Abstract
It has previously been shown that the Shewanella putrefaciens W3-18-1 strain produces remarkably high current in microbial fuel cells (MFCs) and can form magnetite at 0°C. To explore the underlying mechanisms, we developed a genetic manipulation method by deleting the restriction-modification system genes of the SGI1 (Salmonella genome island 1)-like prophage and analyzed the key genes involved in bacterial respiration. W3-18-1 has less respiratory flexibility than the well-characterized S. oneidensis MR-1 strain, as it possesses fewer cytochrome c genes and lacks the ability to oxidize sulfite or reduce dimethyl sulfoxide (DMSO) and timethylamine oxide (TMAO). W3-18-1 lacks the hydrogen-producing Fe-only hydrogenase, and the hydrogen-oxidizing Ni-Fe hydrogenase genes were split into two separate clusters. Two periplasmic nitrate reductases (NapDAGHB and NapDABC) were functionally redundant in anaerobic growth of W3-18-1 with nitrate as the electron acceptor, though napDABC was not regulated by Crp. Moreover, nitrate respiration started earlier in W3-18-1 than in MR-1 (with NapDAGHB only) under microoxic conditions. These results indicate that Shewanella putrefaciens W3-18-1 is well adapted to habitats with higher oxygen levels. Taken together, the results of this study provide valuable insights into bacterial genome evolution.
INTRODUCTION
Shewanella strains, most renowned for their dissimilatory metal reduction and potential applications in the bioremediation of heavy metal contamination, have frequently been isolated from redox-stratified environments. Some Shewanella strains, including the best-characterized S. oneidensis MR-1 strain, have been isolated from freshwater environments (1, 2). However, most of the sequenced Shewanella strains were isolated from marine environments and this genus was believed to have a marine origin (3). The dissimilatory metal reduction of Shewanella species has been intensively studied for the potential applications in bioremediation of radioactive waste of groundwater.
Shewanella putrefaciens is a facultative anaerobic Gram-negative psychrophile with relevance to fish spoilage, oil pipeline corrosion, and human infections (4). The type strain of S. putrefaciens, ATCC 8071, was isolated from butter in England during the 1930s. Other strains, W3-18-1, 200, and CN-32, were isolated from marine sediment, a Canadian oil pipeline, and anaerobic shale sandstone, respectively (4). S. putrefaciens strains originally composed a highly heterogeneous phylogenetic group and have been subjected to frequent reclassification (1, 5, 6). A number of novel species, including S. algae, S. baltica, S. frigidimarina, S. pealeana, and S. oneidensis (ATCC 700550T [MR-1]), were initially classified as S. putrefaciens but renamed later. S. putrefaciens and S. algae are also human pathogens associated with septicemia, cellulitis, ear infection, cerebellar abscesses, leg ulcers, osteomyelitis, and arthritis (7).
To date, many Shewanella species have been sequenced, providing a unique opportunity for exploring bacterial genome evolution and adaptation to their specific habitats (4). S. putrefaciens W3-18-1 is particularly interesting because it achieves high current production in microbial fuel cells (8) and reduces metals and forms magnetite at 0°C (9, 10). Here we report an integrated genetics and physiological analysis of the respiration of W3-18-1 in order to provide insights into the underlying mechanisms. Our results indicate that the respiratory flexibility of S. putrefaciens W3-18-1 is lower than that of S. oneidensis MR-1 and that the strain is well adapted to its habitats with higher oxygen levels.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Shewanella putrefaciens W3-18-1 and S. putrefaciens CN-32 were previously isolated from the deep marine sediments underlying the depth of 670 m below the sea level off the Washington state coast (9, 10) and from anaerobic shale sandstone at the depth of 250 m in the Morrison formation of Cerro Negro, New Mexico (4), respectively. Bacterial strains were usually cultured in Luria-Bertani broth (or plates) (supplemented with 15 and 50 μg/ml of gentamicin [Gm] and kanamycin when necessary) and modified M1 minimal medium (1, 11).
Genome sequencing, annotation, and bioinformatics analysis.
The DNA sequencing, assembly, and annotation of the W3-18-1 genome were conducted by Joint Genome Institute (http://genome.jgi-psf.org/she_w/she_w.home.html) (Fig. 1). The orthologs are identified by using bidirectional BLASTP (best hits) for comparisons of W3-18-1 and other Shewanella strains and also on the basis of genome synteny. The paralog(s) of polypeptides was searched by BLAST against the same genome (>70% sequence similarity).
Fig 1.
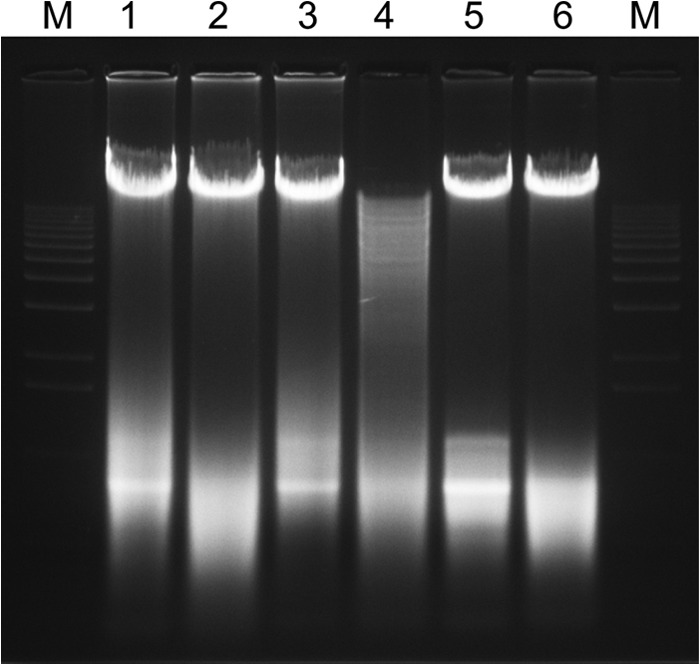
PtsI restriction digestion of the chromosomal DNAs of the wild-type (lane 6), ΔpstI (lane 2), and ΔpstI pstM (lane 4) strains. The DNAs were subjected to electrophoresis on the 1% (wt/vol) agarose gel. M, 1-kb DNA markers. Lanes 1, 3, and 5 are the undigested chromosomal DNAs as the controls.
