Abstract
Influenza A H10N7 virus with a hemagglutinin gene of North American origin was detected in Australian chickens and poultry abattoir workers in New South Wales, Australia, in 2010 and in chickens in Queensland, Australia, on a mixed chicken and domestic duck farm in 2012. We investigated their genomic origins by sequencing full and partial genomes of H10 viruses isolated from wild aquatic birds and poultry in Australia and analyzed them with all available avian influenza virus sequences from Oceania and representative viruses from North America and Eurasia. Our analysis showed that the H10N7 viruses isolated from poultry were similar to those that have been circulating since 2009 in Australian aquatic birds and that their initial transmission into Australia occurred during 2007 and 2008. The H10 viruses that appear to have developed endemicity in Australian wild aquatic birds were derived from several viruses circulating in waterfowl along various flyways. Their hemagglutinin gene was derived from aquatic birds in the western states of the United States, whereas the neuraminidase was closely related to that from viruses previously detected in waterfowl in Japan. The remaining genes were derived from Eurasian avian influenza virus lineages. Our analysis of virological data spanning 40 years in Oceania indicates that the long-term evolutionary dynamics of avian influenza viruses in Australia may be determined by climatic changes. The introduction and long-term persistence of avian influenza virus lineages were observed during periods with increased rainfall, whereas bottlenecks and extinction were observed during phases of widespread decreases in rainfall. These results extend our understanding of factors affecting the dynamics of avian influenza and provide important considerations for surveillance and disease control strategies.
INTRODUCTION
In March 2010, a low-pathogenic avian influenza A H10N7 virus was identified as the cause of increased chicken mortality and decreased egg production in a commercial poultry farm in New South Wales (NSW), Australia (1). Subsequently, infection with the virus was confirmed in two abattoir workers that handled chickens originating from the same farm (1). Sequencing and phylogenetic analyses of the hemagglutinin (HA) genes showed that these viruses were identical and were most likely derived from influenza viruses that are predominant in North American wild aquatic birds (1), suggesting that these viruses were introduced into Australia by aquatic birds. A second incident of H10N7 in poultry occurred in Queensland (Qld) in 2012 on a mixed chicken and domestic duck farm. Virus was isolated only from chickens, but the domestic ducks sampled were seropositive for H10N7. Due to the lack of H10 subtype virus data from aquatic birds in Australia, the route of introduction of these poultry outbreaks and the prevalence of H10N7 viruses in wild birds within Australia have not been well understood.
The vast global literature on avian influenza viruses has shown that interspecies transmission of H5, H7, and H9 subtype avian influenza viruses into terrestrial poultry and mammalian hosts, including humans, has occurred sporadically (2, 3), whereas interspecies transmission of the H10 subtype, as observed in Australia, has been rare. Prior to this, only one instance of H10 subtype virus infection of humans, which occurred in two infants in Egypt, has been reported worldwide (http://www.paho.org/english/ad/dpc/cd/eid-eer-07-may-2004.htm). Previously reported avian influenza outbreaks in Australian poultry have been caused by H7 subtype viruses, including H7N3 and H7N4 in 1994 and 1997, respectively (4); however, at the time of these outbreaks, similar viruses were not reported in wild birds. More recently, during November 2012, the first highly pathogenic avian influenza A H7N7 virus outbreak in over 15 years was reported in commercial layers in NSW, Australia (5). The source of this latest outbreak remains to be determined.
The delineation of the avian influenza viruses into geographically distinct virus lineages (viz., Eurasian and North American lineages) has been attributed to the geographical separation of avian hosts between the American and Eurasian landmasses (6–8). Historical as well as recent surveys of influenza virus in Australian wild aquatic birds have indicated that the Australian avian influenza viruses, including subtypes H5, H7, and H9, form viral lineages that are distinct from those circulating in Eurasia and North America (9, 10). Australian influenza virus lineages do, however, share a more recent common ancestor with divergent Eurasian lineages, indicating periodic introduction of the Eurasian lineage, followed by periods of establishment in Australian wild waterfowl (10). Bird migration data suggest that aquatic birds of the Charadriiformes family (e.g., shorebirds and waders) migrate from Eurasia and North America to Australia along the East Asian or West Pacific flyways (11); however, confirmed introductions of the North American avian influenza virus lineages into Australian wild birds have been rare (12). Members of the Anseriformes (e.g., ducks and geese) in Australia generally limit their movement to movement within Australia, with occasional spread to Papua New Guinea and New Zealand (13, 14).
Intercontinental virus transmission and reassortment between Eurasian and North American influenza virus lineages have been periodically observed (15, 16); however, their establishment in the introduced region has rarely been recorded (6). Intercontinental gene flow between divergent lineages can lead to long-lasting effects on the local avian influenza virus population structure, with eventual extinction of the endemic lineages (6). For example, the Eurasian H6 virus lineage that was first introduced into North American wild waterfowl in about 1980 rapidly replaced the North American H6 viruses. Reports since 2000 have shown that the introduced H6 viruses reassorted with circulating North American lineages, crossed species barriers, and caused widespread outbreaks in poultry (17, 18). The effect of repeated introduction of Eurasian lineages and the recent introduction of the North American lineage on the population structure of avian influenza virus lineages in Australia is poorly understood. This is largely due to the lack of continuous influenza surveillance in aquatic birds or poultry in Australia. Most Australian wild waterfowl data arise from surveys carried out in isolated locations during limited time periods (19–22). Two recent publications did, however, report on a recent (2005 to present) ongoing surveillance study of influenza in wild waterfowl and shorebirds across various parts of Australia, but predominantly Victoria (Vic) and NSW (10, 23).
