Abstract
Host cells activate innate immune signaling pathways to defend against invading pathogens. To survive within an infected host, viruses have evolved intricate strategies to counteract host immune responses. Herpesviruses, including herpes simplex virus type 1 (HSV-1), have large genomes and therefore have the capacity to encode numerous proteins that modulate host innate immune responses. Here we define the contribution of HSV-1 tegument protein VP16 in the inhibition of beta interferon (IFN-β) production. VP16 was demonstrated to significantly inhibit Sendai virus (SeV)-induced IFN-β production, and its transcriptional activation domain was not responsible for this inhibition activity. Additionally, VP16 blocked the activation of the NF-κB promoter induced by SeV or tumor necrosis factor alpha treatment and expression of NF-κB-dependent genes through interaction with p65. Coexpression analysis revealed that VP16 selectively blocked IFN regulatory factor 3 (IRF-3)-mediated but not IRF-7-mediated transactivation. VP16 was able to bind to IRF-3 but not IRF-7 in vivo, based on coimmunoprecipitation analysis, but it did not affect IRF-3 dimerization, nuclear translocation, or DNA binding activity. Rather, VP16 interacted with the CREB binding protein (CBP) coactivator and efficiently inhibited the formation of the transcriptional complexes IRF-3–CBP in the context of HSV-1 infection. These results illustrate that VP16 is able to block the production of IFN-β by inhibiting NF-κB activation and interfering with IRF-3 to recruit its coactivator CBP, which may be important to the early events leading to HSV-1 infection.
INTRODUCTION
Herpes simplex virus type 1 (HSV-1) is a double-stranded DNA virus that undergoes productive replication in the nucleus of infected cells. The linear genome is packaged into a nucleocapsid that is released into the cytoplasm upon fusion of the viral and host cell membranes. Also released are the preformed tegument proteins, which play important roles in counteracting host defenses and stimulating viral gene expression. The tegument protein VP16 acts to stimulate immediate-early (IE) gene expression through the recruitment of general transcription factors and RNA polymerase II to the IE promoters (1), launching the temporal cascade of gene expression.
VP16 is an abundant 65-kDa virion phosphoprotein that is synthesized late in infection and subsequently packaged into virions (2). VP16 delivered by the infecting virions acts during the earliest stages of infection to stimulate transcription of the viral IE genes, thereby facilitating the onset of the lytic program of viral gene expression (3). Intensive studies have shown that the C-terminal portion of VP16 is a potent transcriptional activation domain and that VP16 is targeted to the TAATGARATTC consensus sequence found in IE promoters through interactions with the host factors Oct-1 and host cell factor. Mutations that inactivate the transcriptional activation function of VP16 result in reduced levels of IE gene expression during low-multiplicity infection and a greatly increased particle-to-PFU ratio, indicating that transactivation by VP16 increases the probability that cells infected with a single virus particle will enter the lytic cycle (4, 5). Such transactivation-deficient mutants can, however, be propagated in tissue culture, demonstrating that the activation function of VP16 is not essential for virus replication and assembly. In contrast, certain VP16 mutations prevent production of infectious progeny virus. For example, it has been demonstrated that the ts mutation in HSV-2 VP16 (ts 2203) is lethal, as are some in-frame linker insertion mutations in the HSV-1 VP16 gene (6). The ts 2203 mutation blocks virus assembly, arguing that VP16 plays an essential role in this process. Weinheimer et al. provided additional evidence supporting a role for VP16 in virion maturation by demonstrating that an HSV-1 VP16 null mutant (8MA) displayed a severe defect in virus assembly during infection of noncomplementing cells (7).
The innate immune system is the first line of defense in response to virus infection. Besides Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in the endosome and cytoplasm, respectively, RNA helicases such as retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA-5) are able to recognize characteristic patterns of invading pathogens and induce the production of type I interferons (IFNs), potent antiviral molecules (8, 9). In HSV-1-infected macrophages, MDA-5 was shown to be the primary mediator of HSV recognition using small interfering RNA knockdown (10). Expression of type I IFN genes has been found to be regulated by the so-called enhanceosome, constituted by the transcription factors IFN regulatory factors 3 and 7 (IRF-3/7), NF-κB, and ATF/c-Jun (11). Upon recognition of viral RNA species, RIG-I interacts with the mitochondrial antiviral signaling protein (MAVS; also known as IPS-1, VISA, and CARDIF) in the mitochondrial membrane. This leads to the phosphorylation and activation of both IRF-3 and IRF-7 by IKKε and TBK1 (12). Upon secretion, IFN binds to specific IFN receptors in an autocrine or paracrine manner and activates the JAK/STAT pathway. This leads to the formation of the IFN-stimulated gene factor 3 (ISGF3) transcription complex, which drives the expression of antiviral genes, such as protein kinase R (PKR), Mx GTPases, and others, for establishing an antiviral state in infected and neighboring noninfected cells (13, 14).
The transcriptional factors IRF-3 and IRF-7 play important roles in virus-induced type I interferon gene activation following virus infection (15, 16). Virus-induced C-terminal phosphorylation of IRF-3 promotes cytoplasmic-to-nuclear translocation, DNA binding, association with CREB binding protein (CBP)/p300 histone acetyltransferases, and transactivation of downstream target genes. IRF-3 possesses a restricted DNA binding site specificity and interacts with CBP/p300 coactivators, while IRF-7 has a broader DNA binding specificity that contributes to its capacity to stimulate delayed-type I IFN gene expression (17).
To survive within an infected host, viruses have evolved intricate strategies to counteract host immune responses. HSV-1 has a large genome and therefore has the capacity to encode numerous proteins that modulate host innate immune responses. Our previous studies demonstrated that HSV-1 tegument protein US11 is a novel antagonist of the IFN-β pathway and downregulates the Rig-like receptor (RLR) signaling pathway via direct interactions with both RIG-I and MDA-5 (18). In this study, we defined the contribution of HSV-1 tegument protein VP16 in the inhibition of IFN-β production. Our results indicated that VP16 efficiently inhibited the Sendai virus (SeV)-induced expression of endogenous IFN-β. Additionally, VP16 blocked both SeV infection-induced and tumor necrosis factor alpha (TNF-α)-induced activation of the NF-κB promoter and expression of NF-κB-dependent genes through interaction with p65. Coexpression analysis demonstrated that VP16 selectively blocked IRF-3-mediated but not IRF-7-mediated transactivation. Repression of IRF-3-mediated transcription by VP16 correlated with the capacity of VP16 to compete with IRF-3 in vivo for recruitment of the coactivator CBP in the context of HSV-1 infection.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
HEK 293T cells, HeLa cells, and Vero cells were grown in Dulbecco's modified minimal essential medium (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) as described previously (18, 19). The wild-type (WT) HSV-1 F strain virus and SeV were propagated and titers were determined as described previously (18). For UV inactivation, WT HSV-1 was exposed to short-wave UV light for 2 h prior to infection. Infections with UV-inactivated viruses were based on titers before UV irradiation.
Rabbit antisera against IRF-3-S396 were described previously (20). The protease inhibitor mixture cocktail, mouse anti-Myc (isotype IgG1), and anti-Flag (isotype IgG2b) monoclonal antibodies (MAbs) were purchased from CST (Boston, MA). Mouse anti-hemagglutinin (anti-HA) MAb (isotype IgG2b) was purchased from Roche (Mannheim, Germany). Mouse monoclonal IgG1 and IgG2b isotype control antibodies were purchased from eBioscience Inc. (San Diego, CA). Rabbit anti-IRF-3 polyclonal antibody (PAb), mouse anti-CBP MAb, and mouse anti-VP16 MAb were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Human recombinant TNF-α was purchased from Biovision (San Francisco, CA).
Plasmid construction.
All enzymes used for cloning procedures were purchased from TaKaRa (Dalian, China) except for T4 DNA ligase (New England BioLabs, MA). To construct VP16-HA and VP16-Flag, the VP16 gene was amplified from the HSV-1 genome as described in our previous study (21) and cloned into the BamHI and EcoRI sites of the pCMV-HA vector and pCMV-Flag (Beyotime, Shanghai, China), respectively. Additionally, the plasmids pVP16(1-210)-Flag, pVP16(1-410)-Flag, and pVP16(1-410)-HA were constructed similarly. Commercial reporter plasmids included NF-κB-Luc (Stratagene, La Jolla, CA) and pRL-TK (Promega). Gift plasmids included (PRDIII-I)4-Luc (22), pcDNA3.1-Flag-TBK1 and pcDNA3.1/Zeo-MAVS (23), pcDNA3.1-Flag-IKKi (24), pEF-Flag-MDA-5 and pEF-Flag-RIG-IN (25), Flag-IRF-3/5D and Flag-IRF-7/6D (26), pCAGGS-NS1 (27), pCMV-p65-Flag (28), Myc-IRF-3/5D, and Myc-IRF3(1-197), Myc-IRF3(1-394), and IFN-β promoter reporter plasmid IFN-β-Luc (29).
RNA isolation and real-time quantitative reverse transcription-PCR (qRT-PCR).
The cells were harvested, duplicate wells were pooled, and RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) as described in our previous studies (18, 30). RNA was quantified based on absorbance at an optical density at 260 nm, and 2 μg of DNase-treated RNA was reverse transcribed using OmniScript (Sigma-Aldrich, St. Louis, MO) and oligo(dT) primers (Invitrogen, Carlsbad, CA). The resulting cDNA product was amplified using the ABI Prism 7700 sequence detector in the presence of 2× SYBR green master mix (Applied Biosystems, Foster City, CA) and 0.4 μM (each) forward and reverse primers, resulting in the amplification products. The abundance of each mRNA was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression and compared to that in untreated cells to calculate the relative induction. The denaturing and annealing/extension conditions for each cycle of PCR were 95°C for 15 s and 60°C for 1 min, respectively. The primer sets used for each gene of interest were as follows: for GAPDH, 5′-TGACCTCAACTACATGGTTTACATGT-3′ and 5′-AGGGATCTCGCTCCTGGAA-3′; for IFN-β, 5′-AAACTCATGAGCAGTCTGCA-3′ and 5′-AGGAGATCTTCAGTTTCGGAGG-3′; for interleukin-6 (IL-6), 5′-GGATTCAATGAGGAGACTTGCC-3′ and 5′-ACAGCTCTGGCTTGTTCCTCAC-3′; for IL-8, 5′-CAACACAGAAATTATTGTAAAGCTTTCT-3′ and 5′-GAATTCTCAGCCCTCTTCAAAAA-3′; for ISG54, 5′-ACGGTATGCTTGGAACGATTG-3′ and 5′-AACCCAGAGTGTGGCTGATG-3′; for ISG56, 5′-AAGGCAGGCTGTCCGCTTA-3′ and 5′-TCCTGTCCTTCATCCTGAAGCT-3′.
