector (Invitrogen) by using the restriction enzymes HindIII and KpnI or XhoI. pcDNA3-PDGFβR was generated by subcloning the 3.32-kb open reading frame of PDGFβR (accession no. NM_002609.3) into the KpnI/EcoRI sites of pcDNA3. HIP1 and HIP1r deletion and point mutants were generated using a QuikChange XL multisite-directed mutagenesis kit (Stratagene). The pcDNA3.1HIP1-IRES-GFP construct was described previously (9) and was used as the substrate to generate the HIP1/HPM(4xYF) mutant in the IRES-GFP vector.
The HIP1r and MycHis-tagged HIP1a (previously referred to as full-length HIP1) constructs were reported previously (9, 26). The MycHis-tagged HIP1b construct was generated by reverse transcription-PCR (RT-PCR) with HEK293T cell RNA as the starting material. A 1.2-kb fragment was generated using the following primers: CCG GCG GCC GCG CCT CGG TCA TGG ATG TGA GCA AG (forward) and CAG TTC TGC CCG CAG GAA TTC ACA C (reverse). The resulting fragment was digested with NotI and EcoRI to generate overhangs at the 5′ and 3′ ends, respectively. This fragment was ligated into NotI- and EcoRI-digested pcDNA3.1-HIP1a.
Immunoprecipitation.
Ten micrograms of a combination of HIP1-Myc or HIP1r (untagged) and EGFR-V5 or EGFRVIII-V5 cDNA constructs in pcDNA3 was coexpressed in HEK293T cells by using the Superfect reagent (Qiagen). At 24 h posttransfection, cells were extracted in lysis buffer containing protease and phosphatase inhibitors (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, protease inhibitors [Roche], 30 mM sodium pyrophosphate, 50 mM NaF, 100 μM sodium orthovanadate), cleared of unbroken cells by centrifugation, and diluted to a protein concentration of 0.5 to 2.0 mg/ml. For immunoprecipitation, 500 μl of lysate was first precleared for 1 h at 4°C with protein G-Sepharose beads (GE Healthcare) and then incubated for 2 h with rotation at 4°C with 50 μl of a 50-50 slurry of anti-Myc–agarose (Sigma) or first incubated with antibody (UM410 or anti-EGFR antibody) followed by protein G beads (GE Healthcare) in lysis buffer containing protease and phosphatase inhibitors. The pellets were washed three times with lysis buffer, resuspended in Laemmli buffer, boiled for 5 min, centrifuged, and then separated by SDS-PAGE for Western blot analysis. For immunoprecipitation of HIP1 without an epitope tag, 10 μl of the anti-HIP1 UM410 antibody was used (26). For immunoprecipitation of HIP1r without an epitope tag, 10 μl of the anti-HIP1r UM374 antibody was used (9).
Western blotting.
Whole-cell lysates (WCL) or immune precipitates in Laemmli buffer were separated in 6% or 10% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were probed with the following antibodies: mouse monoclonal anti-HIP1 4B10 and anti-HIP1r 1C5, rabbit polyclonal anti-HIP1 UM410 and UM323 and anti-HIP1r UM374, rabbit monoclonal anti-Myc (1:5,000; Cell Signaling), monoclonal antiphosphotyrosine (4G10; Millipore) (1:1,000), monoclonal anticlathrin (TD.1; Sigma) (1:250), polyclonal anti-PDGFβR (1:2,000; Pharmingen), and mouse monoclonal anti-V5 (1:5,000; Invitrogen). Anti-green fluorescent protein (anti-GFP) and antitubulin polyclonal antibodies were purchased from Cell Signaling (1:5,000). Blots were then incubated with horseradish peroxidase (HRP)-conjugated mouse or rabbit secondary antibodies (1:5,000; GE Healthcare) and developed using chemiluminescence (Pierce).
Mass spectrometry.
Protein extracts from 100 10-cm dishes of EGF-stimulated HeLa cells (97 mg of starting material) were immunoprecipitated with the UM410 anti-HIP1 antibody. The 120-kDa HIP1 band (6% PAGE) was excised, and an in-gel tryptic digestion was performed prior to matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis. The Mascot server (Matrix Sciences) was used to compare our spectra to those in the Swiss-Prot database, and HIP1 tryptic polypeptides were the majority identified.
Confocal immunofluorescence microscopy.
HeLa cells were cultured in 6-well plates with coverslips and transfected the following day with HIP1-Myc. Cells on coverslips were stimulated with 1 μg/ml EGF-555 (Invitrogen) at 4°C for 30 min, and then the medium was changed to 37°C for the indicated times. Cells were fixed for 20 min in 4% paraformaldehyde at 4°C, blocked, and stained with the antibodies listed above. Images were collected using a Zeiss LSM 510-Meta laser scanning confocal microscope with a 63× water immersion objective. Colocalization was calculated in ImageJ (NIH) by using the JACoP plug-in to calculate Manders coefficients, using automated thresholding. Statistical analyses were performed using GraphPad Prism software.
Apoptosis assays.
COS-7 cells growing on coverslips were transfected with the HIP1(r), HIP1(r)/HPM(4xYF), HIP1(r)/HPM(2xYF), or HIP1/ΔANTH construct and fixed with 4% formaldehyde at 24 h posttransfection. Cells were permeabilized with 0.1% Triton X-100 and blocked with 5% milk in phosphate-buffered saline (PBS)–0.1% Triton for 1 h. Cells were then incubated with anti-Myc monoclonal antibody or anti-HIP1r (1C5 monoclonal antibody) to identify transfected cells (1:100). Bound antibodies were visualized with goat anti-mouse IgG–Alexa Fluor 594 (Invitrogen). Coverslips were mounted on glass slides by using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI). Cells expressing the HIP1 or HIP1r constructs were scored for apoptosis by nuclear morphology. At least 100 cells were counted for each sample, and transfections were performed in triplicate.
5′RACE analysis.
The 5′ rapid amplification of cDNA ends (5′RACE) was performed using a GeneRacer kit (Invitrogen). All procedures were performed according to the manufacturer's suggestions. Total RNA (5 μg) isolated from mouse brain and spleen was used as the starting material for analysis. PCR-amplified products generated in the last step of the 5′RACE procedure were separated in a 1% agarose gel in 0.5× Tris-borate-EDTA (TBE) running buffer. Bands of interest were extracted from the agarose by use of a QIAquick gel extraction kit (Qiagen) according to the manufacturer's protocol. Purified products were subcloned into the pCR 4-TOPO vector for sequencing. Sequence analysis was performed using the UCSC BLAT Search Genome program (http://www.genome.ucsc.edu/cgi-bin/hgBlat). RNAs for 5′RACE were originally isolated from mouse brain and spleen because these two tissues had previously displayed high HIP1 protein levels together with a protein doublet by Western blotting (28, 29). The exon 1a-containing transcript (Hip1a) has been used consistently by our laboratory, and the human homologue was the isoform used for the initial phosphorylation studies displayed in Fig. 1 to 5 of this study. The mouse HIP1 sequences were used to identify the two forms in human 293T cells by use of RT-PCR.
Fig 1.
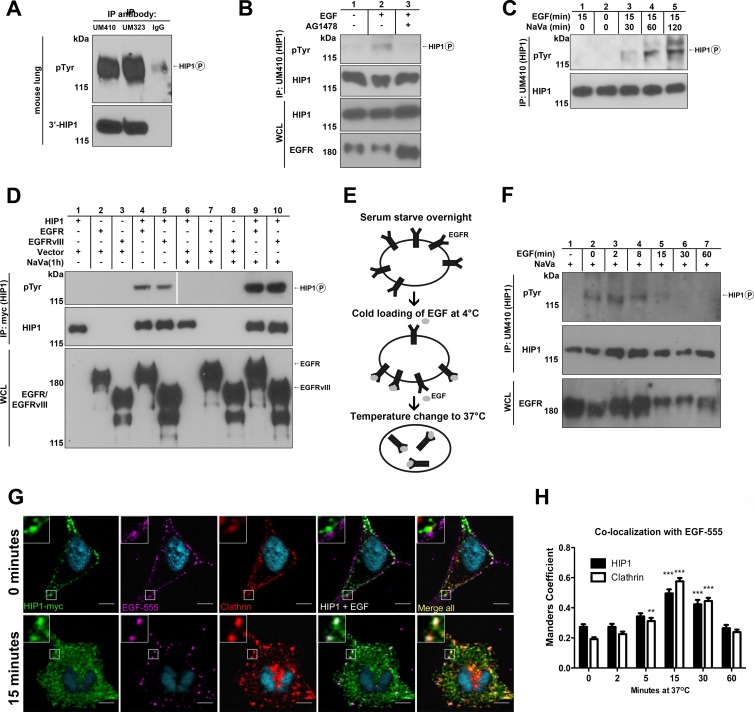
HIP1 association with and phosphorylation mediated by EGFR and EGFRvIII. (A) HIP1 was immunoprecipitated (IP) from mouse lung tissue by use of anti-HIP1 polyclonal antibodies UM410 and UM323. Western blotting with antiphosphotyrosine monoclonal antibody 4G10 (Millipore) showed a HIP1-sized phosphotyrosine band. (B) Fibroblasts were serum starved for 24 h and then stimulated with EGF (100 ng/ml) for 15 min prior to extract preparation. One set of cells was treated with 1 μM AG1478 (also known as tyrphostin) for 30 min prior to addition of EGF. HIP1 was immunoprecipitated from 2 mg of HIP1-transformed fibroblast WCL (23) by use of anti-HIP1 polyclonal antibody UM410. Antiphosphotyrosine monoclonal antibody 4G10 was used to detect phosphorylated HIP1 in the immunoprecipitates. Anti-HIP1 monoclonal antibody 4B10 was used to detect HIP1. (C) Cells were treated with sodium orthovanadate (NaVa) at a final concentration of 2 mM for different periods, and EGF was added to the NaVa-containing medium for the last 15 min of each period, prior to harvest. HIP1 was immunoprecipitated and analyzed as described for panel B. The protein that migrated slower than (above) HIP1 did not plateau at 60 min. The identity of this band is unknown, as it did not comigrate with HIP1 (bottom panel). (D) HEK293T cells were cotransfected with HIP1-Myc and EGFR-V5 or EGFRvIII-V5 and lysed 24 h after transfection. HIP1-Myc was immunoprecipitated from 1 mg of lysate by use of anti-Myc antibody-conjugated agarose beads (Sigma). Anti-V5 antibodies (Invitrogen) were used to detect EGFR. (E) Diagram illustrating cold-load stimulation, a strategy for precise temporal analysis of endocytic processes. (F) HIP1 tyrosine phosphorylation was assayed at several time points for up to 60 min after initiation of endocytosis. Cells were starved, and then EGFR was “loaded” with 100 nM EGF for 1 h at 4°C. The cold medium was then exchanged for medium prewarmed to 37°C. This temperature change allowed for receptor internalization to proceed. (G) HIP1-Myc-transfected HeLa cells were treated with 1 μM EGF-555 in the “cold-load stimulation” experimental paradigm. Cells were then fixed, stained for HIP1 (anti-Myc; Cell Signaling) and clathrin (X22; AbCAM) to mark clathrin-coated vesicles, and imaged using a 1-μm slice thickness on a Zeiss confocal microscope. (H) Quantification of EGF colocalization with HIP1 and clathrin over time. ***, P < 0.0001; **, P < 0.001 (n = 37 cells per time point over 3 experiments).
