Abstract
The aim of this longitudinal study was to determine and compare the prevalences and genotypic profiles of antimicrobial-resistant (AR) Salmonella isolates from pigs reared in antimicrobial-free (ABF) and conventional production systems at farm, at slaughter, and in their environment. We collected 2,889 pig fecal and 2,122 environmental (feed, water, soil, lagoon, truck, and floor swabs) samples from 10 conventional and eight ABF longitudinal cohorts at different stages of production (farrowing, nursery, finishing) and slaughter (postevisceration, postchill, and mesenteric lymph nodes [MLN]). In addition, we collected 1,363 carcass swabs and 205 lairage and truck samples at slaughter. A total of 1,090 Salmonella isolates were recovered from the samples; these were isolated with a significantly higher prevalence in conventionally reared pigs (4.0%; n = 66) and their environment (11.7%; n = 156) than in ABF pigs (0.2%; n = 2) and their environment (0.6%; n = 5) (P < 0.001). Salmonella was isolated from all stages at slaughter, including the postchill step, in the two production systems. Salmonella prevalence was significantly higher in MLN extracted from conventional carcasses than those extracted from ABF carcasses (P < 0.001). We identified a total of 24 different serotypes, with Salmonella enterica serovar Typhimurium, Salmonella enterica serovar Anatum, Salmonella enterica serovar Infantis, and Salmonella enterica serovar Derby being predominant. The highest frequencies of antimicrobial resistance (AR) were exhibited to tetracycline (71%), sulfisoxazole (42%), and streptomycin (17%). Multidrug resistance (resistance to ≥3 antimicrobials; MDR) was detected in 27% (n = 254) of the Salmonella isolates from the conventional system. Our study reports a low prevalence of Salmonella in both production systems in pigs on farms, while a higher prevalence was detected among the carcasses at slaughter. The dynamics of Salmonella prevalence in pigs and carcasses were reciprocated in the farm and slaughter environment, clearly indicating an exchange of this pathogen between the pigs and their surroundings. Furthermore, the phenotypic and genotypic fingerprint profile results underscore the potential role played by environmental factors in dissemination of AR Salmonella to pigs.
INTRODUCTION
Salmonella is a major bacterial food-borne pathogen causing infection in both humans and animals (1). In the United States, Salmonella is responsible for the highest number of food-borne related illnesses, with a reported 1.4 million illnesses, 15,000 hospitalizations, and deaths of more than 500 people each year (2). It is important to note that the actual incidence of salmonellosis is estimated to be 38 times the number of reported cases (3). Emergence and persistence of antimicrobial-resistant (AR) food-borne pathogens due to routine use of antimicrobials for therapeutic, preventive, and growth purposes in conventional swine production are a major public health concern (4). Multidrug-resistant (MDR) Salmonella strains, exhibiting resistance to third-generation cephalosporins, have been reported in commercial pigs (4–7). With growing consumer concerns over AR in bacterial pathogens from commercial swine that are given antimicrobials for prophylaxis and treatment, the demand for antimicrobial-free (ABF) and organic products has increased over the past decade (8). However, previous studies have highlighted the occurrence of MDR Salmonella in both ABF and organic food animal production despite the apparent absence of antimicrobial selection pressure (7, 9, 10).
The prevalence of Salmonella in swine farms in the United States ranges from 1.4 to 33% (11–13). North Carolina is the second-largest pork-producing state in the United States next to Iowa, with a 14.4% contribution to the national inventory (14). Studies in swine production systems have been conducted in North Carolina to report Salmonella prevalence (6, 7, 9). Interestingly, a higher Salmonella prevalence (16 to 29%) has been reported in the swine farm environment than in fecal samples within the same farm (12). Salmonella prevalence at the processing plant ranges from 0% to 77% (15, 16). In these studies, higher prevalence of Salmonella at slaughter was believed to be due to transportation stress, cross-contamination, and the hygienic condition of the slaughter facility. Phenotypic and genotypic analyses have shown that the environment and preslaughter handling, such as transport and lairage, play a significant role in the dissemination of this pathogen in pigs (15, 17–19). It is quite evident that the environment plays a crucial role as a reservoir in transmission of AR pathogens to pigs all along the production chain, either directly or indirectly (6, 12, 17).
Genotyping of Salmonella using pulsed-field gel electrophoresis (PFGE) has been found to be effective in epidemiological studies for identifying different environmental factors as important in contributing to the dynamics of pathogen transmission (6, 14, 20). However, there is a paucity of information regarding the role of the environment in dissemination of AR Salmonella at farm versus slaughter in ABF and conventional systems. To the authors' knowledge, no longitudinal study has been conducted along the entire production chain from farrowing to slaughter to compare the prevalence, antimicrobial resistance, and genotypic diversity of Salmonella among swine reared in ABF and conventional production systems and their environment. The objectives of this study were to (i) determine Salmonella prevalence and serotype distribution in swine and their environment in two distinct swine production systems at farm and slaughter, (ii) compare the AR profiles of isolates from swine and their environment, and (iii) evaluate the genotypic diversity and/or similarity among Salmonella isolates from swine and their environment along the production chain.
MATERIALS AND METHODS
Study design and sample source.
In this longitudinal study design, a total of eight cohorts of ABF and 10 cohorts of conventionally raised pigs were sampled in eight ABF and 30 conventional farms in North Carolina. In the conventional systems, pig cohorts flowed through 3 different farms at different stages of production (i.e., 10 cohorts times 3 farms per cohort = 30 farms). The conventional farms belonged to two different large-scale companies, while the ABF farms were owned by individual swine producers. In the conventional system, pigs were reared indoors and followed an all-in-all-out (AIAO) production system. The purpose of an AIAO system is to reduce disease transmission from one growth stage to another (21), and in order to do so, the pigs were grouped together based on age, weight, and production stage and moved from one location to another at the end of each stage of production (i.e., farrowing, nursery, and finishing stages). Trucks were used to transport pigs from one farm to the next in line. Trucks that ferried pigs were washed and cleaned before they arrived on a farm to load pigs. In the ABF production system, pigs were housed outdoors on agricultural land and had access to the ambient environment. Pigs under the ABF production system were given nonpelleted feed, while the conventionally reared pigs were provided the pelleted form.
All the stages of the pig life cycle under the ABF production system were at the same location but involved rotation to different pastures. The conventionally raised pigs were given antimicrobials for growth, prophylaxis, and therapeutic purposes, whereas ABF pigs raised to slaughter age were not given antimicrobials for any purpose; that is, in keeping with ethical standards, any ABF pig requiring treatment with antimicrobials for bacterial infection was provided such care and subsequently removed from the herd.
Sample size was calculated based on type I (α = 0.05) and type II (β = 0.20) allowable errors, and it was estimated that 27 to 35 pigs needed to be sampled to detect a statistically significant difference in the proportion of Salmonella-positive pigs in the two production systems. We purposely selected healthy pigs at the farrowing farm with the aim of sampling the same cohort of 35 pigs at slaughter.
Sampling on farms.
During each sequential visit, samples were collected from the ABF and conventional pig cohorts and their environment. Sampling was carried out from October 2008 to December 2010 at various stages of production, including once at farrowing (7 to 10 days old), twice each at the nursery (4 and 7 weeks of age) and finishing (16 and 26 weeks of age) stages, and finally once at slaughter. During the farrowing stage, a cohort of 35 healthy piglets per farm (4 piglets/sow) was selected and ear tagged for identification; subsequently, sampling followed the same cohort of pigs at different sampling stages during farm and slaughter stages. Fresh fecal samples (10 g) were collected from piglets using sterile fecal loops (Webster Veterinary, Devens, MA) and from their respective sows using sterile gloves to aid in the determination of the transmission of Salmonella from sows to piglets at birth. Similarly, fecal samples were collected from the ear-tagged pigs twice at each of the nursery and finishing stages using gloved hands. Environmental samples were also collected at every stage of sampling to determine the role played by the environment as a reservoir and in the transmission of Salmonella to/from and among the pigs. Environmental sampling at ABF and conventional farms consisted of five samples each of water, feed, soil, and barn floor swabs. All of the ABF farm environmental samples were collected outdoors, whereas the conventional environmental samples were collected indoors, except soil samples, which were collected from outside the barns. In addition to these environmental samples, lagoon (repository of wastewater draining from the barns) and interfarm truck samples were collected only at conventional farms. Since trucks form an integral part of the pig environment, we sampled the four corners and the center of the truck floor by swabs presoaked with buffered peptone water (BPW; Difco, Becton, Dickinson, Sparks, MD). Similarly, the barn floor swab samples from conventional farms and the inside of hoop structures in ABF farms were collected. Overall, we collected a total of 2,889 fecal (ABF, 1,239; conventional, 1,650), 450 feed (ABF, 200; conventional, 250), 450 floor (ABF, 200; conventional, 250), 449 soil (ABF, 199; conventional, 250), 448 water (ABF, 198; conventional, 250), 245 lagoon (only conventional), and 80 interfarm truck (only conventional) samples from eight ABF and 30 (representing 10 cohorts of pigs) conventional farms and their environment. Samples were transported to the laboratory on ice at 4°C and processed immediately upon arrival.