Transfection and dual luciferase reporter (DLR) assay.
HEK 293T cells were cotransfected with reporter plasmids, such as IFN-β-Luc, NF-κB-Luc, (PRDIII-I)4-Luc, and ISRE-Luc and internal control plasmid pRL-TK, with or without expression plasmids, as indicated, by standard calcium phosphate precipitation (31, 32). At 24 h posttransfection, cells were infected with 100 HA units (HAU) of SeV ml−1 for 16 h or treated with IFN-β or TNF-α, and then luciferase assays were performed with a dual-specific luciferase assay kit (Promega, Madison, WI) as described in our previous studies (18, 30).
Coimmunoprecipitation assay and Western blot analysis.
Coimmunoprecipitation (co-IP) assays and Western blot (WB) analysis were performed as described in our previous studies (18, 19). Briefly, HEK 293T cells (∼5 × 106) were cotransfected with 10 μg of each of the indicated expression plasmids carrying Flag, Myc, or HA tags. For virus infection, the cells were then infected with HSV-1 for an additional 16 h at 20 h posttransfection and lysed on ice with 1 ml of lysis buffer. For each IP, a 0.5-ml aliquot of lysate was incubated with 0.5 μg of the anti-HA MAb, anti-Flag MAb, anti-CBP MAb, or nonspecific mouse monoclonal antibody and 30 μl of a 1:1 slurry of protein A/G Plus-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) for at least 4 h or overnight at 4°C. The beads were washed four times with 1 ml of lysis buffer containing 500 mM NaCl and then subjected to WB analysis. All IP assays were repeated at least two times, and similar data were obtained.
Native PAGE.
Native polyacrylamide gel electrophoresis (PAGE) was performed using ReadyGels (7.5%; Bio-Rad) as described in our previous study (18). In brief, the gel was prerun with 25 mM Tris and 192 mM glycine, pH 8.4, with 1% deoxycholate (DOC) in the cathode chamber for 30 min at 40 mA. Samples in native sample buffer (10 μg protein, 62.5 mM Tris-Cl [pH 6.8], 15% glycerol, and 1% DOC) were size fractionated by electrophoresis for 60 min at 25 mA and transferred to nitrocellulose membranes for WB analysis.
Immunofluorescence assays.
Immunofluorescence assays were performed as described in our previous studies (18, 21, 30). Briefly, HeLa cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; 0.137 M NaCl, 0.003 M KCl, 0.008 M Na2HPO4, 0.001 M NaH2PO4; pH 7.4) for 20 min, washed three times with PBS, and permeabilized with 0.5% Triton X-100 in PBS for 10 min. The cells were rinsed with PBS and then incubated with PBS containing 5% bovine serum albumin (BSA) for 20 min at room temperature. Subsequently, the cells were incubated with rabbit anti-IRF-3 PAb (diluted 1:500) or with mouse anti-HA MAb (diluted 1:2,000) for 2 h at 37°C, followed by incubation with tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG (Pierce) or fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Sigma-Aldrich) in PBS containing 0.5% BSA for 1 h at 37°C. After each incubation step, cells were washed extensively with PBS. Samples were analyzed using fluorescence microscopy (Zeiss, Germany).
ELISA for IFN-β.
An enzyme-linked immunosorbent assay (ELISA) to quantify secreted IFN-β was carried out on culture supernatants collected from infected cells. Medium was collected and centrifuged to remove cell debris. Fifty microliters of cleared supernatant or the IFN-β standard was used in duplicate for detection of IFN-β by using a human IFN-β ELISA kit (PBL IFN Source, Piscataway, NJ) as described in our previous study (18).
EMSA.
Whole-cell extracts were prepared at 40 h posttransfection with 5 μg of expression plasmid, as indicated for individual experiments. Cells were washed in phosphate-buffered saline and lysed in 10 mM Tris-Cl (pH 8.0), 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluroide, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml aprotinin, 0.5 μg/ml chymostatin, and 0.25 μM microcystin. Equivalent amounts of whole-cell extract (20 μg) were assayed for IRF-3 binding in a gel shift analysis using biotin-labeled double-stranded oligonucleotide corresponding to the PRDI-PRDIII region of IFN-β promoter, 5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′. Complexes were formed by incubating the probe with 20 μg of each whole-cell extract in the presence or absence of the indicated antibodies. The binding mixture (20 μl) contained 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 50 mM NaCl, 2 mM DTT, 5% glycerol, 0.5% NP-40, 10 μg/μl of BSA, 62.5 μg/ml of poly(dI-dC) was added to reduce nonspecific binding. To demonstrate the specificity of protein-DNA complex formation, a 500-fold molar excess of unlabeled wild-type or mutated (mut) oligonucleotide corresponding to the IFN-stimulated response element (IRE) region of the RANTES promoter (WT, 5′-CTATTTCAGTTTTCTTTTCCGTTTTGTG-3′, mut, 5′-CTATTTCAGTAAACTAAACCGTTTTGTG-3′) was added to the cell extract before adding labeled probe. After 20 min of incubation with the probe, extracts were loaded on a 5% polyacrylamide gel (60:1 cross-link) prepared in 0.5× Tris-borate-EDTA. Protein-oligonucleotide complexes were electroblotted to a nylon membrane and detected using a Light Shift chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Pierce). After incubation in blocking buffer for 15 min at room temperature, the membrane was incubated with streptavidin-horseradish peroxidase conjugate for 30 min at room temperature. The membrane was then incubated with chemiluminescent substrate for 5 min before analysis in a Fluochem HD2 imaging system (Alpha Innotech).
RESULTS
VP16 represses SeV-mediated production of IFN-β and activation of the NF-κB promoter induced by SeV infection or TNF-α treatment.
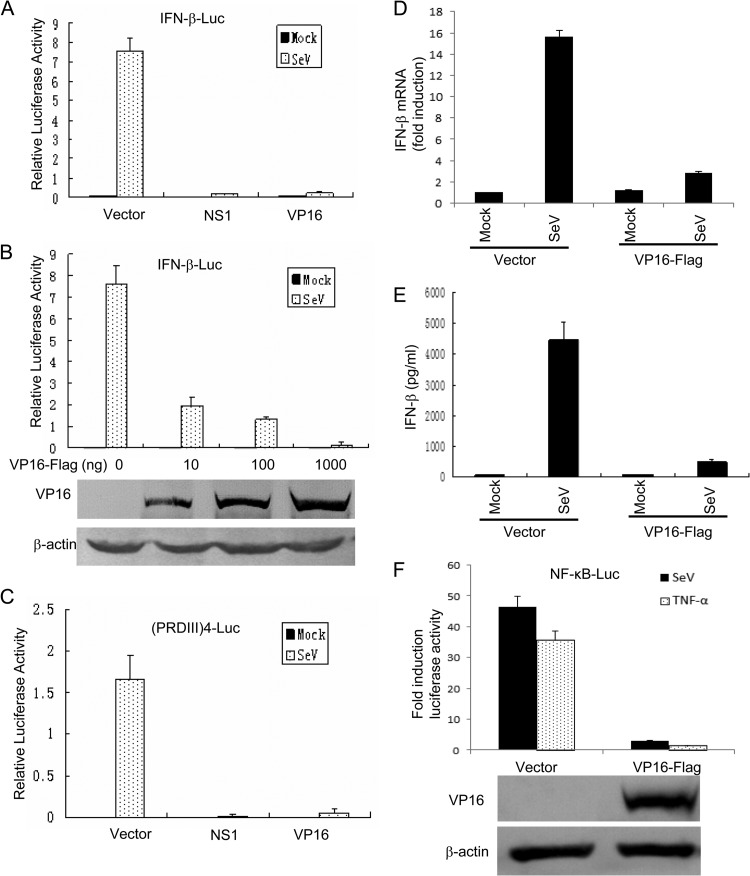
To determine the ability of tegument protein VP16 to inhibit SeV-mediated activation of IFN-β gene transcription, a Flag-tagged VP16 expression plasmid was cotransfected into HEK 293T cells together with an IFN-β promoter construct. SeV infection resulted in strong induction of IFN-β reporter activity (Fig. 1A). In contrast, ectopic expression of VP16 significantly inhibited SeV-mediated activation of the IFN-β promoter, similar to the positive-control influenza A virus NS1 protein (Fig. 1A). Additionally, VP16 inhibited IFN-β promoter activity in a dose-dependent manner (Fig. 1B); the expression of the VP16 protein was confirmed by Western blot analysis (Fig. 1B).
Fig 1.
VP16 inhibits SeV-induced IFN-β production and the activation of the NF-κB promoter induced by SeV or TNF-α treatment. (A and C) HEK 293T cells were transfected with 500 ng of the IFN-β promoter reporter plasmid pIFN-β-luc (A) or p(PRDIII-I)4-Luc (C), together with Renilla luciferase plasmid pRL-TK (50 ng) and pCMV-Flag empty vector or plasmids encoding the indicated viral proteins (1,000 ng). At 24 h after transfection, cells were mock infected or infected with 100 HAU ml−1 SeV as indicated and luciferase activity was measured 16 h postinfection. (B) Results of an experiment similar to that shown in panel A, except an increased amount of VP16-Flag expression plasmid was used, as indicated. The expression of VP16 was analyzed by Western blotting using anti-Flag and anti-β-actin (as a control) MAbs. Data are the relative luciferase activity, with standard deviations of three independent experiments performed in duplicate. (D) HEK 293T cells were transfected with pCMV-Flag empty vector or VP16-Flag expression plasmid. At 24 h posttransfection, cells were mock infected or infected with 100 HAU ml−1 SeV for 16 h and total RNA was analyzed by qRT-PCR. GAPDH levels were quantified for normalization of the data. The fold change in gene expression was determined by comparison to the mock-treated samples. The data represent means + standard deviations of three replicates. (E) Medium from infected cells in shown in panel D was isolated and analyzed by ELISA for IFN-β secretion as described in Materials and Methods. The data represent means + standard deviations of three replicates. (F) HEK 293T cells were transfected with the NF-κB promoter reporter plasmid NF-κB-luc, together with pRL-TK and pCMV-Flag empty vector or plasmid pVP16-Flag. At 24 h posttransfection, cells were treated or not with 10 ng/ml recombinant human TNF-α and incubated for an additional 6 h or infected with SeV for 16 h. NF-κB-driven luciferase activity was determined as described for panel A.