Fig 5.
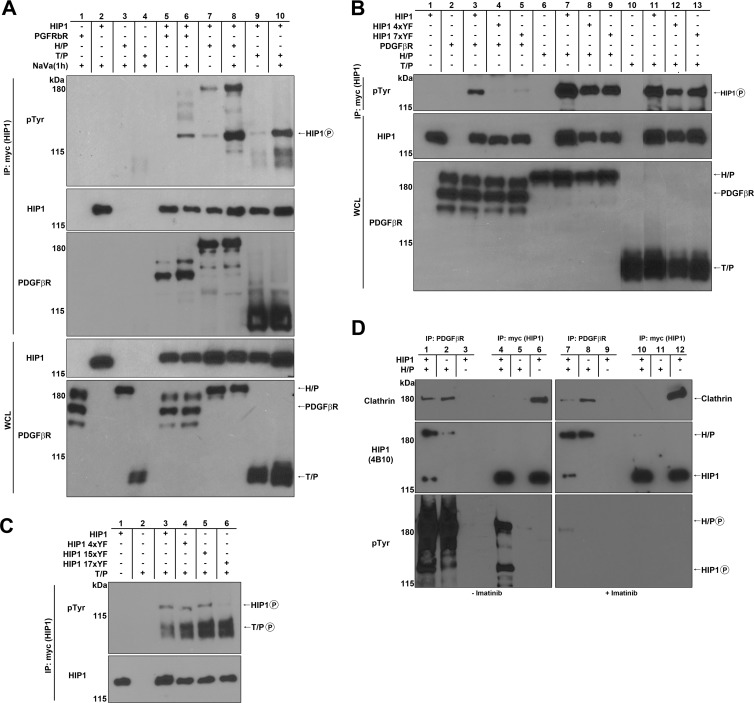
HIP1 association with and phosphorylation mediated by PDGFβR and its oncogenic derivatives. (A to D) HEK293T cells were cotransfected with HIP1-Myc and RTKs. HIP1-Myc was immunoprecipitated from 1 mg of HEK293T whole-cell lysate by use of anti-Myc beads. Phosphorylation was detected with antiphosphotyrosine antibody 4G10, and clathrin association was detected with anticlathrin antibody TD.1 (Sigma). Anti-PDGFβR (BD Pharmingen) was used to detect PDGFβR, H/P, and T/P. (A) Cells were cotransfected with HIP1-Myc and either PDGFβR, H/P, or T/P. HIP1 phosphorylation was observed in these cotransfections, and the phosphorylation was enhanced when NaVa was added to cells 1 h before lysis. (B) The HPM Y-F mutants (HIP1 4xYF and 7xYF) were not phosphorylated by PDGFβR but were phosphorylated by H/P and T/P. Lanes 10 and 11 were spliced and rearranged from the same gel for presentation purposes. (C) T/P requires amino acids 281 and 475 in HIP1 for HIP1 phosphorylation, as the HIP1 15xYF mutant had only these two tyrosines intact. (D) In contrast to HIP1, H/P readily interacted with clathrin when H/P was coexpressed with HIP1. Extracts were immunoprecipitated with anti-PDGFβR (lanes 1 to 3 and 7 to 9) to directly precipitate H/P rather than HIP1. Extracts were immunoprecipitated with anti-Myc (lanes 4, 5, 10, and 11) to directly precipitate Myc-HIP1 rather than H/P. The extracts in the panels on the right were derived from imatinib-treated cells to inhibit H/P phosphorylation, as demonstrated in the lower panels.
RESULTS
HIP1 is phosphorylated by EGFR and its oncogenic derivative EGFRvIII.
Having previously described physical (25) and functional (9) interactions between HIP1 and EGFR, we hypothesized that HIP1 may be phosphorylated by the EGFR tyrosine kinase. In fact, previous reports using the Dictyostelium HIP1r homologue, Hip, have found that Hip migrates as a protein doublet that collapses upon treatment of extracts with calf intestine phosphatase (30, 31). In support of this hypothesis, mouse lung tissue extracts contained a tyrosine-phosphorylated protein that comigrated with HIP1 and was immunoprecipitated by either of two distinct anti-HIP1 antibodies but not by control nonimmune serum (Fig. 1A). The same result was obtained for HIP1-transformed fibroblasts (23) when the cells were treated with EGF (Fig. 1B, lane 2). This observed phosphorylation was likely dependent on EGFR activation, as the phospho-“HIP1” band was not detected when the EGF-stimulated cells were also treated with the EGFR tyrosine kinase inhibitor AG1478 (Fig. 1B, lane 3).
We observed that the tyrosine phosphorylation of HIP1 was enhanced in HIP1-transformed fibroblasts when cells were incubated with the protein tyrosine phosphatase inhibitor NaVa prior to harvest (32). After 30 min of incubation with NaVa, increased HIP1 phosphorylation (Fig. 1C, lane 3 versus lane 1) was observed, and it reached a maximum after 60 min (Fig. 1C, lanes 4 and 5 versus lane 3). These data demonstrate that HIP1 is phosphorylated in cells and tissues and that its phosphorylation is opposed by tyrosine phosphatase activity in cells.
We further tested the idea that HIP1 is an EGFR tyrosine kinase substrate by coexpressing Myc-tagged HIP1 and V5-tagged EGFR in HEK293T cells in the presence and absence of NaVa, immunoprecipitating them with anti-Myc antibody, and probing with the antiphosphotyrosine antibody 4G10 to determine the HIP1 phosphorylation status. Incubation with NaVa for 1 h prior to harvest was sufficient to fully maximize HIP1 phosphorylation in these cells (Fig. 1D, lane 4 versus lane 9). As expected, HIP1 was not phosphorylated by the EGFR kinase-inactive K721M point mutant, even in the presence of the phosphatase inhibitor (data not shown).
Because HIP1 is expressed at high levels in many cancers (26) and can directly transform cells (23), we examined whether HIP1 can serve as a substrate for an N-terminally truncated oncogenic mutant of EGFR, EGFRvIII. This mutant is expressed in tumor cells from 60 to 70% of patients with glioblastoma multiforme and has a 267-amino-acid deletion in its N terminus which leads to constitutive kinase activity and slowed degradation (33–36). Indeed, we found that HIP1 is also a substrate for EGFRvIII (Fig. 1D, lane 5 versus lane 10). The level of phosphorylation of HIP1 by EGFRvIII was similar to that induced by wild-type EGFR (Fig. 1D, lanes 5 and 10 versus lanes 4 and 9).
To explore the chronology of HIP1 phosphorylation by EGFR after ligand stimulation, we turned to a timed, “cold-load” ligand stimulation method (37) in NIH 3T3 cells (illustrated in Fig. 1E). This method allows for coordinated internalization of receptors following incubation of the receptors with ligand at temperatures that are not permissive to internalization. In this experiment, no HIP1 phosphorylation was observed during starvation, even though vanadate was present (Fig. 1F, lane 1). In contrast, strong phosphorylation of HIP1 was observed at the end of the EGF load at 4°C (Fig. 1F, lane 2 versus lane 1). HIP1 phosphorylation remained high 8 min after the shift to 37°C. The HIP1 phosphotyrosine signal began to diminish at 15 min and was absent by 60 min, indicating that HIP1 is transiently phosphorylated on tyrosines in the early phases of EGFR endocytosis.