Sampling at slaughter facilities.
Conventional pigs were transported to a large-scale slaughter plant (9,000 pigs/day) which had a blast-chilling facility (−30°C) to quickly freeze the carcasses. The ABF pigs were transported to two smaller-scale slaughter plants (250 pigs/day), each of which had an overnight chilling facility (4°C) to freeze the carcasses. These smaller-scale plants slaughtered only ABF pigs. At slaughter, we collected carcass swabs from the same cohort of pigs at two stages, specifically, the postevisceration and postchilling stages. At the postevisceration stage, we collected samples of mesenteric lymph node (MLN) from the pigs. Carcass swab samples were collected by wiping at three different positions (jowls, belly, and ham) on each carcass using the USDA-recommended method (22). Environmental samples from the floor of the truck transporting the pigs to the processing plant and lairage floor samples were collected and processed for Salmonella isolation. A total of 455 MLN (ABF, 184; conventional, 271), 454 postevisceration carcass swab (ABF, 182; conventional, 272), 454 postchill carcass swab (ABF, 199; conventional, 255), 130 lairage floor swab (ABF, 80; conventional, 50), and 75 truck floor swab (ABF, 35; conventional, 40) samples were collected. Samples were transported to the laboratory on ice and processed within 3 h of collection.
Salmonella isolation and confirmation.
Isolation and confirmation were performed as previously described (7, 9). Briefly, the samples were preenriched by adding 90 ml of buffered peptone water (BPW) (Difco, Becton, Dickinson) to cups containing either fecal or environment samples, whereas 30 ml of BPW was added to each bag containing either MLN or carcass swabs. Before the MLN was cut into small pieces with a sterilized blade, the outside surface was cleaned with alcohol and flamed to avoid cross-contamination. Preenriched samples were mixed thoroughly and incubated at 37°C for 24 h. After incubation, 100 μl of preenriched BPW suspension from each sample was transferred to 9.9 ml of Rappaport-Vassiliadis (RV) broth (Difco, Becton, Dickinson) and incubated at 42°C for 24 h. A loopful (10 μl) of enriched RV suspension was streaked onto a xylose lactose Tergitol (XLT4) selective agar plate (Difco, Becton, Dickinson) and incubated at 37°C for 24 h. To determine the phenotypic and genotypic diversity of Salmonella within a positive sample, we selected three black-colored colonies from XLT4 and characterized them biochemically by stabbing into triple sugar iron (TSI) and urea agar slants (Difco, Becton, Dickinson). Biochemical testing was interpreted from the TSI and urea agar slants; colonies with positive TSI and negative urea tests were confirmed as Salmonella isolates. Further confirmation of Salmonella was performed by PCR amplification of a targeted Salmonella-specific invasive (invA) gene (23). The confirmed Salmonella isolates were appropriately labeled and stored in brucella broth (Difco, Becton, Dickinson) at −80°C for further characterization.
Salmonella serotyping.
All Salmonella isolates (n = 1,090) were serotyped using one of three methods. Initially, a multiplex PCR was performed to scan the entire isolate set to identify Salmonella enterica serovar Typhimurium using published primers and protocols (24, 25). The template DNA for this multiplex PCR was purified using a Qiagen DNeasy blood and tissue kit (Qiagen, Germany) according to the manufacturer's protocol. In the second approach, pulsed-field gel electrophoresis (PFGE) fingerprint profiles were generated for a subset of 86 isolates, and the serotypes were identified by matching their fingerprint profiles with a database of previously confirmed Salmonella serotypes (26–28). The remaining isolates (n = 684) were sent to the National Veterinary Services Laboratories (NVSL) for traditional phenotypic serotyping.
Antimicrobial susceptibility testing.
All the confirmed Salmonella isolates (n = 1,090) from pigs and the environment were tested against a panel of 15 antimicrobials by the broth microdilution method (Trek Diagnostic Systems, Inc., Cleveland, OH). The panel of antimicrobials tested, along with their respective concentration ranges increasing 2-fold, included amikacin (AMI; 0.5 to 64 μg/ml), ampicillin (AMP; 1 to 32 μg/ml), amoxicillin-clavulanic acid (AUG; 0.5 to 32/16 μg/ml), ceftriaxone (AXO; 0.25 to 64 μg/ml), cefoxitin (FOX; 0.5 to 32 μg/ml), ceftiofur (TIO; 0.25 to 8 μg/ml), chloramphenicol (CHL; 2 to 32 μg/ml), ciprofloxacin (CIP; 0.015 to 2 μg/ml), gentamicin (GEN; 0.25 to 16 μg/ml), kanamycin (KAN; 8 to 64 μg/ml), nalidixic acid (NAL; 0.5 to 32 μg/ml), sulfisoxazole (FIS; 16 to 256 μg/ml), streptomycin (STR; 32 to 64 μg/ml), trimethoprim-sulfamethoxazole (SXT; 0.12/2.38 to 4/76 μg/ml), and tetracycline (TET; 4 to 32 μg/ml). Briefly, 10 μl of bacterial culture (adjusted to a 0.5 McFarland standard) was transferred to 11 ml of Mueller-Hinton broth. Using the Sensititre semiautomated system (Trek Diagnostic Systems, Inc., Cleveland, OH), 50 μl of Mueller-Hinton broth was distributed to each well in a 96-well Sensititre CMV1AGNF plate (Trek Diagnostic Systems, Inc., Cleveland, OH). The plates were sealed and incubated at 37°C for 24 h. The Escherichia coli ATCC 25922 strain was used for quality control. The MICs were recorded, and breakpoints were determined based on Clinical and Laboratory Standards Institute (CLSI) recommendations where available (29). National Antimicrobial Resistance Monitoring System (NARMS) consensus breakpoints were used where CLSI breakpoints were indeterminate (5). Those isolates exhibiting resistance to three or more antimicrobials were considered multidrug resistant (MDR).
PFGE analysis.
A total of 340 Salmonella isolates representing different sampling stages, types of samples, serotypes, and AR profiles (see Fig. 4) from pigs (ABF, 36; conventional, 100) and environment (ABF, 32; conventional, 172) were genotyped by PFGE using the CDC's PulseNet protocol (30). Briefly, Salmonella isolates were grown overnight on LB agar plates. The culture cells were added to cell suspension buffer (CSB), and the concentration was adjusted between optical densities (OD) of 0.48 to 0.52 using a Dade MicroScan turbidity meter. The OD-adjusted bacterial cell suspension (400 μl) was lysed using proteinase K (20 mg/ml), and intact genomic DNA was digested with 50 U of XbaI (Roche) restriction enzyme in agarose-embedded plugs. The restriction fragments were separated by electrophoresis in 0.5× Tris-borate-EDTA (TBE) buffer and 1% ultrapure agarose (SeaKem gold agarose; Lonza, ME) for 18 h at 14°C in a PFGE CHEFF DR III (Bio-Rad) using pulse times of 2.2 to 63.8 s. The XbaI-digested Salmonella enterica serovar Braenderup H9812 strain was used as the reference DNA marker. Gels were stained with ethidium bromide (10 mg/ml) for 30 min in 400 ml of reagent grade water, followed by two washings with NanoPure water, and photographed under UV light. The PFGE images were analyzed by BioNumerics software version 6.1 (Applied Maths, Belgium). Clonal relationships among these isolates were determined using the unweighted-pair group method using average linkages (UPGMA), with band position tolerance and optimization of 1.5% each.
Fig 4.

Dendrogram showing genotypic similarity among Salmonella isolates from ABF systems at various stages of production. aAM, amikacin; AMP, ampicillin; AUG, amoxicillin-clavulanic acid; AXO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; FIS, sulfisoxazole; FOX, cefoxitin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; TIO, ceftiofur. bA2 and A3, ABF cohort.
Statistical analysis.
Statistical analysis was carried out using STATA version 12.1 (Stata Corp., College Station, TX). Descriptive analysis comparing farm types (ABF versus conventional) consisting of unique longitudinal cohorts (which were nested within each farm type) was carried out before pursuing multivariable analyses. Each farm type, along with the stage of production and type of sample collected for isolation of Salmonella, was also considered for descriptive analysis before forcing these variables for multivariable modeling. Contingency table analyses without adjustments for clustering by cohort/farm type were carried out using likelihood ratio (LR) χ2 test statistics for each of the variable types and used to examine their association with Salmonella prevalence. The LR χ2 test for Salmonella prevalence was also carried out for the source of sampling and stages of production. Separate multivariable analyses for pigs versus their environment were carried out using the logistic regression procedure (XTLOGIT) with either random effects (RE) or generalized estimating equation (GEE) models. The XTLOGIT procedure was used instead of XTMELOGIT (multilevel hierarchical logistic regression) because of problems achieving convergence in XTMELOGIT given the high numbers of zero cells in the ABF farm type. The main effects of farm type, stage of production, and sample type, along with their 2-way and 3-way interaction terms, were tested. The final full factorial RE or GEE model was constructed for both main effects and their interaction terms. The final significant model (all variable sets, P < 0.05) was selected based on the associations of these variables and their interaction terms with the prevalence of Salmonella. The same procedure was repeated by forcing cohorts (farm types) for robust variance estimation and compared. From the final model, marginal predictions were obtained for the proportion of positive Salmonella isolates, and these were estimated with 95% confidence intervals. The marginal means were plotted using final predictions from the full factorial RE and GEE models for (i) Salmonella prevalence among pigs by farm type and different stages of production, (ii) Salmonella prevalence in environmental samples by farm type and different stages of production, and (iii) Salmonella prevalence in environmental samples by farm type and different stages of production accounting for sample type differences.