The activation of IFN-β gene transcription depends on synergistic interactions among IRFs, NF-κB, and other transcription factors that bind to distinct regulatory domains in the promoter. To examine the role of VP16 in inhibition of SeV-mediated activation of IRFs, we measured expression of the luciferase reporter gene driven by tandem IRF binding sites from the IFN-β promoter [(PRDIII-I)4-Luc]. SeV infection resulted in strong induction of (PRDIII-I)4-Luc reporter activity (Fig. 1C). However, coexpression of VP16 significantly inhibited the activation of the (PRDIII-I)4 reporter (Fig. 1C). To further determine the role of the VP16 protein in the inhibition of SeV-induced endogenous IFN-β production, both IFN-β mRNA accumulation and protein secretion were measured, by qRT-PCR and ELISA, respectively. As expected, mRNA and protein levels of endogenous IFN-β were strongly upregulated by SeV infection (Fig. 1D and E), whereas VP16 significantly reduced the accumulation of endogenous IFN-β mRNA and IFN-β secretion (Fig. 1D and E, respectively). Additionally, the activation of the NF-κB-Luc reporter induced by SeV infection was inhibited with VP16 coexpression (Fig. 1F). Similarly, coexpression of VP16 also blocked the TNF-α-induced activation of the NF-κB-Luc reporter (Fig. 1F). Taken together, these results demonstrated that VP16 expression dramatically reduced SeV-mediated production of endogenous IFN-β and blocked NF-κB promoter activation induced by SeV or TNF-α treatment.
The transcriptional activation domain of VP16 is not responsible for inhibiting SeV-mediated activation of the IFN-β promoter.
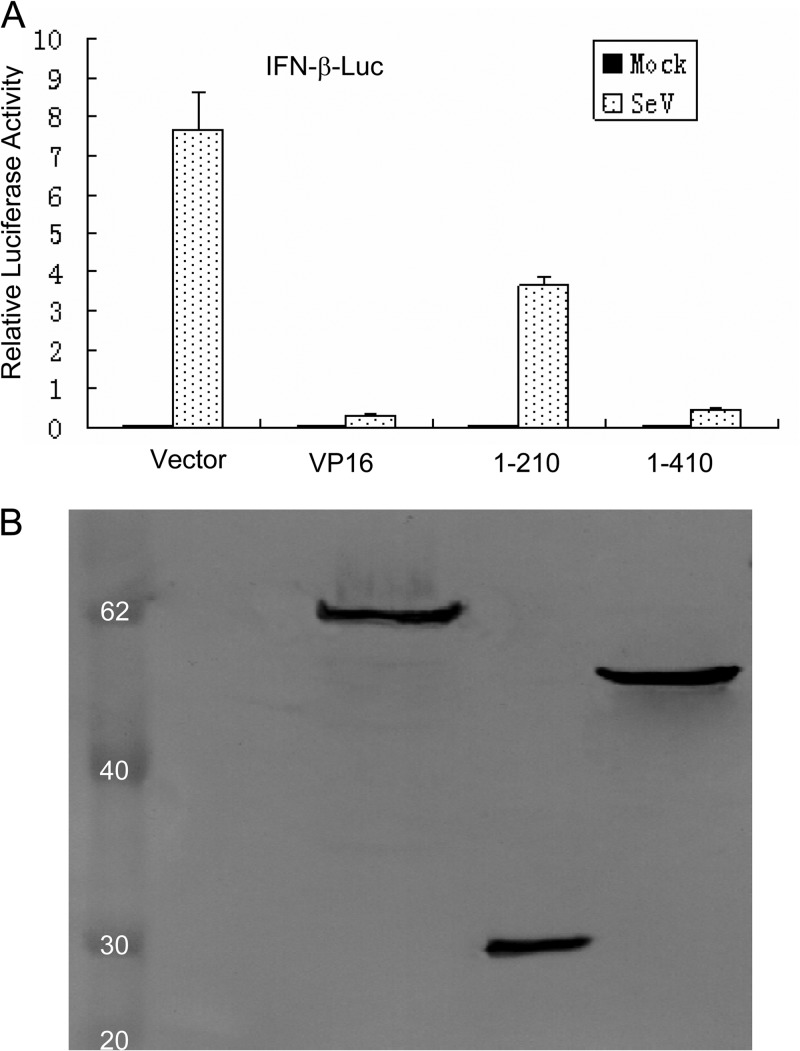
Like other transcription factors, VP16 contains a carboxy-terminal transcriptional activation domain (residues 411 to 490) and a central conserved core (residues 49 to 385), which is sufficient for VP16-induced complex formation (33, 34). To determine which region of VP16 was responsible for the inhibition of virus-mediated activation of the IFN-β promoter, a series of VP16 expression plasmids, including VP16-HA, VP16(1-210)-Flag, and VP16(1-410)-Flag, were constructed in our study. SeV infection significantly activated the IFN-β promoter; however, overexpression of full-length VP16-HA blocked the activation of the IFN-β promoter, consistent with VP16-Flag data (Fig. 2A and 1A), indicating that the presence of different small tags in the VP16 fusion proteins did not affect the inhibitory activity of VP16. The expression of VP16(1-410)-Flag, which did not contain the transcriptional activation domain, blocked SeV-mediated activation of the IFN-β promoter, while the expression of VP16(1-210)-Flag only partially inhibited SeV-induced activation of the IFN-β promoter (Fig. 2A). The expression of the VP16 fusion proteins from the recombinant plasmids was detected at their expected molecular mass (Fig. 2B). These results suggest that the carboxy-terminal transcriptional activation domain of VP16 is not responsible for inhibiting SeV-mediated IFN-β production.
Fig 2.
The C-termimal transcription activation domain of VP16 is not responsible for inhibition of SeV-mediated activation of the IFN-β promoter. (A) Luciferase assays in HEK 293T cells were performed as described for Fig. 1A to measure the activation of the IFN-β promoter following SeV infection in the presence of full-length and deletion mutants of VP16, including VP16-HA, VP16(1-210)-Flag, and VP16(1-410)-Flag. Data are the relative luciferase activity, with standard deviations of three independent experiments performed in duplicate. (B) The expression of these VP16 deletion mutants was verified by Western blotting using mouse anti-Flag or anti-HA MAbs.
VP16 represses IFN-β promoter activity at the IRF-3 level.
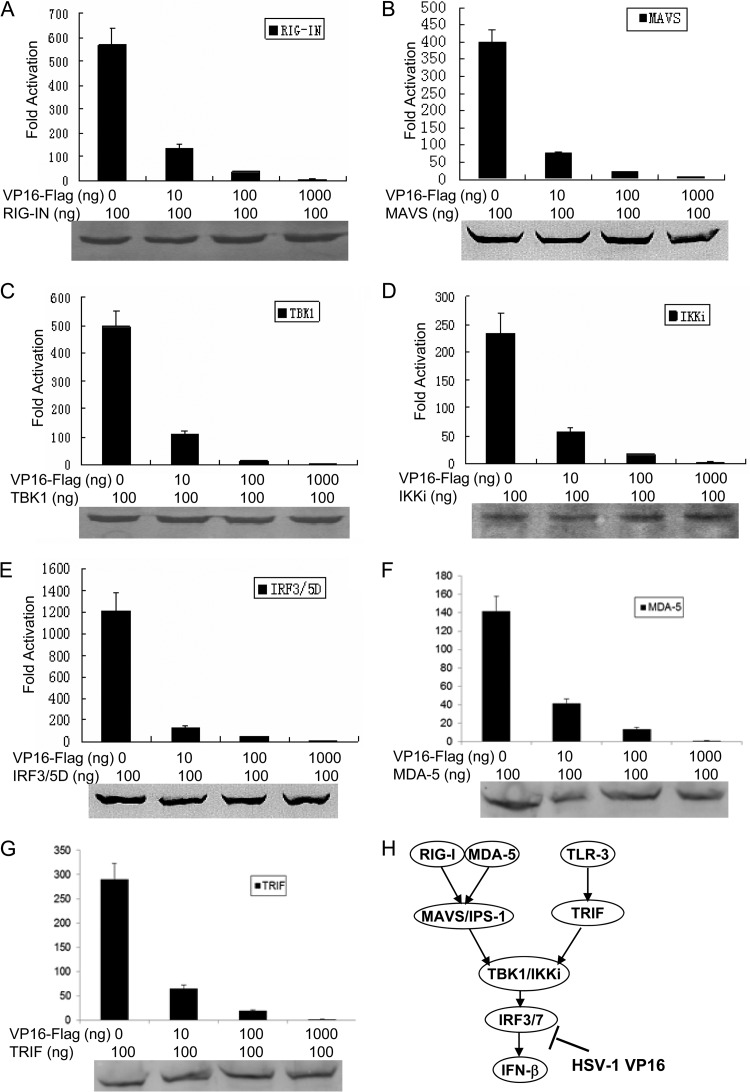
In order to examine at which stage the VP16 protein inhibits the IFN-β production pathway, we used different stimuli downstream of RIG-I or MDA-5 to induce the IFN-β reporter in 293T cells. We overexpressed RIG-IN (a constitutively active variant containing only the amino-terminal CARD of RIG-I), MAVS, TBK1, IKKi, IRF-3/5D (a phosphomimetic form of IRF-3), MDA-5, and TRIF (a downstream adaptor of TLR-3) and analyzed the activation of the IFN-β reporter in the presence of increasing amounts of VP16. As a result, ectopic expression of RIG-IN significantly activated the IFN-β promoter (Fig. 3A). VP16 expression significantly inhibited RIG-IN-mediated activation of the IFN-β promoter in a dose-dependent manner (Fig. 3A). Similarly, when MAVS, IKKi, TBK1, IRF-3/5D, MDA-5, or TRIF signaling components were used to activate IFN-β promoter activity, the VP16 protein suppressed IFN-β promoter activation, and these results were observed even at a higher protein expression level (Fig. 3B, C, D, E, F, and G). Additionally, the expression of all adaptor proteins, including RIG-IN, MAVS, IKKi, TBK1, IRF-3/5D, MDA-5, and TRIF, was confirmed by Western blotting (Fig. 3). A sketch map of the IFN-β pathway mediated by RIG-I, MDA-5, or TLR-3 is shown in Fig. 3H. Collectively, these results suggest that the VP16 protein likely acts at the IRF-3 level in the IFN-β production signaling axis.