To visualize where HIP1 is located when it is being phosphorylated during EGFR activation and endocytosis in the cell, we used the same “cold-load” stimulation protocol followed by confocal microscopy (Fig. 1E). We found that HIP1 and clathrin were recruited to the plasma membrane in the earliest stages of EGF internalization. Both were observed lining the internal portion of the plasma membrane, while EGF remained on the external portion of the surface of the cell (Fig. 1G, top row). Since we saw significant phosphorylation of HIP1 at time zero when the cells were cold loaded with EGF, the RTK phosphorylation of HIP1 located at the plasma membrane is likely very robust. As EGF was internalized, colocalization of HIP1 and clathrin with EGF-containing vesicles peaked at 15 min (Fig. 1G, bottom row). Manders coefficient analysis of EGF colocalization with HIP1 indicated that there was a significant increase in the proportion of EGF colocalizing with HIP1 over time (Fig. 1H) (P < 0.001; one-way analysis of variance [ANOVA]). Post hoc analysis using the Newman-Keuls multiple-comparison test also showed that EGF association with HIP1 increased significantly between 0 min and 15 min (Fig. 1H). This time span approximately reflects the period during which we observed transient HIP1 tyrosine phosphorylation (Fig. 1F). Collectively, these data indicate that HIP1 is recruited to clathrin-coated pits as they form in response to EGFR activation and continues to associate with these structures as they internalize and become clathrin-coated vesicles.
Since HIP1 was phosphorylated in the presence of active EGFR as well as its oncogenic mutant EGFRvIII, we investigated whether HIP1 was phosphorylated in the presence of other activated normal RTKs, such as ErbB2, that contribute to tumorigenesis in a variety of cancers. ErbB2, also known as HER2/neu, is an oncogenic member of the EGFR family that lacks ligand binding capability but is activated upon dimerization with itself or other ligand-bound EGFR family members (38). Although HIP1 coimmunoprecipitated with ErbB2 in cotransfected cells, HIP1 was not readily phosphorylated by activated ErbB2, even after incubation of the cells with NaVa for 1 h to enhance detection of phosphorylated tyrosine (data not shown). Similarly, insulin-like growth factor 1 receptor (IGF-1R) failed to induce HIP1 phosphorylation at detectable levels, even after incubation with NaVa, despite the observation that activated IGF-1R readily bound HIP1 (data not shown).
HIP1 phosphorylation in the presence of EGFR and EGFRvIII requires the HIP1 USH domain.
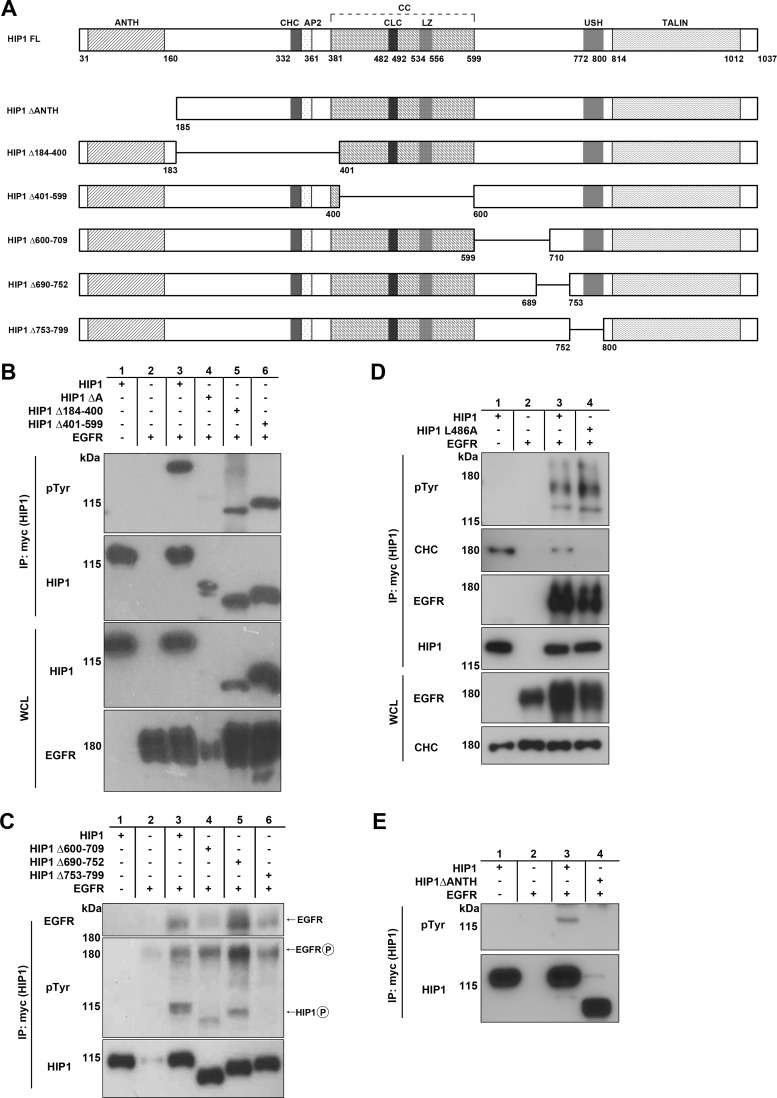
To identify the locations of phosphorylated tyrosine residues in HIP1, we tested a series of HIP1 deletion mutants for the ability to serve as kinase substrates. We used a previously constructed deletion mutant, HIP1ΔANTH (26), and constructed five new mutants (HIP1Δ184-400, HIP1Δ401-599, HIP1Δ600-709, HIP1Δ690-752, and HIP1Δ753-799) that span the HIP1 sequence (Fig. 2A). The HIP1Δ753-799 mutant has a deletion of the upstream helix (USH) domain, which is required for a conformational shift in the presence of actin that prevents clathrin binding (39). These Myc-tagged mutants were then coexpressed with EGFR to determine which domain(s) was necessary for tyrosine phosphorylation. Tyrosine phosphorylation of all the deletion mutants was intact (Fig. 2B, lanes 5 and 6, and C, lanes 3 to 5), with the exception of HIP1ΔANTH (Fig. 2B, lane 4) and HIP1Δ753-799 (Fig. 2C, lane 6). Both of these mutants lack two HIP1 regulatory regions, i.e., the ANTH and USH domains (26, 39).
Fig 2.
The HIP1 ANTH domain and amino acids 753 to 799 are required for HIP1 phosphorylation mediated by EGFR. (A) Schematic of HIP1 deletion mutations as they relate to known HIP1 domains. Domains: ANTH, AP180 N-terminal homology; CHC, clathrin heavy chain binding; AP2, clathrin adaptor protein 2 binding; CC, coiled coil; CLC, clathrin light chain binding; LZ, leucine zipper; USH, upstream helix; TALIN, TALIN homology. (B to E) HEK293T cells were cotransfected with EGFR-V5 and either wild-type HIP1 or the deletion mutants, all of which were Myc tagged. Immunoprecipitation was performed with 1 mg lysate and anti-Myc beads, and phosphorylation was detected with antiphosphotyrosine antibody 4G10. (B) HIP1Δ184-400 and HIP1Δ401-599 were phosphorylated by EGFR, whereas the ΔANTH mutant was not. (C) HIP1Δ600-709 and HIP1Δ690-752 were phosphorylated by EGFR, and HIP1Δ753-799 was not. All mutants interacted with EGFR, as evidenced by their coimmunoprecipitation with EGFR. (D) Phosphorylation of HIP1 by EGFR does not require HIP1 binding to clathrin. The top panel shows phosphorylation of HIP1L486A (lane 3). Lane 4 contained the wild-type HIP1 control. Although the binding mutation is in the clathrin light chain binding site, we consistently observed a loss of binding to the entire clathrin triskelion, as represented by the blot for the clathrin heavy chain in this case. (E) The HIP1ΔANTH mutant is not phosphorylated by EGFR. Data are representative of three independent experiments.
The deleted amino acids in the HIP1Δ753-799 mutant (also called HIP1ΔUSH) do not include tyrosine residues that can serve as substrates for phosphorylation (Fig. 2A); however, since this mutant has a deletion of the USH domain, a sequence that is necessary for a conformational shift in the presence of clathrin that inhibits actin binding (39), it was first considered likely that the lack of phosphorylation was due to loss of binding to EGFR during clathrin-mediated endocytosis. However, the binding of HIP1Δ753-799 to EGFR was found to be similar to that of wild-type HIP1 (Fig. 2C, lane 6 versus lane 3). These data suggest that the USH domain allows for full HIP1 phosphorylation through either conformational changes that promote activation of the RTKs or inhibition of phosphatases. However, because vanadate did not restore phosphorylation of the HIP1Δ753-799 mutant (Fig. 3B), the inhibition of phosphatases is an unlikely explanation for the lack of phosphorylation.
Fig 3.
