Abstract
We identified a new subgroup of koala retrovirus (KoRV), named KoRV-J, which utilizes thiamine transport protein 1 as a receptor instead of the Pit-1 receptor used by KoRV (KoRV-A). By subgroup-specific PCR, KoRV-J and KoRV-A were detected in 67.5 and 100% of koalas originating from koalas from northern Australia, respectively. Altogether, our results indicate that the invasion of the koala population by KoRV-J may have occurred more recently than invasion by KoRV-A.
TEXT
When an exogenous retrovirus integrates into the genome of germ line cells, the provirus becomes permanently embedded into the host genome as an endogenous retrovirus (ERV), resulting in vertical transmission to offspring (1). Although most ERVs are replication defective due to mutations and deletions, endogenous koala retroviruses (KoRVs) are replication competent and can be horizontally transmitted as an exogenous virus. Tarlinton et al. reported that KoRV proviruses are absent in the genomes of some populations of koalas in southern Australia but present in those of koalas in northern Australia (2). These observations indicate that the invasion by KoRV into the koala genome occurred recently and suggest that KoRV is a unique retrovirus in the midst of becoming an endogenous retrovirus (2).
Koala retrovirus belongs to the genus Gammaretrovirus in the family Retroviridae, and it is closely related to Gibbon ape leukemia virus (GALV) (3, 4). Notably, incidences of leukemia and lymphoma, which are the most common neoplastic diseases caused by gammaretrovirus infections, are extremely high in koalas. It has been reported that 3 to 5% of wild koalas and up to 80% of captive koalas die due to these diseases in Australia (3, 5–8). Similarly, captive koalas in zoological parks in Japan suffer from leukemia and lymphoma at a relatively high rate (approximately 10%). A correlation between these neoplastic diseases and the plasma viral load of KoRV, the suspected causative agent, has been reported (6, 7). In addition, many koalas suffer from opportunistic infectious diseases, such as chlamydiosis and cryptococcosis, indicating that KoRV may also induce immunosuppression (3, 8–10). To clarify the relationship between KoRV and these diseases, it is necessary to accumulate more epidemiological and virological data.
KoRV infection is mediated by Pit-1, a sodium-dependent phosphate symporter that also functions as a receptor for feline leukemia virus (FeLV) subgroup B (FeLV-B) and GALV (11–13). Previously, we succeeded in isolating several KoRV strains (OJ-1 to OJ-5) from Queensland koalas reared in Kobe Municipal Oji Zoo (Hyogo, Japan) (14). By utilizing an interference assay, we found that one KoRV isolate (strain OJ-4) did not interfere completely with FeLV-B, indicating that the isolate may contain an additional gammaretrovirus or a KoRV variant that utilizes a different receptor than Pit-1 (data not shown). In this study, we identified a new KoRV subgroup in the isolate and found that this subgroup, named KoRV-J, uses thiamine transport protein 1 (THTR1), which is known to be a receptor for FeLV-A.