Electron donor and acceptor utilization assays.
Biolog dye was used to monitor sulfite and hydrogen utilization in modified M1 minimal medium. Electron acceptor utilization was assayed in the modified M1 medium supplemented with 50 mM sodium lactate as the electron donor and carbon source and the electron acceptors to be tested. The bacterial cultures were incubated under anaerobic or microoxic conditions (without shaking to limit aeration). Nitrate and the nitrite concentrations were measured by using a standard colorimetric method (12).
Genetic manipulation.
The two-step protocol of selection (antibiotics resistance for the single crossover) and counterselection (sucrose sensitivity for the double crossover) was applied for in-frame deletion of a specific gene(s) by using suicide vector pDS3.0 (R6K replicon, sacB, Gmr)-based constructs carrying a fusion of upstream and downstream sequences of target genes (see Tables S1 and S2 in the supplemental material) as previously described (13). The suicide vector was introduced into Shewanella by mating using Escherichia coli WM3064 as the donor strain.
RNA extraction and RT-PCR analysis of gene transcription.
Total RNA was extracted by using RNAiso Plus (TaKaRa) and an RNAprep Pure Cell/Bacteria kit (Tiangen Biotech [Beijing] Co., Ltd.), and RNA was further purified using DNase I treatment. The integrity of RNA was evaluated by agarose (0.8%) gel electrophoresis. The RNA concentration and purity were measured on a spectrophotometer (Nanodrop Technologies, Wilmington, DE). To prepare cDNA, 2 μg of total RNA was reverse transcribed (RT) using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa) and a TIANscript RT kit (Tiangen Biotech [Beijing] Co., Ltd.) according to the manufacturer's protocol. The PCR thermal cycles were as follows: 5 min at 95°C for cDNA denaturation followed by 27 to 30 cycles of 40 s at 95°C, 40 s at 51°C, and 30 s at 72°C. A final elongation step was performed for 10 min at 72°C. RT-PCR products were separated on a 0.8% agarose gel containing ethidium bromide and visualized by UV light and Bio-Rad Image software. The data presented represent relative mRNA levels normalized to 16S rRNA transcript levels, and the value of the control was set to 1. All the experiments described were performed in triplicate to obtain means and standard deviations (SD). The PCR products were also sequenced to confirm amplification of target genes. The primers used are listed in Table S2 in the supplemental material.
RESULTS
Establishment of genetic manipulation in strain W3-18-1.
S. putrefaciens W3-18-1 has a genome size of 4,708 kb and harbors a single circular chromosome with a predicted total of 4,237 open reading frames (ORFs). Among them, about 700 genes, many of which encode bacteriophages, lateral flagella, and bacterial microcompartments, are absent in the MR-1 genome (Table 1). Unlike MR-1 and CN-32, W3-18-1 is recalcitrant to genetic manipulations, which may be due to DNA restriction mediated by prophage-borne restriction-modification systems and CRISPR (clustered regularly interspaced short palindromic repeat) elements. Five prophage elements and a few degenerate phages are present in the W3-18-1 genome (Table 1). The phage (Sputw3181_4072-4090) shares many genes with SGI1 (Salmonella genome island 1) of pathogenic Salmonella enterica serovar Typhimurium DT120 (14). We noted that the SGI1 island of W3-18-1 encodes a PstI-like restriction-modification system. Consistently, its chromosomal DNA could not be digested by commercial PstI endonuclease (New England BioLabs), probably due to the methylation of recognition sites (Fig. 2). To facilitate genetic manipulation, we deleted the putative endonuclease (encoded by Sputw3181_4075) and DNA methylase (designated pstM; Sputw3181_4074) genes. The pstM deletion resulted in sucessful digestion of chromosomal DNA of strain W3-18-1ΔpstIΔpstM by PstI, confirming that PstM mediated methylation of DNA.
Table 1.