We generated genomic sequence data from the H10 viruses collected from poultry on NSW and Qld farms in March 2010 and June 2012, respectively, and phylogenetically analyzed them with newly generated partial sequence data from wild waterfowl H10 subtype viruses collected from 2007 to 2011 in NSW and Vic as part of the ongoing wild bird surveillance program (Table 1). This phylogenetic analysis identified the transmission pathway of the North American H10 virus and helps to provide an understanding of the overall prevalence of the H10 subtype in Australia. We also conducted large-scale phylogenetic analyses of all available avian influenza virus sequence data from Australia to understand the population structure of avian influenza viruses. Our aims were also to understand the effects of repeated introduction of Eurasian lineage viruses and the apparent recent introduction of the North American viruses on the population structure of avian influenza viruses in Australia and to identify research gaps in avian influenza surveillance in the region.
Table 1.
Influenza A H10 viruses isolated from poultry and duck feces in Australia
| Virus | Isolation date | Source |
|---|---|---|
| A/Chicken/Sydney/D808/2010 | March 2010 | Chicken, NSW |
| A/Chicken/Sydney/D809/2010a | March 2010 | Chicken, NSW |
| A/Chicken/Sydney/D891/2010a | March 2010 | Chicken, NSW |
| A/Chicken/Queensland/1/2012 | June 2012 | Chicken, Qld |
| A/Chicken/Queensland/2/2012a | June 2012 | Chicken, Qld |
| A/Chicken/Queensland/3/2012a | June 2012 | Chicken, Qld |
| A/Wild Bird/NSW/TM106907/2007a | September 2007 | Duck droppings, NSW |
| A/Wild Bird/NSW/2338/2009a | June 2009 | Duck droppings, NSW |
| A/Wild Bird/NSW/6526/2011a | September 2011 | Duck droppings, NSW |
| A/Wild Bird/AUS/471/2009a | December 2009 | Duck droppings, Vic |
| A/Wild Bird/AUS/475/2009a | December 2009 | Duck droppings, Vic |
| A/Wild Bird/Victoria/1426/2009a | April 2009 | Duck droppings, Vic |
| A/Wild Bird/Victoria/1434/2009a | April 2009 | Duck droppings, Vic |
| A/Wild Bird/Victoria/1443/2009a | April 2009 | Duck droppings, Vic |
The genomes of these viruses were partially sequenced.
MATERIALS AND METHODS
Surveillance.
Virological surveillance for avian influenza viruses was conducted in wild birds as part of an ongoing national avian influenza surveillance program (see references 10 and 23 for more information). H10 samples under study were collected from coastal wetlands around Newcastle, NSW, and around Melbourne, Vic. The samples from the H10N7 virus-infected chickens from the mixed chicken/duck farm in Qld in 2012 were kindly provided by Biosecurity Qld, Department of Agriculture, Fisheries and Forestry, Archerfield, Qld.
Virus isolation and sequencing.
Influenza viruses were isolated and passaged in Madin-Darby canine kidney (MDCK) cells in Dulbecco's modified Eagle's medium (DMEM) in the presence of antibiotics as previously described (24). RNA extraction was performed using a QIAamp viral RNA minikit (Qiagen) according to the manufacturer's protocol. Reverse transcription-PCR (RT-PCR) for all the H10 influenza virus genes was carried out using a MyTaq one-step RT-PCR kit (Bioline) with gene-specific primers (primer sequences are available upon request), and PCR products were purified and set up for sequencing reactions as previously described (25). Sequencing was performed using a BigDye Terminator cycle sequencing kit (Applied Biosystems), and the sequences were run on ABI 3500xl genetic analyzer (Applied Biosystems).
Virus sequence data.
To investigate the evolutionary origins of the H10 viruses detected in chickens from the NSW and Qld farms, we sequenced the full and partial genomes of 6 representative viruses detected from both locations and the HA genes from 8 H10 subtype viruses of various or unknown neuraminidase (NA) types isolated from wild waterfowl in Australia (Table 1). To identify the evolutionary history and infer the timeline of introduction, we analyzed the surface protein genes of these viruses with all publicly available H10 HA and N7 NA sequences.
To investigate gene flow and the long-term evolutionary dynamics of Australian avian influenza viruses, we also generated data sets for all internal gene segments of multiple subtypes, including the polymerase basic 2 (PB2), polymerase basic 1 (PB1), polymerase acidic (PA), nucleoprotein (NP), matrix (M), and nonstructural (NS) genes that were derived from wild aquatic birds on the Australian continent over a 31-year period (1975 to 2006) (see Fig. S1 in the supplemental material), and combined them with all available full-length sequence data for influenza A viruses from terrestrial and aquatic birds in Eurasia and North America. For the Eurasian and North American viruses, we removed duplicate sequences and those with 100% similarity to viruses from the same region and same year, while ensuring that representative virus sequences of each monophyletic clade were included in all subsequent analyses.
Phylogenetic analysis.
Multiple-sequence alignments for each data set were conducted using the MAFFT program (26) and optimized manually using the Se-Al program (http://tree.bio.ed.ac.uk/software/seal/). The best-fit evolutionary model for each data set was estimated using the JModeltest program (27). Phylogenetic trees and bootstrap supports for each gene data set were estimated using the best-fit evolutionary model and the maximum likelihood (ML) method in PhyML, v3.0, software (28).
