Abstract
The coronavirus nucleocapsid (N) protein plays a multifunctional role in the virus life cycle, from regulation of replication and transcription and genome packaging to modulation of host cell processes. These functions are likely to be facilitated by interactions with host cell proteins. The potential interactome of the infectious bronchitis virus (IBV) N protein was mapped using stable isotope labeling with amino acids in cell culture (SILAC) coupled to a green fluorescent protein-nanotrap pulldown methodology and liquid chromatography-tandem mass spectrometry. The addition of the SILAC label allowed discrimination of proteins that were likely to specifically bind to the N protein over background binding. Overall, 142 cellular proteins were selected as potentially binding to the N protein, many as part of larger possible complexes. These included ribosomal proteins, nucleolar proteins, translation initiation factors, helicases, and hnRNPs. The association of selected cellular proteins with IBV N protein was confirmed by immunoblotting, cosedimentation, and confocal microscopy. Further, the localization of selected proteins in IBV-infected cells as well as their activity during virus infection was assessed by small interfering RNA-mediated depletion, demonstrating the functional importance of cellular proteins in the biology of IBV. This interactome not only confirms previous observations made with other coronavirus and IBV N proteins with both overexpressed proteins and infectious virus but also provides novel data that can be exploited to understand the interaction between the virus and the host cell.
INTRODUCTION
For positive-strand RNA viruses, the viral genomic RNA-binding protein (often referred to as a capsid or nucleocapsid [N] protein) plays a key role in virus biology, with functions in encapsidation and assembly, and for many viruses contributes toward the regulation of viral RNA synthesis (e.g., see reference 1). These proteins may also interact with host cell proteins both to facilitate their activities related to the virus life cycle and possibly to disrupt host cell signaling pathways (e.g., see reference 2). Coronaviruses and arteriviruses are positive-strand RNA viruses with common genome replication and expression strategies that encode an N protein that has multiple functions in the virus life cycle (3–8). Many of these functions may be mediated by subcellular localization (9–11) and multiple interactions with host cell proteins.
The coronavirus N proteins show high sequence homology within each genus but divergent homology between genera (12). This is illustrated by the conservation of functional regions within N proteins, such as RNA-binding motifs and membrane (M) protein binding domains (13). However, the location of such sites in the primary structure of the protein differs between genera. The N protein is best known as a viral structural protein, whose function is to bind the RNA genome to form a helical ribonucleoprotein (RNP) in mature virions (14). The N-terminal region of N protein has been implicated in RNA binding for infection bronchitis virus (IBV) (15–17), severe acute respiratory syndrome coronavirus (SARS-CoV) (18), mouse hepatitis virus (MHV) (19), and human coronavirus OC43 (20), as have additional sites in region 2 of MHV (21) and region 3 of IBV (22). Structural data suggested conserved tertiary structures between these viruses which may be involved in RNA binding (23). Phosphorylation has been shown to play a role in the discrimination of viral and nonviral RNA in IBV N protein (15, 24), with glycogen synthase kinase 3 (GSK3) being the acting kinase for SARS-CoV and MHV N protein. IBV N protein is postulated to bind RNA via a lure-and-lock mechanism (15, 16).
Aside from the structural functions, N protein has been implicated in several other processes. The SARS-CoV N protein has been shown to alter the activities of the AP-1, Akt, Erk, Jnk, and p38 mitogen-activated protein kinase signaling pathways as well inducing a block in the G0/G1 phase of the cell cycle (25–27). The association of N protein with elements of the viral genome as well as cellular RNA-binding factors strongly suggests a role in the transcription of subgenomic mRNAs. N protein has been shown to be essential for the efficient rescue of a number of infectious clones of coronavirus genomes, including IBV (28). While replication and transcription can occur in the absence of N protein, the efficiency of both processes requires N protein to be provided either in cis or in trans (4, 6, 29).
N protein colocalizes with cytoplasmic replication complexes at early time points postinfection in both transmissible gastroenteritis coronavirus and MHV and does so in a dynamic manner (30, 31), which agrees with the suggested involvement of the coronavirus N protein in virus replication (29). In a wide range of coronaviruses and arteriviruses, the N protein also localizes to the nucleolus (9, 32–34), a subnuclear structure whose key role is in ribosome biogenesis, but with additional roles in cell cycle regulation and the response to cell stress (35). Some variable degree of nuclear localization may also be observed, and many positive-strand RNA virus RNA-binding proteins have been shown to be located in the cytoplasm and nucleolus (for a review, see reference 36).
The interaction of coronavirus N proteins with host cell factors have not been subjected to extensive study, and therefore, the interactome of a coronavirus N protein was investigated through the combination of stable isotope labeling with amino acids in cell culture (SILAC)-based mass spectrometry (MS) and green fluorescent protein (GFP)-nanotrap pulldown methodologies. IBV N protein was chosen because both its biophysical properties (15–17, 24, 37–39) and subcellular localization to the cytoplasm and nucleolus (40–44) have been extensively characterized, as have the changes in the host cell proteome as a result of infection with IBV (45, 46).
MATERIALS AND METHODS
Expression and harvesting of EGFP and EGFP-N in 293T cells.
293T cells were cultured in SILAC-Dulbecco's modified Eagle medium (DMEM) lacking arginine (R) and lysine (K) supplemented with versions of these amino acids containing R0K0 (for enhanced GFP [EGFP] expression) and R6K4 (R = 13C-6, K = D-4) (for EGFP-IBV N expression). Cells were maintained in SILAC medium supplemented with 10% dialyzed fetal bovine serum (FBS) and 5% penicillin-streptomycin for at least 5 cell divisions to ensure >95% labeling of cells. For the passage of cells, trypsinization was avoided, with cell dissociation buffer (phosphate-buffered saline [PBS] based; Invitrogen) being used instead. Calcium phosphate was used to transfect cells with plasmid DNA. Per 10-cm2 dish, 500 μl of 2× HEPES-buffered saline and 1.5 mM Na2HPO4 (in which the pH was adjusted to 7) was added dropwise to a solution of 61 μl of 2 M CaCl2 and 10 μg of DNA (diluted in nuclease-free water); this solution was then added to 293T cells.
Lysis and immunoprecipitation of GFP-fusion proteins were performed using GFP-trap beads (Chromotek) that consist of a single-domain anti-GFP antibody conjugated to an agarose bead matrix. Cell pellets were incubated for 30 min with 200 μl lysis buffer, 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, and 1× EDTA-free protease inhibitor (Roche). The lysate was cleared by centrifugation and diluted 5-fold with dilution buffer comprising 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 1× EDTA-free protease inhibitor (Roche). The GFP-trap beads were equilibrated with ice-cold dilution buffer and then incubated with diluted cell lysate for 2 h at 4°C on a rotary mixer, followed by centrifugation at 2,700 × g for 2 min. The bead pellet was washed once with dilution buffer, followed by a single wash in buffer comprising 10 mM Tris-HCl, pH 7.5, 300 mM NaCl (concentrations of 150 mM and 500 mM were also tested [see below]), 0.5 mM EDTA, and 1× EDTA-free protease inhibitor (Roche). After centrifugation of the GFP-trap beads at 2,700 × g and removal of the wash buffer, the beads were resuspended in 2× SDS-sample buffer and boiled for 10 min to elute bound proteins. All stages of the processes were conducted on ice or at 4°C.
LC-MS/MS.
Protein samples generated by GFP-trap immunoprecipitations were separated by one-dimensional SDS-PAGE (4 to 12% bis-Tris Novex minigel; Invitrogen). The resulting separated proteins were cut from the gel in 10 slices and subjected to in-gel digestion with trypsin. Trypsin-digested peptides were separated using an Ultimate U3000 nanoflow liquid chromatography (LC) system (Dionex Corporation) consisting of a solvent degasser, micro- and nanoflow pumps, a flow control module, a UV detector, and a thermostated autosampler. A sample volume of 10 μl (comprising 2 μg) was loaded at a constant flow rate of 20 μl/min onto a PepMap C18 trap column (0.3 mm by 5 mm; Dionex Corporation). After trap enrichment, peptides were eluted onto a PepMap C18 nanocolumn (75 μm by 15 cm; Dionex Corporation) with a linear gradient of 5 to 35% solvent B (90% acetonitrile with 0.1% formic acid) over 65 min at a constant flow rate of 300 nl/min. The high-pressure liquid chromatography system was coupled to an LTQ Orbitrap XL instrument (Thermo Fisher Scientific Inc.) via a nanoelectrospray ion source (Proxeon Biosystems). The spray voltage was set to 1.2 kV, and the temperature of the heated capillary was set to 200°C. Full-scan MS survey spectra (m/z 335 to 1,800) in profile mode were acquired in the Orbitrap instrument with a resolution of 60,000 after accumulation of 500,000 ions. The five most intense peptide ions from the preview scan in the Orbitrap instrument were fragmented by collision-induced dissociation (normalized collision energy, 35%; activation Q, 0.250; activation time, 30 ms) in the LTQ instrument after the accumulation of 10,000 ions. Maximal filling times were 1,000 ms for the full scans and 150 ms for the MS/MS scans. Precursor ion charge state screening was enabled, and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 90 s and a relative mass window of 10 ppm. The lock mass option was enabled for survey scans to improve mass accuracy. The data were acquired using Xcalibur software.
Peptide quantification.
Quantification was performed with the MaxQuant program, version 1.0.7.4 (47), and was based on a two-dimensional centroid of the isotope clusters within each SILAC pair. The generation of the peak list, SILAC- and extracted ion current-based quantitation, calculation of the posterior error probability, calculation of a false discovery rate based on search engine results, peptide-to-protein group assembly, and data filtration and presentation were carried out using the MaxQuant program. The derived peak list was searched with the Mascot search engine (version 2.1.04; Matrix Science, London, United Kingdom) against a concatenated database combining 80,412 proteins from the International Protein Index (IPI) human protein database, version 3.6 (forward database), and the reversed sequences of all proteins (reverse database).
Data deposition.