Specific genetic loci of S. putrefaciens W3-18-1 that are absent in S. oneidensis MR-1
| Locus | Length (kb) | Gene(s) | Predicted function(s) |
|---|---|---|---|
| Sputw3181_0088-0096 | 10.4 | Cytochrome bo oxidase genes cyoABCDE | Electron transfer and energy transduction |
| Sputw3181_0197-0204 | 9.1 | Anion transporter, fumarase, and fumarate reductase genes frdABCD | Nutrient uptake and fumarate reduction |
| Sputw3181_0305-0330 | 27 | Degenerate phage elements, type I restriction-modification system | |
| Sputw3181_0341-0367 | 27.8 | Drugs efflux genes, metal resistance genes | Toxin and heavy metal resistancea |
| Sputw3181_0408-0445 | 82.4 | Bacterial microcompartment operon, including vitamin B12-independent diol dehydratase genes, and phosphotransferase system genes | 1,2-Propanediol utilization and sugar uptake |
| Sputw3181_0454 −0493 | 35.8 | Lateral flagellum operon | Cell motility and colonization |
| Sputw3181_0554-0559 | 8.6 | Tannase/feruloyl esterase, outer member porin, and fumarate reductase-like flavoprotein genes | |
| Sputw3181_0862-0868 | 8.5 | Potassium ion-transporting ATPase operon and two-component regulatory system genes | Osmotic stress response |
| Sputw3181_0872-0873 | 4.1 | Nitric oxide reductase gene norZ and regulatory gene norR | Denitrification and detoxification of nitric oxide |
| Sputw3181_1077-1183 | 108.6 | SXT/R391 ICE (integrating conjugative element)-like prophage inserted into prfC gene | Mobile efflux pumpsa |
| Sputw3181_1380-1395 | 4.3 | Acylneuraminate cytidylyltransferase and sugar nucleotidyltransferase genes | Utilization of amino and nucleotide sugars; lipopolysaccharide O antigen biosynthesis and flagellin glycosylation |
| Sputw3181_1667-1670 | 7.2 | Cobaltochelatase (CobN), siderophore receptor, and tolQ genes | Porphyrin metabolism |
| Sputw3181_1944-1966 | 32.8 | l-Arabinose and polymer utilization operon | Uptake and degradation of L-arabinose and arabinan |
| Sputw3181_2014-2044 | 31.3 | Aldo/keto reductase and glutathione S-transferase genes | —a |
| Sputw3181_2102-2107 | 4.6 | Periplasmic nitrate reductase operon napDABC (nap-α) | Dissimilatory nitrate reduction and anaerobic respiration |
| Sputw3181_2184-2212 | 37.7 | CRISPR elements | Plasmid and phage restriction |
| Sputw3181_2399 | 2.5 | Retron- and RNA-directed DNA polymerase gene | —a |
| Sputw3181_2877-2921 | 37.5 | Phi phage element | |
| Sputw3181_2930-2954 | 24.1 | Mu phage element and arsenate reductase genes | Arsenate resistance |
| Sputw3181_2974-2977 | 6.0 | Aldehyde oxidoreductase, 4Fe-4S ferredoxin, and decaheme cytochrome genes | Carbon source utilization and respiration |
| Sputw3181_3133-3136 | 3.5 | Sodium ion-translocating oxaloacetate decarboxylase genes | Energy transduction |
| Sputw3181_3204-3212 | 7.2 | Mercury resistance operon | Mercury resistance |
| Sputw3181_3245-3248 | 3.6 | Cyanide-insensitive terminal oxidase genes cioAB | Aerobic respiration |
| Sputw3181_3326-3332 | 4.9 | Mlt-interacting MipA family proteins/OmpV, MipA, and two-component system | Osmotic stress response and envelope biogenesis |
| Sputw3181_3435-3437 | 3.0 | Radical S-adenosylmethionine (SAM) domain protein genes | |
| Sputw3181_3472-3473 | 2.6 | NnrS family protein and nitrite reductase NrfA-like cytochrome C552 genes | Nitrite reduction and detoxification |
| Sputw3181_3508-3513 | 9.7 | Tetrathionate reductase and two-component sensory kinase-response regulator genes | Tetrathionate reduction |
| Sputw3181_3902-3909 | 11.8 | Pseudomonas Cup type IV pilus operon | Biofilm formation |
| Sputw3181_3982-3996 | 20.9 | Proline biosynthesis and sodium/proline symporter genes | Proline uptake and osmotic stress response |
| Sputw3181_4067-4090 | 20.7 | Prophage genes, similar to SGI1 element from Salmonella, PstI restriction and modification genes | Modification and restrictiona |
The indicated loci are absent in the Shewanella putrefaciens CN-32 strain.
Fig 2.
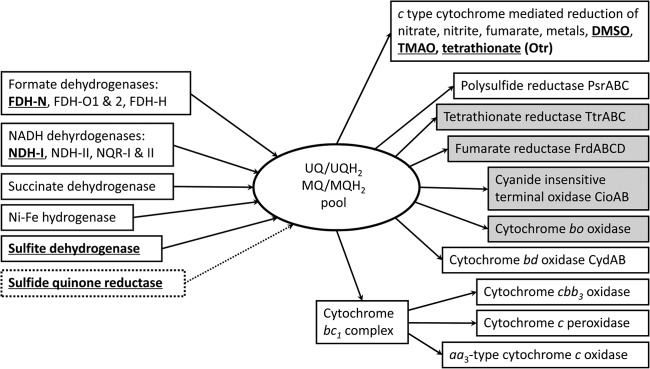
Comparison of respiratory chain components between W3-18-1 and MR-1. The highlighted items are present in MR-1 but absent in W3-18-1. The shaded boxes indicate items present in W3-18-1 but not in MR-1. The broken box indicates that the gene is mutated. UQ/UQH2 and MQ/MQH2 represent ubiquinone/ubiquinol and menaquinone/menaquinol, respectively. See Table S4 in the supplemental material for more details.
There are several transposons and insertion sequences in W3-18-1, most of which are present in multiple copies. The IS4 family insertion sequences (sputw3181_1472, -3732, and -3852) and Tn5 family transposons (sputw3181_1767, -1850, -2145, -2150, and -3364) are present in W3-18-1 but not other sequenced Shewanella strains. The transposon-disrupted genes include ifcA (sputw3181_3363), rtxB (sputw3181_3731), and pilN (sputw3181_3851). The ifcA gene encodes a Fe(III)-induced flavocytochrome c3, while rtxB is part of an operon involved in biofilm and pellicle formation in MR-1 (SO_4317-4321) (15). The pilN gene encodes the pilus assembly protein for the type IV pili (pilM to -Q) involved in cell adhesion and twitching motility of bacteria (16). These genetic differences may have contributed to the phenotypic variations between W3-18-1 and CN-32 revealed by previous studies (8, 17).
Respiratory chains, c-type cytochromes, and respiratory flexibility.