Relaxed phylogenies and estimation of population dynamics.
To estimate the rates of nucleotide substitution and the time of the most recent common ancestors (TMRCA), we used a Bayesian Markov chain Monte Carlo (MCMC) method implemented in the program BEAST, v1.7.4 (29). Each gene data set was analyzed with the codon-based SRD06 nucleotide substitution model (30), the constant-coalescent-population model, and an uncorrelated lognormal relaxed-clock model that allows evolutionary rates to vary along branches within a lognormal distribution (31). For each data set, three to five independent Bayesian MCMC runs were conducted for 20 million to 30 million generations sampled to produce 10,000 trees. Convergence of the runs was confirmed using the Tracer program, v1.4 (http://tree.bio.ed.ac.uk/software/tracer/), and effective sample size values of >500 indicated a sufficient level of sampling. The results of the multiple runs were then combined using the LogCombiner program, v1.7.4 (29). Mean evolutionary rates and divergence times were calculated using the TreeAnnotator program, v1.7.4 (29), after the removal of an appropriate burn-in (10 to 20% in most cases), and phylogenetic trees were visualized with the FigTree program, v1.1.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Climate data.
To understand the effects of regional climate change on the evolution of Australian avian influenza viruses, we acquired high-quality spatial rainfall data sets spanning from 1900 to 2012 (32) and rainfall data from the Bureau of Meteorology of Australia (http://www.bom.gov.au/climate/). We assembled data sets for each of the six states of Australia (NSW, Qld, South Australia [SA], Tasmania [Tas], Vic, and Western Australia [WA]) and from the Northern Territory (NT). We also obtained data from six regions representing distinct climatologic regions (see Fig. S2 in the supplemental material).
Nucleotide sequence accession numbers.
All newly generated sequences were deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database under accession numbers EPI397842 to -397864 and EPI464596 to -464604.
RESULTS
Emergence of viruses with North American H10 virus HA in Australia.
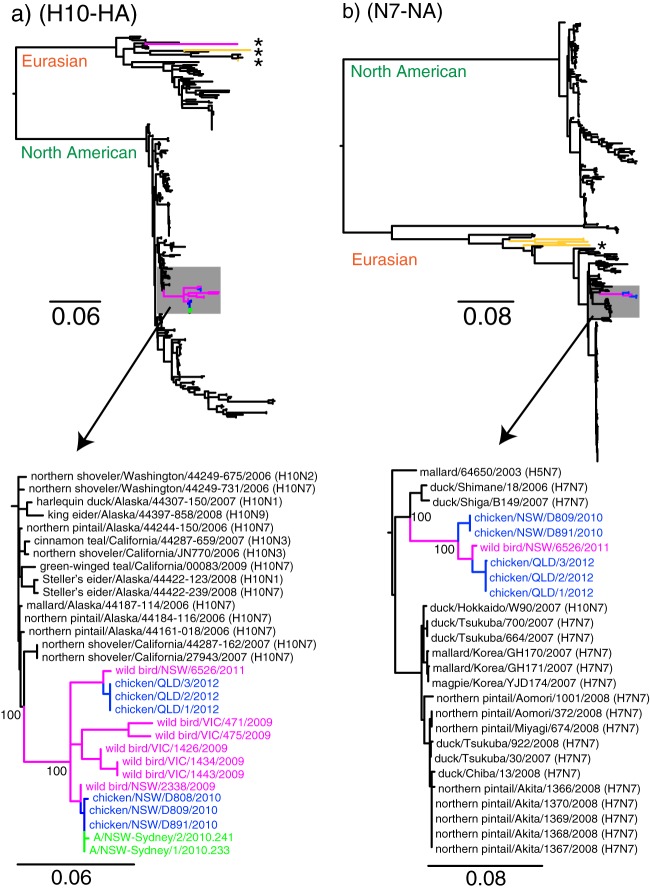
The HA and NA genes of influenza A H10N7 viruses obtained from chickens and abattoir workers in NSW, chickens from the farm in Qld, and wild ducks in NSW and Vic were analyzed with all publicly available H10 subtype HA and N7 subtype NA gene sequences derived from viruses from multiple hosts and continents. The final data sets contained 282 and 393 sequences for the HA and NA genes, respectively. The ML phylogeny of the H10 virus HA genes (Fig. 1a) showed that except for the early wild bird isolate from 2007 (A/Wild Bird/NSW/TM106907/2007), all other newly sequenced isolates, including the human, chicken, and wild waterfowl sequences from Australia, formed a monophyletic lineage (termed AUS/2009-12). The 2007 isolate belonged to the Eurasian lineage, whereas the recent HA genes were all derived from the North American H10 lineage and were most closely related to viruses isolated from aquatic wild birds along the westernmost coastal states of the United States, including, Alaska, Washington, and California, during 2006 and 2007. A/Wild Bird/NSW/TM106907/2007 isolate was the first H10N7 virus reported from Oceania prior to detection of such isolates in poultry from the NSW and Qld farms; however, the H10 HA sequences of one virus of the H10N8 subtype (A/Shearwater/Australia/2/1972) and one of the H10N4 subtype (A/Mallard/New Zealand/223-6/2005) were publicly available. The H10 HA genes of these viruses belonged to the Eurasian H10 lineage and therefore are distantly related to the HA gene of AUS/2009-12 sequences.