LC-MS/MS data from this project for the IBV N protein were deposited in the Proteomics Identifications (PRIDE) database using the PRIDE converter tool (48, 49) and also the IntAct database (50, 51) and were deposited through the IntAct database to the IMEx data resource and assigned the identifier IM-15828.
Bioinformatic analysis.
Ingenuity pathway analysis (IPA; Ingenuity Systems) and STRING analysis were used to analyze the cellular protein data sets and to group proteins into similar functional classes and examine potential interactions. IPA networks were generated using data sets containing gene identifiers and corresponding expression values, which were uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB). Focus genes were overlaid onto a global molecular network developed from information contained in the IPKB. Networks of these focus genes were then algorithmically generated on the basis of their connectivity. Graphical representations of the molecular relationships between genes/gene products were generated. Genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature or from canonical information stored in the IPKB. Human, mouse, and rat orthologs of a gene are stored as separate objects in the IPKB but are represented as a single node in the network. STRING analysis was performed using version 9 (52), and STRING has been found to be a robust noncommercial algorithm for predicting protein-protein interactions (53).
Immunoblot analysis.
Polyvinylidene difluoride (PVDF) membranes (Millipore) were activated in 100% methanol and equilibrated by immersion in SDS-PAGE transfer buffer (25 mM Tris-HCl, 192 mM glycine, 20% [vol/vol] methanol). Transfer between the SDS-polyacrylamide gel and the membrane was performed using a Bio-Rad semidry blotting system according to the manufacturers' instructions. Transfers were performed at 15 V for 60 min. After transfer, PVDF membranes were blocked in 10% (wt/vol) skimmed milk powder (Marvel) made up in Tris-buffered saline containing Tween 20 (TBS-T; 50 mM Tris-HCl, pH 8.3, 150 mM NaCl, 0.5% [vol/vol] Tween 20) and incubated for 60 min. Blocking reagent was discarded, and primary antibody in TBS-T containing 5% (wt/vol) skimmed milk powder was added to the membrane and agitated for 60 min. Unbound antibody was removed by washing in TBS-T. Horseradish peroxidase-conjugated secondary antibody in TBS-T containing 5% (wt/vol) skimmed milk powder was then added to the membrane and the membrane was agitated for 60 min. Unbound antibody was removed by washing in PBS-Tween 20. Protein-antibody complexes were visualized using an enhanced chemiluminescence (ECL) system (Amersham Biosciences).
Ribosome cosedimentation.
Ribosome cosedimentation was performed on 293T cells. At 2 h prior to harvest, cells were treated with 100 μg/ml cycloheximide. A total of 5 × 106 cells were used for each experiment. These cells were resuspended in 1 ml of cytoplasmic lysis buffer (10 mM Tris-HCl [pH 7.4]. 2.5 mM MgCl2, 100 mM NaCl, 0.5% [vol/vol] NP-40 alternative) and incubated on ice for 5 min before passing the sample through a 25-gauge needle three times. The samples were then centrifuged at 1,500 × g for 2 min and then 1,500 × g for 15 min, with the supernatant retained at each stage. Sucrose gradients (5% and 20%) were made up in 10 mM Tris-HCl, pH 7.4, 100 mM NaCl, and 20 mM EDTA and passed through a 22-μm-pore-size filter. These were then used to generate 12-ml 5 to 20% sucrose gradients using a Hoefer 15 gradient maker. The cytoplasmic fraction (200 μl) was then layered over the gradient and centrifuged at 287,000 × g in a swinging-bucket rotor for 5 h at 4°C. Fractions (1 ml) were collected from the bottom of the tube by needle puncture.
Virus infection.
Cells of the Vero cell line (an African green monkey kidney-derived epithelial cell line) were maintained in DMEM supplemented with 10% fetal calf serum (FCS). IBV Beaudette US, a Vero cell-adapted isolate of Beaudette CK (28), was propagated in Vero cells. All cell culture experiments in this study were conducted in the absence of antibiotic or antifungal agents and were performed on actively replicating subconfluent cells (i.e., cells not undergoing contact inhibition). At 60% confluence, cells were infected with IBV and then incubated for 1 h at 37°C, after which the cells were incubated in maintenance medium; cells were then processed for analysis at 12 h postinfection.
Cell viability assay.
Cell viability/cytotoxicity during siRNA treatment was assessed using a colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Vero cells were seeded at a density of 3.8 × 103 (for 40 to 50% confluence) cells/well in clear 96-well microplates at 24 h prior to small interfering RNA (siRNA) treatment, following which 24 mg MTT powder was dissolved in 10 ml DMEM (plus 10% [vol/vol] FBS) at 37°C to make a 10 mM solution and the solution was filtered through a 0.2-μm-pore-size filter. The medium was aspirated, and the wells were washed with PBS. The absorbance at between 425 and 570 nm was then measured on a Dynex plate reader. A dose-response curve was then produced to assess cell viability.
siRNA-mediated knockdown.
For siRNA-mediated knockdown of target mRNAs, the following approach was used: two sequences for the same target gene were used alongside a negative control (Qiagen). The lyophilized oligomers were resuspended in nuclease-free water to make a 10 μM stock solution, and aliquots were further diluted to provide 10 nM working stocks. Lipofectamine 2000 was used (according to the manufacturer's instructions) to transfect Vero cells with siRNA. For a typical 24-well plate transfection, cells were seeded at a density of 3 × 104 in antibiotic-free DMEM at 24 h prior to transfection to achieve 30 to 40% confluence. Prior to transfection, the growth medium was replaced with 400 μl fresh antibiotic-free DMEM and 10 pmol of each siRNA pair (20 pmol for the negative control) and the DMEM and siRNA were mixed with 50 μl well Opti-MEM medium (Invitrogen) at room temperature for 5 min. In parallel, 1 μl/well Lipofectamine 2000 was mixed with 50 μl/well Opti-MEM medium at room temperature for 5 min. The dilute oligomers and the dilute Lipofectamine 2000 were then mixed gently and incubated at room temperature for 20 min. The oligomer-Lipofectamine 2000 complexes were then added to each well (100 μl/well) dropwise to give a final siRNA concentration of 20 pmol total siRNA/100 μl Opti-MEM medium. Control wells of reagent only and medium only were included to assess any background reactivity.
Confocal imaging.
Cells were fixed using a 4% (wt/vol) solution of paraformaldehyde made up in PBS and incubated for 10 min at 20°C. Following fixation, cells were made permeable with 0.2% (vol/vol) Triton X-100 made up in PBS for 10 min at 20°C. Primary antibody was added in PBS containing 1% FBS, and primary and secondary antibodies were tested to ensure no cross-reactivity. Slides were imaged on an LSM 510 META confocal microscope (Carl Zeiss). All images were captured using either a ×40 or a ×63 objective and a digital zoom factor of 1 to 4 within the software. The rainbow feature of the software was used to ensure that images were taken wherever possible within the linear range (unless stated otherwise).
RESULTS
To investigate the cellular interactions of the IBV N protein, a SILAC-based quantitative proteomic approach coupled to an immunoprecipitation strategy based on expression of an EGFP-N fusion protein in cells was used (Fig. 1A). Expressing the target protein as an EGFP-fusion protein has been shown to improve sensitivity and allow discrimination of specific from nonspecific interactions with the target protein (54, 55).
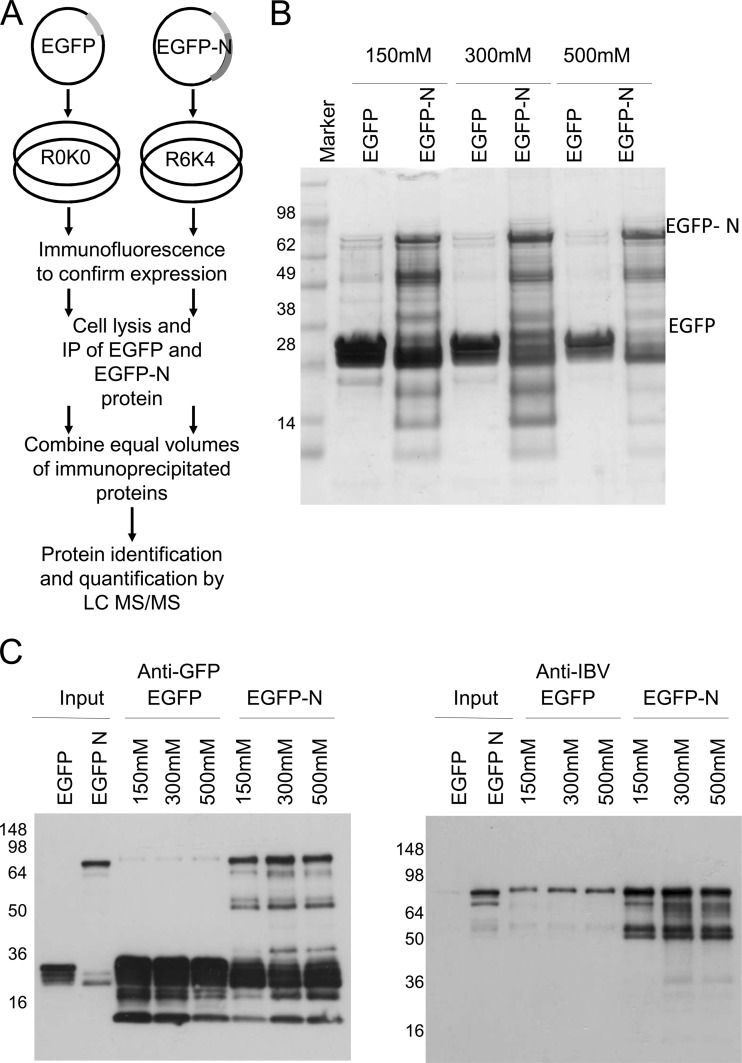
Fig 1.