Based on the CXXCH motif and comparative genomic analysis, a total of 32 c-type cytochrome genes were identified in W3-18-1, while 42 and 55 are present in S. oneidensis MR-1 and S. piezotolerans WP3, respectively (18, 19, 20). W3-18-1 does not harbor unique c-type cytochrome genes that are absent in other sequenced Shewanella genomes. Both W3-18-1 and MR-1 could grow anaerobically with fumarate as the electron acceptor (21), but only the S. putrefaciens strains (W3-18-1 and CN-32) contain the four-subunit integral-membrane fumarate reductase complex genes frdABCD (Table 1 and Fig. 2) among the sequenced Shewanella strains. FrdABCD does not contain heme groups or contribute to the proton gradient (22). In addition, W3-18-1 lacks the gene clusters encoding sulfite hydrogenase SorAB, octaheme tetrathionate reductase Otr, timethylamine oxide (TMAO) reductase, dimethyl sulfoxide (DMSO) reductase, and the secondary metal reductase MtrDEF (see Tables S3 and S4 in the supplemental material). A number of in-frame deletion mutants have been generated in W3-18-1 and MR-1 for experimentally testing the cellular functions of c-type cytochromes and other respiratory genes as described below.
Nitrate reduction.
Like several other Shewanella strains, W3-18-1 contains two periplasmic nitrate reductase operons, nap-α (napDABC) and nap-β (napDAGHB), and two nitrite reductase paralog (pentaheme cytochrome NrfA) genes (Table 1). In contrast, S. oneidensis MR-1 harbors only napDAGHB whereas the denitrifier S. denitrificans OS217 contains only napDABC. It has been proposed that nap-α is involved in denitrification and redox balance and nap-β is involved in nitrate ammonifcation in Shewanella (23). To test this hypothesis, we generated the in-frame deletion mutants of nap-β and nap-α operons. Our results showed that both single mutants grew on nitrate at rates similar to that seen with the wild-type strain (Fig. 3). Nonetheless, nitrate reduction was abolished in the double mutant, suggesting that both nap operons were functional in the nitrate reduction in W3-18-1. W3-18-1 harbors the nitric oxide (NO) reductase gene norZ and regulatory gene norR, which may be involved in NO detoxification (21).
Fig 3.
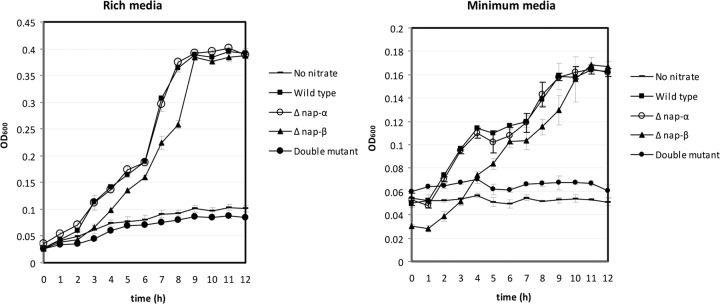
Bacterial growth (optical density at 600 nm [OD600]) of the S. putrefaciens W3-18-1 wild-type strain, nap-α in-frame deletion mutant, nap-β mutant, and double mutant of nap-α and nap-β on nitrate (2 mM) in the rich (1% [wt/vol] tryptone and 0.5% [wt/vol] yeast extract) and modified M1 minimum media supplemented with 50 mM sodium lactate as the electron donor and carbon source. The wild-type strain grown in media without nitrate was used as the control. Error bars represent SD.
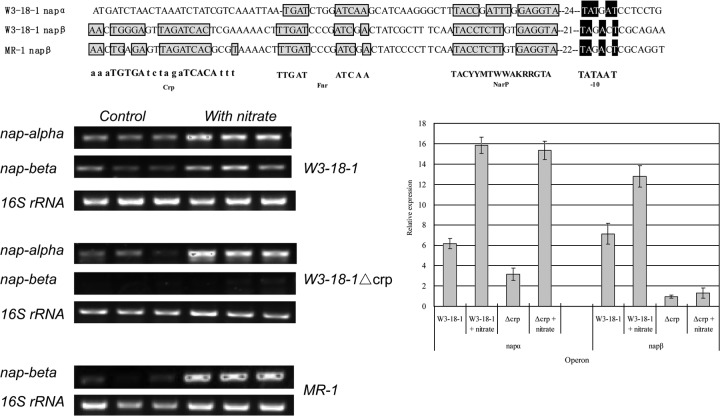
In MR-1, anaerobic respiration is mainly regulated by cyclic AMP (cAMP) and the cAMP receptor protein (Crp), and the crp mutant is deficient in anaerobic growth with, and reduction of, Fe(III), Mn(IV), nitrate, nitrite, fumarate, and DMSO (24). It has been previously noted that the consensus binding sequence of Crp is absent in the upstream region of the nap-α (napDABC) operon in some Shewanella species, though it is present upstream of nap-β (napDAGHB) and nrfA (encoding nitrite reductase) in those species as well as MR-1 (25). This is also the case for W3-18-1 (see Fig. 5). Thus, we monitored the time course of dissimilatory nitrate reduction (represented by the changes in nitrate and nitrite levels in the media) of W3-18-1 and MR-1 under micoaerobic conditions (cultivation without shaking to limit aeration [Fig. 4]). The results showed that nitrate respiration started earlier in W3-18-1 than in MR-1 (with NapDAGHB only). In addition, the level of transcription of napA-α, as indicated by RT-PCR, was still high in the crp null mutant W3-18-1 Δcrp (Fig. 5). Furthermore, the anaerobic growth of the W3-18-1 Δcrp mutant with nitrate as the electron acceptor was not completely abolished, in contrast to what was shown in the MR-1 crp mutant (24), albeit the growth of the Δcrp Δnap-α double mutant was totally abolished (see Fig. S1 in the supplemental material). Together, these results suggested that the expression of the nap-α operon was truly independent of the presence of Crp, allowing for an earlier transcription for nitrate respiration when oxygen supply is limited.