RESULTS
Salmonella prevalence in pigs and the environment at farms.
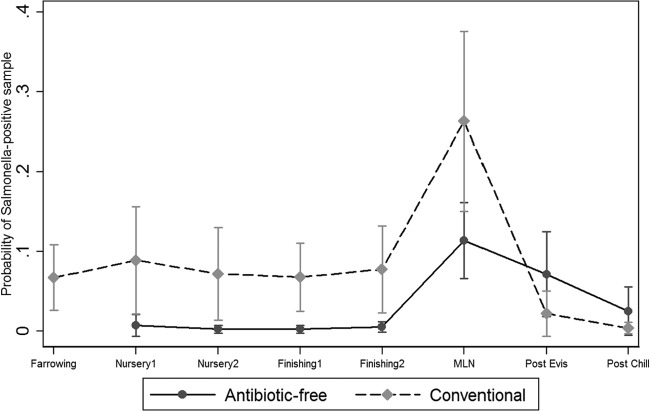
A total of 1,090 Salmonella isolates were isolated from all the samples collected in the study population. The overall proportion of samples that were positive for Salmonella was higher in the conventional production system, both in pigs (66/1,650, 4%) and the environment (156/1,325, 11.7%), than in ABF pigs (2/1,239, 0.2%) and the environment (5/797, 0.6%). The multivariable analysis using logistic regression generated the final significant model (all variables, P < 0.05), which was selected based on the associations of these variables and their interaction terms with the estimated prevalence of Salmonella and plotted along with 95% confidence intervals (Fig. 1, 2, and 3). The breakdown of Salmonella isolates from pigs and the environment samples by farm, type of farm, and stage of production are highlighted in Tables 1 and 2, respectively. Overall, there were statistically significant differences (P < 0.05) between the proportions of samples positive for Salmonella in ABF (2/1,239, 0.2%) and conventional (66/1,650, 4%) pigs at the following different sampling stages: farrowing (ABF, 0%; conventional, 6.7%), nursery 1 (ABF, 0.7%; conventional, 8.8%,), nursery 2 (ABF, 0.2%; conventional, 7.2%), finishing 1 (ABF, 0.2%; conventional, 6.7%), and finishing 2 (ABF, 0.5%; conventional, 7.7%) (Fig. 1).
Fig 1.
Salmonella prevalence among pigs at farm and slaughter. MLN, mesenteric lymph node.
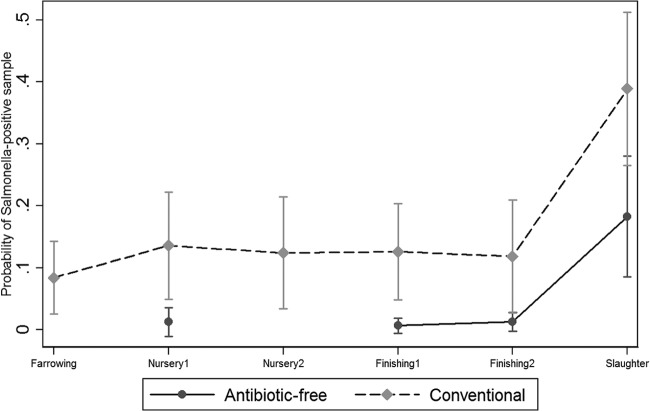
Fig 2.
Salmonella prevalence in the environment at farm and slaughter.
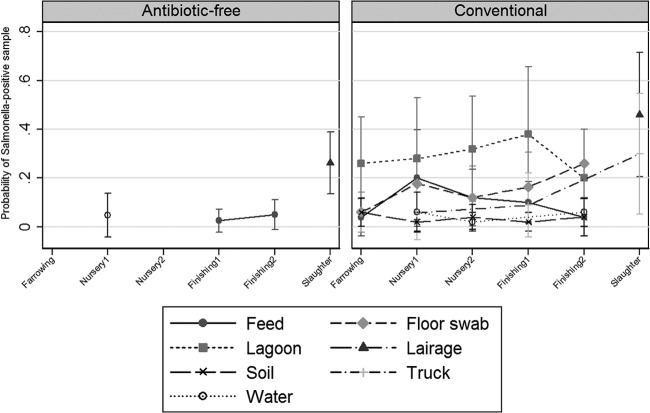
Fig 3.
Salmonella prevalence among environmental samples at farm and slaughter.
Table 1.
Breakdown of Salmonella isolates by farm and farm type
| Farma | Farm type | Cohortb | Frequencyc (n) | % positive |
|---|---|---|---|---|
| ABF 1 | ABF | A1 | 394 | 5.9 |
| ABF 2 | ABF | A2 | 333 | 5.1 |
| ABF 3 | ABF | A3 | 341 | 5.2 |
| ABF 4 | ABF | A4 | 342 | 5.2 |
| ABF 5 | ABF | A5 | 366 | 5.6 |
| ABF 6 | ABF | A6 | 344 | 5.2 |
| ABF 7 | ABF | A7 | 365 | 5.6 |
| ABF 8 | ABF | A8 | 232 | 3.5 |
| CONV 1d | Conventional | C1 | 388 | 5.9 |
| CONV 2d | Conventional | C2 | 362 | 5.5 |
| CONV 3 | Conventional | C3 | 121 | 1.84 |
| CONV 4 | Conventional | C3 | 171 | 2.6 |
| CONV 5 | Conventional | C3 | 72 | 1 |
| CONV 6 | Conventional | C4 | 195 | 2.9 |
| CONV 7 | Conventional | C4 | 119 | 1.8 |
| CONV 8 | Conventional | C4 | 67 | 1 |
| CONV 9 | Conventional | C5 | 69 | 1.1 |
| CONV 10 | Conventional | C5 | 70 | 1.1 |
| CONV 11 | Conventional | C5 | 286 | 4.4 |
| CONV 12 | Conventional | C6 | 69 | 1.1 |
| CONV 13 | Conventional | C6 | 67 | 1 |
| CONV 14 | Conventional | C6 | 239 | 3.6 |
| CONV 15 | Conventional | C7 | 37 | 1 |
| CONV 16 | Conventional | C7 | 127 | 1.9 |
| CONV 17 | Conventional | C7 | 215 | 3.3 |
| CONV 18d | Conventional | C8 | 210 | 3.2 |
| CONV 19d | Conventional | C8 | 185 | 2.8 |
| CONV 20 | Conventional | C9 | 194 | 2.9 |
| CONV 21 | Conventional | C9 | 67 | 1 |
| CONV 22 | Conventional | C9 | 122 | 1.9 |
| CONV 23d | Conventional | C10 | 183 | 2.8 |
| CONV 24d | Conventional | C10 | 197 | 3 |
ABF 1 to 8, antimicrobial-free farms owned by individual farmers; CONV 1 to 24, conventional farms owned by two different companies.
A1 to A8, ABF cohorts; C1 to C10, conventional cohorts.
Number of samples collected. Includes fecal, environmental, and slaughter samples.
The total number of conventional farms sampled was 30. The indicated farms were sampled at three different locations; since the name of the farm is the same, the locations appear as a single farm. Therefore, there are only 24 farms listed in the table.
Table 2.
Breakdown of Salmonella isolates by production/processing stage from ABF and conventional production systems
| Production stagea | Frequencyb (n) | % positive |
|---|---|---|
| Farrowing | 1,112 | 16.9 |
| Nursery 1 | 1,026 | 15.6 |
| Nursery 2 | 974 | 14.8 |
| Finishing 1 | 986 | 14.9 |
| Finishing 2 | 913 | 13.8 |
| Slaughter | 672 | 10.21 |
| Postevisceration | 454 | 6.9 |
| Postchilling | 454 | 6.7 |
Farrowing includes fecal samples from sows and piglets and environmental samples; nursery 1 and 2 and finishing 1 and 2 include fecal and environmental samples; slaughter includes mesenteric lymph node, lairage, and truck swabs.
Total number of samples collected from pigs and the environment.