Fig 3.
VP16 inhibits activation of the IFN-β promoter at the IRF-3 level. HEK 293T cells were transfected with plasmids expressing RIG-IN (A), MAVS (B), TBK1 (C), IKKi (D), IRF-3/5D (E), MDA-5 (F), or TRIF (G) together with pIFN-β-luc and the Renilla luciferase reporter plasmid pRL-TK. At 36 h after transfection, luciferase assays were performed. Data are expressed as the fold induction, with standard deviations of three independent experiments performed in duplicate. The expression of all adaptor proteins, including RIG-IN, MAVS, IKKi, TBK1, IRF-3/5D, MDA-5, and TRIF, was confirmed by Western blotting. (H) Schematic of the IRF signaling pathway mediated by both RLR (RIG-I and MDA-5) or TLR-3 and leading to IFN-β production. Both TLR-3- and RLR-mediated IFN-β production was inhibited by HSV-1 VP16.
VP16 does not prevent the nuclear translocation and phosphorylation of IRF-3 induced by SeV infection.
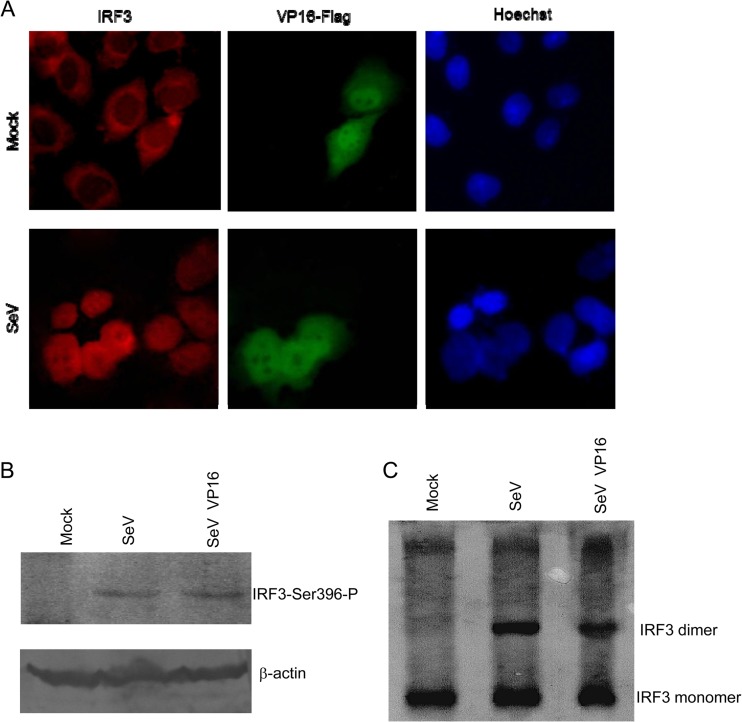
In response to cellular stimulation, activation of IRF-3 is achieved by phosphorylation of IRF-3. Upon phosphorylation, IRF-3 dimerizes and translocates to the nucleus, where it can form a complex with CBP/p300 and act as a transcription factor. The holocomplex can then bind to the IRF-3 sites in the IFN-β promoter region eventually to activate the transcription of the IFN-β gene. As the aforementioned results indicated that VP16 might repress IFN-β promoter activation at the IRF-3 level, we subsequently investigated whether VP16 inhibited SeV-induced IRF-3 phosphorylation and nuclear translocation. First, in an indirect immunofluorescence assay, nuclear translocation of IRF-3 was analyzed in HeLa cells transfected or not with the VP16-Flag expression plasmid and infected with SeV to stimulate IRF-3 translocation from the cytoplasm to the nucleus. In uninfected cells, IRF-3 localized exclusively to the cytoplasm and the expression of VP16, which localized in the cytoplasm and nucleus, especially in the nucleus, did not affect the expression or localization of IRF-3 (Fig. 4A). In contrast, in SeV-infected cells, endogenous IRF-3 translocated into the nucleus (Fig. 4A). Similarly, SeV-induced IRF-3 translocation was not affected in the presence of full-length VP16 (Fig. 4A). These results indicated that VP16 did not block IRF-3 nuclear translocation induced by SeV infection.
Fig 4.
VP16 does not block the nuclear translocation and dimerization of IRF-3. (A) VP16 does not block the nuclear translocation of IRF-3 induced by SeV infection. HeLa cells were transfected with a VP16-Flag expression plasmid or pCMV-Flag vector. At 24 h posttransfection, cells were mock infected or infected with 100 HAU ml−1 SeV for 8 h. Cells were stained with mouse anti-Flag MAb and rabbit anti-IRF3 PAb. FITC-conjugated goat anti-mouse (green) and TRITC-conjugated goat anti-rabbit (red) antibodies were used as the secondary antibodies. Cell nuclei (blue) were stained with Hoechst 33258. The images were obtained by fluorescence microscopy using a 40× objective. (B and C) VP16 does not inhibit SeV-induced IRF-3 phosphorylation and dimerization. HEK 293T cells were transfected with VP16-Flag expression plasmid. At 24 h posttransfection, cells were then mock infected or infected with 100 HAU ml−1 SeV for 8 h. The protein extracts were subjected to SDS-PAGE (B) or native PAGE (C) and subsequent immunoblotting with anti-phospho-IRF3 (Ser396) (B) or anti-IRF3 (C) antibody. Whole IRF-3 and β-actin (as a loading control) were also detected.
Generally, Ser 396 is targeted in vivo for phosphorylation following virus infection and plays an essential role in IRF-3 activation (35). Therefore, the phosphorylation state of IRF-3 following virus infection was evaluated by Western blotting using the phospho-specific IRF-3 (S396) antibody. SeV infection induced the accumulation of Ser 396 phosphorylation (Fig. 4B, lane 2), while SeV-induced Ser 396 phosphorylation was not inhibited by VP16 (Fig. 4B, lane 3). IRF-3 dimer formation is a consequence of IRF-3 phosphorylation. Next, we tested whether the dimerization of IRF-3 induced by SeV could be inhibited by VP16. We found that IRF-3 dimerization was not reduced in SeV-infected cells expressing VP16 protein (Fig. 4C). Taken together, these results demonstrate that VP16 does not prevent the phosphorylation and dimerization of IRF-3 induced by SeV infection.
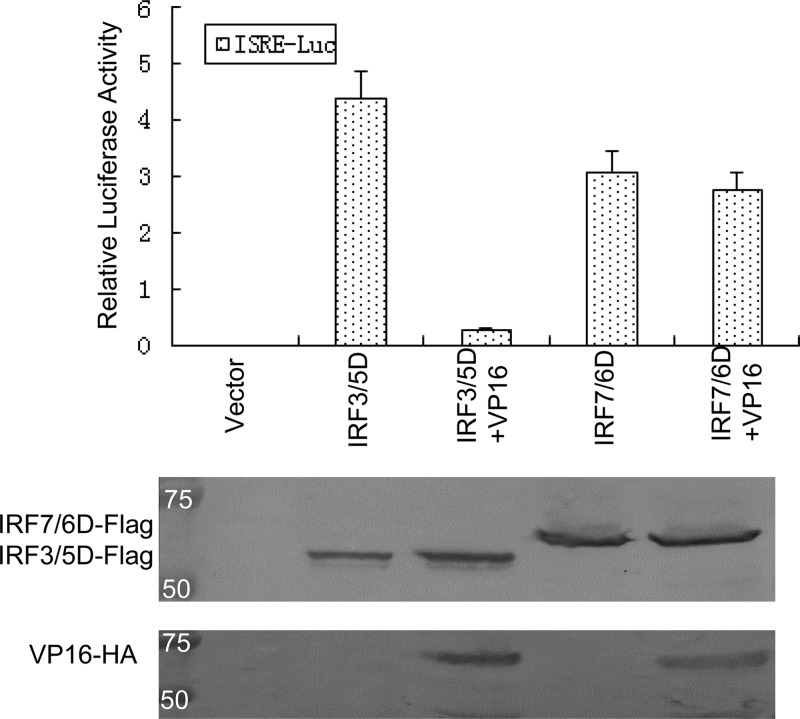
VP16 represses IRF-3-mediated but not IRF-7-mediated activation.
We next determined whether VP16 repressed IRF-3- or IRF-7-mediated transcriptional activity. A transient-expression assay was performed using the IFN-stimulated response element (ISRE) promoter construct together with Flag-IRF-3/5D (the constitutively active form of IRF-3), Flag-IRF-7/6D (the constitutively active form of IRF-7), and/or VP16 expression plasmids. Cotransfection of the constitutively active form of IRF-3/5D or IRF-7/6D easily activated the ISRE promoter up to 410-fold or 300-fold, respectively, and overexpression of VP16 reduced the IRF-3/5D-mediated transactivation more than 15-fold (Fig. 5). However, coexpression of VP16 had essentially no effect on the inducible expression of the ISRE promoter induced by IRF-7/6D (Fig. 5). Additionally, the expression of all expression plasmids, including Flag-IRF-3/5D, Flag-IRF-7/6D, and VP16-Flag, was confirmed by Western blotting (Fig. 5). These results demonstrate that VP16 represses IRF-3-mediated but not IRF-7-mediated transcriptional activation.
Fig 5.