Fig 1.
Phylogenetic relationship of the hemagglutinin (a) and neuraminidase (b) gene segments of H10N7 subtype viruses isolated from poultry (blue), abattoir workers (green), and wild birds (pink) in New South Wales, Queensland, and Victoria, Australia, since 2007. Branches leading to H10 and N7 subtype viruses previously isolated from Australia are highlighted in yellow with an asterisk.
The H10 HA genes of the viruses from the NSW chicken farm and the associated human cases in 2010 were monophyletic and clustered most closely with A/Wild Bird/NSW/2338/2009, whereas the H10 HA genes of the viruses detected on the Qld chicken/duck farm in 2012 were most closely related to A/Wild Bird/NSW/6526/2011. The observed genetic differences between the chicken H10 viruses isolated from the NSW and Qld farms (blue viruses in Fig. 1a) and their relatedness to wild waterfowl isolate sequences from 2009 and 2011, respectively (pink viruses in Fig. 1a), may suggest that the H10N7 viruses that caused the 2010 and 2012 incidents were independently introduced into chickens from waterfowl. However, considering the lack of clinical symptoms in ducks infected with the H10N7 virus and the absence of ongoing active influenza surveillance in poultry in Australia, we cannot exclude the possibility that domestic ducks are the source of infection of chickens.
The N7 virus NA gene of AUS/2009-12 sequences from chickens and wild waterfowl also clustered in a monophyletic lineage (Fig. 1b) similar to that of the HA gene; however, these viruses were derived from the Eurasian lineage. In particular, the NA genes of AUS/2009-12 were most closely related to those of H7N7 viruses isolated from ducks in Japan during 2006 and 2007. The sequences of the N7 NA genes of four H7N7 subtype viruses previously reported from Australia (collected during the 1976 and 1985 high-pathogenicity avian influenza [HPAI] virus outbreaks in Vic) (yellow branches with asterisks in Fig. 1b and 2b) were distantly related to the new sequence data, suggesting that the viruses detected on the NSW and Qld farms were a new reassortant. Taken together, these results show that while the HA and NA genes of the viruses detected on the NSW and Qld farms were derived from Australian aquatic birds, they were not typical of avian influenza viruses normally detected in Australia. However, due to the close proximity of domestic ducks to chickens in both the NSW and Qld farms—a large duck breeder farm is located 1.5 km from the NSW farm, and the Qld farm contained both chickens and ducks—the possibility that domestic ducks played an intermediary role cannot be excluded. The close phylogenetic relationship of the genes for the H10 viruses with viruses detected across multiple continents highlights the possible interaction of migratory birds from multiple continents along the Western Pacific flyway.
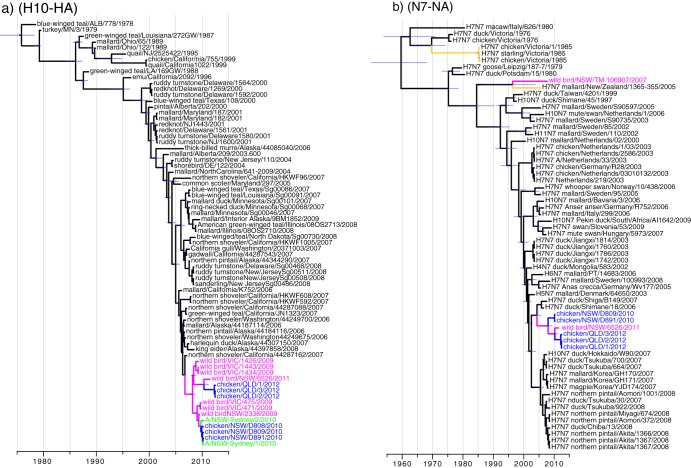
Fig 2.
Dated phylogenies of H10 virus subtype HA and N7 virus subtype NA regions showing the time of the most recent common ancestor and the divergence times of Australian H10N7 viruses. Tree branches and virus names are highlighted using the same color scheme described in the legend to Fig. 1.
Recent introduction of H10 HA and N7 NA gene segments.
The detection of North American H10 virus HA genes over 3 years in Australian wild waterfowl suggests that the North American lineage may now be endemic in aquatic waterfowl in Australia. To infer the timeline of introduction of the H10N7 viruses into Australia, we estimated the TMRCA for influenza A H10N7 viruses isolated from humans, poultry, and wild birds in Australia using the Bayesian relaxed-clock models (31). The TMRCA of the AUS/2009-12 H10 HA genes was estimated to be early 2008 (95% Bayesian confidence interval [CI], June 2007 to June 2008), whereas the time of divergence of this lineage from the North American lineage was estimated to be early 2007 (Fig. 2a). These results indicate that the introduction of the North American lineage into Australian wild waterfowl occurred at least 2 years prior to the first detection of the virus in poultry in 2010. The TMRCA of the AUS/2009-12 N7 NA lineage was similarly estimated to be June 2007 (95% CI, 2006 to 2008), whereas divergence of these viruses from the Eurasian lineage was estimated to be from 2005 to 2007 (Fig. 2b).