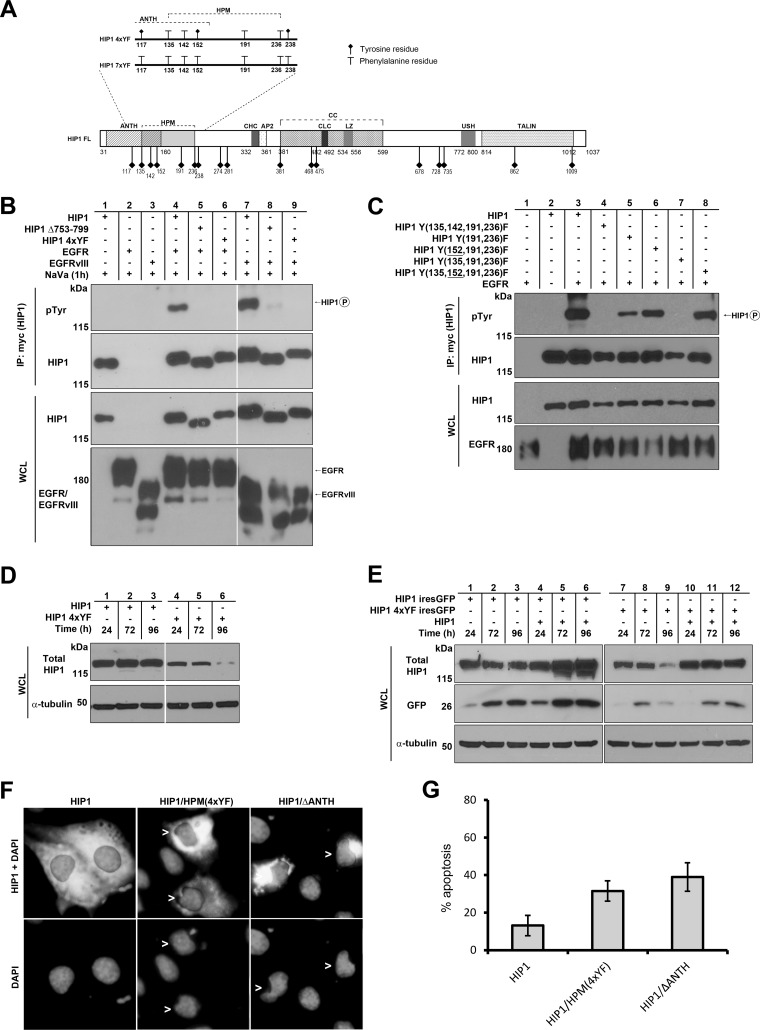
Identification of tyrosine residues in HIP1 required for EGFR-mediated phosphorylation of HIP1. (A) Schematic of HIP1 Y-to-F mutations. The diagram also displays the locations of all 17 tyrosine residues (diamond-headed stalks) within the domains of HIP1. The key mutants that were not phosphorylated by wild-type RTKs are illustrated in the magnified area (HIP1 4xYF and 7xYF). Abbreviations are as described in the legend to Fig. 2. HPM, HIP1 phosphorylation motif. (B and C) HEK293T cells were cotransfected with EGFR-V5 and either Myc-tagged wild-type HIP1 or Myc-tagged HIP1 Y-to-F point mutants. Immunoprecipitation was performed with anti-Myc beads, and phosphorylation was detected with antiphosphotyrosine antibody 4G10. (B) The USH-deficient mutant (HIP1Δ753-799) and the HPM Y-to-F mutants (HIP1 4xYF) were not phosphorylated by EGFR or EGFRvIII. (C) Y152 of HIP1 inhibits EGFR phosphorylation. Phosphorylation-resistant mutants (lanes 5 and 7) were phosphorylation sensitive when the Y152F point mutation was included (lanes 6 and 8). The levels of EGFR were increased in the presence of wild-type HIP1 but not in the presence of phosphorylation-resistant mutants (bottom panel). (D) HEK293T cells were transfected with Myc-tagged HIP1 or the HIP1/HPM(4xYF) mutant and assayed for HIP1 levels by Western blotting with anti-Myc antibody over a 96-hour period. (E) Low GFP levels that were associated with cotransfected HIP1/HPM(4xYF) (lanes 7 to 9) were rescued by cotransfection of HIP1/HPM(4xYF)-ires-GFP with wild-type HIP1 (lanes 10 to 12). Wild-type HIP1 also increased GFP levels (lanes 4 to 6) compared to those in cells transfected with wild-type HIP1-ires-GFP (lanes 1 to 3). (F) Cos-7 cells were transfected with Myc-tagged wild-type HIP1, HIP1/HPM(4xYF), and HIP1/ΔANTH DNA constructs. Cells were stained with mouse monoclonal anti-Myc antibody (cytoplasmic staining) to allow for analysis of transfected cells and with DAPI to show nuclear morphology. Cells were scored at 24 h posttransfection and deemed apoptotic if nuclear condensation or fragmentation was observed (arrowheads). These images are representative of the overall results, where the HIP1-transfected cells were found more frequently with smooth, nonblebbed nuclei and contained large regions of HIP1-expressing cytoplasm. In comparison, the mutant cells displayed more condensed cytoplasm, and the nuclei were less frequently intact. (G) Apoptotic cells in the three different transfections described for panel F were quantitated. The experiment was performed on three separate occasions, and data were averaged. Error bars denote standard deviations. At least 100 cells from each sample in each experiment were scored for apoptosis by two blinded investigators (A.A.W. and A.C.), according to nuclear morphology. The percentages of apoptotic cells were compared to the baseline cell death frequency (15%) of surrounding HIP1-negative cells in each experiment.
Because the conformation of the HIP1 USH domain is known to be altered upon clathrin binding (39), we tested if clathrin binding was required for HIP1 phosphorylation. A point mutation (HIP1 L486A) that prevents HIP1 interaction with clathrin was generated (40). Although this mutation abolished HIP1 binding to clathrin, as expected, no effect on tyrosine phosphorylation was observed (Fig. 2D). These findings indicate that the HIP1 USH domain influences EGFR-mediated tyrosine phosphorylation of HIP1 in a manner independent of its ability to interact with clathrin and that clathrin binding is not required for HIP1 phosphorylation.
HIP1 phosphorylation in the presence of EGFR requires tyrosine residues in the ANTH domain.
As mentioned above, the HIP1ΔANTH mutant, which lacks the phospholipid-binding ANTH domain as well as four tyrosine residues, was not phosphorylated in the presence of EGFR. This mutant exhibited low protein expression due to its known proapoptotic activity (26) (Fig. 2B, lane 4). To confirm that this mutant was not phosphorylated, we repeated the immunoprecipitation experiment with 15 times the amount of lysate used in the experiment with wild-type HIP1 and EGFR. Even under these exaggerated conditions, the HIP1ΔANTH mutant was not phosphorylated (Fig. 2E). To determine which tyrosine(s) within the ANTH domain is phosphorylated, a series of point mutants were constructed by converting each or all of the four tyrosines in the ANTH domain to phenylalanine (Y117F, Y135F, Y142F, and Y152F mutants) (Fig. 3A; Table 1). All of these mutants were readily phosphorylated, indicating that although the lack of the ANTH domain inhibited EGFR-mediated phosphorylation, the tyrosine residues in the HIP1 ANTH domain are not completely necessary for phosphorylation. Because the ANTH domain deletion mutant is unable to transform cells (23), these data raise the hypothesis that phosphorylation may be required for transformation.
Table 1.
Phosphorylation of HIP1 and its mutants by EGFRa
| Construct | Phosphorylation |
|---|---|
| HIP1a | +++ |
| HIP1b | + |
| HIP1a ΔANTH | − |
| HIP1a Δ753-799 | − |
| HIP1a 2xYF (135, 142) | ++ |
| HIP1a 2xYF (135, 191) | +++ |
| HIP1a 2xYF (142, 191) | + |
| HIP1a 2xYF (191, 236) | ++ |
| HIP1a 2xYF (862, 1009) | +++ |
| HIP1a 3xYF (117, 135, 142) | + |
| HIP1a 3xYF (117, 191, 236) | + |
| HIP1a 3xYF (135, 191, 236) | + |
| HIP1a 3xYF (142, 191, 236) | +/− |
| HIP1a 4xYF (117, 135, 191, 236) | +/− |
| HIP1a 4xYF (117, 142, 191, 236) | +/− |
| HIP1a 4xYF (135, 142, 191, 236) | − |
| HIP1a 1xYF (152) | ++++ |
| HIP1a 3xYF (152, 191, 236) | ++++ |
| HIP1a 4xYF (135, 152, 191, 236) | ++++ |
| HIP1a 4xYF (142, 152, 191, 236) | ++ |
| HIP1a 4xYF (117, 135, 142, 152) | +++ |
| HIP1a 5xYF (135, 142, 152, 191, 236) | ++ |
| HIP1a 6xYF (117, 135, 142, 152, 191, 236) | ++ |
| HIP1a 7xYF (117, 135, 142, 152, 191, 236, 238) | − |
293T cells were cotransfected with HIP1 or its mutants together with EGFR and assayed for HIP1 phosphorylation by coimmunoprecipitation with anti-Myc beads and antiphosphotyrosine Western blot analysis. +++, full phosphorylation; ++, moderately diminished phosphorylation; +, severely diminished but detectable phosphorylation; −, no detectable phosphorylation; +++++, hyperphosphorylation. The presence of the Y152F mutation is indicated in bold.
To determine which of the HIP1 tyrosine residues were phosphorylated by EGFR, we initially performed mass spectrometry on HIP1 immunoprecipitates from EGF-stimulated HeLa cells. We found that a fraction of the HIP1 127-144 tryptic peptide contained phosphotyrosines at positions 135 and/or 142. Two of 23 peptides were phosphorylated, and there was 81% total HIP1 sequence coverage. This finding, however, did not rule out the possibility that HIP1 phosphorylation occurred in other areas of the HIP1 protein, due to the low sensitivity of mass spectrometry for tyrosine-phosphorylated peptides. Thus, we employed a computerized prediction model as a complementary approach to evaluate phosphorylation sites in HIP1. Using the NetPhos 2.0 prediction algorithm, we predicted that positions 135 and 191, near the HIP1 ANTH domain, were most likely to serve as tyrosine kinase substrates.