VP16 represses IRF-3- but not IRF-7-mediated activation. HEK 293T cells were cotransfected with Flag-IRF3/5D or Flag-IRF-7/6D, ISRE reporter plasmid ISRE-Luc, Renilla luciferase reporter plasmid pRL-TK and expression plasmid VP16-HA, or pCMV-HA empty vector. At 36 h after transfection, DLR luciferase assays were performed. Data are expressed as the relative luciferase activity, with standard deviations of three independent experiments performed in duplicate.
VP16 associates with both IRF-3 and p65 and blocks the expression of NF-κB-dependent genes.
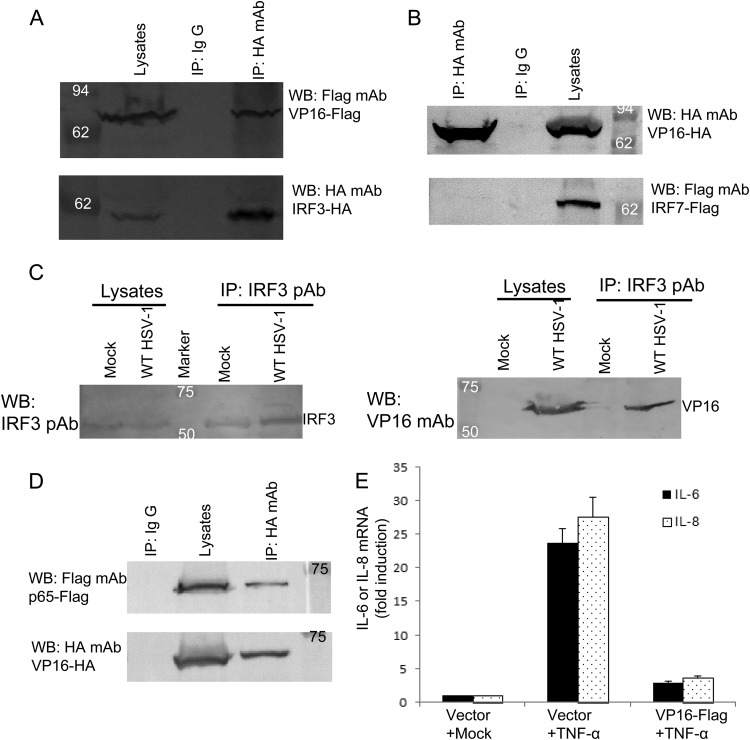
To examine the possibility that VP16 associates with IRF-3, coimmunoprecipitation experiments were performed using HEK 293T cells cotransfected with Flag-tagged VP16 and HA-tagged IRF-3, as described in our previous studies (18, 19). We found that VP16 was efficiently coimmunoprecipitated with IRF-3 by the anti-HA MAb (Fig. 6A), but not by nonspecific mouse monoclonal antibody IgG (Fig. 6A). IRF-3 and IRF-7, which are highly homologous, play an important role in the virus-induced type I IFN gene activation following virus infection (15). Next, we tested whether VP16 also interacted with IRF-7. The coimmunoprecipitation analysis demonstrated that VP16 did not interact with Flag-tagged IRF-7 (Fig. 6B), which was consistent with the above conclusion that VP16 has no effect on IRF-7 signaling.
Fig 6.
VP16 interacts with IRF-3 and blocks the expression of NF-κB-dependent genes through interaction with p65. (A and B) VP16 associates with IRF-3 but not IRF-7. HEK 293T cells (∼5 × 106) were cotransfected with expression plasmids VP16-Flag and HA-IRF-3 or plasmids VP16-HA and Flag-IRF-7. At 36 h posttransfection, cells were lysed and the samples were then subjected to immunoprecipitation assays (IP) using anti-HA MAb or nonspecific mouse monoclonal antibody (IgG2b). Cell lysates and immunoprecipitated proteins were separated in denaturing 10% polyacrylamide gels and transferred to nitrocellulose membranes. The transferred proteins were probed with anti-HA and anti-Flag MAbs. (C) VP16 interacts with endogenous IRF-3 in HSV-1-infected cells. HEK 293T cells were infected with WT HSV-1 at a multiplicity of infection of 1 for 16 h. The cells were then lysed, and the extracts were subjected to immunoprecipitation using anti-IRF-3 PAb or control IgG. Precipitates were analyzed by Western blotting using anti-VP16 MAb. (D) VP16 interacts with NF-κB subunit p65. HEK 293T cells were cotransfected with expression plasmids VP16-HA and p65-Flag. At 36 h posttransfection, immunoprecipitation assays was performed using anti-HA MAb as described for panel A. (E) HEK 293T cells were transfected with pCMV-Flag empty vector or VP16-Flag expression plasmid. At 24 h posttransfection, the cells were treated with TNF-α (15 ng/ml) for 4 h, and total RNA was analyzed by qRT-PCR. GAPDH levels were quantified for normalization of the data. The fold change in gene expression of IL-6 or IL-8 was determined by comparison to the mock-treated samples. The data represent means + standard deviations of three replicates.
The goal of the subsequent experiment was to determine whether VP16 interacted with endogenous IRF-3 in the context of HSV-1 infection. In WT HSV-1-infected cells, the VP16 protein was easily immunoprecipitated by IRF-3 when we used anti-IRF-3 PAb (Fig. 6C), but not by control antibody IgG (data not shown). In contrast, in mock-infected cells, no VP16 protein was immunoprecipitated by IRF-3 (Fig. 6C). Collectively, these data suggest that the VP16 protein interacts with endogenous IRF-3 during HSV-1 infection.
It is well established that activation of the IFN-β promoter requires not only activation by IRF-3 but also NF-κB. We showed that VP16 blocks the activation of the NF-κB promoter induced by SeV infection or TNF-α treatment (Fig. 1F). Next, we analyzed the NF-κB subunit p65 for its potential interaction with VP16. VP16-HA and Flag-p65 expression plasmids were cotransfected into HEK 293T cells for co-IP analysis. As expected, the ectopic expression of p65 was efficiently coimmunoprecipitated by VP16 when we used the anti-HA MAb (Fig. 6D). To further determine if VP16 also blocks the expression of NF-κB-dependent genes that are not IRF-3 dependent, the mRNA induction of two inflammatory chemokines (IL-6 and IL-8) was analyzed by qRT-PCR. We found that mRNA levels of endogenous IL-6 and IL-8 were strongly induced by TNF-α treatment (Fig. 6E), whereas coexpression of VP16 significantly reduced the accumulation of endogenous mRNAs of both IL-6 and IL-8 (Fig. 6E). Taken together, these results suggest that VP16 also blocks the expression of NF-κB-dependent genes, likely by binding to p65.
The carboxy terminus of IRF-3 is required for interaction with the amino terminus of VP16.
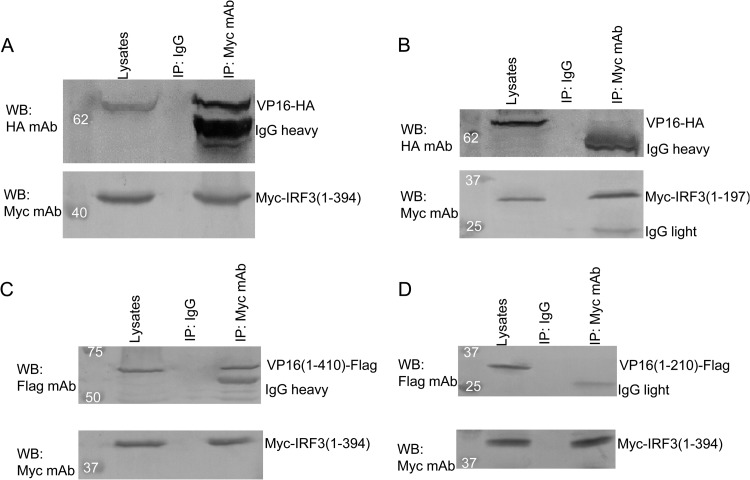
To further determine which region of IRF-3 binds to VP16, IRF-3 mutants, including Myc-IRF3(1-197), which contains the N-terminal DNA binding domain but lacks the C-terminal IRF association domain, and Myc-IRF3(1-394), which contains the C-terminal IRF association domain, were tested in co-IP analyses. In HEK 293T cells cotransfected with VP16-HA and either Myc-IRF3(1-197) or Myc-IRF3(1-394), VP16 was easily immunoprecipitated with IRF3(1-394) by anti-Myc MAb (Fig. 7A), but not with IRF3(1-197) or control antibody IgG (Fig. 7B), indicating that the C-terminal region of IRF-3 is required for interaction with VP16.
Fig 7.
The carboxy terminus of IRF-3 is required for interaction with the amino terminus of VP16. (A and B) HEK 293T cells were transfected with plasmids VP16-HA and pMyc-IRF3(1-394) (A) or pMyc-IRF3(1-197) containing the N-terminal DNA binding domain (B), respectively. (C and D) HEK 293T cells were transfected with plasmids pMyc-IRF3(1-394) and pVP16(1-410)-Flag or pVP16(1-210)-Flag, respectively. At 24 h posttransfection, immunoprecipitation with anti-Myc MAb or nonspecific mouse monoclonal antibody (IgG2b) and Western blot analysis were performed as described for Fig. 6A.
Additionally, VP16(1-410)-Flag was successfully immunoprecipitated by anti-Myc MAb from cells expressing VP16(1-410)-Flag and Myc-IRF3(1-394) (Fig. 7C), while no detectable VP16(1-210)-Flag was coimmunoprecipitated by anti-Myc MAb from cells expressing VP16(1-210)-Flag and Myc-IRF3(1-394) (Fig. 7D), further demonstrating that the N-terminal amino acids (aa) 1 to 410, but not the C-terminal transcriptional activation domain, of the VP16 protein are responsible for interacting with IRF-3. Taken together, these data suggest that the N-terminal aa 1 to 410 of VP16 are required for binding to IRF-3.
VP16 blocks the interaction between IRF-3 and the CBP coactivator in the context of HSV-1 infection.