On the basis of these analyses, it is not certain if reassortment of the Eurasian and North American lineage viruses occurred within Australia or along the flyway before they were introduced intro Australia. However, the lack of detection of the North American H10 viruses elsewhere in Asia (e.g., Japan, where the N7 precursors were identified) suggests that the mixing may have occurred in Australia. A thorough investigation of virus prevalence and the migratory patterns of birds that arrive in Australia may provide additional insights into the introduction pathways and the most likely location for the reassortment of viruses.
Introduced H10N7 continues to reassort with endemic viruses.
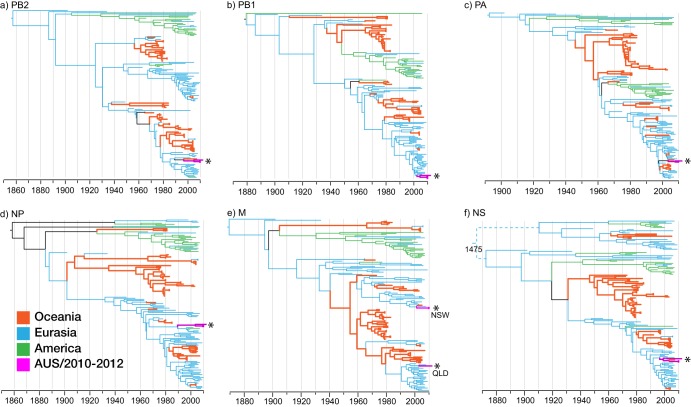
Analysis of all 6 internal gene segments of the H10 viruses from the NSW and Qld farms showed that they were all derived from the Eurasian lineage; however, these viruses were not closely related to the viruses previously detected in Australia. Furthermore, the H10 viruses from the NSW and Qld farms differed in their M gene sequences, showing that they were derived from independent lineages that were cocirculating in aquatic birds (Fig. 3e). Taken together, these results suggest that the introduced H10N7 viruses may be continuously reassorting with introduced and endemic viruses in waterfowl to generate new genome constellations.
Fig 3.
Phylogenetic relationship of the PB2 (a), PB1 (b), PA (c), NP (d), M (e), and NS (f) protein genes of influenza viruses of all subtypes isolated from Oceania (orange), Eurasia (blue), and the Americas (green). H10 viruses isolated from Australia from 2009 to 2012 (pink) are highlighted with an asterisk. Phylogenies with virus names are presented in Fig. S3 in the supplemental material.
Evolutionary dynamics of avian influenza virus lineages in Australia.
To understand the long-term evolutionary dynamics of influenza viruses circulating in wild aquatic waterfowl in Australia, we used the Bayesian relaxed-clock method to analyze the internal gene segments (PB2, PB1, PA, NP, NS, and M genes) of all influenza virus subtypes. We included all publicly available and newly generated sequence data from aquatic birds in Australia and New Zealand collected over the past 40 years as well as representative sequences from the American and Eurasian influenza virus gene pool (Fig. 3). The phylogenies of the internal gene segments showed that until the early 1980s, the majority of viruses detected in Australia formed major monophyletic lineages in each of the internal gene segments (orange branches in Fig. 3), even though these viruses were isolated from a range of different locations around Oceania (see Fig. S1 in the supplemental material). The times of divergence of the major lineages indicate that these viruses likely circulated in Australian waterfowl in the region for at least 10 to 30 years (orange branches in Fig. 3). Interestingly, during the early 1980s, the PB2, PB1, and MP genes of this lineage became extinct, while an evolutionary bottleneck was observed for the PA, NP, and NS genes. Since the extinction of the major Australian avian influenza viruses in the 1980s, a number of repeated introductions from the Eurasian influenza virus gene pool were observed; however, none of these viruses persisted for more than a few years. While the persistence of some lineages for several years suggests that the Australian waterfowl were capable of maintaining an independent influenza virus lineage over a number of years, the period since the early 1980s may represent a significant transition in the dynamics of avian influenza virus lineages in Australia.
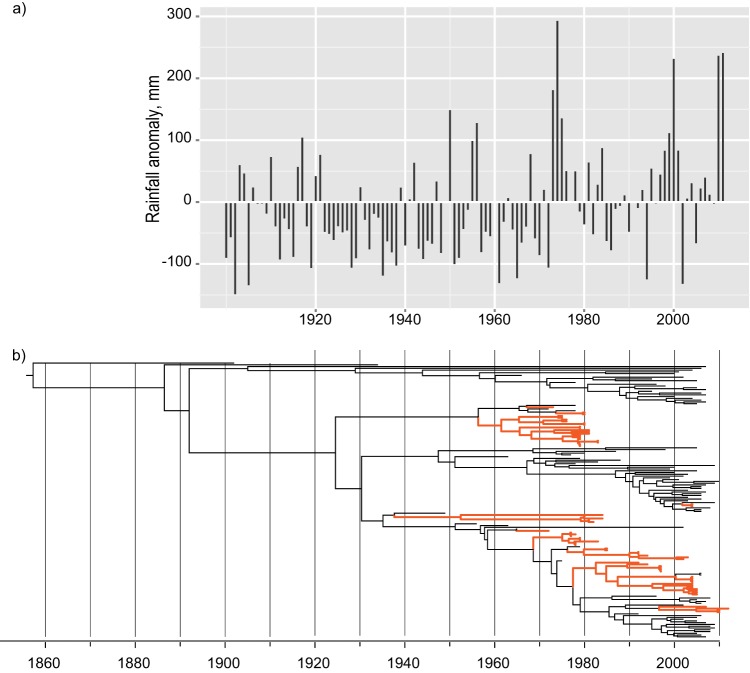
Weather patterns, especially rainfall, are known to affect the behavior of Anseriformes in Australia (33). To deduce any such affects on the observed dynamics of Australian avian influenza viruses, we plotted the observed deviations in rainfall over the last hundred years in Australia. We found that during the early 1980s and mid-2000s, when we observed a marked decrease in avian influenza virus diversity (Fig. 3 and 4b), the average rainfall across Australia was below average (Fig. 4a; see Fig. S2 in the supplemental material). In particular, the lowest recorded rainfall across a large region in Australia was observed during April 1982 and February 1983, resulting in one of the most severe and widespread droughts recorded in Australian history (www.bom.gov.au/climate/drought/livedrought.shtml). Conversely, the period prior to the early 1980s, when a monophyletic lineage of viruses was observed in multiple parts of the country, represented a time when above-average rainfall was observed across multiple states in Australia for several years (Fig. 4a).