(A) Strategy for analyzing cellular interacting partners of IBV N protein via immunoprecipitation of an EGFP-N protein expressed in cells labeled by SILAC. Immunoprecipitations (IPs) were carried out using GFP-trap beads to minimize nonspecific binding. Labeling by SILAC was employed, as it allowed less stringent wash conditions to be employed, as contaminating proteins should be present in roughly equal amounts in both samples. Proteins showing an increased abundance with EGFP-N as the bait versus EGFP as the control bait are more likely to represent true interactions. Various stages in this process are highlighted. (B) Silver-stained SDS-polyacrylamide gel showing EGFP and EGFP-N pulldowns at various salt concentrations (indicated). (C). Immunoblot analysis of EGFP and EGFP-N pulldowns at various salt concentrations (indicated) to test the effects of buffer conditions on protein binding. Numbers to the left of the gels are molecular masses (in kilodaltons).
Optimization of immunoprecipitation conditions.
From previous experience investigating proteome changes in IBV-infected cells using various model cell lines (Vero [primate-derived] and DF1 [avian-derived] cells) (45, 46), a human cell line was used in this study to provide cellular proteins with potential interaction with N protein. This was due to the superior annotation of the human genome allowing protein identification. 293T cells were also chosen because of their efficiency in taking up exogenous plasmid DNA by calcium phosphate (>45% by fluorescence-activated cell sorting; data not shown) compared to that of Vero or DF1 cells, thus allowing higher exogenous protein expression, which could be monitored using the EGFP tag. IBV N protein was expressed in 293T cells as part of an EGFP fusion protein (EGFP-N), as previously described (41–44), and the GFP-trap system was then applied to allow isolation of potential interaction partners from the cell lysates.
Various concentrations of NaCl (150, 300, and 500 mM) were tested in the wash buffer to determine the optimum binding conditions. Immunoprecipitated samples were analyzed by SDS-PAGE, and proteins were visualized by silver staining (Fig. 1B). EGFP alone gave heavy staining between the 28- and 38-kDa marker bands, with a few faint bands being present at different molecular masses under the 150 mM NaCl wash condition. These products became less apparent as the concentration of NaCl was increased. In contrast, immunoprecipitation from the EGFP-N samples indicated a large number of heavily stained products from a low molecular mass (14 kDa) upwards. A densely staining band at approximately 75 kDa was identified as the EGFP-N fusion protein. At either a 150 mM or a 300 mM NaCl concentration, silver-stained bands in the EGFP-N samples did not show significant decreases in their intensity. However, at 500 mM NaCl, intensity differences became more apparent. Immunoblot analysis using either an anti-EGFP antibody or an anti-IBV antibody confirmed the expression and immunoprecipitation of EGFP and EGFP-N, respectively (Fig. 1C). Note that the species visualized migrating between the 98- and 64-kDa markers in the anti-GFP and anti-IBV EGFP samples was present in other pulldown samples, and we postulate that it represents nonspecific binding of a host protein by the GFP-trap, with the apparent high level of enrichment leading to antibody cross-reactivity. Taken together, a 300 mM NaCl concentration was used in the wash buffer, as this appeared to yield a decrease in nonspecific binding in the EGFP pulldown samples while maintaining staining patterns in the EGFP-N samples. Further, SILAC coupled to LC-MS/MS was used to discriminate between background and specific interactions with the N moiety.
LC-MS/MS analysis.
SILAC-labeled samples (R0K0 EGFP, R6K4 EGFP-N) were immunoprecipitated, and equal volumes were combined and analyzed by LC-MS/MS to identify and quantify target proteins (45, 56, 57). Out of the 245 cellular proteins identified, 232 were also quantified. This data set and the associated mass spectra were uploaded to the PRIDE repository (49). For many quantitative proteomics analyses, an arbitrary 2-fold cutoff has been used to select potential true versus nonspecific interactions. When applied to this data set, 142 cellular proteins met this criterion (Table 1). These proteins were used in downstream analysis and also deposited with the IntAct database.
Table 1.
SILAC ratios of proteins identified from the IBV N protein immunoprecipitation experimenta
| Gene name | Binding ratio | No. of peptides | Sequence coverage |
|---|---|---|---|
| RPL19 | 15.78 | 3 | 18.4 |
| RPL35 | 14.98 | 5 | 30.1 |
| CAPRIN1 | 14.32 | 9 | 12.8 |
| G3BP-2 | 14.02 | 11 | 29.5 |
| RPL31 | 13.99 | 3 | 27.3 |
| RPL13 | 13.45 | 2 | 9 |
| RPS18 | 13.20 | 16 | 69.7 |
| RPS3A | 12.23 | 14 | 48.9 |
| G3BP-1 | 12.21 | 14 | 38.8 |
| ALYREF | 11.94 | 2 | 6.8 |
| RPS9 | 11.89 | 3 | 9.3 |
| RPS19 | 11.55 | 14 | 62.1 |
| RPS16 | 10.67 | 13 | 63 |
| RPS5 | 10.26 | 11 | 42.6 |
| RPL24 | 9.81 | 4 | 21.4 |
| RPS8 | 9.76 | 8 | 35.9 |
| RPL26 | 9.59 | 8 | 48.4 |
| RPL22 | 9.54 | 4 | 48.4 |
| YBX1 | 9.47 | 9 | 43 |
| RPS11 | 9.45 | 10 | 54.4 |
| RPS10 | 9.39 | 8 | 33.9 |
| RPS17 | 9.07 | 7 | 65.2 |
| RPS25 | 9.07 | 8 | 46.4 |
| SFRS4 | 8.94 | 2 | 3.2 |
| RPS3 | 8.92 | 18 | 69.1 |
| RPL4 | 8.74 | 7 | 19.7 |
| RPS14 | 8.54 | 7 | 40.4 |
| RPS26 | 8.40 | 4 | 37.4 |
| SFRS3 | 8.30 | 3 | 24.4 |
| RPS23 | 8.15 | 5 | 29.4 |
| RPS12 | 8.02 | 8 | 63.6 |
| RPL15 | 8.00 | 4 | 21.6 |
| RPS13 | 7.97 | 8 | 45 |
| HIST1H1D | 7.79 | 5 | 17.2 |
| RPS29 | 7.72 | 5 | 29.6 |
| GSK3A | 7.60 | 5 | 13 |
| RPS15A | 7.60 | 8 | 46.9 |
| RPL13A | 7.45 | 3 | 16.7 |
| RPS15 | 7.43 | 4 | 22.1 |
| RPL23 | 7.24 | 3 | 27.1 |
| RPL22L1 | 7.19 | 5 | 51.6 |
| RPL7 | 7.03 | 7 | 25.7 |
| RPL18A | 6.96 | 4 | 22.2 |
| RPL10 | 6.94 | 7 | 35.2 |
| RPS7 | 6.88 | 8 | 47.4 |
| RPL18A | 6.78 | 4 | 27.8 |
| RPS4 | 6.61 | 14 | 48.3 |
| NCL | 6.58 | 22 | 29.7 |
| RPL21 | 6.38 | 2 | 11.2 |
| RPL17 | 6.33 | 5 | 32.1 |
| RPS24 | 6.33 | 2 | 20.3 |
| DDX5 | 6.29 | 14 | 23.8 |
| RPL8 | 6.26 | 3 | 13.4 |
| HNRNPA0 | 6.26 | 4 | 17.7 |
| c14orf166 | 5.91 | 3 | 11.9 |
| RPL3 | 5.83 | 4 | 8.9 |
| RBM3 | 5.76 | 2 | 19.1 |
| USP10 | 5.55 | 5 | 9.1 |
| RPL27A | 5.51 | 3 | 31.1 |
| XRCC6 | 5.51 | 15 | 28.1 |
| DDX21 | 5.48 | 13 | 21.1 |
| RPL9 | 5.42 | 5 | 28.6 |
| NONO | 5.42 | 12 | 25.3 |
| SFRS1 | 5.34 | 6 | 29.8 |
| RPL27 | 5.19 | 2 | 22.9 |
| SFPQ | 5.19 | 10 | 17.5 |
| HNRNPD | 5.15 | 9 | 29.9 |
| RPL10A | 5.15 | 7 | 36.5 |
| RPL23A | 5.00 | 3 | 17 |
| DDX1 | 4.99 | 13 | 24.5 |
| XRCC5 | 4.97 | 12 | 19.7 |
| HNRNPA/B | 4.95 | 8 | 31.6 |
| SSB | 4.89 | 7 | 20.1 |
| RPL5 | 4.79 | 6 | 26.6 |
| HNRNPA1 | 4.69 | 16 | 43 |
| GSK3B | 4.69 | 5 | 15.9 |
| C22orf28 | 4.60 | 11 | 27.5 |
| RPL0 | 4.53 | 7 | 28.4 |
| RPL14 | 4.41 | 3 | 15 |
| SFRS2 | 4.37 | 2 | 10.9 |
| HNRNPQ | 4.35 | 13 | 26.5 |
| CSDA | 4.34 | 6 | 25.8 |
| RPL30 | 4.34 | 6 | 58.3 |
| DDX3X | 4.32 | 10 | 18.3 |
| RPL11 | 4.31 | 6 | 33.1 |
| DDX17 | 4.31 | 16 | 22.8 |
| DHX9 | 4.13 | 32 | 29.8 |
| FAM98A | 4.10 | 5 | 12.1 |
| HNRNPA3 | 4.07 | 6 | 25.7 |
| PABP1 | 4.05 | 34 | 48.1 |
| DHX30 | 4.01 | 13 | 12.8 |
| HNRNPU | 3.96 | 21 | 25.2 |
| FUS | 3.92 | 4 | 10.8 |
| SNRPD3 | 3.85 | 3 | 22.9 |
| RPL12 | 3.84 | 6 | 48.5 |
| SERBP1 | 3.82 | 10 | 33.1 |
| BLM | 3.77 | 2 | 1.6 |
| RPSA | 3.77 | 10 | 38 |
| HNRNPR | 3.77 | 5 | 9 |
| HNRNPA2/B1 | 3.75 | 15 | 45 |
| NPM1 | 3.57 | 10 | 37.1 |
| RPLP1 | 3.56 | 2 | 28.9 |
| LARP1 | 3.45 | 28 | 31.5 |
| RBM14 | 3.40 | 2 | 3.6 |
| SFRS7 | 3.39 | 2 | 8.8 |
| YTHDF2 | 3.38 | 5 | 10.2 |
| GNB2L1 | 3.33 | 13 | 50.1 |
| HNRNPG | 3.30 | 2 | 6.6 |
| IGF2BP1 | 3.29 | 20 | 40.2 |
| PTBP1 | 3.27 | 11 | 26.8 |
| PABP4 | 3.25 | 30 | 41.5 |
| DHX36 | 3.23 | 6 | 7 |
| MOV10 | 3.21 | 3 | 3.6 |
| DHX57 | 3.18 | 2 | 1.7 |
| H1FX | 3.11 | 3 | 21.1 |
| ATXN2L | 3.09 | 3 | 2.8 |
| FMR1 | 3.05 | 3 | 4.9 |
| DHX15 | 3.01 | 2 | 2.9 |
| HNRNPH1 | 3.01 | 6 | 19.5 |
| HNRNPF | 2.98 | 3 | 10.6 |
| H2AFX | 2.97 | 4 | 37.9 |
| HNRNPK | 2.95 | 12 | 35.6 |
| ILF2 | 2.91 | 3 | 10.3 |
| DDX6 | 2.80 | 2 | 4.3 |
| LRRC59 | 2.78 | 3 | 11.4 |
| HNRNPM | 2.69 | 10 | 17.3 |
| HDLBP | 2.68 | 9 | 8.9 |
| HNRNPH3 | 2.54 | 2 | 8.4 |
| PARP1 | 2.50 | 7 | 8.6 |
| ELAV1 | 2.36 | 5 | 16.4 |
| HNRNPC | 2.36 | 5 | 19.6 |
| ILF3 | 2.35 | 13 | 16.3 |
| MYL6 | 2.35 | 2 | 16.6 |
| RPS27A | 2.30 | 5 | 42.3 |
| ZCCHC3 | 2.29 | 2 | 6.4 |
| HNRPDL | 2.24 | 5 | 11.7 |
| LRPPRC | 2.15 | 12 | 11 |
| MYH9 | 2.13 | 34 | 22.5 |
| IGF2BP3 | 2.11 | 12 | 25.9 |
| HNRNPL | 2.07 | 2 | 4.1 |
| SND1 | 2.05 | 2 | 2.5 |
| RENT1 | 2.03 | 20 | 22 |
Ratios are organized from the potential strongest interaction to the weakest interaction. Shown are the gene name of the protein, binding ratio (EGFP-N/EGFP), number of different peptides used to identify the protein, and, from this value, the sequence coverage (which is calculated by dividing the number of amino acids observed by LC-MS/MS by the protein amino acid length). Proteins identified to have one peptide and/or a binding ratio of less than 2.00 have been excluded.