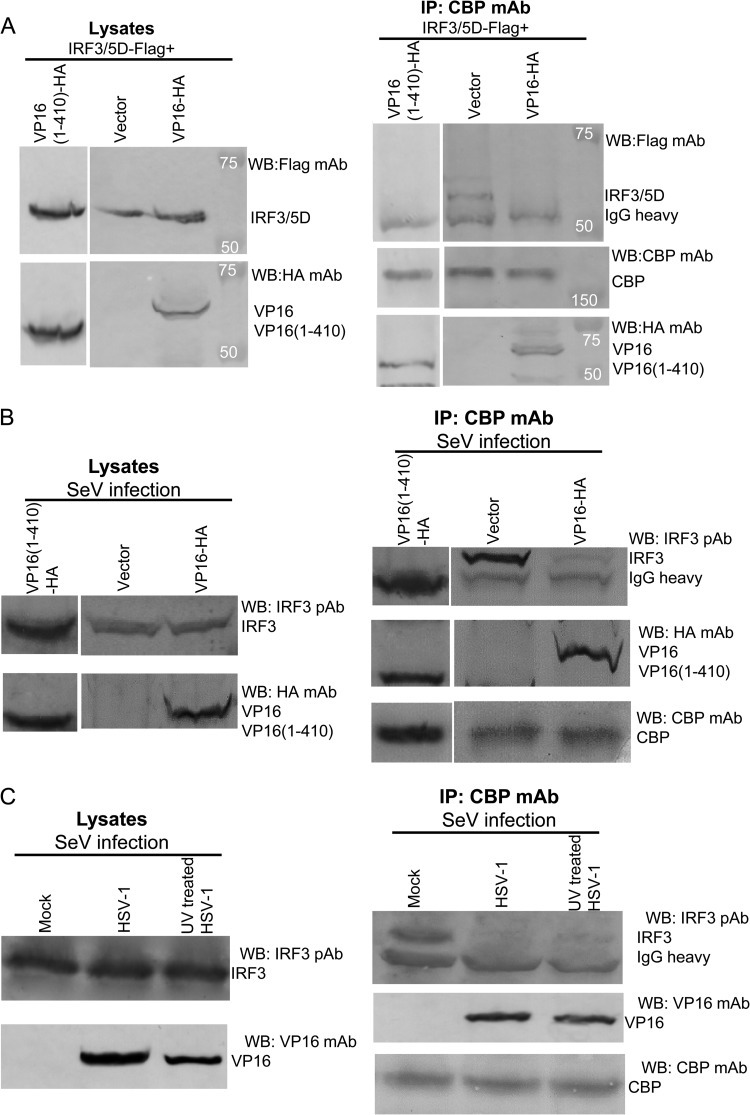
Next, we explored the mechanisms involved in the inhibition of IRF-3-mediated transactivation by VP16. It has been shown that the histone acetyltransferase coactivators CBP and p300 associate with the C-terminally phosphorylated form of IRF-3 (11, 36, 37), but not with IRF-7 (17). Since VP16 repressed IRF-3-mediated but not IRF-7-mediated transactivation, we next examined whether VP16 could interact with the CBP coactivator in vivo and block the association between IRF-3 and the CBP coactivator. Flag-IRF-3/5D and VP16-HA or VP16(1-410)-HA, or an empty vector, were transiently cotransfected into HEK 293T cells. After immunoprecipitation of endogenous CBP from whole-cell extracts with anti-CBP MAb, WB analysis with anti-HA MAb revealed that VP16 coimmunoprecipitated with CBP (Fig. 8A, lane 3, third panel), but not with control antibody IgG (data not shown). WB analysis of the same membrane with anti-Flag MAb indicated that in the absence of VP16, IRF-3/5D strongly associated with CBP (Fig. 8A, lane 2, first panel), while coexpression of VP16 blocked the interaction between IRF-3/5D and the CBP coactivator (Fig. 8A, lane 3, first panel). Similarly, aa 1 to 410 of VP16, a region that lacks the activation domain but retains the ability to block IFN gene expression, immunoprecipitated with CBP and blocked the interaction between IRF-3/5D and the CBP coactivator (Fig. 8A, lane 1).
Fig 8.
VP16 inhibits the interaction between endogenous IRF-3 and CBP in the context of HSV-1 infection. (A) HEK 293T cells (∼5 × 106) were cotransfected with plasmids Flag-IRF-3/5D and VP16-HA or VP16(1-410)-HA, or empty vector, respectively. At 36 h posttransfection, cells were lysed and the clarified supernatants were subjected to immunoprecipitation assays (IP) using anti-CBP MAb or nonspecific mouse monoclonal antibody (IgG1). CBP, IRF-3/5D, and VP16 were detected by Western blotting using anti-CBP, anti-Flag, or anti-HA MAbs, respectively. (B) HEK 293T cells were transfected with plasmid VP16-HA or VP16(1-410)-HA, or empty vector, respectively. At 20 h posttransfection, cells were infected with SeV for another 16 h. Then, the cells were lysed and subjected to immunoprecipitation assays as described for panel A. Additionally, rabbit anti-IRF-3 PAb was used for detection of endogenous IRF-3 expression. (C)HEK 293T cells were first mock infected or infected with wild-type HSV-1 (multiplicity of infection of 1) or UV inactivated with HSV-1 (multiplicity of infection of 10) for 12 h and then infected with SeV for another 12 h. The cells were lysed, and the clarified supernatants were subjected to immunoprecipitation assays using an anti-CBP MAb as in panel B.
Following the overexpression studies, SeV infection-induced interaction of endogenous IRF-3 and CBP coactivator was also analyzed in HEK 293T cells with or without VP16 expression or of the truncated VP16(1-410)-HA. After SeV infection, CBP was immunoprecipitated with anti-CBP MAb, and the immunoprecipitate was analyzed for the presence of IRF-3 by using IRF-3 PAb. Endogenous IRF-3 coimmunoprecipitated easily with CBP in SeV-infected cells (Fig. 8B, lane 2, first panel), but not in uninfected cells (data not shown). However, in virus-infected cells expressing VP16, the amount of IRF-3 that coimmunoprecipitated together with CBP was dramatically decreased (Fig. 8B, lane 3, first panel). Additionally, VP16 was successfully immunoprecipitated by anti-CBP MAb from cells expressing VP16-HA (Fig. 8B, lane 3, second panel), but not in the cells without VP16 expression (Fig. 8B, lane 2, second panel). Similarly, in SeV-infected cells expressing truncated VP16(1-410)-HA, the amount of IRF-3 that coimmunoprecipitated together with CBP was significantly reduced (Fig. 8B, lane 1). Taken together, these data suggest that the aa 1 to 410 region of VP16, which lacks the activation domain but retains the ability to block IFN-β gene expression, is enough to block the interaction between endogenous IRF-3 and the CBP coactivator in virus infection.
To further investigate whether VP16 has the same effect during HSV-1 infection, HEK 293T cells were first mock infected or infected with wild-type HSV-1 or UV-inactivated HSV-1 for 12 h and then infected with SeV for another 12 h prior to co-IP analysis. In mock-infected cells, endogenous IRF-3 coimmunoprecipitated easily with CBP after SeV infection (Fig. 8C, lane 1). However, in HSV-1-infected cells, the amount of IRF-3 that coimmunoprecipitated together with CBP was dramatically decreased because of the expression of VP16 in HSV-1 infection (Fig. 8C, lane 2). Similarly, in UV-inactivated HSV-1-infected cells, the interaction between endogenous IRF-3 and CBP was significantly reduced due to the expression of VP16 in incoming virions (Fig. 8C, lane 3). These results demonstrate that HSV infection disrupts the association between IRF-3 and CBP and suggest that this effect is due to VP16. Although we cannot definitively demonstrate that the inhibitory effect is due to virion-associated VP16, the data are fully consistent with this hypothesis.
VP16 blocks IRF-3–CBP–DNA complex formation.
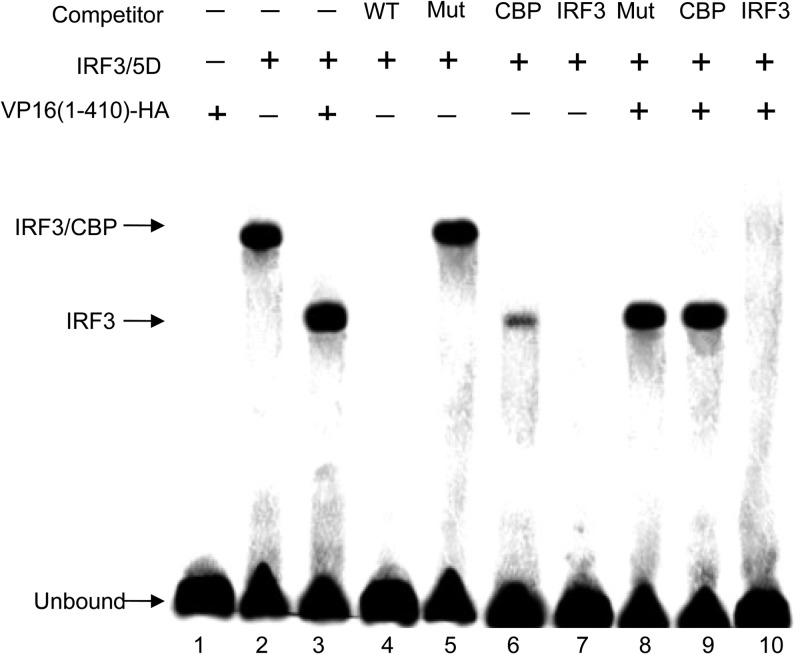
To further support the inhibition of IRF-3-mediated IFN activation with specific protein DNA interactions by VP16, IRF-3/5D and/or VP16 was transiently expressed in HEK 293T cells, and the DNA binding capacity of IRF-3/5D was determined by EMSA using a PRDIII-PRDI oligonucleotide from the IFN-β gene (Fig. 9). In cells expressing IRF-3/5D, a new protein-DNA complex was easily identified by EMSA (Fig. 9, lane 2) that contained IRF-3 and the CBP coactivator, as confirmed by EMSA using anti-CBP Ab and anti-IRF-3 Ab (Fig. 9, lanes 6 and 7), which was consistent with findings in a previous report (38). Because addition of anti-CBP Ab could abrogate the formation of the IRF-3–CBP–DNA complex and strongly inhibited IRF-3–DNA complex formation (Fig. 9, lane 6), while formation of either the slower-migrating IRF-3–CBP–DNA complex or faster-migrating IRF-3–DNA complex was fully abrogated by adding anti-IRF-3 Ab (Fig. 9, lane 7). Strikingly, this slower-migrating IRF-3–CBP–DNA complex disappeared when the aa 1 to 410 region of VP16 was coexpressed with IRF-3/5D (Fig. 9, lane 3) and was replaced by a faster-migrating protein-DNA complex (Fig. 9, lane 3). Additionally, this faster-migrating protein-DNA complex contained IRF-3, as confirmed by EMSA analysis with anti-IRF-3 Ab (Fig. 9, lane 10), but did not contain CBP, since the addition of the anti-CBP Ab did not inhibit complex formation (Fig. 9, lane 9). Competition with excess WT ISREs from the IFN-β promoter successfully depleted the binding of the complex (Fig. 9, lane 4), whereas mutated ISRE failed to compete for IRF-3/5D binding activity (Fig. 9, lanes 5 and 8). Additionally, the aa 1 to 410 region of VP16 alone did not bind to the PRDIII-PRDI site (Fig. 9, lane 1). Taken together, these results indicate that VP16 is able to block the formation of the IRF-3–CBP–DNA complex but does not affect the independent DNA binding activity of IRF-3.