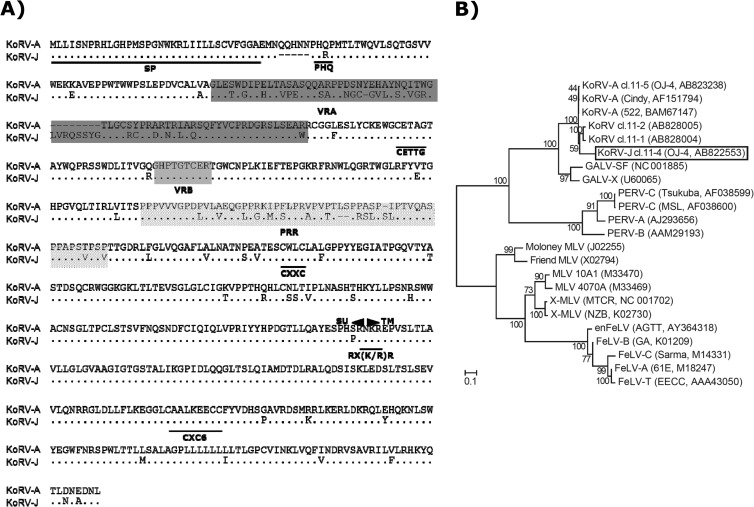
To investigate the genetic variability of the KoRV env of strain OJ-4 (14), full-length env was amplified by PCR using Pfu Ultra II (Stratagene) with a primer set corresponding to the 3′ terminus of pol and the 3′ untranslated region (GenBank accession number AF151794) (Table 1). Genomic DNA of HEK293T cells chronically infected with KoRV strain OJ-4 (14) was used as a PCR template. The PCR fragments were subcloned into pCR Blunt II TOPO (Invitrogen), and the resulting env clones were then digested for restriction enzyme mapping. The clones whose restriction patterns were different from that of the original KoRV env (pcindy) (3) were subsequently subjected to sequencing analysis. We identified four unique KoRV env sequences (clones 11-1, 11-2, 11-4, and 11-5). Two of the clones (clones 11-4 and 11-5) were functional in the pseudotype virus infection assay, described below. The nucleotide identity of clone 11-5 env with that of pcindy was 99.7% (data not shown). The amino acid sequence alignment of pcindy (KoRV subgroup A [KoRV-A]) and clone 11-4 (KoRV-J) is shown in Fig. 1A. The identity of amino acid sequences between the two subgroups was 87.6%, and the variability was mainly found in variable region A (VRA), which is the major determinant of receptor usage (17), and the proline-rich region (PRR) of the envelope glycoprotein (Env). The CETTG motif, which is involved in fusion activity (18), was disrupted by mutations in both KoRV-A and KoRV-J. KoRV-J had a replacement of Gln with Arg in the PHQ motif, which is important for the fusion activity (19–22). Other important motifs that influence the functions of Env and/or the viral infectivity were highly conserved between the two subgroups.
Table 1.
Primers used in this study
| Assay | Target | Orientation | Sequence | Position (nucleotides) | Amplicon size (bp) |
|---|---|---|---|---|---|
| Cloning | KoRV env | Forward | 5′-ATGCTTCTCATCTCAAACCC-3′ | 1–20 | 1,983 |
| Reverse | 5′-TTAAAGGTTATCGGCGTTGT-3′ | 1964–1983 | |||
| Differential PCR | KoRV pol | Forward | 5′-CCTTGGACCACCAAGAGACTTTTGA-3′ | 2710–2734 | 1,051 |
| Reverse | 5′- TCAAATCTTGGACTGGCCGA-3′ | 3741–3760 | |||
| KoRV-A | Forward | 5′-TATGAACATGCTTATAATCAGATCAC-3′ | 6242–6267 | 723 | |
| Reverse | 5′-ATAATGACTTGCCCTGGAGTTGCG-3′ | 6940–6964 | |||
| KoRV-J | Forward | 5′-AAATGGCTGTGGGGTGCTTTATAGTCAG-3′ | 303–330 | 547 | |
| Reverse | 5′-TAGGGAGCGTGGGGTAGTGACTGGTG-3′ | 824–849 | |||
| Real-time PCR | KoRV-A | Forward | 5′-AACTATGAACATGCTTATAATCAGATCACTT-3′ | 6239–6269 | 101 |
| Reverse | 5′-GGGCACACGTAGAACTGGGA-3′ | 6320–6339 | |||
| KoRV-J | Forward | 5′-TGCTTTATAGTCAGGTCGGCAG-3′ | 317–338 | 101 | |
| Reverse | 5′-CTGAGATTGAGCCAGTCTGTTCC-3′ | 395–417 | |||
| Koala β-actin | Forward | 5′-GAGACCTTCAACACCCCAGC-3′ | 373–392 | 111 | |
| Reverse | 5′-GTGGGTCACACCATCACCAG-3′ | 464–483 |
Fig 1.