Fig 4.
Correlation of rainfall and avian influenza virus diversity in Australia. (a) Bars represent deviations in annual rainfall observed between 1900 and 2012 from an average observed during the same period in Australia. The average anomaly in rainfall for each state in Australia and the major climatologic regions are provided in Fig. S2 in the supplemental material. (b) Phylogenetic tree showing the relationship of the PB2 gene of avian influenza viruses isolated in Oceania (orange).
DISCUSSION
Through the phylogenetic analyses of H10N7 subtype viruses isolated from farmed poultry in 2010 and 2012 and from wild waterfowl in Australia, we have shown that the H10N7 viruses that caused infection in chickens were also present in wild waterfowl in Vic and NSW, suggesting that the initial introduction of the North American H10 virus occurred in aquatic birds. In addition, our data show that the North American H10 subtype viruses may have developed endemicity in the Australian wild bird population and are continuing to reassort with other circulating avian influenza virus lineages, resulting in new gene constellations such as those detected in farmed poultry in Qld in 2012. The occurrence of poultry outbreaks 2 years apart on farms that are approximately 1,000 km apart may indicate the endemic nature of the North American H10 subtype viruses introduced into Australian wild waterfowl. However, movements of domestic poultry from the affected farm in NSW and the area in Qld where the second farm studied is located are known to occur. Therefore, it is not possible to rule out the possibility that the incursion of the virus on both farms was associated with the presence of infection in domestic ducks and movements of domestic poultry.
Domestic ducks have also been implicated in the transmission of H7 subtypes in at least two HPAI outbreaks in Australia (4, 34). They are also the group mostly found to harbor low-pathogenicity avian influenza (LPAI) virus in Australia, with 70% of all LPAI viruses reported between 1976 and 2012 being in domestic duck farms or in mixed enterprises containing both chickens and ducks (35). The Qld farm was a mixed enterprise where both species were infected with H10N7 (35). Therefore, an alternative hypothesis is that domestic ducks may have acquired the North American H10 viruses from wild waterfowl and then transmitted these viruses to chickens at the farms, a transmission pathway that has consistently been observed in other countries (36, 37). Further investigation is warranted to understand the genomic relationships of an HPAI H7N7 virus outbreak on a free-range chicken farm in NSW in November 2012 (http://www.oie.int/wahis_2/public/wahid.php/Reviewreport/Review?reportid=12612) with the H10N7 viruses reported from wild birds and poultry in this study, especially as they share the same NA type.
Our study also provides evidence on the long-term evolutionary dynamics of avian influenza viruses on the Australian continent, previously a gap in knowledge of the global dynamics of avian influenza viruses. Evolutionary analyses of all available internal gene segments of multiple influenza virus subtypes from waterfowl provided crucial insight into the dynamics of avian influenza viruses in the region, suggesting that large-scale changes in weather may be an important determinant of waterfowl behavior, thereby affecting the dynamics of avian influenza viruses in the region (33, 38).
The severe drought that was observed across a wide region in Australia during the 1980s appears to have played an important role in limiting avian influenza virus diversity in the region. However, the effect of regional variation in weather and the periodicity of such changes on the evolutionary dynamics of avian influenza viruses in Australia overall is not clear. For example, when most states in Australia experienced drought conditions during the early 1980s, above-average rainfall was recorded in the state of Western Australia (see Fig. S2 in the supplemental material). Enhanced disease surveillance coupled with a better understanding of Australian water bird movements on the local, regional, and continental scales is required to better understand the ecology of avian influenza viruses in Australia.
Extreme climatic conditions may also prove to be a barrier for the establishment and spread of new virus lineages following introduction by the Charadriiformes that migrate into Australia. The ecology of Australian Anseriformes differs significantly from that of Anseriformes in the Northern Hemisphere. While clear and established patterns of seasonal migration are observed in the Northern Hemisphere, movement of Australian Anseriformes is more dynamic and is dependent on habitat, food, rainfall, and breeding requirements (33). Furthermore, the majority of birds that have been known to carry influenza virus (e.g., ducks) are not migratory in the region. Of the 656 of all bird species that are regularly observed in Australia, only about 90 species regularly move between Asia and Australia (14). Therefore, if migratory birds are, in fact, bringing the viruses into Australia, the interface between nonmigratory bird species (e.g., ducks) and migratory bird species during weather extremes may be an important site for the implementation of enhanced avian influenza surveillance in Australia.