Fig 9.
The effect of VP16 on IRF-3 DNA binding activity, determined by EMSA analysis. HEK 293T cells were transfected with plasmid Flag-IRF-3/5D with or without the VP16(1-410)-HA expression plasmid for 36 h, and an EMSA was performed with whole-cell extracts (20 mg) derived from the cells. The biotin-labeled probe corresponds to the PRDI-PRDIII (5′-GAAAACTGAAAGGGAGAAGTGAAAGTG-3′) motif of the IFN-β promoter. The anti-CBP MAb (shown as CBP) and anti-IRF-3 PAb (shown as IRF3) were added as indicated to demonstrate the presence of CBP and IRF-3 in the protein-DNA complex, indicated by the arrows. The bands composed of unbound oligonucleotides are also indicated by the arrows. For oligonucleotide competition, a 500-fold molar excess of unlabeled WT or mutated (Mut) oligonucleotide corresponding to the ISRE region of the RANTES promoter (WT, 5′-CTATTTCAGTTTTCTTTTCCGTTTTGTG-3′; Mut, 5′-CTATTTCAGTAAACTAAACCGTTTTGTG-3′) was added as indicated above the lanes.
VP16 diminishes activation of ISREs and ISGs downstream of IFN-β.
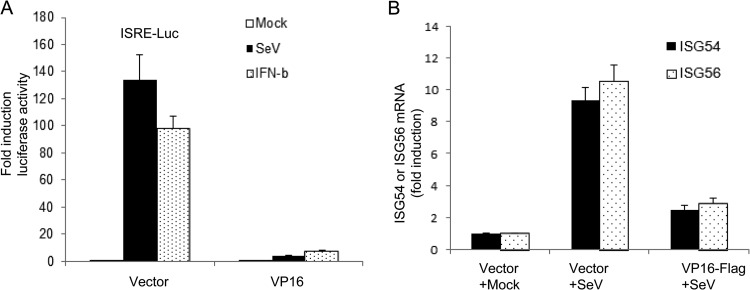
The binding of type I IFNs, including IFN-α and IFN-β, to the IFNAR (IFN-α receptor) initiates a signaling cascade that leads to the activation of ISREs and the induction of more than 300 ISGs (39). To determine the effect of VP16 on downstream IFN signaling, the well-established ISRE-luciferase reporter assay was applied to assess whether VP16 inhibited the ISRE promoter activity activated by SeV infection or IFN-β treatment. HEK 293T cells were transfected with the ISRE-Luc or RL-TK (internal control) reporter construct in the presence or absence of VP16-Flag. At 24 h posttransfection, cells were treated with IFN-β or infected with SeV, and a luciferase assay was performed at 16 h posttreatment. SeV infection or IFN-β treatment significantly induced the activation of the ISRE promoter by about 130-fold or 90-fold, respectively (Fig. 10A), whereas the activity was significantly abolished in the presence of VP16 (Fig. 10A). Furthermore, to determine the effects of VP16 on the activation of ISGs, qRT-PCR was performed to measure the mRNA levels of ISG54 and ISG56. The mRNA levels of ISG54 and ISG56 were easily activated by SeV infection (Fig. 10B), whereas coexpression of VP16 significantly reduced the mRNA levels of ISG54 and ISG56 (Fig. 10B). Collectively, these data demonstrate that VP16 also diminishes the activation of ISREs and ISGs downstream of IFN-β.
Fig 10.
VP16 inhibits SeV- or IFN-β-induced ISRE activation downstream of IFN-β. (A) VP16 inhibits SeV- or IFN-β-induced ISRE promoter activity. The experiments were performed as described for Fig. 1A, except that the ISRE reporter plasmid ISRE-Luc was used here. (B) HEK 293T cells were transfected with VP16-HA expression plasmid or pCMV-HA vector. At 24 h posttransfection, cells were then mock infected or infected with 100 HAU ml−1 SeV for 8 h, and total RNA was analyzed by qRT-PCR. GAPDH levels were quantified for normalization of the data. The fold change in gene expression of ISG54 or ISG56 was determined by comparison to the mock-treated samples. The data represent means + standard deviations of three replicates.
DISCUSSION
Innate immunity represents an ancient, conserved defense mechanism that rapidly responds to pathogen invasion. Upon virus infection, IFNs mediate antiviral effects through the upregulation of a diverse set of ISGs. However, to ensure their survival, viruses have evolved strategies to overcome these cellular responses. Thus, early events that occur between a virus and a cell dictate the fate of both parties. HSV-1 is an extremely successful human pathogen that has evolved multiple immune evasion strategies that allow it to exist for the lifetime of its host. The first HSV protein shown to block IRF-3 activation and IFN production was ICP0 (40, 41). However, its importance and activity remain controversial, due to different in vitro experimental approaches (42–45). The virion host shutoff protein (vhs) is a RNase that degrades both viral and cellular mRNAs (46). vhs from HSV-2 suppresses the expression of TLRs and RLRs, blocks IRF-3 activation, and inhibits IFN and ISG induction (47). Conversely, HSV-1 vhs activity on ISG mRNAs is only observed in the absence of ICP0 (40), consistent with an inability to directly affect IRF-3 activation (48). While ICP34.5 is best known for its ability to inhibit the IFN-inducible kinase PKR, it also dampens IFN production by binding and sequestering TBK-1, the kinase that phosphorylates IRF-3 (49). Recently, we showed that another tegument protein, US11, blocks IRF-3 activation and IFN production through disruption of the RLR pathway (18). While the IE protein ICP27 has been implicated in blocking IFN signal transduction cascades (50, 51), viruses lacking ICP27 induce higher levels of activated IRF-3 and cytokines, including IFN, in monocytic cells than does wild-type virus (43). Similarly, removal of the viral kinase US3 increases IRF-3 activation and TLR-3 and IFN levels in infected monocytic cells (52). Here, we now add the tegument protein VP16 to the long list of IFN-modulating proteins in HSV-1.
To search further for other HSV-1 genes that block IFN-β production, a screening of HSV-1 open reading frames for their abilities to block SeV-mediated activation of the IFN-β promoter was performed. Interestingly, HSV-1 tegument protein VP16 was found to significantly inhibit SeV-induced IFN-β production, based on mRNA level and protein secretion, and its transcriptional activation domain was not responsible for this inhibition activity. The previous evidence demonstrated that VP16 plays an indispensable role in formation of infectious virions (2), and UL48, which encodes the VP16 protein, is an essential gene (3), so there is no way to make a truly negative VP16 null virus. Additionally, VP16 was demonstrated to block the TNF-α-induced activation of the NF-κB promoter and expression of NF-κB-dependent genes through interaction with NF-κB subunit p65. We further showed that VP16 specifically blocked the transactivation activity of IRF-3, but not IRF-7, through interaction with IRF-3 but not IRF-7 as detected by co-IP analysis. Since VP16 did not block the nuclear translocation, phosphorylation, or dimerization of IRF-3 induced by SeV infection, we speculated that VP16 might interfere with the interaction between IRF-3 and its coactivator, CBP. As expected, VP16 was demonstrated to interact with the CBP coactivator and efficiently inhibit the formation of transcriptionally competent IRF-3–CBP complexes in the contexts of both live and UV-inactivated HSV-1 infection. Taken together, these data indicate that VP16 abrogates IFN-β production by inhibiting NF-κB and blocking IRF-3 from recruiting its coactivator, CBP, which may be important for the early events leading to HSV-1 infection.
Transcriptional repressors may function by one of several mechanisms: (i) displacement of activators from their DNA binding sites; (ii) masking of transcriptional activation domains; (iii) interference with the assembly of the basal transcription machinery; (iv) recruitment of multiprotein complexes with chromatin-modifying activities to specific sites on DNA (53, 54). Since VP16 has been shown to lack significant DNA binding activity (34), it is unlikely to function by a competition or displacement mechanism. Our data demonstrate that VP16 does not function by blocking the nuclear translocation, phosphorylation, or dimerization of IRF-3 induced by SeV infection, suggesting that VP16 prevents the recruitment of CBP to transcriptionally active promoters, based on data showing that in the presence of VP16, IRF-3 is able to bind DNA in vitro but does not form a high-molecular-weight complex with CBP (Fig. 9). In contrast, VP16 is able to physically associate with CBP, based on co-IP analysis. The fact that IRF-7-mediated activation is not blocked by VP16 is consistent with this interpretation, since IRF-7 does not associate with CBP/p300 (17).
The CBP/p300 transcriptional coactivators are important regulators of many cellular processes and have tumor-suppressing activity (55–57). CBP and p300 proteins interact with a variety of activators, such as NF-κB (58). VP16 may block the induction of the NF-κB promoter, probably by binding p65 and sequestering CBP.