Genetic analysis of KoRV-A and KoRV-J. (A) Comparison of amino acid sequences of KoRV subgroups A and J. Dots indicate the same amino acids. VRA, VRB, and PRR are shaded in gray. Abbreviations: SP, signal peptide; PHQ, PHQ motif; CETTG, CETTG motif; CXXC, CXXC motif; SU, surface unit; TM, transmembrane; RX(K/R)R, furin cleavage site; CXC6, CXC6 motif. (B) Maximum likelihood tree of the entire amino acid sequences of env genes of KoRV isolates and other gammaretroviruses. Numbers at the nodes indicate the percentage of rapid bootstrap values (1,000 replicates). Amino acid sequences used for the analyses were retrieved from the GenBank database, and the accession numbers are shown in parentheses. Abbreviations: X-MLV, xenotropic MLV; enFeLV, endogenous FeLV.
Phylogenetic analysis of env using the maximum likelihood approach (23) revealed that KoRV isolates (Cindy [3], 522 [24], and KoRV-J [clone 11-4]) and GALV clustered together, but they were distinct from the cluster that consists of FeLVs, murine leukemia viruses (MLVs), and porcine endogenous retroviruses (PERVs) (Fig. 1B). A similar topology of the phylogenetic tree was obtained using the transmembrane region of env (data not shown). KoRVs and GALVs are distantly related to PERVs. Similarities of the Env amino acids among the KoRV-A isolates (Cindy, 522, and OJ-4 [clone 11-5]) were shown to be high, and the degree of diversity between KoRV-A and KoRV-J was less than those of the FeLV and PERV subgroups (data not shown).
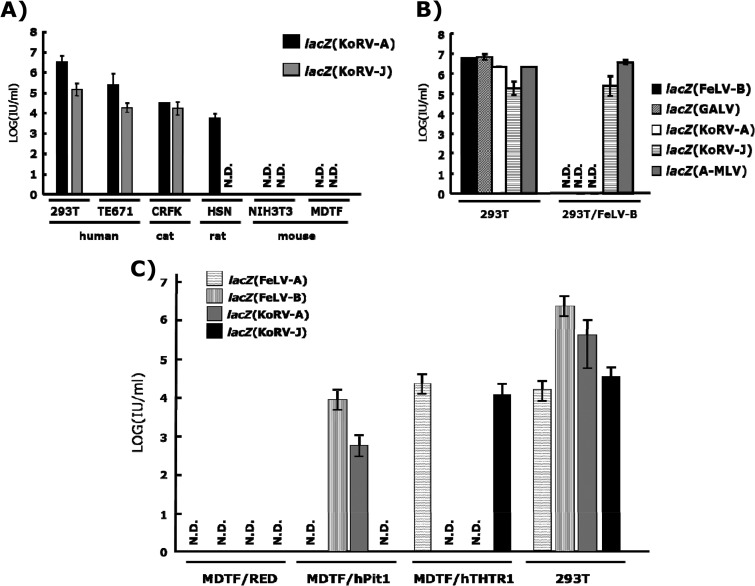
We next performed a LacZ pseudotype assay to characterize biological properties of KoRV-J Env. The env expression plasmids, termed pFBFeLV-A (25), pFBFeLV-B (26), and pFBASALF (16), were used for the expression of Envs of FeLV-A (strain Glasgow-1), FeLV-B (strain Gardner-Arnstein), and amphotropic MLV (A-MLV, strain 4070), respectively. KoRV-A (clone 11-5), KoRV-J (clone 11-4), and GALV env genes were cloned into pFB envelope expression plasmids to produce pFBKoRV-A, pFBKoRV-J, and pFBGALV, respectively (25). The GALV env was cloned from the genomic DNA of TELCeB/GAF cells (26). These env expression plasmids were transfected into TELCeB6 cells, which express large amounts of MLV core particles, and an MFGnlsLacZ vector to produce pseudotype viruses (16). After selection of the transfectants with 50 μg/ml of phleomycin, virus supernatants were harvested from the pooled phleomycin-resistant cell populations, filtered through 800-nm Millipore filters, and used immediately in LacZ pseudotype assays. Titration of LacZ pseudotypes was performed as described previously (27). The LacZ pseudotype virus bearing the KoRV-J Env [lacZ(KoRV-J)] could infect HEK293T cells, although the titer was approximately 10-fold lower than that for lacZ(KoRV-A) (Fig. 2A). These data suggest that the KoRV-J Env is functional. Interestingly, substitutions in the PHQ motif and deletions in the vicinity of the motif did not influence the fusion activity.