Bioinformatic analysis of the interactome data set.
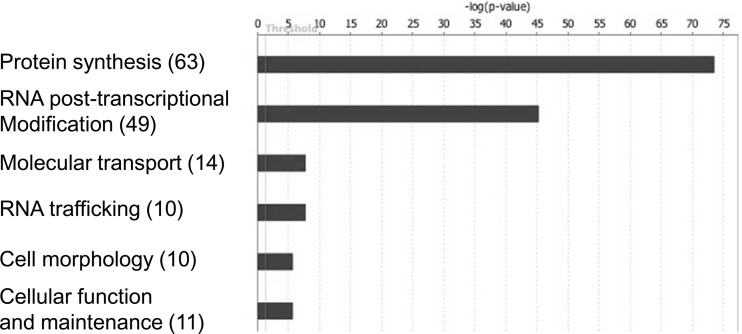
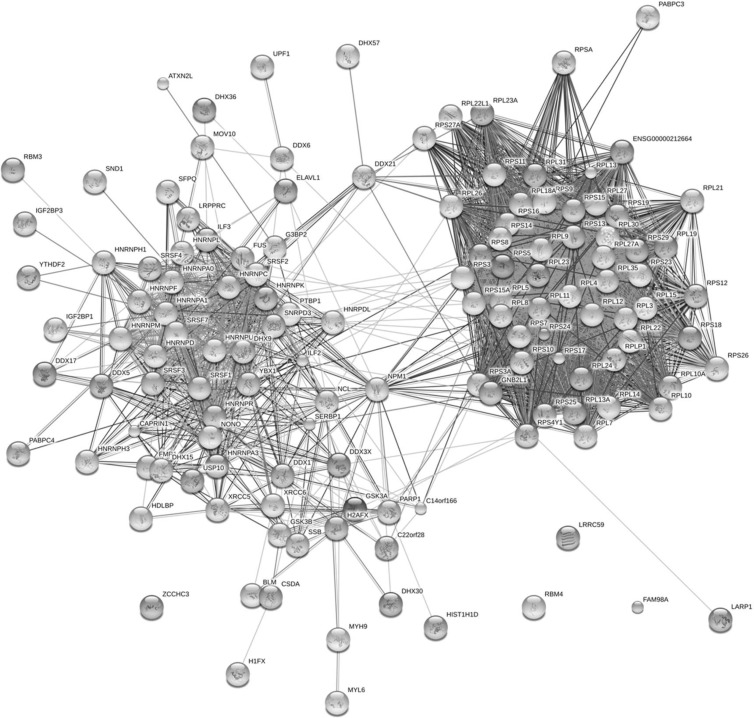
Ingenuity pathway analysis was used to investigate whether the identified proteins grouped into distinct functional clusters. This revealed that the identified proteins were mainly involved in protein synthesis and RNA posttranscriptional modification (Fig. 2). Independent analysis of the potential network interactions using STRING revealed clustering of proteins into those involved in translation (Fig. 3, right) and RNA processing/modification (Fig. 3, left). Many proteins were associated with the large (28 proteins) and small (21 proteins) ribosomal subunits, as well as 16 hnRNP proteins with other groups, including the DDX and DHX RNA helicases and splicing factors and the kinases GSK3α and GSK3β.
Fig 2.
Bioinformatic analysis of the EGFP-N protein interactome using ingenuity pathway analysis showing the data organized into functional groupings in the cell. Numbers in parentheses indicate the number of proteins identified in each grouping.
Fig 3.
STRING (version 9.0) analysis of the cellular proteins identified by the SILAC pulldown approach to be potentially interacting with IBV N protein. Proteins group into two large units: those associated with translation (right) and those involved in RNA modification/processing (left).
The efficiency of binding to EGFP-N over that to EGFP does not reflect the abundance of the proteins in the cellular proteome.
One hypothesis that could explain the data set was that the degree of enrichment of binding to EGFP-N merely reflected the abundance of the specific protein in the cellular proteome; i.e., the most abundant cellular proteins would be overrepresented. To investigate this, we compared the EGFP-N data set with data in the PaxDb: Protein Abundance across Organisms database, which ranks proteins according to their abundance in various human cell types on the basis of spectral counting data. The eight most abundant proteins in human cells at the time of analysis were recorded as APOA2, RBP4, APOC2, ALB, TTR, APOA1, APOC1, and APOC3; however, none of these were part of the 142 proteins associated with EGFP-N. RPL19 was the most enriched protein out of the 142, and this is ranked the 446th most abundant out of 12,797 cellular proteins. RENT1, which was the least enriched protein out of the 142 selected, is ranked 1,564 out of 12,797. Between these two proteins were proteins that occurred with different frequencies. For example, GSK3α and GSK3β are ranked the 3,755th and 2,962nd most abundant out of 12,797 proteins, respectively, and G3BP-2 is ranked 1,568 out of 12,797. Therefore, there is no apparent correlation between the abundance of a protein in a cell and its association with N protein.
Validation of SILAC immunoprecipitation results by immunoblotting.
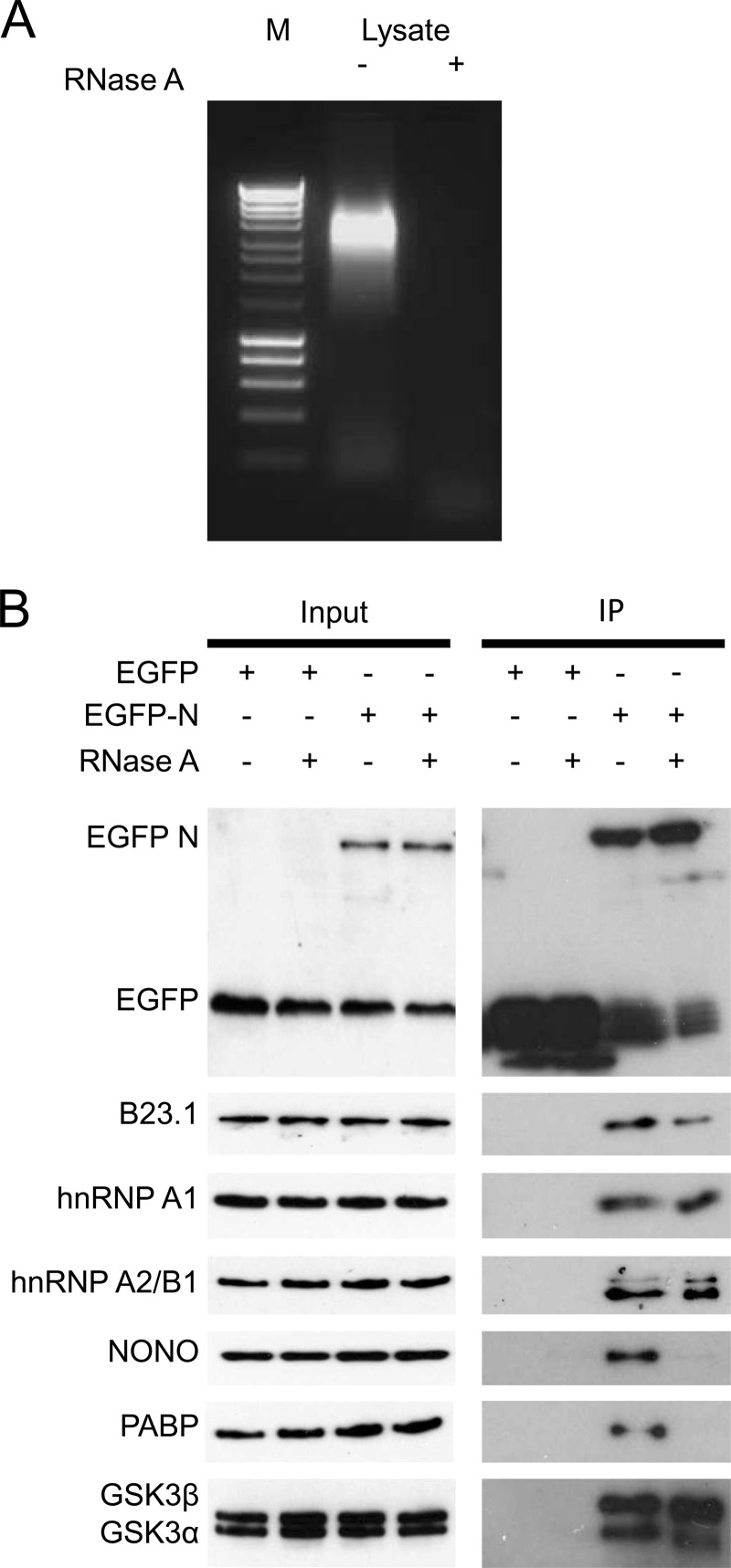
To increase confidence in the SILAC EGFP-N pulldown results, the experiment was repeated in the absence of label and immunoblotting was used to confirm the identity of N-interacting cellular proteins using specific antibodies (Fig. 4). As N protein is a known RNA-binding protein (24), analysis of the EGFP-N protein input lysate (to be used in the pulldown assays) indicated the presence of RNA (Fig. 4A), and this interaction may also determine an association with some cellular proteins. Examples of proteins used for validation from the data set were taken from Table 1. All of the proteins were confirmed to be interacting with the EGFP-N protein, with the interaction with NONO and poly(A)-binding protein (PABP) being potentially mediated by RNA (Fig. 4B). Other proteins, such as nucleolin (NCL), have previously been shown to interact with native IBV N protein in an RNA-independent manner (40). No identified proteins were shown to interact with EGFP (Fig. 3B). There is also some evidence from the EGFP blot that some EGFP may be cleaved from EGFP-N, and this may be through the action of caspase (58).