Fig 5.
Expression of napA-α and napA-β of S. putrefaciens W3-18-1, the crp deletion mutant (W3-18-1Δcrp), and S. oneidenesis MR-1 (harboring only the nap-β operon) under microoxic conditions. Bacteria were cultivated in the modified M1 minimal media (8 ml of bacterial culture in the 15-ml culture tubes incubated without shaking for enhanced aeration). The blank represents the used culture media without bacterial inoculation. The presented data represent the samples collected at 6 h of incubation. The three lanes on the left represent the controls without nitrate, and the three on the right represent the treatments supplemented with 2 mM sodium nitrate. Error bars represent SD. (MR-1 sequence alignment adapted from reference 25.)
Fig 4.
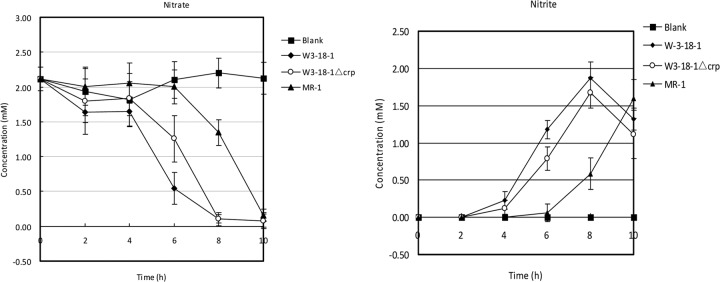
Nitrate reduction of S. putrefaciens W3-18-1, the crp deletion mutant (W3-18-1Δcrp), and S. oneidenesis MR-1 under microoxic conditions. Bacteria were cultivated in the modified M1 minimal media supplemented with 2 mM sodium nitrate (8 ml of bacterial culture in the 15-ml culture tubes incubated without shaking for enhanced aeration). The blank represents the used culture media without bacterial inoculation. Error bars represent SD.
Hydrogen utilization.
W3-18-1 lacks the Fe-only hydrogenase (HydAB) gene cluster that enables S. oneidensis MR-1 to use protons as electron acceptors in the absence of an external electron acceptor, resulting in hydrogen production (26). On the other hand, Ni-Fe hydrogenase is involved in the utilization of hydrogen as the electron donor in S. decolorationis S12 and MR-1 (27, 28). In most sequenced Shewanella strains, Ni-Fe hydrogenase is encoded by a single operon (e.g., SO_2089 to SO_2099 in MR-1), which is split into two separate gene clusters (Sputw3181_1919-1924 and Sputw3181_2173-2178) in W3-18-1. To examine utilization of hydrogen, Biolog dye and a previously described method (27) were used. Dye color changes were observed in both W3-18-1 and MR-1 cultures supplemented with hydrogen via a syringe compared to the results seen with the control experiments performed with nitrogen gas (Table 2) that resulted in only slight color changes, indicative of hydrogen utilization as an electron donor in both strains.
Table 2.
Electron donors and acceptors utilized by Shewanella putrefaciens W3-18-1 and S. oneidensis MR-1 for respirationa
| Chemical | Electron donor or acceptor | S. putrefaciens W3-18-1 | S. oneidensis MR-1 | Enzyme |
|---|---|---|---|---|
| Fumarate | Acceptor | + | + | Fumarate reductase |
| Nitrate | Acceptor | + | + | Nitrate reductase |
| Nitrite | Acceptor | + | + | Nitrite reductase |
| Ferric citrate | Acceptor | + | + | Metal reductase |
| Dimethyl sulfoxide (DMSO) | Acceptor | − | + | DMSO reductase |
| Timethylamine oxide (TMAO) | Acceptor | − | + | TMAO reductase |
| Thiosulfate | Acceptor | + | + | Polysulfide reductase |
| Tetrathionate | Acceptor | + | + | Tetrathionate reductase |
| Hydrogen ion (proton) | Acceptor | − | + | Fe-only hydrogenase |
| Hydrogen gas | Donor | + | + | Ni-Fe hydrogenase |
| Sulfite | Donor | − | + | Sulfite dehydrogenase |
+ and − indicate that utilization of an electron donor or acceptor was and was not observed, respectively.
Deficiency in DMSO and TMAO reduction in W3-18-1.
Most Shewanella strains, including MR-1, can use trimethylamine oxide (TMAO) and dimethyl sulfoxide (DMSO) as electron acceptors under anoxic conditions (29). These species harbor the TMAO reductase gene cluster (SO_1228 to SO_1233 in MR-1), but W3-18-1 lacks the pentaheme c TorC and MtrAD-like decaheme c proteins (SO_1427 and SO_4360, involved in reduction of DMSO). The torF gene (SO_4694) and torECAD are positively coregulated by the transcriptional regulator TorR (30). These genes are absent in W3-18-1 (see Tables S3 and S4 in the supplemental material). Accordingly, W3-18-1 respired neither TMAO nor DMSO in our assays (Table 2).
Oxidation and reduction of sulfur compounds.
MR-1 harbors several c-type cytochrome genes that are probably involved in sulfur compound metabolisms, such as reduction of thiosulfate and/or other sulfur-containing molecules (11). We noted that two types of tetrathionate reductases, tetrathionate reductase (TTR) and octaheme tetrathionate reductase (OTR), exist in the Shewanella strains. W3-18-1 harbors the tetrathionate reductase structural genes ttrA, ttrB, and ttrC as well as ttrS and ttrR encoding the two-component regulatory system (Table 1; see also Table S4 in the supplemental material). The gene products are homologous to TtrRSBCA in Salmonella Typhimurium LT2, which are responsible for respiring tetrathionate (31). On the other hand, MR-1 harbors the octaheme tetrathionate reductase encoded by SO_4144 (32), which is also present in many other Shewanella strains. OTR can also reduce nitrite and hydroxylamine (33). Several Shewanella strains, including S. baltica strains MR-4 and ANA-3, contain both tetrathionate reductases. Both W3-18-1 and MR-1 could grow on tetrathionate (20 mM) under anaerobic condition (Table 2). Our assays showed that the growth of MR-1 was better than that of W3-18-1 under thiosulfate-reducing conditions, probably due to the psrABC gene cluster (34).