Fig 2.
Receptor usage of KoRV-A and KoRV-J. (A) Host ranges of KoRV subgroups. lacZ pseudotypes were inoculated into the indicated cells, and titers of the viruses were determined in a LacZ assay. Data were obtained from three independent experiments, and the values are expressed as means ± 1 standard deviation (SD). N.D., not detected. (B) Interference assay of KoRV subgroups. lacZ pseudotypes were inoculated onto naive HEK293T cells and HEK293T cells chronically infected with FeLV-B strain Gardner-Arnstein, and titers of the viruses were determined in a LacZ assay. Data were obtained from three independent experiments, and the values are expressed as means ± 1 SD. (C) Infectivity of KoRV subgroups in MDTF cells expressing human Pit-1 (MDTF/hPit1) and human THTR1 (MDTF/hTHTR1). cDNAs of human Pit1 and THTR1 were cloned into a RetroQDsRed monomer vector (Clontech) to produce pRetro-hTHTR-DsRed. The resultant plasmid was transfected into MDTF cells, selected by puromycin, and subjected to a lacZ pseudotype infection assay. lacZ pseudotypes were inoculated onto MDTF/hPit1, MDTF/hTHTR1, and MDTF cells transfected with an empty expression vector (MDTF/RED) and HEK293T cells. Titers of the viruses were determined in a LacZ assay. Data were obtained from three independent experiments, and the values are expressed as means ± 1 SD.
We then examined the host cell ranges of KoRV-A and -J by using target cells derived from human (HEK293T and TE671), cat (CRFK), rat (HSN), and mouse (NIH 3T3 and MDTF) cells (Fig. 2A). Consistent with previous reports (11, 24, 28), lacZ(KoRV-A) showed the same host cell range as FeLV-B and GALV, which utilize Pit-1 as a receptor. On the other hand, lacZ(KoRV-J) could not infect rat HSN cells (Fig. 2A) but efficiently infected HEK293T cells chronically infected with FeLV-B (Fig. 2B). In addition, the stable expression of human Pit-1 rendered MDTF cells susceptible to lacZ(KoRV-A), but not to lacZ(KoRV-J) (Fig. 2C). These data clearly show that KoRV-J utilizes an unidentified receptor distinct from Pit-1. To further characterize the receptor usage of KoRV-J, we conducted a receptor interference assay using six gammaretroviruses that utilize different receptors, namely, FeLV-A, -B, or -C, RD-114 virus, xenotropic murine leukemia virus (X-MLV), and A-MLV (Table 2). We found that lacZ(KoRV-J) interfered with FeLV-A on FEA cells (feline fibroblasts). The receptor for FeLV-A is known to be THTR1 (29, 30). As shown in Fig. 2C, lacZ(KoRV-J) and lacZ(FeLV-A) infected MDTF cells expressing human THTR1 (hTHTR1) but not naive MDTF cells. These data indicate that KoRV-J utilizes THTR1 as a receptor.
Table 2.
Interference between gammaretroviruses
| Virus | Receptor interference between the two viruses in the indicated cell typea |
||||||
|---|---|---|---|---|---|---|---|
| FeLV-A in FEA | FeLV-B in HEK293T | FeLV-C in HEK293T | A-MLV in TE671 | X-MLV in TE671 | RD-114 in TE671 | KoRV OJ-4 in 293T | |
| lacZ(KoRV-A) | − | + | − | − | − | − | + |
| lacZ(KoRV-J) | + | − | − | − | − | − | + |
| lacZ(FeLV-A) | + | − | − | − | − | − | + |
| lacZ(FeLV-B) | − | + | − | − | − | − | + |
+, interference observed; −, no interference.