Further empirical Y-to-F mutagenesis in various combinations of the remaining 13 tyrosine residues (there are 17 residues in total, 4 of which are in the ANTH domain) in HIP1 was then carried out in order to experimentally identify the putative site(s) of HIP1 phosphorylation (Table 1). Guided by the NetPhos 2.0 program, initial Y-to-F mutations were made simultaneously at several of the most likely positions based on this program (Y135F, Y191F, Y468F, Y475F, Y678F, Y862F, and Y1009F) (41). Notably, no mutant with only one Y-to-F mutation displayed decreased tyrosine phosphorylation (Table 1), suggesting that HIP1 phosphorylation occurs at multiple tyrosines.
We narrowed down the necessary tyrosines to those in the Y135,142,191,236F mutant (Fig. 3A, HIP1 4xYF), which was not phosphorylated in the presence of EGFR (Fig. 3B, lane 6 versus lane 4) or EGFRvIII (Fig. 3B, lane 9 versus lane 7). Even when the cells were maximally stimulated with EGF (100 ng/ml for 15 min after 24 h of serum starvation), we were unable to observe significant phosphorylation of this HIP1 Y135,142,191,236F 4xYF mutant (data not shown). In sum, we have identified a patch of tyrosine residues close to the N terminus of HIP1, in addition to the ANTH and USH domains of HIP1 (HIP1ΔANTH and HIP1Δ753-799), that are required for EGFR-mediated phosphorylation. For reference, we have designated this 101-amino-acid motif that contains the four required tyrosine residues the “HIP1 phosphorylation motif” (HPM).
Although the double or triple Y-to-F mutations of the HPM were still phosphorylated, they were less phosphorylated than wild-type HIP1 (Table 1; Fig. 3C, lane 5 versus lane 3). Addition of the Y152F mutation to HIP1 double or triple Y-to-F HPM mutants corrected the diminished EGFR-mediated phosphorylation, indicating that the Y152 residue inhibits the phosphorylation of HIP1 (Fig. 3C, lane 5 versus lane 6 and lane 7 versus lane 8). This raises the possibility that the Y152 residue dampens tyrosine phosphorylation of HIP1 by acting as a pseudosubstrate for RTKs.
HPM tyrosine-to-phenylalanine mutations may convert HIP1 into a proapoptotic protein.
Whether phosphorylation of HIP1 by RTKs is required for HIP1 function is not known, so we considered the use of HPM mutants to probe for a role of phosphorylation in the function of HIP1. First, the ability of the phosphorylation-deficient HIP1 HPM mutant to associate with clathrin was found to be intact; however, it was noted that the steady-state protein level of the HIP1/HPM(4xYF) mutant was significantly lower than that of wild-type HIP1 at 48 h posttransfection (data not shown). This reduced expression of the HIP1 phosphorylation-deficient mutant suggested that the HIP1/HPM(4xYF) mutant may exhibit proapoptotic activity similar to that described previously for the HIP1/ΔANTH mutant (26). Consistent with the low level of the HIP1 phosphorylation-deficient HPM mutant compared to wild-type HIP1, we noted that the “half-lives” of transfected HIP1/HPM(4xYF) and another phosphorylation resistant mutant [HIP1(7xYF)] (Fig. 3A) were indeed shorter than that of transfected wild-type HIP1 (Fig. 3D, lanes 4 to 6 versus lanes 1 to 3, and data not shown).
One alternative explanation to the proapoptotic possibility of a shortened half-life of the HIP1 HPM mutant is that the mutant protein is less stable than the wild type. To determine whether promotion of cell death could explain our observations, we expressed the inert protein GFP in tandem with wild-type HIP1 or HIP1/HPM(4xYF), using a HIP1-ires-GFP or HIP1/HPM(4xYF)-ires-GFP DNA construct, and probed for GFP expression via Western blotting (Fig. 3E, lanes 1 to 3 versus lanes 7 to 9). Like the expression of HIP1/HPM(4xYF) protein, expression of GFP in HIP1/HPM(4xYF)-ires-GFP-transfected cells was significantly lower than that in HIP1-ires-GFP-transfected cells at 72 h and 96 h posttransfection. Since the GFP signal in cells transfected with the wild-type HIP1-ires-GFP construct remained steady at 96 h posttransfection, the decrease in GFP signal at the 96-h time point in concert with the decrease of the HIP1/HPM(4xYF) protein level in HIP1/HPM(4xYF)-transfected cells suggests that there was death of the transfected cells rather than abnormal degradation of GFP caused by coexpression with the HIP1/HPM(4xYF) protein. Coexpression of wild-type HIP1 with the HIP1/HPM(4xYF) mutant was able to partially rescue the lower levels of GFP (Fig. 3E, lanes 10 to 12 versus lanes 7 and 8). This suggests that the HIP1/HPM(4xYF) mutant behaves as a dominant negative mutant that opposes prosurvival activities of wild-type HIP1.
Apoptosis was examined directly in HIP1-transfected cells by using morphological criteria as previously described (35) (Fig. 3F). At 24 h posttransfection, 31% of cells transfected with HIP1/HPM(4xYF) and 38% of cells transfected with HIP1/ΔANTH were apoptotic (Fig. 3G). These frequencies were significantly higher than the baseline apoptotic frequency, which was 13% for wild-type HIP1-transfected cells (Fig. 3G) (P < 0.01 by the Pearson chi-square test). These data suggest that cell death is associated with the HIP1/HPM(4xYF) mutant, which explains, at least in part, the decreased half-life of this mutant and the coexpressed GFP (Fig. 3D and E).
Also, the suggested proapoptotic effects of the HIP1/HPM(4xYF) mutant make it difficult to interpret EGFR half-life experiments. The lower panel of Fig. 3C demonstrates that EGFR levels increased in the presence of wild-type HIP1 (lane 3 versus lane 1). In contrast, this was not the case for the HPM mutants (Fig. 3C, lanes 4 to 7). The decreased levels of EGFR may have been due to possible proapoptotic effects of the HPM mutants.
HIP1r is also phosphorylated in the presence of EGFR and requires the conserved tyrosine residues in the HPM motif for phosphorylation.
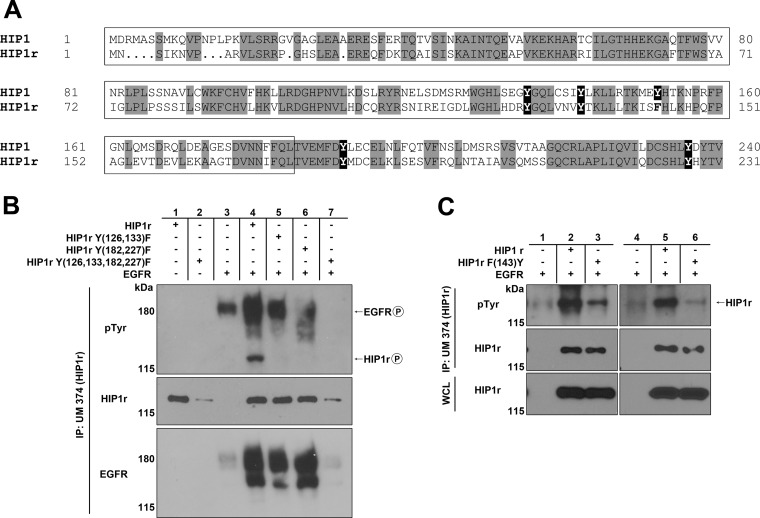
HIP1r is the only known mammalian relative of HIP1, and this protein compensates for HIP1 deficiency in vivo (13). Consistent with this functional and physical overlap, we found that the four key tyrosines of the HIP1 HPM (Y135, Y142, Y191, and Y236) are conserved in HIP1r (Y126, Y133, Y182, and Y227) (Fig. 4A). Note that the tyrosine in the HPM sequence at HIP1 position 152, which is unnecessary for HIP1 phosphorylation and appears to inhibit phosphorylation, is a phenylalanine (homologous position 143) in HIP1r (Fig. 4A). With this amino acid conservation in mind, we examined EGFR-mediated phosphorylation of HIP1r to determine whether the homologous Y-to-F mutants in HIP1r showed reduced phosphorylation. The 2× HIP1r Y-to-F mutants, with mutations in each of the two halves of the HPM, were not phosphorylated at detectable levels. The HIP1r HPM/4xYF mutant, in which tyrosines 126, 133, 182, and 227 were converted to phenylalanine, was consistently expressed at much lower levels (Fig. 4B, lanes 5 to 7 versus lane 4). The reason for this low level of expression is not known. However, even when the amount of extract was increased 10-fold, phosphorylation of the HIP1r 4xYF mutant was not detected. Additionally, when the phenylalanine at position 143 in HIP1r was converted to tyrosine to mimic the inhibitory Y152 residue in HIP1, HIP1r phosphorylation was similarly inhibited (Fig. 4C, lane 3 versus lane 2 and lane 6 versus lane 5). These HIP1r data further support the notion that this tyrosine (position 152 in HIP1 and position 143 in mutated HIP1r) inhibits EGFR-mediated phosphorylation of HIP1 and HIP1r. Whether this tyrosine and its surrounding sequence might inhibit RTK phosphorylation of other substrates is an important question for future research.