The overall Salmonella prevalence in the environmental samples on conventional farms (156/1,325, 11.7%) was higher than that in the ABF farms (5/797, 0.62%). At both the farrowing and nursery 1 stages, all the environmental samples from the ABF production system were negative for Salmonella (Fig. 2). Overall, the marginal mean predictions for Salmonella in the conventional farm environment were significantly higher than those in the ABF farm environment at nursery 1 (ABF, 1.2%; conventional, 13.5%), finishing 1 (ABF, 0.6%; conventional, 12.5%), and finishing 2 (ABF, 1.2%; conventional, 11.8%). On the conventional farms, Salmonella was successfully recovered from water, soil, feed, floor swabs, lagoons, and truck samples. Among all the environmental samples, the Salmonella mean prediction was higher in lagoons than in other environmental samples (Fig. 3); on the other hand, on ABF farms, only water (nursery 1, 4.8%) and feed (finishing 1, 0.6%; finishing 2, 1.2%) samples were positive for Salmonella (Fig. 3).
Salmonella prevalence in carcasses and the environment at slaughter.
Overall, the proportion of positive samples for Salmonella was significantly higher in MLN from conventional carcasses than in MLN from ABF carcasses (P < 0.001). However, the prevalence of Salmonella in postevisceration and postchill carcass swabs was higher in ABF carcasses than conventional carcasses. There was a statistically significant difference between the postevisceration swabs (P = 0.008) of ABF carcasses and conventional carcasses. The marginal prediction for Salmonella was highest in MLN (ABF, 11.3%; conventional, 26.3%), postevisceration swabs (ABF, 7.1%; conventional, 2.2%), and postchill swabs (ABF, 2.5%; conventional, 0.39%) (Fig. 1). Salmonella isolates were also obtained from the slaughter environment. Overall, the prevalence of Salmonella was higher in the conventional slaughter environment (38.8%) than in ABF environmental samples (18.6%), with the highest marginal predictions in lairage (ABF, 26.2%; conventional, 46%) and conventional truck (30%) samples. On the other hand, no ABF truck samples tested positive for Salmonella.
Identification and distribution of Salmonella serotypes.
Three different methods were used to identify the different serotypes. S. Typhimurium (n = 320) isolates were identified by using multiplex PCR, and other serotypes were identified by fingerprint profile matching and by traditional phenotypic serotyping at NSVL. We identified 24 Salmonella serotypes among the ABF and conventional pigs and the environment at farm and slaughter (Table 3). The ABF and conventional production systems had certain unique Salmonella serotypes which were unevenly distributed in each of the respective production systems at farm and slaughter. Certain serotypes, including Salmonella enterica serovar Anatum, Salmonella enterica serovar Infantis, and S. Typhimurium, were isolated from both production systems. The predominant Salmonella serotypes in the ABF system on the farm were S. Anatum (pigs, 60%; environment, 21.4%), Salmonella enterica serovar Give (pigs, 40%; environment, 42.8%), and S. Typhimurium (pigs, 0; environment, 21.4%). At slaughter, S. Anatum (carcass, 10.4%; environment, 28.5%) and S. Infantis (carcass, 39.5%; environment, 60.3%) were the predominant serotypes. S. Give was identified only in the ABF system at farm and slaughter. At ABF slaughter, we identified specific serotypes which were not found at the farm level, such as S. Infantis, S. Braenderup, Salmonella enterica serovar Derby, Salmonella enterica serovar Inverness, Salmonella enterica serovar Muenchen, Salmonella enterica serovar Newport, and Salmonella enterica serovar London (Table 3). The conventional system had a greater variety of serotypes at farm and slaughter (Table 3). On the farm, the major serotypes identified were S. Typhimurium (pigs, 28.5% of isolates; environment, 35% of isolates), S. Infantis (pigs, 16.4%; environment, 13.8%), S. Anatum (pigs, 15.8%; environment, 12%), and Salmonella enterica serovar Rissen (pigs, 3%; environment, 8.8%). The S. Rissen serotype was reported for the first time in pigs in the United States. At conventional slaughter, the major serotypes were S. Typhimurium (carcasses, 37%; environment, 30%), S. Derby (carcasses, 35.5%; environment, 1%), and S. Infantis (carcasses, 6.5%; environment, 48.4%).
Table 3.
Distribution of Salmonella serotypes from pigs and the environmental samples at farm and slaughtera
| Serotype identified | No. (%) of each type of isolate with each serotype |
|||||||
|---|---|---|---|---|---|---|---|---|
| Farm |
Slaughter |
|||||||
| ABF |
Conventional |
ABF |
Conventional |
|||||
| Pigs (n = 5) | Environment (n = 14) | Pigs (n = 189) | Environment (n = 439) | Carcasses (n = 86) | Environment (n = 63) | Carcasses (n = 197) | Environment (n = 97) | |
| S. Agona | 0 | 0 | 0 | 0 | 1 (1.1) | 0 | 1 (0.5) | 0 |
| S. Anatum | 3 (60) | 3 (21.4) | 30 (15.8) | 53 (12) | 9 (10.4) | 18 (28.5) | 6 (3) | 15 (15.4) |
| S. Braenderup | 0 | 0 | 0 | 1 (0.2) | 3 (3.4) | 0 | 0 | 0 |
| Salmonella enterica serovar Cerro | 0 | 0 | 12 (6.3) | 3 (0.6) | 0 | 0 | 0 | 0 |
| S. Derby | 0 | 0 | 4 (2.1) | 31 (7) | 5 (5.8) | 0 | 70 (35.5) | 1 (1) |
| S. Give | 2 (40) | 6 (42.8) | 0 | 0 | 3 (3.4) | 0 | 0 | 0 |
| S. Heidelberg | 0 | 0 | 0 | 17 (3.8) | 0 | 0 | 0 | 0 |
| S. Infantis | 0 | 0 | 31 (16.4) | 61 (13.8) | 34 (39.5) | 38 (60.3) | 13 (6.5) | 47 (48.4) |
| S. Inverness | 0 | 0 | 0 | 2 (0.4) | 13 (15.1) | 0 | 0 | 0 |
| Salmonella enterica serovar Johannesburg | 0 | 0 | 0 | 1 (0.2) | 0 | 0 | 5 (2.5) | 0 |
| S. London | 0 | 0 | 0 | 3 (0.6) | 0 | 3 (4.7) | 0 | 2 (2) |
| Salmonella enterica serovar Mbandaka | 0 | 0 | 2 (1) | 2 (0.4) | 0 | 0 | 3 (1.5) | 1 (1) |
| S. Muenchen | 0 | 0 | 3 (1.5) | 0 | 11 (12.7) | 0 | 0 | 0 |
| S. Newport | 0 | 0 | 0 | 1 (0.2) | 3 (3.4) | 0 | 0 | 0 |
| S. Ohio | 0 | 0 | 20 (10.5) | 24 (5.4) | 0 | 0 | 6 (3) | 1 (1) |
| S. Ouakam | 0 | 0 | 23 (12.1) | 41 (9.3) | 0 | 0 | 8 (4) | 0 |
| S. Rissen | 0 | 0 | 6 (3.1) | 39 (8.8) | 0 | 0 | 1 (0.5) | 0 |
| Salmonella enterica serovar Rough_O:r:1,5 | 0 | 0 | 0 | 0 | 0 | 1 (1.5) | 9 (4.5) | 0 |
| Salmonella enterica serovar Schwarzengrund | 0 | 0 | 0 | 2 (0.4) | 0 | 0 | 0 | 0 |
| Salmonella enterica serovar Senftenberg | 0 | 0 | 4 (2.1) | 2 (0.4) | 0 | 0 | 1 (0.5) | 0 |
| S. Typhimurium | 0 | 3 (21.4) | 54 (28.5) | 154 (35) | 1 (1.1) | 3 (4.7) | 73 (37) | 30 (30.1) |
| S. Typhimurium Var 5 | 0 | 2 (14.2) | 0 | 0 | 0 | 0 | 1 (0.5) | 0 |
| 6,7, nonmotile | 0 | 0 | 0 | 2 (0.4) | 0 | 0 | 0 | 0 |
| III_44:z4,z32:- | 0 | 0 | 0 | 0 | 3 (3.4) | 0 | 0 | 0 |
n = 1,090.
Antimicrobial resistance profile of Salmonella.
The overall MIC distribution and prevalence of AR Salmonella isolates from pigs and their environment at different stages on farm and at slaughter are represented in Table 4. A total of 1,090 Salmonella isolates were tested (ABF, n = 168; conventional, n = 922) against a panel of 15 antimicrobials. AR was higher in conventional isolates (resistant, 80%; pansusceptible, 20%) than ABF isolates (resistant, 27%; pansusceptible, 73%). Overall, Salmonella isolates exhibited a wide spectrum of AR, with the highest frequency of resistance to TET (70.6%), followed by FIS (41.6%) and STR (17.3%). In addition, Salmonella isolates exhibited resistance to β-lactams, including cephalosporins, with the highest frequency of resistance to AMP (12.1%), FOX (4.4%), and AXO and TIO (4% each). All the isolates from both production systems were susceptible to AMI and CIP. A “squashtogram” was generated both to illustrate and to compare resistance and MIC distributions of Salmonella isolates from pigs and the environment in the conventional production system (Table 5). Salmonella isolates from pigs and the environment exhibited similar AR profiles and MIC distributions for predominant antimicrobials, with the exception of TET. Most of the environmental isolates which were resistant to TET had a MIC of either 16 μg/ml (0.4%) or 32 μg/ml (78%); on the other hand, the isolates from pigs had MICs of 32 μg/ml (1%) and >32 μg/ml (79%). We observed the highest frequency of resistance in conventional isolates to TET (pigs, 80.3%; environment, 78.3%), followed by FIS (pigs, 56%; environment, 43.4%), STR (pigs, 27.7%; environment, 14.5%), and AMP (pigs, 13.9%; environment, 14.1%).