Fig 4.
Immunoblotting confirmation of the interactions of identified cellular proteins with EGFP-N protein. (A) Agarose electrophoresis analysis of a small aliquot of the input lysate (to be used in the pulldown assay) in the absence (−) and presence (+) of RNase. Lane M, a 100-bp DNA ladder. (B) The pulldown experiment was repeated in the presence and absence of RNase and immunoblot analysis of selected cellular proteins.
Interaction of IBV N with the small ribosomal subunit.
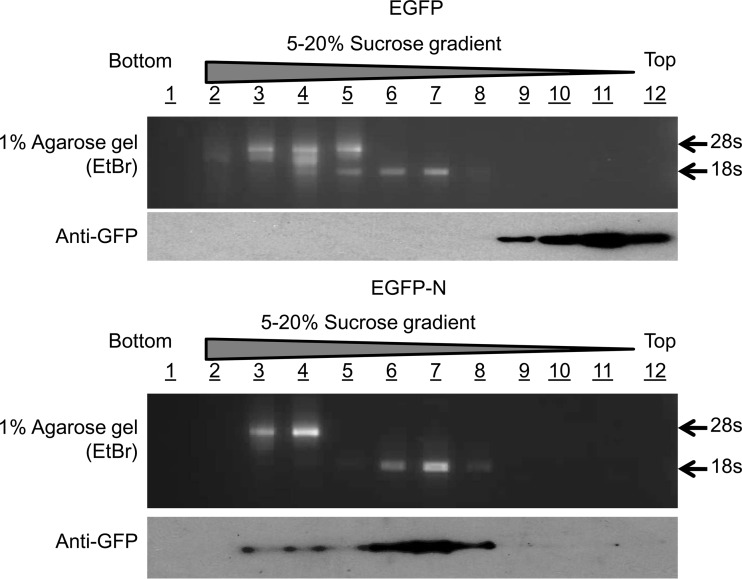
As over a third of the identified cellular proteins were ribosomal proteins, a potential interaction between N protein and ribosomal proteins was suggested. To investigate whether EGFP-N protein sedimented with either or both of the ribosomal subunits, 28S and 18S rRNAs were used to show the migration of the large (60S) and small (40S) ribosomal subunits, respectively (Fig. 5). In EGFP-expressing control cells, EGFP sedimented at the top of the gradient in fractions 9 to 12 and was most concentrated in fraction 11, while the ribosomal subunits occupied fractions 2 to 7. In contrast, EGFP-N protein was the most concentrated in fraction 7, the least concentrated in fraction 8, and at an intermediate level in fraction 6, a pattern duplicated by the 18S rRNA. This suggested a preference for one of the ribosomal subunits. The small ribosomal subunit is associated with translation initiation.
Fig 5.
Cosedimentation of EGFP-N protein with the small ribosomal subunit. Cosedimentation was performed on a 5 to 20% sucrose gradient in the presence of EDTA to induce separation of the large and small ribosomal subunits. The migration of these subunits was determined by extraction of RNA from the fractions and running of these on a 1% agarose gel containing ethidium bromide (EtBr), allowing visualization of the 18S and 28S rRNAs. Migration of the EGFP control or EGFP-N protein was determined by immunoblotting using an anti-GFP antibody.
Interaction of the EGFP-N protein with G3BP.
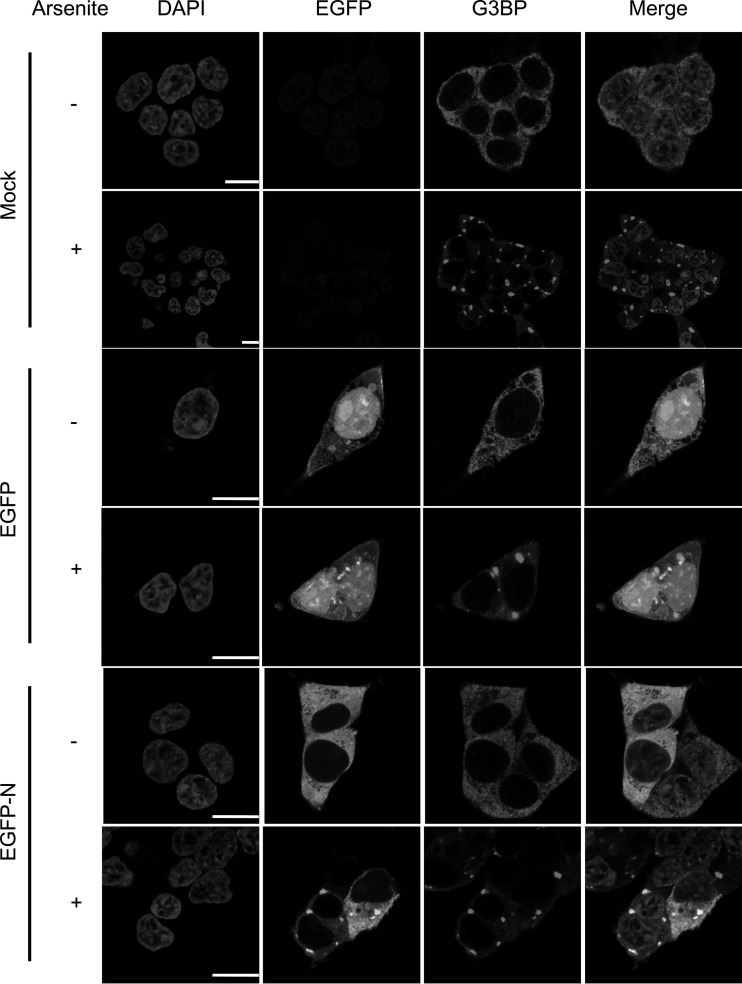
Three proteins with some of the highest potential binding ratios were caprin-1, G3BP-1, and G3BP-2 (Table 1) and are involved in the formation of cytoplasmic stress granules (59, 60). Indirect immunofluorescence confocal microscopy was used to investigate whether the EGFP-N protein colocalized with G3BP. This provided an independent validation of the interaction between these proteins separate from immunoblotting. To induce stress granule formation and, hence, G3BP, cells were treated with sodium arsenite (Fig. 6) and G3BP was used as a stress granule marker. In cells lacking stress granules, G3BP staining was diffuse and cytoplasmic, whereas in arsenite-stressed cells, G3BP was concentrated in punctate cytoplasmic foci. In EGFP-expressing cells, arsenite treatment successfully induced stress granule formation, with granules showing no enrichment of EGFP. In contrast, in EGFP-N protein-expressing cells, arsenite treatment caused EGFP-N protein to relocalize to G3BP-rich foci (Fig. 6, yellow signal), suggestive of an interaction between this protein and EGFP-N protein.
Fig 6.
Indirect immunofluorescent confocal microscopy showing colocalization of EGFP-N protein with the stress granule marker in G3BP in arsenite-stressed 293T cells. Cells were mock treated (−) or treated (+) with 0.5 mM sodium arsenite for 1 h prior to fixation. Nuclei are colored blue (DAPI [4′,6-diamidino-2-phenylindole]), EGFP is in green, G3BP is in red, and a merge image is presented. Colocalization is suggested by a yellow signal. Bars, 10 mm.
Potential relocalization and association of cellular proteins with N protein in IBV-infected cells.