W3-18-1 lacks the monoheme cytochrome c genes of SO_0714, SO_0716, and SO_0717 in MR-1. SO_0715 and SO_0716 encode the periplasmic sulfite dehydrogenase subunits SorA and SorB, respectively. SorAB could oxidize sulfite (SO32−) and transfer the electrons to cytochrome c552, which could be oxidized by the terminal cytochrome c oxidase. To experimentally test the oxidation of sulfite (2 to 4 mM), Biolog dye was used to monitor aerobic respiration by MR-1 and W3-18-1 in the minimal medium supplemented by sulfite. Respiration (dye color change) was observed in MR-1 but not in W3-18-1 (Table 2), which was indicative of active sulfite oxidation and/or detoxification by MR-1. In addition, W3-18-1 was more sensitive to higher (>16 mM) levels of sulfite than MR-1 in the modified M1 minimum medium (supplemented with 50 mM pyruvate as the carbon source) and LB broth. These results indicated that MR-1 could utilize sulfite as an electron donor under aerobic condition.
Dissimilatory metal reduction.
Previous studies showed that W3-18-1 exhibited a lower capability of metal oxide reduction than several other Shewanella strains (8). The metal-reductase-containing locus is highly diverse among the sequenced Shewanella genomes (4). W3-18-1 lacks the mtrFED genes encoding the secondary metal reductase as shown in S. oneidensis MR-1. Furthermore, the allele of omcA1 of MR-1 (SO_1779, decaheme c) encodes an 11-heme cytochrome (designated UndA) in W3-18-1 and S. baltica OS223 (4). In addition, W3-18-1 lacks the orthologs for cytochromes encoded by SO_1413 (split tetraheme flavocytochrome), SO_1427 (decaheme c), SO_2930 (diheme c), SO_2931 (diheme c), SO_3300 (split tetraheme flavocytochrome), SO_4360 (decaheme c), SO_3623 (split tetraheme flavocytochrome), SO_4570 (monoheme c), and SO_4572 (triheme c) in MR-1 (see Table S3 in the supplemental material). Moreover, W3-18-1 grew only with Fe(III), Mn(IV), and Se(IV) whereas S. putrefaciens CN-32 could grow with all of six metal ions and metalloids tested, including Co(III), Cr(VI), and As(V) (8). We noted that similar sets of the c-type cytochromes are present in both W3-18-1 and CN-32 strains whereas the sputw3181_3363 (ifcA) locus is disrupted by a transposon (sputw3181_3364) in W3-18-1 as aforementioned. The ifcA gene encodes the ferric ion-induced flavocytochrome c3 (Ifc3), a soluble periplasmic low-potential heme c cytochrome involved in Fe(III) respiration and modulation of fumarate reduction rates in S. frigidimarina NCIMB 400 (35, 36). The ΔmtrD, ΔmtrF, and ΔifcA1 mutants of MR-1 had an even greater capacity to reduce the solid-phase iron, a combination of geothite, hematite, and nanoparticles of hydrous ferric oxide (37). Moreover, deletion of mtrD, mtrF, ifcA, SO_0714, sorB, SO_0717, SO_1427 (dmsC), SO_2930 (diheme c), SO_3300, and otr resulted in higher current density in microbial fuel cells (MFC) (37).
The NapC/NirT family tetraheme CymA is required for Fe(III), fumarate, nitrate, and arsenate respiration (38, 39) and is responsible for electron transfer from the quinol pool to periplasmic and outer-membrane-bound reductases. We generated an in-frame cymA deletion mutant in W3-18-1 and compared the reduction of ferric citrate of this mutant to that of the wild-type strain. The results showed that deletion of cymA led to a significant decrease in the rate of reduction of ferric citrate (Table 2), suggesting CymA plays a role similar to that of its ortholog in MR-1 and Shewanella sp. ANA-3 strains.
DISCUSSION
The respiratory chains of Shewanella strains are highly diversified and could utilize a series of electron acceptors for respiration (Fig. 2). S. putrefaciens W3-18-1 has less respiratory flexibility than S. oneidensis MR-1, as it possesses fewer cytochrome c genes, resulting in the inability to reduce DMSO and TMAO or oxidize sulfite. In most Shewanella strains, there are two functionally redundant dissimilatory periplasmic nitrate reductases encoded by nap-α and nap-β operons; OS217 harbors only the former and MR-1 encodes the latter (23). Our results demonstrated that these seemingly redundant respiratory-chain components may be differentially expressed and function under the different environmental conditions, particularly with respect to oxygen levels and availability of other electron acceptors. These subtly different gene clusters may have enhanced the ecological fitness of Shewanella strains (23).