Table 4.
MIC distribution (squashtogram) of Salmonella isolates from all swine samplesa
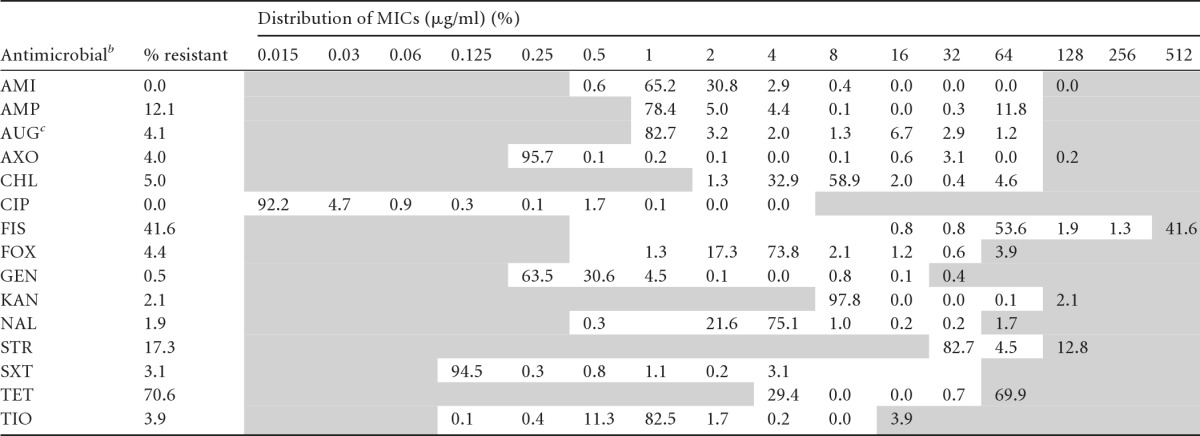
n = 1,090. Areas with white backgrounds indicate the range of dilutions tested for each antimicrobial. Shaded areas fall outside the range of tested concentrations. The vertical bars indicate the CLSI or NARMS consensus breakpoints for resistance (R versus I and S combined). Numbers in the shaded areas on the right indicate the percentages of isolates with undetermined MICs known to be greater than the highest concentrations measured on the broth microdilution plates.
AMI, amikacin; AMP, ampicillin; AUG, amoxicillin-clavulanic acid; AXO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; FIS, sulfisoxazole; FOX, cefoxitin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; TIO, ceftiofur.
The MIC represents the first antibiotic (of two).
Table 5.
Comparison of resistance and the MIC distribution (squashtogram) for Salmonella isolates from the conventional production system at farm and slaughtera
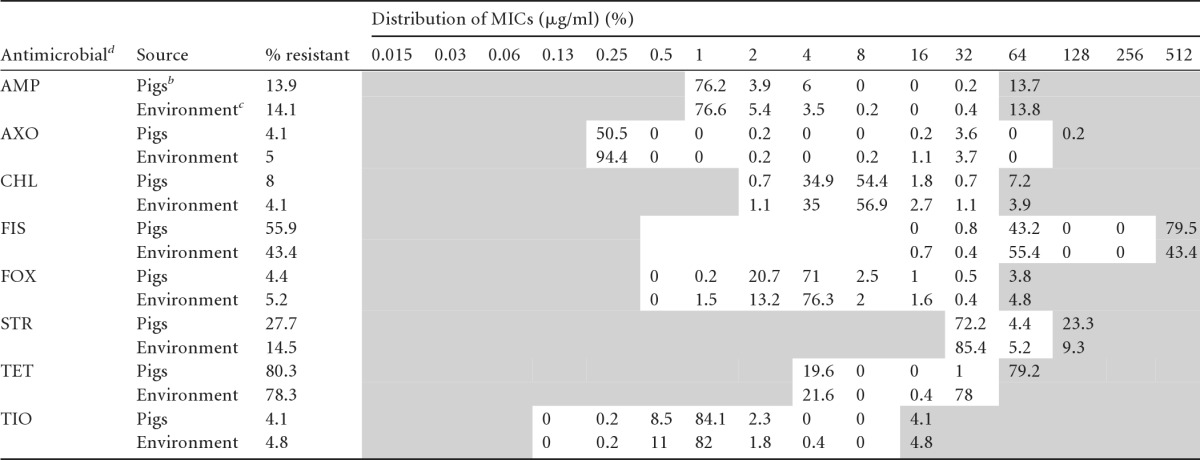
Pigs, n = 386; environment, t n = 536. Areas with white backgrounds indicate the range of dilutions tested for each antimicrobial. Shaded areas fall outside the range of tested concentrations. The vertical bars indicate the CLSI or NARMS consensus breakpoints for resistance (R versus I and S combined). Numbers in the right-side shaded areas indicate the percentages of isolates with undetermined MICs known to be greater than the highest concentrations measured on the broth microdilution plates.
Pigs includes isolates from conventional pig fecal samples (n = 189) at the farm and carcass samples (n = 197) at slaughter.
Environment includes isolates from conventional farm (n = 439) and slaughter (n = 97) environments.
AMI, amikacin; AMP, ampicillin; AXO, ceftriaxone; CHL, chloramphenicol; FIS, sulfisoxazole; FOX, cefoxitin; STR, streptomycin; TET, tetracycline; TIO, ceftiofur.
Distribution and association of MDR patterns with Salmonella serotypes.
We observed a higher frequency of MDR isolates from the conventional system at various stages of production. Conventional isolates had different MDR patterns (27.5%, 254/922) associated with various serotypes. The most common MDR patterns, associated serotypes, and distributions are presented in Table 6. FIS STR TET (n = 72) was the predominant MDR pattern that we found on farms (pigs, 3%; environment, 21%) and at slaughter (carcasses, 75%; environment, 1%), associated with the serotype S. Derby. Two major MDR patterns were associated with S. Anatum, namely, AMP AUG AXO FOX TIO TET (n = 25), which was found only at the farm level (pigs, 44%; environment, 56%), and AMP AUG AXO FOX TIO (n = 5), which was identified only with farm (40%) and slaughter (60%) environmental isolates. S. Typhimurium was associated with five major MDR patterns, with the most common MDR pattern, AMP CHL FIS STR TET (n = 41), a pentaresistant pattern common to S. Typhimurium DT104, found at both farm (pig, 12%; environment, 41%) and slaughter (carcasses, 39%; environment, 7%). FIS SXT TET (n = 25) was observed in farm and slaughter environments (32%) and carcasses (36%). FIS STR TET (n = 18) was found only at slaughter in carcasses (61%) and the environment (39%). The Salmonella serotypes S. Anatum and S. Typhimurium with MDR patterns highlighting β-lactams, including cephalosporins (AMP AUG AXO FOX TIO TET), were found only at the farm level. Salmonella enterica serovar Heidelberg had a specific MDR pattern (KAN STR TET) that we found only in the environment (100%). In the ABF system, we found only one isolate with an MDR pattern (AMP CHL FIS STR TET) associated with S. Typhimurium isolated from a carcass swab at slaughter.
Table 6.
Distribution of Salmonella serotypes associated with predominant MDR patterns in conventional production systems
| Serotype (n) | Predominant MDR patterna (n) | No. (%) of each type of isolate with each pattern |
|||
|---|---|---|---|---|---|
| Farm |
Slaughter |
||||
| Pigs | Environment | Carcasses | Environment | ||
| S. Anatum (103) | AMP AUG AXO FOX TIO TET (25) | 11 (44) | 14 (56) | 0 | 0 |
| AMP AUG AXO FOX TIO (5) | 0 | 2 (40) | 0 | 3 (60) | |
| S. Typhimurium (311) | AMP AUG AXO FOX TIO TET (7) | 3 (43) | 4 (57) | 0 | 0 |
| AMP FIS NAL STR TET (13) | 3 (23) | 8 (61) | 1 (8) | 0 | |
| AMP CHL FIS STR TET (41) | 5 (12) | 17 (41) | 16 (39) | 3 (7) | |
| FIS SXT TET (25) | 0 | 8 (32) | 9 (36) | 8 (32) | |
| FIS STR TET (18) | 0 | 0 | 11 (61) | 7 (39) | |
| S. Derby (106) | FIS STR TET (72) | 2 (3) | 15 (21) | 54 (75) | 1 (1) |
| S. Heidelberg (17) | KAN STR TET (11) | 0 | 11 (100) | 0 | 0 |
AMP, ampicillin; AUG, amoxicillin-clavulanic acid; AXO, ceftriaxone; CHL, chloramphenicol; FIS, sulfisoxazole; FOX, cefoxitin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; TET, tetracycline; TIO, ceftiofur.