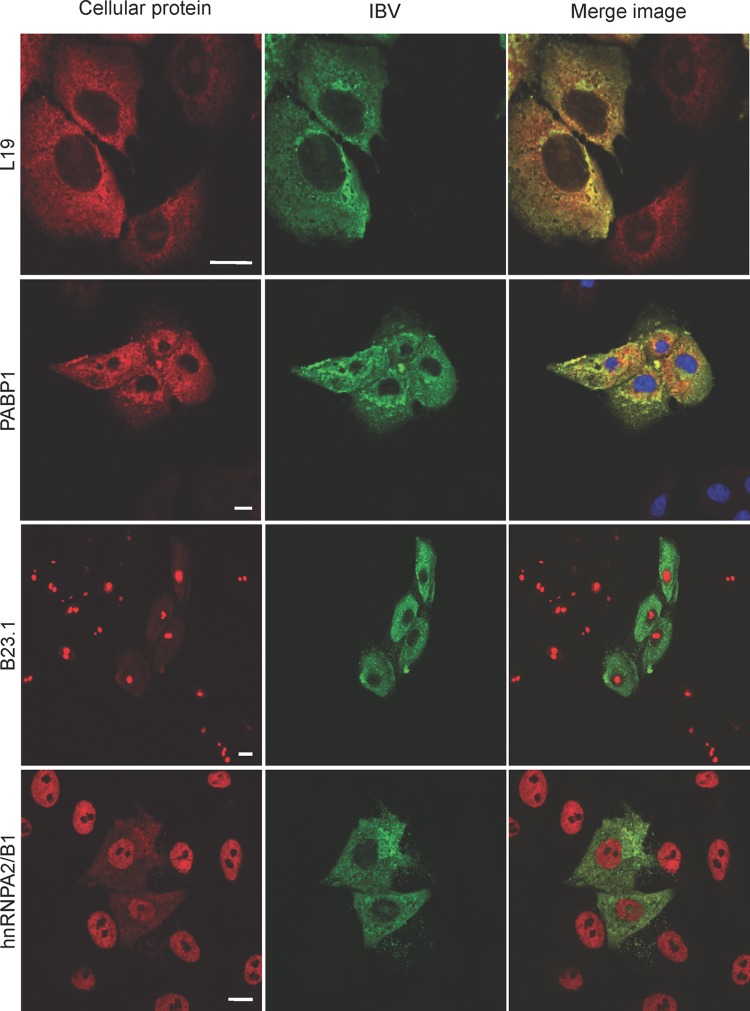
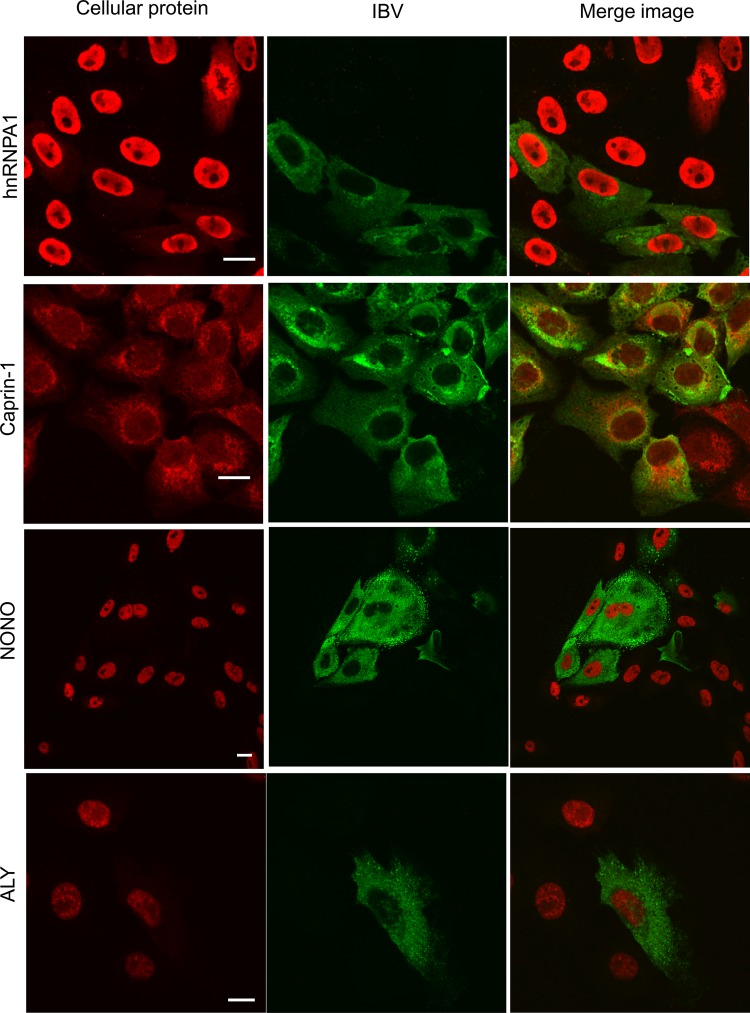
To investigate whether any of the cellular proteins that interacted with N protein were either redistributed in IBV-infected cells or colocalized with N protein, Vero cells were infected with IBV and subcellular localization was determined at 12 h postinfection (Fig. 7). Selection of cellular proteins was on the basis of a potential association and also the availability and avidity of antibodies. The ribosomal protein L19 (shown in red) localized to both the cytoplasm and the nucleolus both in bystander cells and also in infected cells, as would be predicted for a ribosomal protein. Additionally, the data indicated that L19 colocalized with N protein in infected cells (green), as shown by the yellow signal in the merged image. PABP (red) colocalized with N protein (green) in infected cells (yellow in the merge image). Both B23.1 (a nucleolar protein, indicated in red) and hnRNPA2/B1 (a nuclear protein, indicated in red) localized to the nucleolus and nucleus, respectively, in bystander cells. However, in infected cells (green), B23.1 (red) and hnRNPA2/B1 (red) localized not only to the nucleolus and nucleus but also to the cytoplasm, as shown by the red signal in the cytoplasm. Colocalization was not possible to investigate, given the difference in signal intensity in the cytoplasm between B23.1/hnRNPA2/B1 and N protein. In contrast, no relocalization or colocalization with N protein was observed with hnRNPA1, despite capturing the signal for hnRNPA1 beyond the linear range (i.e., red-saturated pixels). Likewise, no relocalization or colocalization with N protein was observed with caprin-1, NONO, or ALY in IBV-infected cells.
Fig 7.
Indirect immunofluorescent confocal microscopy analysis of the localization of selected cellular proteins that potentially interact with N protein in IBV-infected cells. Cellular proteins are shown in red, IBV-infected cells are shown in green, and a merged image is presented. Colocalization is suggested by a yellow signal. Bars, 10 mm.
Potential importance of the cellular proteins that associate with N protein in the viral life cycle.
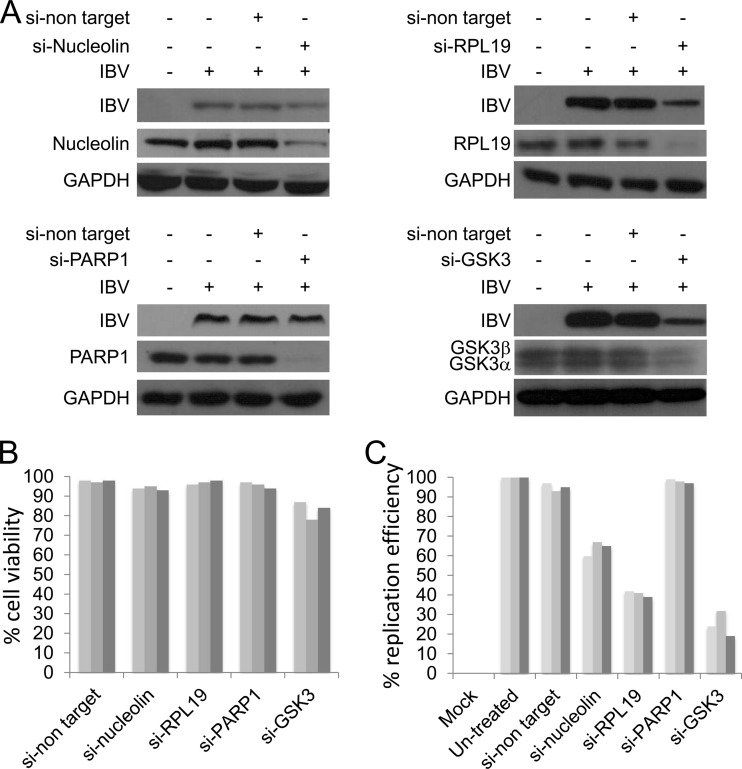
Several approaches have been used to investigate the importance of cellular proteins in a virus life cycle that interact with viral proteins; generally, these involved ablation or partial ablation of the cellular protein using siRNA (e.g., for IBV [61]). To investigate whether the identified cellular proteins that interacted with EGFP-N were involved in the replication of IBV, Vero cells were transfected with two specific siRNAs per mRNA directed toward selected cellular targets, prior to infection with IBV. The cellular targets were chosen on the basis of demonstrated siRNA knockdown and potential association with the N protein, including proteins of high and low probability. Four target proteins that matched these criteria were chosen: RPL19, GSK3α/GSKβ, nucleolin, and procyclic acidic repetitive protein 1 (PARP1). These proteins had EGFP-to-EGFP-N binding ratios of 15.78, 7.60/4.69, 6.58, and 2.5, respectively. In addition, cells were transfected with a nontargeting siRNA pair. Western blotting indicated that the abundance of all four proteins was decreased in the presence of the specific siRNA but not the nontargeting siRNA (Fig. 8A). However, the efficiency of ablation was different, with RPL19, PARP1, and GSK3 being the most efficiently depleted. Western blotting indicated that in cells depleted of nucleolin, RPL19, GSK3α, and GSK3β, the abundance of N protein decreased compared to that for the control (e.g., nontargeting siRNA-treated cells) or untreated cells (Fig. 8A). The observed decreased abundance of N protein was not due to any gross effects on cell viability (Fig. 8B), although there was an approximately 15 to 20% greater reduction in cell viability in GSK3-depleted cells than in the other treated cells. Given the decrease in the abundance of N protein in infected cells in which the abundance of nucleolin, RPL19, and GSK3 was decreased, we hypothesized that synthesis of viral RNA may be affected. To test this hypothesis, quantitative reverse transcription-PCR (qRT-PCR) using a specific primer pair to identify the genomic RNA was used to investigate the abundance of this molecule under the different conditions (Fig. 8C). The data indicated that the amount of genomic RNA was decreased in infected cells depleted of nucleolin, RPL19, or GSK3 but not in infected cells depleted of PARP1 or in untreated cells (Fig. 8C).