It has been argued (6) that the flow of influenza virus genes between continents affects the dynamics of the endemic virus populations due to competition, where the introduced lineage becomes predominant while the preexisting lineage eventually becomes extinct. In our scenario, due to the geographical proximity to Eurasia followed by the weather-induced near extinction of local virus diversity, we propose that Australia acts as a sink population for avian influenza virus lineages, thereby contributing less to the global changes in the influenza virus population dynamics than is seen elsewhere, such as in Europe or America.
While our study clearly indicates multiple incursions of avian influenza virus lineages into Australian waterfowl, there appears to be a barrier to the introduction of lineages that are endemic in poultry in Southeast Asia, such as the highly pathogenic avian influenza H5N1 virus and low-pathogenic H9N2 virus lineages. The reasons for this are unknown, but further studies to understand the infectivity and/or pathogenicity of these viruses in the different bird species that migrate between the regions may provide some insight. If such an introduction was to appear, it is most likely to occur through the northern- and northwestern-most areas of Australia, where the climatic barriers that determine waterfowl densities are very different from those that determine the densities of the populations sampled in this study. Intensive virological surveillance in these regions to detect any incursions of novel influenza viruses into Australia remains important.
Supplementary Material
ACKNOWLEDGMENTS
The samples from the Qld chicken outbreak in 2012 that yielded the H10N7 isolate were kindly provided by Biosecurity Qld, Department of Agriculture, Fisheries and Forestry, Archerfield Qld.
The laboratories in the Australian states of Victoria and New South Wales are supported by their state governments, and the collection of wild aquatic bird samples in New South Wales is supported by the University of Newcastle. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing. Wild bird surveillance in New South Wales and Victoria is funded by the Department of Agriculture, Forestry and Fisheries and the National Health and Medical Research Council of Australia. This study was also supported by Singapore Ministry of Education Academic Research Fund grant MOE2011-T2-2-049 (to D.V.); a career development award under National Institute of Allergy and Infectious Diseases contract HHSN266200700005C (to G.J.D.S.); and the Duke-NUS Signature Research Program, funded by the Agency for Science, Technology and Research, Singapore, and the Ministry of Health, Singapore (to D.V., Y.C.F.S., M.F. and G.J.D.S.).
Footnotes
Published ahead of print 17 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03437-12.
REFERENCES
- 1. Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng Y-M, Iannello P, Barr IG, Dwyer DE, Ratnamohan M, McPhie K, Selleck P. 2012. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg. Infect. Dis. 18:814–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander DJ. 2000. A review of avian influenza in different bird species. Vet. Microbiol. 74:3–13 [DOI] [PubMed] [Google Scholar]
- 3. Peiris JSM. 2009. Avian influenza viruses in humans. Rev. Sci. Tech. 28:161–174 [DOI] [PubMed] [Google Scholar]
- 4. Selleck PW, Gleeson L, Hooper P, Westbury H, Hansson E. 1997. Identification and characterisation of an H7N3 influenza A virus from an outbreak of virulent avian influenza in Victoria. Aust. Vet. J. 75:289–292 [DOI] [PubMed] [Google Scholar]
- 5.Department of Primary Industries 15 November 2012. Avian influenza confirmed at Hunter Valley egg farm. Media release. Department of Primary Industries, New South Wales Government, Sydney, NSW, Australia: www.dpi.nsw.gov.au/aboutus/news/all [Google Scholar]
- 6. Bahl J, Vijaykrishna D, Holmes EC, Smith GJD, Guan Y. 2009. Gene flow and competitive exclusion of avian influenza A virus in natural reservoir hosts. Virology 390:289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donis RO, Bean WJ, Kawaoka Y, Webster RG. 1989. Distinct lineages of influenza virus H4 hemagglutinin genes in different regions of the world. Virology 169:408–417 [DOI] [PubMed] [Google Scholar]
- 8. Obenauer JC. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580 [DOI] [PubMed] [Google Scholar]
- 9. Bulach D, Halpin R, Spiro D, Pomeroy L, Janies D, Boyle D. 2010. Molecular analysis of H7 avian influenza viruses from Australia and New Zealand: genetic diversity and relationships from 1976 to 2007. J. Virol. 84:9957–9966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hansbro PM, Warner S, Tracey JP, Arzey KE, Selleck P, O'Riley K, Beckett EL, Bunn C, Kirkland PD, Vijaykrishna D, Olsen B, Hurt AC. 2010. Surveillance and analysis of avian influenza viruses, Australia. Emerg. Infect. Dis. 16:1896–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gill RE, Jr, Piersma T, Hufford G, Servranckx R, Riegen A. 2005. Crossing the ultimate ecological barrier: evidence for an 11000-km long non-stop flight from Alaska to New Zealand and eastern Australia by bar-tailed godwits. Condor 107:1–20 [Google Scholar]