PFGE.
Salmonella isolates (n = 340) from pigs and the environment were genotyped by PFGE. Restriction analyses by XbaI produced on average 10 to 16 bands and distributed the 340 isolates into 58 major clusters consisting of isolates with similar PFGE profiles and another 53 unique PFGE patterns represented by a single isolate each (see Fig. S1 in the supplemental material). Two separate dendrograms representing genotypic similarity within the same flow at different stages of production of the two distinct production systems were created (Fig. 4 and 5). S. Infantis isolated from lairage and carcass swabs originating from ABF pigs of two cohorts (A2 and A3) had 100% similar fingerprint profiles (Fig. 4). Within the conventional production system, we found 100% genotypic similarity among S. Rissen isolates from pig fecal, MLN, and environmental samples, including feed, water, floor swab, and lagoon, at nursery 1, nursery 2, finishing 2, and slaughter representing the same flow (C3) (Fig. 5). Identical fingerprint patterns were detected (cluster 14) (see Fig. S1 in the supplemental material) among S. Infantis isolates from pig and environmental samples of the same flow (C6) at different stages, including farrowing (isolate identifications [ID] S548, S551, S575, S581, and S591), nursery 1 (isolate ID S783 and S789), finishing 1 (isolate ID S963 and S984), and slaughter (isolate ID S1184 and S1195). Furthermore, we found 100% genotypic similarity among S. Infantis (FIS TET pattern) isolates from the conventional production system at farrowing, nursery 1, finishing 1, and slaughter, including slaughter truck samples (cluster 14) (see Fig. S1 in the supplemental material). All of the genotypically similar pansusceptible S. Infantis isolates at slaughter were grouped in respective clusters (clusters 17, 18, 19, and 20) (see Fig. S1 in the supplemental material). The fingerprint profiles of S. Anatum isolated from the ABF and conventional production systems at different stages of production and sample types were grouped into four major clusters (clusters 9, 10, 11, and 12) (see Fig. S1 in the supplemental material). Even though the ABF and conventional slaughter facilities were different, we identified a fingerprint profile (cluster 10) (see Fig. S1 in the supplemental material) associated with S. Anatum highlighting 100% genotypic similarity between the ABF and conventional production systems, which include isolates from pigs and environment samples at farm and slaughter.
Fig 5.

Dendrogram showing genotypic similarity among Salmonella isolates from conventional systems at various stages of production. aAM, amikacin; AMP, ampicillin; AUG, amoxicillin-clavulanic acid; AXO, ceftriaxone; CHL, chloramphenicol; CIP, ciprofloxacin; FIS, sulfisoxazole; FOX, cefoxitin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; TIO, ceftiofur. bMLN, mesenteric lymph node. cC3, conventional cohort.
DISCUSSION
This longitudinal study was conducted to determine and compare AR Salmonella isolates, isolated from pigs and the environment of both ABF and conventional production systems at different stages of production from farm to slaughter, at their phenotypic and genotypic levels. The prevalence of Salmonella in the conventional pigs was significantly higher than that in the ABF pigs at both farm and slaughter, a finding which is in contrast to an earlier study that reported a higher prevalence of Salmonella in an ABF production system (7). The low prevalence of Salmonella in outdoor ABF pigs at the farm in the present study was in accordance with other previous findings (9). We observed an increase in the prevalence of Salmonella in the final stages of production (finishing 1 and 2) in the conventional system, similar to previous reports of higher prevalence of Salmonella among finishing herds. The likely reasons include a previously infected group of pigs at the farm, contaminated transport vehicles, or handling and close contact of pigs during transportation (6, 12, 31). Higher prevalence of Salmonella in pigs during the final stages of production is of greater concern from a public health and food safety perspective. Only a few studies have been conducted to highlight the prevalence of Salmonella in the ABF environment versus that in the conventional farm environment (6, 7, 9, 32). In the ABF farm environment, Salmonella was detected only in three feed and two water samples at nursery 1 and finishing stages, in contrast to the conventional farm, for which positive environmental samples were widely represented by water, soil, feed, floor swabs, lagoon, and truck at different stages of production. A higher number of conventional environmental samples were positive for Salmonella in spite of strict AIAO practices. This highlights the potential role of the farm environment as a reservoir, which is in accordance with studies highlighting the persistence of Salmonella in the farm environment for several months to years (33, 34). Even though the ABF pigs had access to the external environment throughout their production chain, the prevalence of Salmonella in pigs was low, a finding which may be attributed to the absence/low prevalence of Salmonella in the ABF environment.
In our study, we found a higher prevalence of Salmonella in both production systems at slaughter in both pigs and the environment than at the farm. Factors contributing to the increased prevalence of Salmonella at slaughter likely include cross-contamination at the periharvest stage by trucks involved in transfer to the slaughtering facilities, stress experienced by the pigs during transport, and cross-contamination at lairage and at postharvest stages (17, 18, 20). In addition, a previous study highlighted that contaminated feed at the end stage of production has a significant role in dissemination of Salmonella (14). We detected clear evidence of cross-contamination in our study, as shown in clusters 7, 8, 14, 17, 18, and 26 (see Fig. S1 in the supplemental material). The MLN samples from both production systems had a higher prevalence of Salmonella than did fecal samples at the farm, a finding which is in accordance with previous reports suggesting occurrence of Salmonella in the gastrointestinal (GI) tract and lymphatic tissue in carrier pigs (6, 15, 18). Even though the MLN and gut contents are not used for consumption, occurrence of Salmonella in the MLN may act as a reservoir in contaminating carcasses during the postevisceration stage. We also isolated Salmonella from the postevisceration carcasses, which were cleansed with water before they were stored in the chilling facility. This indicates possible cross-contamination during the evisceration process along the slaughter chain. The ABF slaughter facility had overnight chilling of the carcass, whereas the conventional slaughter facility had blast chilling. Blast chilling is preferred to overnight cooling because it improves the meat shelf life and tenderness and it prevents the growth of important food-borne pathogens on the carcass surface (35, 36). Interestingly, we isolated Salmonella from both the systems in postchill swabs irrespective of the chilling facility type. The occurrence of Salmonella in postchill swabs (9) is of critical importance to public health and food safety, as this sample closely represents the final retail product. In this study, we also isolated Salmonella from the floor of trucks which are used to transport conventional and ABF pigs from farm to slaughter. The most significant contribution to positive samples for Salmonella at slaughter was from lairage, where pigs rest for about 2 h before they are slaughtered. It takes less than 2 h for a particular Salmonella serotype to establish in the GI tract of pigs and to be shed in their feces (17, 18). Clusters 7 and 8 (see Fig. S1 in the supplemental material) highlight similar PFGE fingerprint profiles among Salmonella isolates from ABF carcass and lairage swabs. However, the detection of Salmonella in ABF carcass samples, despite the absence of Salmonella at the farm and in transport trucks, clearly highlights the role of lairage and cross-contamination of the carcasses during processing.
Previous studies from Denmark reported various serotypes from organic and outdoor pig farming, including S. Anatum, Salmonella enterica serovar Agona, S. Derby, S. Typhimurium, and S. Newport, also observed in our ABF isolates (32, 37). However, in the ABF system at the farm and slaughter, we identified the S. Give serotype, which is most commonly associated with cattle (38). The likely reason for this serotype was the presence of other animals, including cattle, on the same premises as the pigs in the ABF farms and perhaps outdoor access to pastures which might be an ecological niche. In addition, identification of specific serotypes (Table 3) in ABF carcass and processing plant environmental samples, which were not detected at the farm level, suggests the role of the slaughter environment as a source for cross-contamination. Identification of serotypes, including S. Agona, S. Braenderup, S. Derby, S. Inverness, S. Muenchen, and S. Newport, which were not found in farm and slaughter environments on ABF carcass (Table 3) is attributed to the slaughter robots, including the carcass splitter and other instruments used for processing the carcass as a possible source of contamination during the production chain as described previously (39). We observed some common serotypes in the ABF and conventional production systems, including S. Anatum, S. Infantis, and S. Typhimurium. This is in accordance with a Centers for Disease Control and Prevention (CDC) report of the top four predominant serotypes in swine (40). In the conventional production system, we identified for the first time S. Rissen in pigs and the environment. S. Rissen is one of the top 10 serotypes most commonly isolated from pigs since 2004 in Europe (41) and the most common nonhuman serotype in Asian countries (42). According to the CDC annual report, S. Rissen was isolated from humans (<20 isolates per year) from 1999 to 2007 and there were no reports of its occurrence in food animals in the United States (41, 43). This serotype is uncommon in the United States; it was reported to have entered the United States in late 2008 and early 2009 through imported white pepper, resulting in a human outbreak in northern California and Nevada (44). We identified this serotype in our samples collected in late 2009.