Fig 8.
Investigation of the proviral/antiviral activity of selected cellular proteins that potentially interact with N protein in IBV-infected Vero cells. (A) Representative immunoblots showing the abundance of the target protein and IBV N protein in mock-infected and infected Vero cells either treated with siRNAs targeting the specific mRNA of interest or treated with a nontarget siRNA control. (B) MTT cell viability assay of Vero cells treated with either the nontarget siRNA or the specific siRNA to the mRNAs encoding the selected protein of interest. The experiment was performed three times in triplicate for each condition. (C) qRT-PCR analysis of the abundance of the IBV genomic RNA in either mock-infected cells (mock) or cells treated with the nontarget siRNA or the siRNA specific to the mRNAs encoding the selected protein of interest. Replication efficiency is related to the level observed in untreated cells. si-non target, nontargeting siRNA; si-Nucleolin, siRNA targeting nucleolin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; si-PARP1, siRNA targeting PARP1.
DISCUSSION
This study represents the first work on the use of SILAC-based LC-MS/MS to study the interaction of the coronavirus N protein with host cell proteins and to tentatively identify the cellular interactome of this protein. The SILAC-based LC-MS/MS approach allowed the discrimination between cellular proteins that bound both to EGFP and/or the binding matrix and to N protein. Prior to this work, the sole known interactions confirmed for the IBV N protein were an interaction with the nucleolar protein nucleolin and colocalization (but not necessarily interaction) with fibrillarin (40). Studies carried out with different coronavirus N proteins demonstrated interactions with the kinase GSK3β (62), as well as the A-type hnRNPs (63), which offered the potential for conservation across the Coronaviridae. The approach described herein identified a potential of 142 cellular partners, including a large number of hnRNPs, ribosomal proteins, and the kinases GSK3α and GSK3β, and confirmation of the interaction with nucleolin as well as a novel interaction with B23.1 and with several stress granule marker proteins known to form a heterodimeric complex. N protein is unlikely to interact individually with all of the identified proteins, and these data are most likely to represent those for many complexes of cellular proteins that bind via a number of hub proteins, such as nucleolin (35).
Comparison to other viral proteins where we have used this approach, including the human respiratory syncytial virus (HRSV) NS1 and NS2 proteins (64), indicated that the IBV N data set was unique in terms of highlighting specific proteins and classes of proteins that potentially bound to the N protein. For example, ribosomal proteins were not the top proteins with binding to either HRSV NS1 or NS2. Similar to the analysis of the NS1 cellular interactome (64), the data indicated that the SILAC-based LC-MS/MS approach coupled to the GFP-trap immunoprecipitations could be used to identify protein complexes that bound to the viral protein of interest. Analysis of the related arterivirus PRRSV N protein (65, 79) indicated interaction with translation initiation factors, proteins involved in mRNA stability, hnRNPs, and DEAD RNA helicases. Although PRRSV N protein interacted with some ribosomal proteins, this was not necessarily the repertoire of ribosomal proteins associated with IBV N protein.
Results of repeat pulldown assays with EGFP-N protein confirmed the data in the mass spectrometry-based data set in the absence of label using immunoblotting and also confocal microscopy. Gene depletion analysis on selected targets using siRNA was used to demonstrate the potential functional relevance of cellular proteins in the biology of the virus, although off-target effects could not be ruled out. For example, depletion of GSK-3 in IBV-infected cells resulted in a decrease in the abundance of N protein and a reduction in the amount of genomic RNA compared to the results for the control treated cells. Although cells were generally viable, GSK3α and GSK3β are major cellular kinases and depletion of these molecules also likely affected other cellular pathways. However, GSK3β has previously been shown to be responsible for phosphorylation of the SARS-CoV and MHV N proteins (62). In addition, treatment of IBV-infected cells with lithium chloride (LiCl), which is a GSK3 inhibitor, resulted in decreased amounts of viral RNA and reduced viral titers (66, 67).
Interactions between the coronavirus N protein and hnRNPs have previously been reported for other members of the Coronaviridae, including SARS-CoV and MHV N proteins (68, 69), and have been shown through overexpression/mRNA depletion analysis to potentially be involved in coronavirus replication and transcription (70–72). The discovery that these hnRNP molecules bind N protein in an RNA-independent manner lends credence to the hypothesis that these proteins may be involved in replication and transcription of viral RNA, with at least some of the molecules able to functionally substitute for others in the event of their loss (72).
Ribosomal proteins also represented a large proportion of the cellular proteins identified in this analysis. Very limited data on the role of N protein in translation are available, but in MHV, N protein was found to favor the translation of viral over cellular transcripts, in a mechanism requiring N protein to bind to telomere repeat sequences (73, 74). There was a 4-fold increase in the efficiency of translation for reporter sequences containing MHV leader RNA compared to that for control sequences containing human alpha-globin or reverse complement leader sequence. EGFP-N was also shown to interact with various RNA helicases. One of these, DDX1, has also been shown to interact with IBV and SARS-CoV nonstructural protein 14 (61). Depletion of DDX1 in IBV-infected cells resulted in a reduction of IBV replication (61).
Several of the cellular proteins identified as interacting with the IBV N protein have also shown to be essential cofactors in the replication of other RNA viruses, indicating the importance of recruitment of cellular proteins in the life cycle of viruses. For example, C14orf166, which is involved in the modulation of mRNA transcription by polymerase II, has been shown to interact with the PA subunit of the influenza virus polymerase, and silencing causes a reduction in polymerase activity and a reduction in virus titer (75). DDX17, DDX5, NPM1, and HNRNPM were shown to be required for efficient activity of the influenza virus polymerase (76). The rubella virus capsid proteins binds to PABP and inhibits cellular protein synthesis (77). CAPRIN1 associates with the Japanese encephalitis virus core protein, and this interaction facilitates virus growth as well inhibits stress granule formation (78).
In summary, this study identified and characterized the potential cellular interactome of the coronavirus IBV N protein, using a SILAC-based quantitative proteomic methodology to allow discrimination of specific versus nonspecific interactions during the pulldown process. This interactome both confirms previous observations made with other coronavirus and IBV N proteins with both overexpressed proteins and infectious virus and also provides novel data that can be exploited to understand the interaction between the virus and the host cell.
ACKNOWLEDGMENTS
This work was supported by a BBSRC DTG Ph.D. studentship awarded to J.A.H. to support E.E., a BBSRC PDRA grant, and a Leverhulme Trust Research Fellowship awarded to J.A.H. A.W. was supported by a BBSRC Research Development Fellowship. This study was partially funded by the National Natural Science Foundation of China (no. U0931003/L01) and the National High Technology Research and Development Program (863 Program, no. 2011AA10A208-4) of the Ministry of Science and Technology of China to E.-M.Z.
Footnotes
Published ahead of print 1 May 2013
REFERENCES
- 1.Ilkow CS, Willows SD, Hobman TC. 2010. Rubella virus capsid protein: a small protein with big functions. Future Microbiol. 5:571–584 [DOI] [PubMed] [Google Scholar]
- 2.Ilkow CS, Weckbecker D, Cho WJ, Meier S, Beatch MD, Goping IS, Herrmann JM, Hobman TC. 2010. The rubella virus capsid protein inhibits mitochondrial import. J. Virol. 84:119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuniga S, Sola I, Moreno JL, Sabella P, Plana-Duran J, Enjuanes L. 2007. Coronavirus nucleocapsid protein is an RNA chaperone. Virology 357:215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuniga S, Cruz JL, Sola I, Mateos-Gomez PA, Palacio L, Enjuanes L. 2010. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 84:2169–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurst KR, Koetzner CA, Masters PS. 2009. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 83:7221–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79:6620–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowland RRR, Yoo D. 2003. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: a simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 95:23–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo D, Song C, Sun Y, Du Y, Kim O, Liu HC. 2010. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res. 154:48–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tijms MA, van der Meer Y, Snijder EJ. 2002. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 83:795–800 [DOI] [PubMed] [Google Scholar]
- 10.Lee C, Hodgins D, Calvert JG, Welch S-KW, Jolie R, Yoo D. 2006. Mutations within the nuclear localization signal of the porcine reproductive and respiratory syndrome virus nucleocapsid protein attenuate virus replication. Virology 346:238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pei Y, Hodgins DC, Lee C, Calvert JG, Welch S-KW, Jolie R, Keith M, Yoo D. 2008. Functional mapping of the porcine reproductive and respiratory syndrome virus capsid protein nuclear localization signal and its pathogenic association. Virus Res. 135:107–114 [DOI] [PubMed] [Google Scholar]
- 12.Lapps W, Hogue BG, Brian DA. 1987. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology 157:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enjuanes L, Almazan F, Sola I, Zuniga S. 2006. Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol. 60:211–230 [DOI] [PubMed] [Google Scholar]
- 14.Davies HA, Dourmashkin RR, Macnaughton MR. 1981. Ribonucleoprotein of avian infectious bronchitis virus. J. Gen. Virol. 53:67–74 [DOI] [PubMed] [Google Scholar]
- 15.Spencer KA, Dee M, Britton P, Hiscox JA. 2008. Role of phosphorylation clusters in the biology of the coronavirus infectious bronchitis virus nucleocapsid protein. Virology 370:373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spencer KA, Hiscox JA. 2006. Characterisation of the RNA binding properties of the coronavirus infectious bronchitis virus nucleocapsid protein amino-terminal region. FEBS Lett. 580:5993–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan H, Ooi A, Tan YW, Wang S, Fang S, Liu DX, Lescar J. 2005. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure 13:1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET. 2004. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 43:6059–6063 [DOI] [PubMed] [Google Scholar]
- 19.Nelson GW, Stohlman SA. 1993. Localization of the RNA-binding domain of mouse hepatitis virus nucleocapsid protein. J. Gen. Virol. 74(Pt 9):1975–1979 [DOI] [PubMed] [Google Scholar]
- 20.Huang CY, Hsu YL, Chiang WL, Hou MH. 2009. Elucidation of the stability and functional regions of the human coronavirus OC43 nucleocapsid protein. Protein Sci. 18:2209–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters PS, Parker MM, Ricard CS, Duchala C, Frana MF, Holmes KV, Sturman LS. 1990. Structure and function studies of the nucleocapsid protein of mouse hepatitis virus. Adv. Exp. Med. Biol. 276:239–246 [DOI] [PubMed] [Google Scholar]