Fig 4.
HIP1r requires the HPM for phosphorylation mediated by EGFR. (A) Amino acid alignment of HIP1 and HIP1r N termini. Amino acids in gray are completely conserved. The tyrosines in black are the HPM tyrosines. The tyrosine at position 152 of HIP1 is not conserved in HIP1r, and the amino acid at this position (residue 143) in HIP1r is a phenylalanine. (B and C) HEK293T cells were cotransfected with EGFR-V5 and either wild-type HIP1r or HIP1r Y-to-F point mutants. Immunoprecipitation was performed with anti-HIP1r UM374 polyclonal antibody, and phosphorylation was detected with antiphosphotyrosine monoclonal antibody 4G10. (B) The HIP1r HPM Y-to-F mutants [HIP1r Y(135,142)F and Y(191,236)F] were not phosphorylated by EGFR. (C) The F143Y mutant of HIP1r inhibited phosphorylation of HIP1r. The phosphorylated protein migrating at 120 kDa in the first lane is endogenous HIP1r. Two separate repetitions of these experiments are presented.
The HIP1r/HPM(4xYF) and HIP1r/HPM(2xYF) mutants, which were phosphorylation resistant (Fig. 4B), were also compared to wild-type HIP1r for differences in putative apoptotic activity. We quantitated the number of cells that were morphologically apoptotic at both 12 and 24 h posttransfection. However, unlike the results for HIP1 and HIP1/HPM(4xYF), those for HIP1r and the HIP1r/HPM mutants did not show consistent differences in apoptosis between the wild-type and mutant HIP1r-transfected cells (data not shown). Although the roles of HIP1 and HIP1r in the cell overlap in vivo (14), previous work has shown that, similar to these apoptosis data, HIP1 and HIP1r are not identical, and only HIP1 is able to readily transform cells (23).
HIP1 is phosphorylated in the presence of PDGFβR and its oncogenic derivatives HIP1/PDGFβR (H/P) and TEL/PDGFβR (T/P).
We next asked if HIP1 interacted with and was phosphorylated in the presence of PDGFβR and its oncogenic derivatives. We chose PDGFβR because in addition to the case for EGFR, we have also observed prolongation of the PDGFβR half-life by HIP1 (9). The cDNA for PDGFβR was cotransfected with Myc-tagged HIP1 into 293T cells, and extracts were immunoprecipitated with anti-Myc antibodies. HIP1 phosphorylation mediated by PDGFβR was observed, and it was enhanced by addition of NaVa (Fig. 5A, lane 6 versus lane 5).
Oncogenic derivatives of PDGFβR, such as H/P and T/P, were next tested for the ability to interact with HIP1 and mediate HIP1 phosphorylation. Like EGFRvIII, they exhibited strong phosphorylation of HIP1 (Fig. 5A, top panel, lanes 8 and 10 versus lanes 7 and 9). In contrast to the equivalent phosphorylation of HIP1 mediated by EGFR and EGFRvIII, the ability of H/P and T/P to induce HIP1 phosphorylation was consistently greater than that of wild-type PDGFβR. Having previously found that HIP1 interacts with H/P via the HIP1 region of the fusion oncoprotein (42), and finding in these studies that HIP1 also interacts with the PDGFβR region of the fusion due to its ability to interact with PDGFβR and phosphorylate HIP1 (Fig. 5A, lane 6), we were not surprised by the strong affinity of H/P for HIP1 and the strong phosphorylation of HIP1 by H/P. However, it was an unexpected observation that T/P also induced strong phosphorylation of HIP1 (Fig. 5A, lanes 9 and 10).
Phosphorylation of HIP1 in the presence of PDGFβR but not its oncogenic derivatives requires the HPM tyrosine residues.
We next tested if PDGFβR or its oncogenic derivatives required the HPM tyrosine residues for phosphorylation of HIP1. Although PDGFβR, like EGFR, did not induce phosphorylation of the “HPM” Y-to-F mutants (Fig. 5B, lanes 4 and 5 versus lane 3), to our surprise, the H/P and T/P oncoproteins readily phosphorylated the “HPM” Y-to-F point mutants (Fig. 5B, top panel, lanes 8, 9, 12, and 13). We assayed for HIP1/HPM(4xYF) mutant phosphorylation in the presence of EGFR (n = 17), PDGFβR (n = 6), H/P (n = 5), and T/P (n = 8) several times, in a variety of experiments, and each time we observed HIP1/HPM(4xYF) phosphorylation in the presence of the oncogenes but not the normal RTKs (the difference was statistically significant by two-tailed Fisher's exact chi-square test [P < 0.0001]). The oncogenic translocation products also led to phosphorylation of the HIP1ΔUSH mutant as much as, if not more than, that of wild-type HIP1 (data not shown). In contrast, EGFRvIII was comparable to EGFR in its complete inability to induce phosphorylation of the HIP1/HPM(4xYF) (Fig. 3B, top panel, lane 9) and HIP1ΔUSH (Fig. 3B, top panel, lane 8) mutants.
To determine which tyrosine residues in HIP1 were required for T/P-induced phosphorylation, we generated a tyrosine-free mutant, designated HIP1(17xYF), along with a series of other YF mutants. The HIP1 15xYF mutant, which contains only tyrosine residues 281 and 475, was still phosphorylated, indicating that these residues are required for phosphorylation of HIP1 in the presence of T/P (Fig. 5C, lanes 5 and 6). These data suggest that the specificity of the oncogenic forms of PDGFβR toward HIP1 is altered as a result of the chromosomal translocations. Since the PDGFβR fusion oncoproteins phosphorylate wild-type HIP1 more heavily than normal PDGFβR does (Fig. 5A, lanes 8 and 10 versus lane 6 and lanes 7 and 9 versus lane 5), it is quite possible that tyrosine residues such as residues 281 and 475 are phosphorylated in addition to the HPM tyrosine residues. H/P kinase specificity in these experiments was not analyzed because the additional association of HIP1 with the HIP1 portion of the fusion as well as the PDGFβR portion adds a unique complexity. Using the T/P oncogene for mutagenesis experiments is more generalizable, as HIP1 interacts only with the PDGFβR portion of the fusion. T/P is therefore more representative of the family of PDGFβR fusions that phosphorylate HIP1.
H/P inhibits HIP1 binding to clathrin.
While we did not find significant effects of HIP1 phosphorylation on clathrin binding (data not shown), we noted that when HIP1 was coexpressed with H/P but not with T/P or PDGFβR, there was no detectable HIP1 binding to clathrin in an immunoprecipitation assay for HIP1 (Fig. 5D, lanes 4 and 10). In contrast, clathrin was readily bound by HIP1 in the absence of H/P (Fig. 5D, lanes 6 and 12). When H/P and HIP1 were coexpressed and H/P was instead immunoprecipitated with anti-PDGFβR antibodies to specifically assess clathrin association with H/P rather than HIP1, clathrin was readily detected in the immunoprecipitates (Fig. 5D, lanes 1, 2, 7, and 8 versus lanes 4 and 10). Phosphorylation was not necessary, since treatment with imatinib, a specific inhibitor of H/P, did not affect this pattern of binding (Fig. 5D, lanes 7 to 12 versus lanes 1 to 6). These data suggest that the H/P protein binds clathrin more readily than HIP1 in the same cell, and the more efficient binding may sequester clathrin away from HIP1. An alternative hypothesis is that H/P noncompetitively blocks HIP1 from binding clathrin.
Two isoforms of HIP1 are differentially phosphorylated in the presence of normal RTKs.
In light of the difference in substrate specificity between coexpression with wild-type PDGFβR or its oncogenic fusions, we sought to determine if there were different isoforms of HIP1 that served as better or worse substrates for the RTKs. We surmised that different HIP1 isoforms that have alternative amino termini in the vicinity of the HPM may display altered abilities to be phosphorylated by RTKs. We therefore performed 5′RACE analysis of RNAs isolated from mouse brain and spleen, two tissues that had previously displayed high HIP1 protein levels together with a protein doublet by Western blotting (28, 29). Two transcripts (Hip1a and Hip1b) were identified that had different starting exons, designated exons 1a and 1b (Fig. 6A). The exon 1a-containing transcript (Hip1a) has been used consistently by our laboratory, and the human homologue is the isoform we used for all of the initial phosphorylation studies displayed in Fig. 1 to 5. We used the mouse HIP1 sequences to identify these two forms in human 293T cells by using RT-PCR. Human exon 1b (152 nucleotides) is located ∼99 kb downstream of exon 1a (161 nucleotides), within the 139-kb human intron 1 (Fig. 6A). Using quantitative PCR, we found that the absolute levels of the human HIP1a and HIP1b transcripts in 293T cells were equivalent, with a copy number of ∼80 per ng of total RNA. In both mice and humans, the predicted proteins from both transcripts carry an intact lipid-binding ANTH domain. Furthermore, the N-terminal regions that are different do not contain tyrosine residues (Fig. 6A, HIP1a and HIP1b [see N-terminal sequences and alignment]).
Fig 6.