Salmonella isolates from conventional production had higher AR (80%) than isolates from ABF production (27%). The use of antimicrobials in the conventional system for treatment and growth purposes likely results in a higher prevalence of Salmonella as previously reported (7, 45). Overall, Salmonella isolates of either production system or production stage had the highest frequency of resistance against TET (71%), followed by FIS (42%) and STR (17%). In addition, isolates exhibited resistance to β-lactams, including third-generation cephalosporins. These results are in agreement with previous reports (6, 7, 46). MIC distributions were similar for all the antimicrobials tested except TET, which was highest in Salmonella isolates of pig origin (MIC > 32 μg/ml; resistance, 80%) compared to environmental isolates in the conventional system. The possible reason may be use of tetracyclines as growth promoters administered in feed of growing pigs in our study, which has been reported extensively in the swine industry (47, 48). We detected a higher frequency of MDR isolates in the conventional system (27%) but only a single MDR isolate from the MLN of an ABF carcass exhibiting a pentaresistant pattern of AMP CHL FIS STR TET associated with S. Typhimurium. This result is in contrast with a previous report of higher MDR prevalence in an ABF system (7). In the conventional system, S. Typhimurium was broadly associated with the common MDR pattern of AMP CHL FIS STR TET at farm and slaughter as previously reported (6, 49). This pentaresistant pattern is common to the S. Typhimurium phage type DT104 (50). Identification of this phage type, both at farm and slaughter, is of significant public health concern because this phage type is commonly associated with human food-borne outbreaks worldwide (50, 51). Another important MDR pattern with β-lactams, including third-generation cephalosporins (AXO TIO), was associated with S. Typhimurium and S. Anatum only in conventional pigs and the environment at farm level, as previously reported (6). Emergence of these MDR patterns resistant to β-lactams is of concern because β-lactams (third-generation cephalosporins) are extensively used to treat human clinical Salmonella infections (52).
PFGE was used to genotype a representative subset of Salmonella isolates from pigs and the environment. PFGE is considered the gold-standard test to determine the source of Salmonella in epidemiological studies (12, 15). Therefore, we used this genotyping method to determine whether a similar Salmonella genotype is disseminated from farm to slaughter along the production chain. Based on similar fingerprint profiles, Salmonella isolates in our study were grouped into 58 major clusters. Clustering was consistent with serotypes and resistance patterns reported by a previous study. In addition, we observed fingerprint profile diversity among the same Salmonella serotypes representing different clusters as previously reported (14, 28, 33). Within the conventional production system, 100% genotypic similarity was observed among S. Rissen serotype isolates from pig fecal and environmental samples at different stages of production at farm and slaughter from a single cohort (C3). This result highlights the dissemination of the relatively new S. Rissen serotype in pigs all along the production chain in the United States. It was evident that specific serotypes, including S. Anatum, S. Infantis, S. Typhimurium, Salmonella enterica serovar Ouakam, S. Give, and Salmonella enterica serovar Ohio, were able to persist in the pigs and environment at different stages of production based on phenotypic and genotypic evidence (Table 3; see also Fig. S1 in the supplemental material) (clusters 7, 10, 14, 31, 37, and 42). The identification of genotypically identical S. Infantis from the slaughter environment and carcass samples from the ABF system, which were not observed at the farm level, highlights the importance of the slaughter environment from a food safety perspective. In an epidemiological study, it is difficult to determine the exact mechanism and direction of pathogen transmission between pigs and the environment. However, detection of the same genotype among pigs and environments within distinct production systems clearly suggests the exchange of Salmonella strains.
To summarize, this study demonstrates the presence of AR Salmonella in ABF and conventional production systems at farm, slaughter, and the environment, though at much lower levels in ABF than in conventional systems. The phenotypic and genotypic fingerprint profile results underscore the potential role played by the environment in the persistence and dissemination of transmission of AR Salmonella in the two production systems. We detected MDR isolates throughout all the production stages and the environment in the conventional system, which uses antimicrobials for prophylaxis and growth purposes. The detection of AR Salmonella in ABF pigs and their environment in the absence of selection pressure is a concern. At the phenotypic level, Salmonella isolates from the lairage floor, carcass, and MLN had similar resistance patterns and serotypes, which were not detected at the farm level. This highlights the importance of the farm and slaughter environments as separate but important reservoirs and as a crucial link to determining the dissemination of AR Salmonella among pigs. Future research should focus on environmental factors to develop a better understanding of the molecular epidemiology of this pathogen in the swine production environment and to reduce the burden of AR Salmonella on public health.
Supplementary Material
ACKNOWLEDGMENTS
We thank the swine producers and swine processing plants for providing us access to their facilities.
This work was funded by a U.S. Department of Agriculture National Research Initiative grant (2008-529245), a U.S. Department of Agriculture National Integrated Food Safety Initiative grant (2008-529461), the National Pork Board grant (NPB project number 12-001), and a North Carolina State University Individual Faculty Research and Professional Development Award to Siddhartha Thakur.
Views presented in this article do not necessarily reflect those of the U.S. Food and Drug Administration.
Footnotes
Published ahead of print 21 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01419-13.
REFERENCES
- 1.Lynne AM, Dorsey LL, David DE, Foley SL. 2009. Characterisation of antibiotic resistance in host-adapted Salmonella enterica. Int. J. Antimicrob. Agents 34:169–172 [DOI] [PubMed] [Google Scholar]
- 2.Scallan E, Hoekstra R, Angulo F, Tauxe R, Widdowson M, Roy S, Jones J, Griffin P. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV, Emerging Infections Program FoodNet Working Group 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl 3):S127–S134 [DOI] [PubMed] [Google Scholar]
- 4.Aarestrup FM, Oliver Duran C, Burch DG. 2008. Antimicrobial resistance in swine production. Anim. Health Res. Rev. 9:135–148 [DOI] [PubMed] [Google Scholar]
- 5.NARMS 2009. Executive report. National Antimicrobial Resistance Monitoring System, US FDA, Silver Spring, MD: http://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM268954.pdf [Google Scholar]
- 6.Dorr P, Tadesse D, Zewde B, Fry P, Thakur S, Gebreyes W. 2009. Longitudinal study of Salmonella dispersion and the role of environmental contamination in commercial swine production systems. Appl. Environ. Microbiol. 75:1478–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebreyes W, Thakur S, Morrow W. 2006. Comparison of prevalence, antimicrobial resistance, and occurrence of multidrug-resistant Salmonella in antimicrobial-free and conventional pig production. J. Food. Prot. 69:743–748 [DOI] [PubMed] [Google Scholar]
- 8.USDA 2009. Organic agriculture: organic market overview. US Department of Agriculture, Economic Research Service, Washington, DC: http://www.ers.usda.gov/topics/natural-resources-environment/organic-agriculture/organic-market-overview.aspx#.Ucw9oCIkA7V [Google Scholar]
- 9.Thakur S, Tadesse D, Morrow M, Gebreyes W. 2007. Occurrence of multidrug resistant Salmonella in antimicrobial-free (ABF) swine production systems. Vet. Microbiol. 125:362–367 [DOI] [PubMed] [Google Scholar]
- 10.Ray KA, Warnick LD, Mitchell RM, Kaneene JB, Ruegg PL, Wells SJ, Fossler CP, Halbert LW, May K. 2006. Antimicrobial susceptibility of Salmonella from organic and conventional dairy farms. J. Dairy Sci. 89:2038–2050 [DOI] [PubMed] [Google Scholar]
- 11.Davies PR, Bovee FG, Funk JA, Morrow WE, Jones FT, Deen J. 1998. Isolation of Salmonella serotypes from feces of pigs raised in a multiple-site production system. J. Am. Vet. Med. Assoc. 212:1925–1929 [PubMed] [Google Scholar]
- 12.Rodriguez A, Pangloli P, Richards H, Mount J, Draughon F. 2006. Prevalence of Salmonella in diverse environmental farm samples. J. Food Prot. 69:2576–2580 [DOI] [PubMed] [Google Scholar]
- 13.Barber DA, Bahnson PB, Isaacson R, Jones CJ, Weigel RM. 2002. Distribution of Salmonella in swine production ecosystems. J. Food Prot. 65:1861–1868 [DOI] [PubMed] [Google Scholar]