Genomics analyses also revealed other characteristics of respiratory chains. W3-18-1 and MR-1 share three terminal oxidases, i.e., cbb3-type cytochrome c oxidase (CcoNOQP, a proton-pumping heme-copper oxidase), cytochrome c oxidase COX (complex IV, proton pump), and cytochrome d ubiquinol oxidase CydAB (38, 40). There are two alternative quinol terminal oxidase operons, cyoABCDE and cioAB (41, 42), present in W3-18-1 but absent in MR-1 (Fig. 2; see also Table S4 in the supplemental material). W3-18-1 harbors more terminal oxidases and fewer c-type cytochromes and terminal reductases than MR-1. Moreover, W3-18-1 lacks the anaerobic nitrate-inducible formate dehydrogenase FDH-N, the hydrogen-producing Fe-only hydrogenase, and sulfite dehydrogenase, which are present in MR-1 and many marine Shewanella strains. W3-18-1 does not produce hydrogen in the absence of the Fe-only hydrogenase. These observations suggest that W3-18-1 may also have evolved in and is well adapted to environments with relatively high oxygen tensions. The lower anaerobic respiratory flexibility that resulted from the absence or mutation of respiratory genes, including mtrDEF, ifcA1, and Fe-only hydrogenase genes, may confer the phenotypes of higher current production of W3-18-1 in MFC (17) and its inability to reduce Co(III), Cr(VI), and As(V) (8). S. putrefaciens W3-18-1 is not only suitable for MFC, which could be utilized for electricity generation and wastewater treatment, but also useful for the in situ bioremediation of marine and brackish water environments with higher oxygen tensions and at lower temperatures.
Formate dehydrogenase and NADH dehydrogenase catalyze oxidation of formate and NADH, donating electrons to respiratory chains. The formate dehydrogenase fdh-O and NADH dehydrogenase nqr gene clusters appear to have been duplicated in most Shewanella strains (see Table S4 in the supplemental material). The RNF electron transport complex was thought to be a sodium-translocating NADH dehydrogenase. Only the denitrifier S. denitrificans OS217 harbors a single NQR (NqrABCDEF-2) and lacks many other respiratory chain components such as formate dehydrogenase and hydrogenase. A notable feature of MR-1 is the acquisition of NADH dehydrogenase I operon (ndh-I), composed of nuoA to -N, which is unique to MR-1 among Shewanella strains. This proton-pumping NADH:ubiquinone oxidoreductase (a minimal form of mitochondrion respiratory complex I) may be energetically more efficient under certain conditions such as neutral pH. The ndh-I gene clusters obviously arose from the lateral gene transfer. Such genome divergence could provide valuable insights into bacterial genome evolution and adaptation to their specific niches.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by U.S. Department of Energy (DOE) grant DE-FG02-07ER64383 to J.Z. D.Q. and H.W. were partly supported by the Chinese Academy of Science Grant Y15103-1-401 to D.Q. This work was also supported by ENIGMA under contract DE-AC02-05CH11231 by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy.
Footnotes
Published ahead of print 28 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00619-13.
REFERENCES
- 1.Myers CR, Nealson KH. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319–1321 [DOI] [PubMed] [Google Scholar]
- 2.Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, MacGregor BJ, Ringelberg DB, White DC, Nishijima M, Sano H, Burghardt J, Stackebrandt E, Nealson KH. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705–724 [DOI] [PubMed] [Google Scholar]
- 3.Hau HH, Gralnick JA. 2007. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61:237–258 [DOI] [PubMed] [Google Scholar]
- 4.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JL, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6:592–603 [DOI] [PubMed] [Google Scholar]
- 5.Vogel BF, Jorgensen K, Christensen H, Olsen JE, Gram L. 1997. Differentiation of Shewanella putrefaciens and Shewanella alga on the basis of whole-cell protein profiles, ribotyping, phenotypic characterization, and 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 63:2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziemke F, Höfle MG, Lalucat J, Rosselló-Mora R. 1998. Reclassification of Shewanella putrefaciens Owen's genomic group II as Shewanella baltica sp. nov. Int. J. Syst. Bacteriol. 48:179–186 [DOI] [PubMed] [Google Scholar]
- 7.Khashe S, Janda JM. 1998. Biochemical and pathogenic properties of Shewanella alga and Shewanella putrefaciens. J. Clin. Microbiol. 36:783–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konstantinidis KT, Serres MH, Romine MF, Rodrigues JL, Auchtung J, McCue LA, Lipton MS, Obraztsova A, Giometti CS, Nealson KH, Fredrickson JK, Tiedje JM. 2009. Comparative systems biology across an evolutionary gradient within the Shewanella genus. Proc. Natl. Acad. Sci. U. S. A. 106:15909–15914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray AE, Lies D, Li G, Nealson K, Zhou J, Tiedje JM. 2001. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl. Acad. Sci. U. S. A. 98:9853–9859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stapleton RD, Jr, Sabree ZL, Palumbo AV, Moyer CL, Devol AH, Roh Y, Zhou J. 2005. Metal reduction at cold temperatures by Shewanella isolates from various marine environments. Aquat. Microb. Ecol. 38:81–91 [Google Scholar]
- 11.Beliaev AS, Klingeman DM, Klappenbach JA, Wu L, Romine MF, Tiedje JM, Nealson KH, Fredrickson JK, Zhou J. 2005. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187:7138–7145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Environmental Protection Agency of China 2002. Water and wastewater monitoring methods, 4th ed Chinese Environmental Science Publishing House, Beijing, China [Google Scholar]
- 13.Wan X, VerBerkmoes NC, McCue LA, Stanek D, Connelly H, Hauser LJ, Wu L, Liu X, Yan T, Leaphart A, Hettich RL, Zhou J, Thompson DK. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 186:8385–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, Imberechts H, Mulvey MR. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Windt W, Gao H, Krömer W, Van Damme P, Dick J, Mast J, Boon N, Zhou J, Verstraete W. AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology 152:721–729 [DOI] [PubMed] [Google Scholar]