- 12. Kishida N, Sakoda Y, Shiromoto M, Bai GR, Isoda N, Takada A, Laver G, Kida H. 2008. H2N5 influenza virus isolates from terns in Australia: genetic reassortants between those of the Eurasian and North American lineages. Virus Genes 37:16–21 [DOI] [PubMed] [Google Scholar]
- 13. Dingle H. 2004. The Australo-Papuan bird migration system: another consequence of Wallace's Line. Emu 104:95–108 [Google Scholar]
- 14. Tracey JP, Woods R, Roshier D, West P, Saunders GR. 2004. The role of wild birds in the transmission of avian influenza for Australia: an ecological perspective. Emu 104:109–124 [Google Scholar]
- 15. Ramey AM, Pearce JM, Flint PL, Ip HS, Derksen DV, Franson JC, Petrula MJ, Scotton BD, Sowl KM, Wege ML, Trust KA. 2010. Intercontinental reassortment and genomic variation of low pathogenic avian influenza viruses isolated from northern pintails (Anas acuta) in Alaska: examining the evidence through space and time. Virology 401:179–189 [DOI] [PubMed] [Google Scholar]
- 16. Krauss SL, Obert CA, Franks J, Walker D, Jones K, Seiler P, Niles L, Pryor SP, Obenauer JC, Naeve CW, Widjaja L, Webby RJ, Webster RG. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 3:e167. 10.1371/journal.ppat.0030167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Webby RJ, Woolcock PR, Krauss SL, Walker DB, Chin PS, Shortridge KF, Webster RG. 2003. Multiple genotypes of nonpathogenic H6N2 influenza viruses isolated from chickens in California. Avian Dis 47:905–910 [DOI] [PubMed] [Google Scholar]
- 18. Woolcock PR, Suarez DL, Kuney D. 2003. Low-pathogenicity avian influenza virus (H6N2) in chickens in California, 2000-02. Avian Dis. 47:872–881 [DOI] [PubMed] [Google Scholar]
- 19. Hurt AC, Hansbro P, Selleck P, Olsen B, Minton C, Hampson AW, Barr IG. 2006. Isolation of avian influenza viruses from two different transhemisperic migratory shorebird species in Australia. Arch. Virol. 151:2301–2309 [DOI] [PubMed] [Google Scholar]
- 20. Peroulis I, O'Reiley K. 2004. Detection of avian paramyxoviruses and influenza viruses amongst wild bird populations in Victoria. Aust. Vet. J. 82:79–82 [DOI] [PubMed] [Google Scholar]
- 21. MacKenzie J. 1985. Isolation of avian influenza and paramyxoviruses from wild birds in Western Australia, p 336–339 In Della-Porta AJ. (ed), Veterinary viral diseases: their significance in south-east Asia and the western Pacific. Academic Press, Sydney, NSW, Australia [Google Scholar]
- 22. Mackenzie JS, Edwards E, Holmes R, Hinshaw V. 1984. Isolation of ortho- and paramyxoviruses from wild birds in Western Australia, and the characterization of novel influenza A viruses. Aust. J. Exp. Biol. Med. Sci. 62:89–99 [DOI] [PubMed] [Google Scholar]
- 23. Haynes L, Arzey E, Bell C, Buchanan N, Burgess G, Cronan V, Dickason C, Field H, Gibbs S, Hansbro P, Hollingsworth T, Hurt A, Kirkland P, McCracken H, O'Connor J, Tracey J, Wallner J, Warner S, Woods R, Bunn C. 2009. Australian surveillance for avian influenza viruses in wild birds between July 2005 and June 2007. Aust. Vet. J. 87:266–272 [DOI] [PubMed] [Google Scholar]
- 24. Barr IG, Deng YM, Iannello P, Hurt AC, Komadina N. 2008. Adamantane resistance in influenza A(H1) viruses increased in 2007 in South East Asia but decreased in Australia and some other countries. Antiviral Res. 80:200–205 [DOI] [PubMed] [Google Scholar]
- 25. Deng Y-M, Iannello P, Smith I, Watson J, Barr IG, Daniels P, Komadina N, Harrower B, Wong FYK. 2012. Transmission of influenza A(H1N1) 2009 pandemic viruses in Australian swine. Influenza Other Respi. Viruses 6:e42–e47. 10.1111/j.1750-2659.2012.00337.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 29. Drummond AJH, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shapiro B, Rambaut A, Drummond AJ. 2006. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 23:7–9 [DOI] [PubMed] [Google Scholar]
- 31. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. 10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jones DA, Wang W, Fawcett R. 2009. High-quality spatial climate data-sets for Australia. Aust. Meteorol. Oceanogr. J. 59:233–248 [Google Scholar]
- 33. Kingsford RT, Norman FI. 2002. Australian waterbirds—products of the continent's ecology. Emu 102:47–69 [Google Scholar]
- 34. Selleck PW, Arzey G, Kirkland PD, Reece RL, Gould AR, Daniels PW, Westbury HA. 2003. An outbreak of highly pathogenic avian influenza in Australia in 1997 caused by an H7N4 virus. Avian Dis. 47:806–811 [DOI] [PubMed] [Google Scholar]
- 35. Arzey G. 2013. Low pathogenic avian influenza in Australia and implications, p 4–17 Abstr. Proc. Aust. Vet. Poult. Assoc. Sci. Conf, Sydney, NSW, Australia [Google Scholar]
- 36. Xu X, Subbarao K, Cox NJ, Guo Y. 1999. Genetic characterization of the pathogenic A/Goose/Guangdong/1/1996 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261:15–19 [DOI] [PubMed] [Google Scholar]
- 37. Vijaykrishna D, Bahl J, Riley S, Duan L, Zhang JX, Chen H, Peiris JSM, Smith GJD, Guan Y. 2008. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 4:e1000161. 10.1371/journal.ppat.1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klaasen M, Hoye BJ, Roshier DA. 2010. Identifying crucial gaps in our knowledge of the life-history of avian influenza viruses—an Australian perspective. Emu 111:103–112 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.