- 14.Molla B, Sterman A, Mathews J, Artuso-Ponte V, Abley M, Farmer W, Rajala-Schultz P, Morrow W, Gebreyes W. 2010. Salmonella enterica in commercial swine feed and subsequent isolation of phenotypically and genotypically related strains from fecal samples. Appl. Environ. Microbiol. 76:7188–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botteldoorn N, Heyndrickx M, Rijpens N, Grijspeerdt K, Herman L. 2003. Salmonella on pig carcasses: positive pigs and cross contamination in the slaughterhouse. J. Appl. Microbiol. 95:891–903 [DOI] [PubMed] [Google Scholar]
- 16.De Busser EV, Maes D, Houf K, Dewulf J, Imberechts H, Bertrand S, De Zutter L. 2011. Detection and characterization of Salmonella in lairage, on pig carcasses and intestines in five slaughterhouses. Int. J. Food Microbiol. 145:279–286 [DOI] [PubMed] [Google Scholar]
- 17.Hernández M, Gómez-Laguna J, Luque I, Herrera-León S, Maldonado A, Reguillo L, Astorga RJ. 2013. Salmonella prevalence and characterization in a free-range pig processing plant: tracking in trucks, lairage, slaughter line and quartering. Int. J. Food Microbiol. 162:48–54 [DOI] [PubMed] [Google Scholar]
- 18.Hurd H, Gailey J, McKean J, Rostagno M. 2001. Rapid infection in market-weight swine following exposure to a Salmonella Typhimurium-contaminated environment. Am. J. Vet. Res. 62:1194–1197 [DOI] [PubMed] [Google Scholar]
- 19.Botteldoorn N, Herman L, Rijpens N, Heyndrickx M. 2004. Phenotypic and molecular typing of Salmonella strains reveals different contamination sources in two commercial pig slaughterhouses. Appl. Environ. Microbiol. 70:5305–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebreyes W, Davies P, Turkson P, Morrow W, Funk J, Altier C, Thakur S. 2004. Characterization of antimicrobial-resistant phenotypes and genotypes among Salmonella enterica recovered from pigs on farms, from transport trucks, and from pigs after slaughter. J. Food Prot. 67:698–705 [DOI] [PubMed] [Google Scholar]
- 21.Owsley F, Rodning S, Floyd J. 2013. Scheduling all-in/all-out swine production, vol ANR-0847. Alabama Cooperative Extension System; http://www.aces.edu/pubs/docs/A/ANR-0847/ANR-0847.pdf [Google Scholar]
- 22.USDA 1996. Guidelines for Escherichia coli testing for process control verification in cattle and swine slaughter establishments. USDA, Washington, DC: http://www.fsis.usda.gov/wps/wcm/connect/cebac8d0-954b-4cc2-abcf-205ad4f4ddcd/Guideline_for_Ecoli_Testing_Cattle_Swine_Estab.pdf?MOD=AJPERES. [Google Scholar]
- 23.Oliveira S, Santos L, Schuch D, Silva A, Salle C, Canal C. 2002. Detection and identification of salmonellas from poultry-related samples by PCR. Vet. Microbiol. 87:25–35 [DOI] [PubMed] [Google Scholar]
- 24.Kim S, Frye J, Hu J, Fedorka-Cray P, Gautom R, Boyle D. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 44:3608–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez J, Sota M, Vivanco A, Perales I, Cisterna R, Rementeria A, Garaizar J. 2004. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 42:1734–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou W, Lin WJ, Foley SL, Chen CH, Nayak R, Chen JJ. 2010. Evaluation of pulsed-field gel electrophoresis profiles for identification of Salmonella serotypes. J. Clin. Microbiol. 48:3122–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou W, Lin WJ, Hise KB, Chen HC, Keys C, Chen JJ. 2012. Prediction system for rapid identification of Salmonella serotypes based on pulsed-field gel electrophoresis fingerprints. J. Clin. Microbiol. 50:1524–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liebana E, Guns D, Garcia-Migura L, Woodward MJ, Clifton-Hadley FA, Davies RH. 2001. Molecular typing of Salmonella serotypes prevalent in animals in England: assessment of methodology. J. Clin. Microbiol. 39:3609–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CLSI 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 30.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67 [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez-Buenfil JC, Alvarez-Fleites M, Segura-Correa JC. 2006. Incidence of salmonellosis and identification of serogroups and serotypes in a pig commercial farm in Yucatan. Rev. Latinoam. Microbiol. 48:10–13 [PubMed] [Google Scholar]
- 32.Jensen AN, Dalsgaard A, Stockmarr A, Nielsen EM, Baggesen DL. 2006. Survival and transmission of Salmonella enterica serovar Typhimurium in an outdoor organic pig farming environment. Appl. Environ. Microbiol. 72:1833–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandvang D, Jensen LB, Baggesen DL, Baloda SB. 2000. Persistence of a Salmonella enterica serotype Typhimurium clone in Danish pig production units and farmhouse environment studied by pulsed field gel electrophoresis (PFGE). FEMS Microbiol. Lett. 187:21–25 [DOI] [PubMed] [Google Scholar]
- 34.Baloda SB, Christensen L, Trajcevska S. 2001. Persistence of a Salmonella enterica serovar Typhimurium DT12 clone in a piggery and in agricultural soil amended with Salmonella-contaminated slurry. Appl. Environ. Microbiol. 67:2859–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nesbakken T, Eckner K, Røtterud OJ. 2008. The effect of blast chilling on occurrence of human pathogenic Yersinia enterocolitica compared to Campylobacter spp. and numbers of hygienic indicators on pig carcasses. Int. J. Food Microbiol. 123:130–133 [DOI] [PubMed] [Google Scholar]
- 36.Shackelford SD, King DA, Wheeler TL. 2012. Chilling rate effects on pork loin tenderness in commercial processing plants. J. Anim. Sci. 90:2842–2849 [DOI] [PubMed] [Google Scholar]
- 37.Jensen AN, Lodal J, Baggesen DL. 2004. High diversity of Salmonella serotypes found in an experiment with outdoor pigs. Wageningen J. Life. Sci. 52:109–117 [Google Scholar]
- 38.Edrington TS, Schultz CL, Bischoff KM, Callaway TR, Looper ML, Genovese KJ, Jung YS, McReynolds JL, Anderson RC, Nisbet DJ. 2004. Antimicrobial resistance and serotype prevalence of Salmonella isolated from dairy cattle in the southwestern United States. Microb. Drug Resist. 10:51–56 [DOI] [PubMed] [Google Scholar]
- 39.van Hoek AH, de Jonge R, van Overbeek WM, Bouw E, Pielaat A, Smid JH, Malorny B, Junker E, Löfström C, Pedersen K, Aarts HJ, Heres L. 2012. A quantitative approach towards a better understanding of the dynamics of Salmonella spp. in a pork slaughter-line. Int. J. Food. Microbiol. 153:45–52 [DOI] [PubMed] [Google Scholar]
- 40.CDC 2006. Salmonella surveillance: annual summary, 2005. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 41.Eurosurveillance Editorial Team 2013. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011 has been published. Euro Surveill. 18(15):pii=20449 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20449 [PubMed] [Google Scholar]
- 42.Galanis E, Lo Fo Wong DM, Patrick ME, Binsztein N, Cieslik A, Chalermchikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC, World Health Organization Global Salm-Surv. 2006. Web-based surveillance and global Salmonella distribution, 2000-2002. Emerg. Infect. Dis. 12:381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.CDC 2009. Salmonella annual summary tables. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ncezid/dfwed/pdfs/salmonellaannualsummarytables2009.pdf [Google Scholar]
- 44.CDC 2010. Salmonella Montevideo infections associated with salami products made with contaminated imported black and red pepper—United States, July 2009-April 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1647–1650 [PubMed] [Google Scholar]
- 45.Cromwell GL. 2002. Why and how antibiotics are used in swine production. Anim. Biotechnol. 13:7–27 [DOI] [PubMed] [Google Scholar]
- 46.White DG, Zhao S, McDermott PF, Ayers S, Friedman S, Sherwood J, Breider-Foley M, Nolan LK. 2003. Characterization of integron mediated antimicrobial resistance in Salmonella isolated from diseased swine. Can. J. Vet. Res. 67:39–47 [PMC free article] [PubMed] [Google Scholar]
- 47.Akwar HT, Poppe C, Wilson J, Reid-Smith RJ, Dyck M, Waddington J, Shang D, McEwen SA. 2008. Associations of antimicrobial uses with antimicrobial resistance of fecal Escherichia coli from pigs on 47 farrow-to-finish farms in Ontario and British Columbia. Can. J. Vet. Res. 72:202–210 [PMC free article] [PubMed] [Google Scholar]
- 48.McEwen SA, Fedorka-Cray PJ. 2002. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl 3):S93–S106 [DOI] [PubMed] [Google Scholar]
- 49.Gebreyes W, Thakur S, Davies P, Funk J, Altier C. 2004. Trends in antimicrobial resistance, phage types and integrons among Salmonella serotypes from pigs, 1997-2000. J. Antimicrob. Chemother. 53:997–1003 [DOI] [PubMed] [Google Scholar]
- 50.Helms M, Ethelberg S, Mølbak K, DT104 Study Group 2005. International Salmonella Typhimurium DT104 infections, 1992-2001. Emerg. Infect. Dis. 11:859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Glynn MK, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo FJ. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333–1338 [DOI] [PubMed] [Google Scholar]
- 52.Bradford PA. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.