- 22.Zhou M, Collisson EW. 2000. The amino and carboxyl domains of the infectious bronchitis virus nucleocapsid protein interact with 3′ genomic RNA. Virus Res. 67:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Tong X, Xu X, Li X, Lou Z, Rao Z. 2010. Structures of the N- and C-terminal domains of MHV-A59 nucleocapsid protein corroborate a conserved RNA-protein binding mechanism in coronavirus. Protein Cell 1:688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Gill A, Dove BK, Emmett SR, Kemp FC, Ritchie MA, Dee M, Hiscox JA. 2005. Mass spectroscopic characterisation of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding using surface plasmon resonance. J. Virol. 79:1164–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He R, Leeson A, Andonov A, Li Y, Bastien N, Cao J, Osiowy C, Dobie F, Cutts T, Ballantine M, Li X. 2003. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 311:870–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surjit M, Liu B, Chow VT, Lal SK. 2006. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 281:10669–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surjit M, Liu B, Jameel S, Chow VT, Lal SK. 2004. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 383:13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casais R, Thiel V, Siddell SG, Cavanagh D, Britton P. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359–12369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almazan F, Galan C, Enjuanes L. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 78:12683–12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verheije MH, Hagemeijer MC, Ulasli M, Reggiori F, Rottier PJ, Masters PS, de Haan CA. 2010. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 84:11575–11579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denison MR, Spaan WJ, van der Meer Y, Gibson CA, Sims AC, Prentice E, Lu XT. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowland RR, Kervin R, Kuckleburg C, Sperlich A, Benfield DA. 1999. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 64:1–12 [DOI] [PubMed] [Google Scholar]
- 33.Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. 2001. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 75:9345–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiscox JA, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G. 2001. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75:506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emmott E, Hiscox JA. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep. 10:231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiscox JA. 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan YW, Fang S, Fan H, Lescar J, Liu DX. 2006. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 34:4816–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jayaram H, Fan H, Bowman BR, Ooi A, Jayaram J, Collisson EW, Lescar J, Prasad BV. 2006. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 80:6612–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayaram J, Youn S, Collisson EW. 2005. The virion N protein of infectious bronchitis virus is more phosphorylated than the N protein from infected cell lysates. Virology 339:127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Wurm T, Britton P, Brooks G, Hiscox JA. 2002. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J. Virol. 76:5233–5250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed ML, Dove BK, Jackson RM, Collins R, Brooks G, Hiscox JA. 2006. Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic 7:833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reed ML, Howell G, Harrison SM, Spencer KA, Hiscox JA. 2007. Characterization of the nuclear export signal in the coronavirus infectious bronchitis virus nucleocapsid protein. J. Virol. 81:4298–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cawood R, Harrison SM, Dove BK, Reed ML, Hiscox JA. 2007. Cell cycle dependent nucleolar localization of the coronavirus nucleocapsid protein. Cell Cycle 6:863–867 [DOI] [PubMed] [Google Scholar]
- 44.Emmott E, Dove BK, Howell G, Chappell LA, Reed ML, Boyne JR, You JH, Brooks G, Whitehouse A, Hiscox JA. 2008. Viral nucleolar localisation signals determine dynamic trafficking within the nucleolus. Virology 380:191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emmott E, Rodgers MA, Macdonald A, McCrory S, Ajuh P, Hiscox JA. 2010. Quantitative proteomics using stable isotope labeling with amino acids in cell culture reveals changes in the cytoplasmic, nuclear, and nucleolar proteomes in Vero cells infected with the coronavirus infectious bronchitis virus. Mol. Cell. Proteomics 9:1920–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emmott E, Smith C, Emmett SR, Dove BK, Hiscox JA. 2010. Elucidation of the avian nucleolar proteome by quantitative proteomics using stable isotope labeling with amino acids in cell culture (SILAC) and alteration in the coronavirus infectious bronchitis virus infected cells. Proteomics 10:3558–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- 48.Barsnes H, Vizcaino JA, Eidhammer I, Martens L. 2009. PRIDE converter: making proteomics data-sharing easy. Nat. Biotechnol. 27:598–599 [DOI] [PubMed] [Google Scholar]
- 49.Vizcaino JA, Cote R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. 2009. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9:4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B, Thorneycroft D, Zhang Y, Apweiler R, Hermjakob H. 2007. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 35:D561–D565. 10.1093/nar/gkl958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, Jandrasits C, Jimenez RC, Khadake J, Mahadevan U, Masson P, Pedruzzi I, Pfeiffenberger E, Porras P, Raghunath A, Roechert B, Orchard S, Hermjakob H. 2012. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 40:D841–D846. 10.1093/nar/gkr1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41:D808–D815. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller T, Schrotter A, Loosse C, Helling S, Stephan C, Ahrens M, Uszkoreit J, Eisenacher M, Meyer HE, Marcus K. 2011. Sense and nonsense of pathway analysis software in proteomics. J. Proteome Res. 10:5398–5408 [DOI] [PubMed] [Google Scholar]
- 54.Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, Lamond A. 2008. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183:223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ten Have S, Boulon S, Ahmad Y, Lamond AI. 2011. Mass spectrometry-based immuno-precipitation proteomics—the user's guide. Proteomics 11:1153–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Munday DC, Emmott E, Surtees R, Lardeau CH, Wu W, Duprex WP, Dove BK, Barr JN, Hiscox JA. 2010. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol. Cell. Proteomics 9:2438–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emmott E, Wise H, Loucaides EM, Matthews DA, Digard P, Hiscox JA. 2010. Quantitative proteomics using SILAC coupled to LC-MS/MS reveals changes in the nucleolar proteome in influenza A virus-infected cells. J. Proteome Res. 9:5335–5345 [DOI] [PubMed] [Google Scholar]
- 58.Eleouet JF, Slee EA, Saurini F, Castagne N, Poncet D, Garrido C, Solary E, Martin SJ. 2000. The viral nucleocapsid protein of transmissible gastroenteritis coronavirus (TGEV) is cleaved by caspase-6 and -7 during TGEV-induced apoptosis. J. Virol. 74:3975–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolobova E, Efimov A, Kaverina I, Rishi AK, Schrader JW, Ham AJ, Larocca MC, Goldenring JR. 2009. Microtubule-dependent association of AKAP350A and CCAR1 with RNA stress granules. Exp. Cell Res. 315:542–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, Schrader JW. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27:2324–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. 2010. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J. Virol. 84:8571–8583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu CH, Yeh SH, Tsay YG, Shieh YH, Kao CL, Chen YS, Wang SH, Kuo TJ, Chen DS, Chen PJ. 2009. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J. Biol. Chem. 284:5229–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi ST, Lai MM. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W, Tran KC, Teng MN, Heesom KJ, Matthews DA, Barr JN, Hiscox JA. 2012. The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J. Virol. 86:7777–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jourdan SS, Osorio F, Hiscox JA. 2012. An interactome map of the nucleocapsid protein from a highly pathogenic North American porcine reproductive and respiratory syndrome virus strain generated using SILAC-based quantitative proteomics. Proteomics 12:1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison SM, Tarpey I, Rothwell L, Kaiser P, Hiscox JA. 2007. Lithium chloride inhibits the coronavirus infectious bronchitis virus in cell culture. Avian Pathol. 36:109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J, Yin J, Sui X, Li G, Ren X. 2009. Comparative analysis of the effect of glycyrrhizin diammonium and lithium chloride on infectious bronchitis virus infection in vitro. Avian Pathol. 38:215–221 [DOI] [PubMed] [Google Scholar]
- 68.Luo H, Chen Q, Chen J, Chen K, Shen X, Jiang H. 2005. The nucleocapsid protein of SARS coronavirus has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1. FEBS Lett. 579:2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Zhang X. 1999. The nucleocapsid protein of coronavirus mouse hepatitis virus interacts with the cellular heterogeneous nuclear ribonucleoprotein A1 in vitro and in vivo. Virology 265:96–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sola I, Mateos-Gomez PA, Almazan F, Zuniga S, Enjuanes L. 2011. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 8:237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi KS, Mizutani A, Lai MM. 2004. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 78:13153–13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi ST, Yu GY, Lai MM. 2003. Multiple type A/B heterogeneous nuclear ribonucleoproteins (hnRNPs) can replace hnRNP A1 in mouse hepatitis virus RNA synthesis. J. Virol. 77:10584–10593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tahara SM, Dietlin TA, Bergmann CC, Nelson GW, Kyuwa S, Anthony RP, Stohlman SA. 1994. Coronavirus translational regulation: leader affects mRNA efficiency. Virology 202:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tahara SM, Dietlin TA, Nelson GW, Stohlman SA, Manno DJ. 1998. Mouse hepatitis virus nucleocapsid protein as a translational effector of viral mRNAs. Adv. Exp. Med. Biol. 440:313–318 [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez A, Perez-Gonzalez A, Nieto A. 2011. Cellular human CLE/C14orf166 protein interacts with influenza virus polymerase and is required for viral replication. J. Virol. 85:12062–12066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, Chase G, Martinez-Sobrido L, Schwemmle M, Garcia-Sastre A. 2011. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2(4):e00151–11. 10.1128/mBio.00151-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ilkow CS, Mancinelli V, Beatch MD, Hobman TC. 2008. Rubella virus capsid protein interacts with poly(A)-binding protein and inhibits translation. J. Virol. 82:4284–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, Mori Y, Kamitani W, Matsuura Y. 2013. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with caprin-1 and facilitates viral propagation. J. Virol. 87:489–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jourdan SS, Osorio FA, Hiscox JA. 2012. Biophysical characterisation of the nucleocapsid protein from a highly pathogenic porcine reproductive and respiratory syndrome virus strain. Biochem. Biophys. Res. Commun. 419:137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]