Identification and phosphorylation of two alternative HIP1 isoforms. (A) Diagram of alternative human HIP1 transcripts and their amino acid sequences, designated hHIP1a and hHIP1b. These isoforms differ only in their initial exon, resulting in alternative N-terminal amino acid sequences. (B to E) HEK293T cells were cotransfected with various RTKs and either HIP1a or HIP1b, both of which were tagged with Myc. Immunoprecipitation was performed with anti-Myc beads, and HIP1 phosphorylation was detected with antiphosphotyrosine antibody 4G10. (B) EGFR phosphorylates HIP1b less than HIP1a in the presence or absence of NaVa. (C) EGFRvIII phosphorylates HIP1b to a slightly lesser degree than that for HIP1a in both the presence and absence of NaVa. (D) PDGFβR phosphorylates both HIP1b and HIP1a. (E) H/P and T/P phosphorylate HIP1a and HIP1b equally in both the presence and absence of NaVa.
To determine whether the two different protein products were phosphorylated by the various RTKs used in this study, we overexpressed Myc-tagged versions of the human HIP1 transcripts with the different RTKs in 293T cells. The human exon 1a HIP1 cDNA expressed a protein that migrated slightly slower than the exon 1b HIP1 protein (Fig. 6B, lane 1 versus lane 2). When the isoforms were coexpressed with EGFR, HIP1b was qualitatively phosphorylated less than HIP1a (Fig. 6B, lane 4 versus lane 5 and lane 9 versus lane 10). This difference was observed in the presence or absence of vanadate. A similar but less striking pattern was observed with PDGFβR, as HIP1b was slightly less phosphorylated by PDGFβR than HIP1a (data not shown). In contrast, EGFRvIII (Fig. 6C, lane 1 versus lane 2 and lane 6 versus lane 7) and the PDGFβR fusions (Fig. 6E, lane 4 versus lane 5, lane 7 versus lane 8, lane 10 versus lane 11, and lane 13 versus lane 14) led to equal phosphorylation of HIP1a and HIP1b in the presence or absence of vanadate. These data indicate that as in the case of phosphorylation of the HPM Y-to-F mutants in the presence of normal and neoplastic RTKs, wild-type RTKs display a narrower substrate specificity than that of oncogenic PDGFβR RTKs.
DISCUSSION
In this report, we determined that HIP1 is tyrosine phosphorylated in the presence of EGFR and PDGFβR and that this modification may modulate HIP1's role in cellular survival. These data collectively support the hypothesis that HIP1 is directly phosphorylated by activated RTKs but do not rule out the possibility that there remains an intermediate tyrosine kinase activated by RTKs that then phosphorylates HIP1. To investigate which of the HIP1 tyrosine residues were phosphorylated, we performed extensive tyrosine-to-phenylalanine point mutagenesis and identified four tyrosine residues that are within and just carboxyl to the ANTH domain (Y135, Y142, Y191, and Y236) as necessary for EGFR phosphorylation of HIP1. In support of the significance of these HPM tyrosines, they were completely conserved in HIP1r, and the homologous HPM mutant was also phosphorylation resistant.
To our surprise, the oncogenic versions of the PDGFβR kinase (T/P and H/P) did not require these four tyrosine residues for phosphorylation. Extensive additional point mutagenesis identified tyrosine residues 281 and 475 of HIP1 as required for phosphorylation by the PDGFβR oncoproteins. These data indicate that either multiple domains are phosphorylated at the same time by the oncoproteins or, upon mutagenesis of those tyrosine residues in the HPM that are “normally” used, there is adaptation by the oncoproteins and “normally” unused tyrosine residues are converted to preferred oncogenic kinase substrate sites.
The identification of the HPM as a sequence that is required specifically for membrane-bound RTK phosphorylation of HIP1 suggests a “locale hypothesis” where membrane-bound RTKs have more access to HIP1 sequences than cytoplasmic RTKs. This increased access allows them to modify more of HIP1. An alternative hypothesis is a “priming hypothesis” where phosphorylation of the tyrosine residues in the HPM of HIP1 may be required before the rest of HIP1 serves as a kinase substrate. This priming may not be required by constitutively active oncogenic kinases. Because the oncogenic version of EGFR, EGFRvIII, remains membrane bound (33–36) and did not phosphorylate the HPM Y-F mutants, in contrast to oncogenic PDGFβR, which is primarily cytoplasmic (15), we prefer the “locale hypothesis” over the “priming hypothesis.”
HIP1 is a member of a growing group of oncoproteins that usurp normal endocytic pathways to transform cells by increasing tumor-promoting signals. By altering clathrin trafficking, aberrant endocytic factors such as HIP1 are thought to simultaneously elevate levels of several growth factor receptors (43). The exact mechanism for this is not known, but since HIP1 specifically binds clathrin, we suggest that HIP1 may serve as a clathrin “sponge” that at very high levels competes with a coated pit for its components. This HIP1 “sponge” could prevent or stall endocytosis of activated growth factor receptors by either preventing formation of the endosome or causing abnormal retention of the endosome clathrin coat. Progression to late endosomes and lysosomes is halted, resulting in delayed signal termination. This is a plausible hypothesis, as the pool of clathrin available for endocytosis can be limiting. For instance, we observed that the clathrin-binding HIP1/PDGFβR oncogene competes with HIP1 for clathrin to such an extent that HIP1 binding to clathrin is abrogated when HIP1/PDGFβR is also expressed (Fig. 5D).
Finally, the roles of the EGFR interaction and phosphorylation of HIP1 in cellular transformation are not clear. Although HIP1 itself has been shown to transform cells in an EGFR-dependent fashion (23), this report is the first to demonstrate HIP1 phosphorylation mediated by EGFR. Tyrosine phosphorylation of HIP1 and HIP1r underscores the likelihood that the observed physical interactions between HIP1 or HIP1r and RTKs are functionally significant. Because HIP1 phosphorylation is lost when cells are treated with an EGFR inhibitor (AG1478) and increased when cells are treated with the protein tyrosine phosphatase inhibitor NaVa (Fig. 1), the tyrosine phosphorylation state is clearly transient and is therefore a possible regulatory signal. Furthermore, EGFR inhibitor treatment of HIP1-transformed cells inhibits their transformed phenotype (23). Whether inhibition of HIP1 phosphorylation prevents HIP1's dynamic location in EGF-stimulated cells is of future interest. Additionally, identification of specific tyrosine phosphatases that use HIP1 and HIP1r as substrates and further analysis of the role of the HPM in HIP1 and HIP1r cellular functions in vivo will shed additional light on the biological purpose of HIP1 phosphorylation.
Conclusions.
We report that HIP1 and HIP1r are phosphorylated by EGFR and PDGFβR. We identified a 101-amino-acid HIP1 phosphorylation motif (HPM) that includes a patch of four tyrosine residues that are conserved between HIP1 and HIP1r and are necessary for normal RTK phosphorylation. In contrast, we found that oncogenic PDGFβR mutants do not require the HPM for phosphorylation of HIP1. Within this conserved HPM motif, we identified a tyrosine unique to HIP1 that inhibits HIP1 phosphorylation. Finally, data from a HIP1 mutant with conversion of the four conserved HPM tyrosines to phenylalanine suggest that it is a proapoptotic mutant, indicating that an intact HPM may be necessary for HIP1 to promote cellular survival and transformation.
ACKNOWLEDGMENTS
We are grateful to Linda Pike and Paul Mischel for providing EGFR plasmids and to Sandra Schmid and members of the Ross lab for their technical assistance and intellectual contributions.