IRF-3 is targeted by several different classes of viruses, including paramyxoviruses, herpesviruses, and reovirus, implicating it as the major activation pathway for primary IFN gene expression (59). Kaposi's sarcoma-associated herpesvirus (KSHV) expresses several IRF homologues that act as dominant negative inhibitors by preventing the association of IRF-3 with its coactivators, CBP and p300 (38). Similarly, a viral IRF from KSHV dimerizes with host IRF-7 and inhibits its DNA binding activity (60). Another KSHV protein, the transcription factor K-bZIP, competes with host IRF-3 for binding sites in the IFN-β promoter, thereby blocking promoter activation (61). Alternatively, HSV infected cell protein 0 (ICP0) binds to host IRF-3 and sequesters it together with CBP and p300 in nuclear bodies away from its normal binding sites on host genes (44). In this study, we demonstrated that VP16 efficiently inhibited IRF-3-mediated transactivation by competing for CBP coactivator recruitment. It has been reported that IRF-3 also contributes to virus-induced apoptosis (62). Hence, inhibition of the proapoptotic activities of IRF-3 may also be important for the establishment of persistent lytic infection or latent infection by herpesviruses such as HSV-1.
To date, six HSV-1 proteins have been implicated in inhibiting the production of IFN: ICP0, vhs, ICP34.5, US11, ICP27, and US3 (63). In this study, we demonstrate that the HSV-1 tegument protein VP16 is a novel antagonist of IFN-β production. We have provided evidence that VP16 abrogates the interferon antiviral response by inhibiting NF-κB and blocking IRF-3 to recruit its coactivator, CBP. We now add the tegument protein VP16 to the long list of IFN-modulating proteins of HSV-1. These findings will lead to a better understanding of the mechanisms employed by HSV-1 VP16 to dampen host antiviral signaling and information for the development of therapeutic interventions to modulate HSV-1 pathogenesis.
ACKNOWLEDGMENTS
This work was supported by grants from the Major State Basic Research Development Program of China (973 Program, 2011CB504802 and 2010CB530105), the National Natural Science Foundation of China (81171584 and 81000736), the Program for Changjiang Scholars and Innovative Research Team at Soochow University, and the Jiangsu Provincial Innovative Research Team.
We thank Takashi Fujita, S. Goodbourn, and Yi-Ling Lin for their gifts of plasmids pEF-Flag-RIG-I and pEF-Flag-RIG-IN, pEF-Flag-MDA-5, and Flag-IRF-3/5D, respectively. We thank Karen L. Mossman for critically reviewing the manuscript.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U. S. A. 102:16055–16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mossman KL, Sherburne R, Lavery C, Duncan J, Smiley JR. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roizman B, Knipe DM, Whitley RJ. 2007. Herpes simplex virus, p 2501–2601 In Knipe DM, Howley PM, Griffiths DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 4.Smiley JR, Duncan J. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J. Virol. 71:6191–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tal-Singer R, Pichyangkura R, Chung E, Lasner TM, Randazzo BP, Trojanowski JQ, Fraser NW, Triezenberg SJ. 1999. The transcriptional activation domain of VP16 is required for efficient infection and establishment of latency by HSV-1 in the murine peripheral and central nervous systems. Virology 259:20–33 [DOI] [PubMed] [Google Scholar]
- 6.Ace CI, McKee TA, Ryan JM, Cameron JM, Preston CM. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 63:2260–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinheimer SP, Boyd BA, Durham SK, Resnick JL, O'Boyle DR., II 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001 [DOI] [PubMed] [Google Scholar]
- 9.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, Reis e Sousa C. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397–408 [DOI] [PubMed] [Google Scholar]
- 10.Melchjorsen J, Rintahaka J, Soby S, Horan KA, Poltajainen A, Ostergaard L, Paludan SR, Matikainen S. 2010. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J. Virol. 84:11350–11358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507–518 [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Akira S. 2007. Antiviral signaling through pattern recognition receptors. J. Biochem. 141:137–145 [DOI] [PubMed] [Google Scholar]
- 13.Sarkar SN, Sen GC. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245–259 [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda K, Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658 [DOI] [PubMed] [Google Scholar]
- 16.Lin R, Mamane Y, Hiscott J. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin R, Genin P, Mamane Y, Hiscott J. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 20:6342–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J. Virol. 86:3528–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing J, Wang S, Lin F, Pan W, Hu CD, Zheng C. 2011. Comprehensive characterization of interaction complexes of herpes simplex virus type 1 ICP22, UL3, UL4, and UL20.5. J. Virol. 85:1881–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282:32208–32221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing J, Wu F, Pan W, Zheng C. 2010. Molecular anatomy of subcellular localization of HSV-1 tegument protein US11 in living cells. Virus Res. 153:71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrhardt C, Kardinal C, Wurzer WJ, Wolff T, von Eichel-Streiber C, Pleschka S, Planz O, Ludwig S. 2004. Rac1 and PAK1 are upstream of IKK-epsilon and TBK-1 in the viral activation of interferon regulatory factor-3. FEBS Lett. 567:230–238 [DOI] [PubMed] [Google Scholar]
- 23.Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, Servant MJ, Lin R, Meurs EF, Hiscott J. 2009. Ubiquitin-regulated recruitment of IκB kinase epsilon to the MAVS interferon signaling adapter. Mol. Cell. Biol. 29:3401–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao T, Yang L, Sun Q, Arguello M, Ballard DW, Hiscott J, Lin R. 2007. The NEMO adaptor bridges the nuclear factor-κB and interferon regulatory factor signaling pathways. Nat. Immunol. 8:592–600 [DOI] [PubMed] [Google Scholar]
- 25.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 26.Chang TH, Liao CL, Lin YL. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation. Microbes Infect. 8:157–171 [DOI] [PubMed] [Google Scholar]
- 27.Kochs G, Garcia-Sastre A, Martinez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severa M, Coccia EM, Fitzgerald KA. 2006. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 281:26188–26195 [DOI] [PubMed] [Google Scholar]
- 29.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKε molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 80:6072–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL. 2011. Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J. Virol. 85:11079–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan M, Schallhorn A, Wurm FM. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550 [DOI] [PubMed] [Google Scholar]
- 33.Greaves RF, O'Hare P. 1990. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding protein and target TAATGARAT sequences. J. Virol. 64:2716–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Gong W, Huang CC, Herr W, Cheng X. 1999. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 13:1692–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148–1151 [DOI] [PubMed] [Google Scholar]
- 36.Lin R, Heylbroeck C, Pitha PM, Hiscott J. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ, Jr, Barber GN, Hiscott J. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800–811 [DOI] [PubMed] [Google Scholar]
- 39.Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 95:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melroe GT, DeLuca NA, Knipe DM. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411–8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everett RD, Orr A. 2009. Herpes simplex virus type 1 regulatory protein ICP0 aids infection in cells with a preinduced interferon response but does not impede interferon-induced gene induction. J. Virol. 83:4978–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melchjorsen J, Siren J, Julkunen I, Paludan SR, Matikainen S. 2006. Induction of cytokine expression by herpes simplex virus in human monocyte-derived macrophages and dendritic cells is dependent on virus replication and is counteracted by ICP27 targeting NF-κB and IRF-3. J. Gen. Virol. 87:1099–1108 [DOI] [PubMed] [Google Scholar]
- 44.Melroe GT, Silva L, Schaffer PA, Knipe DM. 2007. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: potential role in blocking IFN-beta induction. Virology 360:305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paladino P, Collins SE, Mossman KL. 2010. Cellular localization of the herpes simplex virus ICP0 protein dictates its ability to block IRF3-mediated innate immune responses. PLoS One 5(4):e10428. 10.1371/journal.pone.0010428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elgadi MM, Hayes CE, Smiley JR. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153–7164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao XD, Rosenthal KL. 2011. Herpes simplex virus type 2 virion host shutoff protein suppresses innate dsRNA antiviral pathways in human vaginal epithelial cells. J. Gen. Virol. 92:1981–1993 [DOI] [PubMed] [Google Scholar]
- 48.Cotter CR, Kim WK, Nguyen ML, Yount JS, Lopez CB, Blaho JA, Moran TM. 2011. The virion host shutoff protein of herpes simplex virus 1 blocks the replication-independent activation of NF-κB in dendritic cells in the absence of type I interferon signaling. J. Virol. 85:12662–12672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verpooten D, Ma Y, Hou S, Yan Z, He B. 2009. Control of TANK-binding kinase 1-mediated signaling by the γ134.5 protein of herpes simplex virus 1. J. Biol. Chem. 284:1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson KE, Knipe DM. 2010. Herpes simplex virus-1 infection causes the secretion of a type I interferon-antagonizing protein and inhibits signaling at or before Jak-1 activation. Virology 396:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson KE, Song B, Knipe DM. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374:487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peri P, Mattila RK, Kantola H, Broberg E, Karttunen HS, Waris M, Vuorinen T, Hukkanen V. 2008. Herpes simplex virus type 1 Us3 gene deletion influences toll-like receptor responses in cultured monocytic cells. Virol. J. 5:140. 10.1186/1743-422X-5-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maldonado E, Hampsey M, Reinberg D. 1999. Repression: targeting the heart of the matter. Cell 99:455–458 [DOI] [PubMed] [Google Scholar]
- 54.Tyler JK, Kadonaga JT. 1999. The “dark side” of chromatin remodeling: repressive effects on transcription. Cell 99:443–446 [DOI] [PubMed] [Google Scholar]
- 55.Kung AL, Rebel VI, Bronson RT, Ch'ng LE, Sieff CA, Livingston DM, Yao TP. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272–277 [PMC free article] [PubMed] [Google Scholar]
- 56.Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T. 1999. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood 93:2771–2779 [PubMed] [Google Scholar]
- 57.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361–372 [DOI] [PubMed] [Google Scholar]
- 58.Suzuki T, Uchida-Toita M, Yoshida M. 1999. Tax protein of HTLV-1 inhibits CBP/p300-mediated transcription by interfering with recruitment of CBP/p300 onto DNA element of E-box or p53 binding site. Oncogene 18:4137–4143 [DOI] [PubMed] [Google Scholar]
- 59.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 60.Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. 2007. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3. J. Virol. 81:8282–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L. 2007. Binding of Kaposi's sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J. Virol. 81:10950–10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heylbroeck C, Balachandran S, Servant MJ, DeLuca C, Barber GN, Lin R, Hiscott J. 2000. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 74:3781–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paladino P, Mossman KL. 2009. Mechanisms employed by herpes simplex virus 1 to inhibit the interferon response. J. Interferon Cytokine Res. 29:599–607 [DOI] [PubMed] [Google Scholar]