- 16.Martin PR, Watson AA, McCaul TF, Mattick JS. 1995. Characterization of a five-gene cluster required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 16:497–508 [DOI] [PubMed] [Google Scholar]
- 17.Bretschger O, Cheung ACM, Mansfeld F, Nealson KH. 2010. Comparative microbial fuel cell evaluations of Shewanella spp. Electoanalysis 22:883–894 [Google Scholar]
- 18.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118–1123 [DOI] [PubMed] [Google Scholar]
- 19.Meyer TE, Tsapin AI, Vandenberghe I, de Smet L, Frishman D, Nealson KH, Cusanovich MA, van Beeumen JJ. 2004. Identification of 42 possible cytochrome C genes in the Shewanella oneidensis genome and characterization of six soluble cytochromes. OMICS 8:57–77 [DOI] [PubMed] [Google Scholar]
- 20.Wang F, Wang J, Jian H, Zhang B, Li S, Wang F, Zeng X, Gao L, Bartlett DH, Yu J, Hu S, Xiao X. 2008. Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One 3:e1937. 10.1371/journal.pone.0001937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsapin AI, Burbaev DS, Nealson KH, Keppen OI. 1995. Investigations of succinate dehydrogenase and fumarate reductase in whole cells of Shewanella putrefaciens (strains MR-1 and MR-7) using electron spin resonance spectroscopy. J. Appl. Magn. Res. 9:509–516 [Google Scholar]
- 22.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. 1999. Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284:1961–1966 [DOI] [PubMed] [Google Scholar]
- 23.Simpson PJL, Richardson DJ, Codd R. 2010. The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiology 156:302–312 [DOI] [PubMed] [Google Scholar]
- 24.Saffarini DA, Schultz R, Beliaev A. 2003. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 185:3668–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart V, Bledsoe PJ, Chen L, Cai A. 2009. Catabolite repression control of napF (periplasmic nitrate reductase) operon expression in Escherichia coli K-12. J. Bacteriol. 191:996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meshulam-Simon G, Behrens S, Choo AD, Spormann AM. 2007. Hydrogen metabolism in Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 73:1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong YG, Guo J, Sun GP. 2008. Identification of an uptake hydrogenase for hydrogen-dependent dissimilatory azoreduction by Shewanella decolorationis S12. Appl. Microbiol. Biotechnol. 80:517–524 [DOI] [PubMed] [Google Scholar]
- 28.Marshall MJ, Plymale AE, Kennedy DW, Shi L, Wang Z, Reed SB, Dohnalkova AC, Simonson CJ, Liu C, Saffarini DA, Romine MF, Zachara JM, Beliaev AS, Fredrickson JK. 2008. Hydrogenase- and outer membrane c-type cytochrome-facilitated reduction of technetium(VII) by Shewanella oneidensis MR-1. Environ. Microbiol. 10:125–136 [DOI] [PubMed] [Google Scholar]
- 29.Gralnick JA, Vali H, Lies DP, Newman DK. 2006. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U. S. A. 103:4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bordi C, Ansaldi M, Gon S, Jourlin-Castelli C, Iobbi-Nivol C, Mejéan V. 2004. Genes regulated by TorR, the trimethylamine oxide response regulator of Shewanella oneidensis. J. Bacteriol. 186:4502–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32:275–287 [DOI] [PubMed] [Google Scholar]
- 32.Mowat CG, Rothery E, Miles CS, McIver L, Doherty MK, Drewette K, Taylor P, Walkinshaw MD, Chapman SK, Reid GA. 2004. Octaheme tetrathinonate reductase is a respiratory enzyme with novel heme ligation. Nat. Struct. Mol. Biol. 11:1023–1024 [DOI] [PubMed] [Google Scholar]
- 33.Atkinson SJ, Mowat CG, Reid GA, Chapman SK. 2007. An octaheme c-type cytochrome from Shewanella oneidensis can reduce nitrite and hydroxylamine. FEBS Lett. 581:3805–3808 [DOI] [PubMed] [Google Scholar]
- 34.Burns JL, DiChristina TJ. 2009. Anaerobic respiration of elemental sulfur and thiosulfate by Shewanella oneidensis MR-1 requires psrA, a homolog of the phsA gene of Salmonella enterica serovar Typhimurium LT2. Appl. Environ. Microbiol. 75:5209–5217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobbin PS, Butt JN, Powell AK, Reid GA, Richardson DJ. 1999. Characterization of a flavocytochrome that is induced during the anaerobic respiration of Fe3+ by Shewanella frigidimarina NCIMB400. Biochem. J. 342:439–448 [PMC free article] [PubMed] [Google Scholar]
- 36.Bamford V, Dobbin PS, Richardson DJ, Hemmings AM. 1999. Open conformation of a flavocytochrome c3 fumarate reductase. Nat. Struct. Biol. 6:1104–1107 [DOI] [PubMed] [Google Scholar]
- 37.Bretschger O, Obraztsova A, Sturm CA, Chang IS, Gorby YA, Reed SB, Culley DE, Reardon CL, Barua S, Romine MF, Zhou J, Beliaev AS, Bouhenni R, Saffarini D, Mansfeld F, Kim B, Fredrickson JK, Nealson KH. 2007. Current production and metal oxide reduction by Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 73:7003–7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers CR, Myers JM. 1997. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179:1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy JN, Saltikov CW. 2007. The cymA gene, encoding a tetraheme c-type cytochrome, is required for arsenate respiration in Shewanella species. J. Bacteriol. 189:2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Oost J, de Noer APN, de Gier JL, Zumft WG, Stouthamer AH, van Spanning RJM. 1994. The heme-copper oxidase family consists of three distinct types of terminal oxidases and is related to nitric oxide reductase. FEMS Microbiol. Lett. 121:1–9 [DOI] [PubMed] [Google Scholar]
- 41.Cotter PA, Chepuri V, Gennis RB, Gunsalus RP. 1990. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172:6333–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cunningham L, Pitt M, Williams HD. 1997. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24:579–591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.