This work was supported by National Cancer Institute grants to T.S.R. (R01 CA82363-03 and R01 CA098730-01) and by a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research. T.S.R. is a Leukemia and Lymphoma Society Scholar.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Kalchman MA, Koide HB, McCutcheon K, Graham RK, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn FC, Wellington C, Metzler M, Goldberg YP, Kanazawa I, Gietz RD, Hayden MR. 1997. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat. Genet. 16:44–53 [DOI] [PubMed] [Google Scholar]
- 2.Wanker EE, Rovira C, Scherzinger E, Hasenbank R, Walter S, Tait D, Colicelli J, Lehrach H. 1997. HIP-I: a huntingtin interacting protein isolated by the yeast two-hybrid system. Hum. Mol. Genet. 6:487–495 [DOI] [PubMed] [Google Scholar]
- 3.Metzler M, Legendre-Guillemin V, Gan L, Chopra V, Kwok A, McPherson PS, Hayden MR. 2001. HIP1 functions in clathrin-mediated endocytosis through binding to clathrin and adaptor protein 2. J. Biol. Chem. 276:39271–39276 [DOI] [PubMed] [Google Scholar]
- 4.Mishra SK, Agostinelli NR, Brett TJ, Mizukami I, Ross TS, Traub LM. 2001. Clathrin- and AP-2-binding sites in HIP1 uncover a general assembly role for endocytic accessory proteins. J. Biol. Chem. 276:46230–46236 [DOI] [PubMed] [Google Scholar]
- 5.Rao DS, Chang JC, Kumar PD, Mizukami I, Smithson GM, Bradley SV, Parlow AF, Ross TS. 2001. Huntingtin interacting protein 1 is a clathrin coat binding protein required for differentiation of late spermatogenic progenitors. Mol. Cell. Biol. 21:7796–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waelter S, Scherzinger E, Hasenbank R, Nordhoff E, Lurz R, Goehler H, Gauss C, Sathasivam K, Bates GP, Lehrach H, Wanker EE. 2001. The huntingtin interacting protein HIP1 is a clathrin and alpha-adaptin-binding protein involved in receptor-mediated endocytosis. Hum. Mol. Genet. 10:1807–1817 [DOI] [PubMed] [Google Scholar]
- 7.Senetar MA, Foster SJ, McCann RO. 2004. Intrasteric inhibition mediates the interaction of the I/LWEQ module proteins Talin1, Talin2, Hip1, and Hip12 with actin. Biochemistry 43:15418–15428 [DOI] [PubMed] [Google Scholar]
- 8.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. 2001. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science 291:1051–1055 [DOI] [PubMed] [Google Scholar]
- 9.Hyun TS, Rao DS, Saint-Dic D, Michael LE, Kumar PD, Bradley SV, Mizukami IF, Oravecz-Wilson KI, Ross TS. 2004. HIP1 and HIP1r stabilize receptor tyrosine kinases and bind 3-phosphoinositides via epsin N-terminal homology domains. J. Biol. Chem. 279:14294–14306 [DOI] [PubMed] [Google Scholar]
- 10.Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. 2001. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291:1047–1051 [DOI] [PubMed] [Google Scholar]
- 11.Metzler M, Li B, Gan L, Georgiou J, Gutekunst CA, Wang Y, Torre E, Devon RS, Oh R, Legendre-Guillemin V, Rich M, Alvarez C, Gertsenstein M, McPherson PS, Nagy A, Wang YT, Roder JC, Raymond LA, Hayden MR. 2003. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 22:3254–3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oravecz-Wilson KI, Kiel MJ, Li L, Rao DS, Saint-Dic D, Kumar PD, Provot MM, Hankenson KD, Reddy VN, Lieberman AP, Morrison SJ, Ross TS. 2004. Huntingtin interacting protein 1 mutations lead to abnormal hematopoiesis, spinal defects and cataracts. Hum. Mol. Genet. 13:851–867 [DOI] [PubMed] [Google Scholar]
- 13.Bradley SV, Hyun TS, Oravecz-Wilson KI, Li L, Waldorff EI, Ermilov AN, Goldstein SA, Zhang CX, Drubin DG, Varela K, Parlow A, Dlugosz AA, Ross TS. 2007. Degenerative phenotypes caused by the combined deficiency of murine HIP1 and HIP1r are rescued by human HIP1. Hum. Mol. Genet. 16:1279–1292 [DOI] [PubMed] [Google Scholar]
- 14.Hyun TS, Li L, Oravecz-Wilson KI, Bradley SV, Provot MM, Munaco AJ, Mizukami IF, Sun H, Ross TS. 2004. Hip1-related mutant mice grow and develop normally but have accelerated spinal abnormalities and dwarfism in the absence of HIP1. Mol. Cell. Biol. 24:4329–4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross TS, Bernard OA, Berger R, Gilliland DG. 1998. Fusion of Huntingtin interacting protein 1 to platelet-derived growth factor beta receptor (PDGFbetaR) in chronic myelomonocytic leukemia with t(5;7)(q33;q11.2). Blood 91:4419–4426 [PubMed] [Google Scholar]
- 16.Grand FH, Burgstaller S, Kuhr T, Baxter EJ, Webersinke G, Thaler J, Chase AJ, Cross NC. 2004. p53-binding protein 1 is fused to the platelet-derived growth factor receptor beta in a patient with a t(5;15)(q33;q22) and an imatinib-responsive eosinophilic myeloproliferative disorder. Cancer Res. 64:7216–7219 [DOI] [PubMed] [Google Scholar]
- 17.Jones AV, Cross NC. 2004. Oncogenic derivatives of platelet-derived growth factor receptors. Cell. Mol. Life Sci. 61:2912–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tefferi A, Vardiman JW. 2008. Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia 22:14–22 [DOI] [PubMed] [Google Scholar]
- 19.Golub TR, Barker GF, Lovett M, Gilliland DG. 1994. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 77:307–316 [DOI] [PubMed] [Google Scholar]
- 20.Carroll M, Tomasson MH, Barker GF, Golub TR, Gilliland DG. 1996. The TEL/platelet-derived growth factor beta receptor (PDGF beta R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF beta R kinase-dependent signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 93:14845–14850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grisolano JL, O'Neal J, Cain J, Tomasson MH. 2003. An activated receptor tyrosine kinase, TEL/PDGFbetaR, cooperates with AML1/ETO to induce acute myeloid leukemia in mice. Proc. Natl. Acad. Sci. U. S. A. 100:9506–9511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oravecz-Wilson KI, Philips ST, Yilmaz OH, Ames HM, Li L, Crawford BD, Gauvin AM, Lucas PC, Sitwala K, Downing JR, Morrison SJ, Ross TS. 2009. Persistence of leukemia-initiating cells in a conditional knockin model of an imatinib-responsive myeloproliferative disorder. Cancer Cell 16:137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao DS, Bradley SV, Kumar PD, Hyun TS, Saint-Dic D, Oravecz-Wilson KI, Kleer CG, Ross TS. 2003. Altered receptor trafficking in Huntingtin interacting protein 1-transformed cells. Cancer Cell 3:471–482 [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Yu W, Cai Y, Ren C, Ittmann MM. 2008. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia 10:847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley SV, Holland EC, Liu GY, Thomas D, Hyun TS, Ross TS. 2007. Huntingtin interacting protein 1 is a novel brain tumor marker that associates with epidermal growth factor receptor. Cancer Res. 67:3609–3615 [DOI] [PubMed] [Google Scholar]
- 26.Rao DS, Hyun TS, Kumar PD, Mizukami IF, Rubin MA, Lucas PC, Sanda MG, Ross TS. 2002. Huntingtin-interacting protein 1 is overexpressed in prostate and colon cancer and is critical for cellular survival. J. Clin. Invest. 110:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. 2011. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J. Clin. Invest. 121:396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradley SV, Smith MR, Hyun TS, Lucas PC, Li L, Antonuk D, Joshi I, Jin F, Ross TS. 2007. Aberrant Huntingtin interacting protein 1 in lymphoid malignancies. Cancer Res. 67:8923–8931 [DOI] [PubMed] [Google Scholar]
- 29.Graves CW, Philips ST, Bradley SV, Oravecz-Wilson KI, Li L, Gauvin A, Ross TS. 2008. Use of a cryptic splice site for the expression of huntingtin interacting protein 1 in select normal and neoplastic tissues. Cancer Res. 68:1064–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Repass SL, Brady RJ, O'Halloran TJ. 2007. Dictyostelium Hip1r contributes to spore shape and requires epsin for phosphorylation and localization. J. Cell Sci. 120:3977–3988 [DOI] [PubMed] [Google Scholar]
- 31.Brady RJ, Damer CK, Heuser JE, O'Halloran TJ. 2010. Regulation of Hip1r by epsin controls the temporal and spatial coupling of actin filaments to clathrin-coated pits. J. Cell Sci. 123:3652–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher JA, Crockett DK, Elenitoba-Johnson KS, Lim MS. 2007. Evaluation of enrichment techniques for mass spectrometry: identification of tyrosine phosphoproteins in cancer cells. J. Mol. Diagn. 9:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grandal MV, Zandi R, Pedersen MW, Willumsen BM, van Deurs B, Poulsen HS. 2007. EGFRvIII escapes down-regulation due to impaired internalization and sorting to lysosomes. Carcinogenesis 28:1408–1417 [DOI] [PubMed] [Google Scholar]
- 34.Hatanpaa KJ, Burma S, Zhao D, Habib AA. 2010. Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 12:675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, Huang CM, Gill GN, Wiley HS, Cavenee WK. 1997. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J. Biol. Chem. 272:2927–2935 [DOI] [PubMed] [Google Scholar]
- 36.Loew S, Schmidt U, Unterberg A, Halatsch ME. 2009. The epidermal growth factor receptor as a therapeutic target in glioblastoma multiforme and other malignant neoplasms. Anticancer Agents Med. Chem. 9:703–715 [DOI] [PubMed] [Google Scholar]
- 37.Parachoniak CA, Park M. 2009. Distinct recruitment of Eps15 via its coiled-coil domain is required for efficient down-regulation of the met receptor tyrosine kinase. J. Biol. Chem. 284:8382–8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepard HM, Brdlik CM, Schreiber H. 2008. Signal integration: a framework for understanding the efficacy of therapeutics targeting the human EGFR family. J. Clin. Invest. 118:3574–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilbur JD, Chen CY, Manalo V, Hwang PK, Fletterick RJ, Brodsky FM. 2008. Actin binding by Hip1 (huntingtin-interacting protein 1) and Hip1R (Hip1-related protein) is regulated by clathrin light chain. J. Biol. Chem. 283:32870–32879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ybe JA, Mishra S, Helms S, Nix J. 2007. Crystal structure at 2.8 A of the DLLRKN-containing coiled-coil domain of huntingtin-interacting protein 1 (HIP1) reveals a surface suitable for clathrin light chain binding. J. Mol. Biol. 367:8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blom N, Gammeltoft S, Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362 [DOI] [PubMed] [Google Scholar]
- 42.Ross TS, Gilliland DG. 1999. Transforming properties of the Huntingtin interacting protein 1/platelet-derived growth factor beta receptor fusion protein. J. Biol. Chem. 274:22328–22336 [DOI] [PubMed] [Google Scholar]
- 43.Mosesson Y, Mills GB, Yarden Y. 2008. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer 8:835–850 [DOI] [PubMed] [Google Scholar]