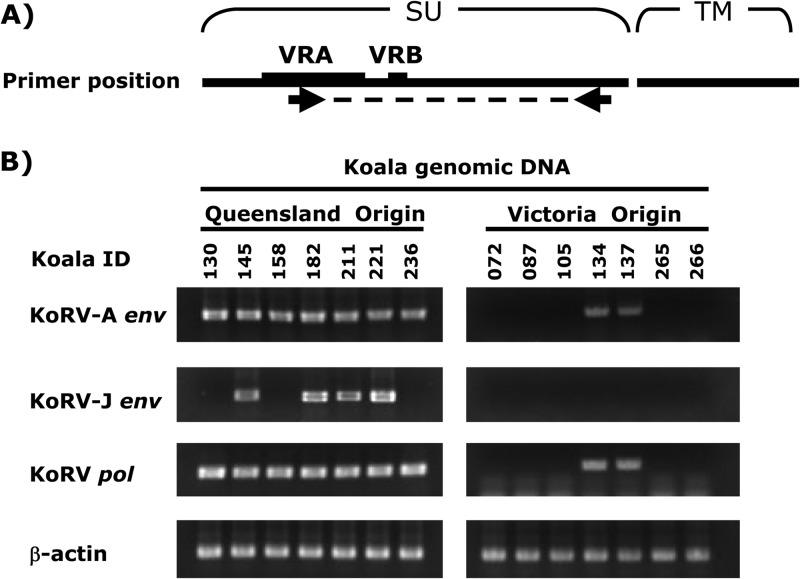
To investigate the prevalence of KoRV subgroups in koalas reared in Japanese zoos, we collected heparinized blood samples from 40 northern koalas (Queensland, New South Wales, and hybrids of Queensland and New South Wales koalas) and 11 Victorian koalas from 9 zoological parks in Japan and performed differential PCR analysis using subgroup-specific primer sets. The forward primer specific to the env of each subgroup and the reverse primer common to all subgroups were used (Fig. 3A; Table 1). The pol gene was detected by a pol-long primer set (6) as a positive control for KoRV infection, and the β-actin primer set was used for the quality check of the genomic DNAs. These genes were amplified by using Ex Taq DNA polymerase (TaKaRa, Ohtsu, Shiga, Japan) under the following cycling conditions: 1 cycle of denaturing (2 min at 94°C), followed by 30 cycles of chain reaction (30 s at 94°C, 30 s at 60°C, and 1 min at 72°C), and an additional extension (4 min at 72°C). KoRV-A was detected in all northern koalas tested and in 4 out of 11 Victorian koalas (Fig. 3B and Table 3), consistent with previous reports that KoRV had become endogenous in koalas in northern Australia (6, 7, 9, 10). In contrast, KoRV-J was detected in 67.5% of northern koalas but not in southern (Victorian) koalas (Fig. 3B and Table 3). These data indicate that the prevalence of KoRV-J is more limited than KoRV-A, and the invasion of KoRV-J into the koala population may have occurred more recently than KoRV-A.
Fig 3.
Prevalence of KoRV subgroups in captive koalas in Japanese zoos. (A) Primer positions. Forward and reverse primers were designed in the VRA and the C terminus of surface unit of Env, respectively (see Table 1). Abbreviations: SU, surface unit; TM, transmembrane; VRB, variable region B. (B) Infection status of KoRV subtypes in koalas in Japanese zoos. Subgroup-specific differential PCR was conducted as described in the text. Representative data are shown.
Table 3.
Prevalence of KoRV subgroups in koalas in Japanese zoos
| Source of animalsa | Infection rateb (%) |
||
|---|---|---|---|
| KoRV-pol | KoRV-A | KoRV-J | |
| Northern koalas | 40/40 (100) | 40/40 (100) | 27/40 (67.5) |
| QLD | 31/31 (100) | 31/31 (100) | 20/31 (64.5) |
| QLD × NSW hybrid | 3/3 (100) | 3/3 (100) | 2/3 (66.7) |
| NSW | 6/6 (100) | 6/6 (100) | 5/6 (83.3) |
| Southern koalas | |||
| VIC | 4/11 (36.4) | 4/11 (36.4) | 0/11 (0) |
| Total | 44/51 (86.3) | 44/51 (86.3) | 27/51 (52.9) |
Animals were obtained from northern or southern Australia. QLD, Queensland; NSW, New South Wales; VIC, Victoria.
The infection rate is the number of positive animals per the total number tested.
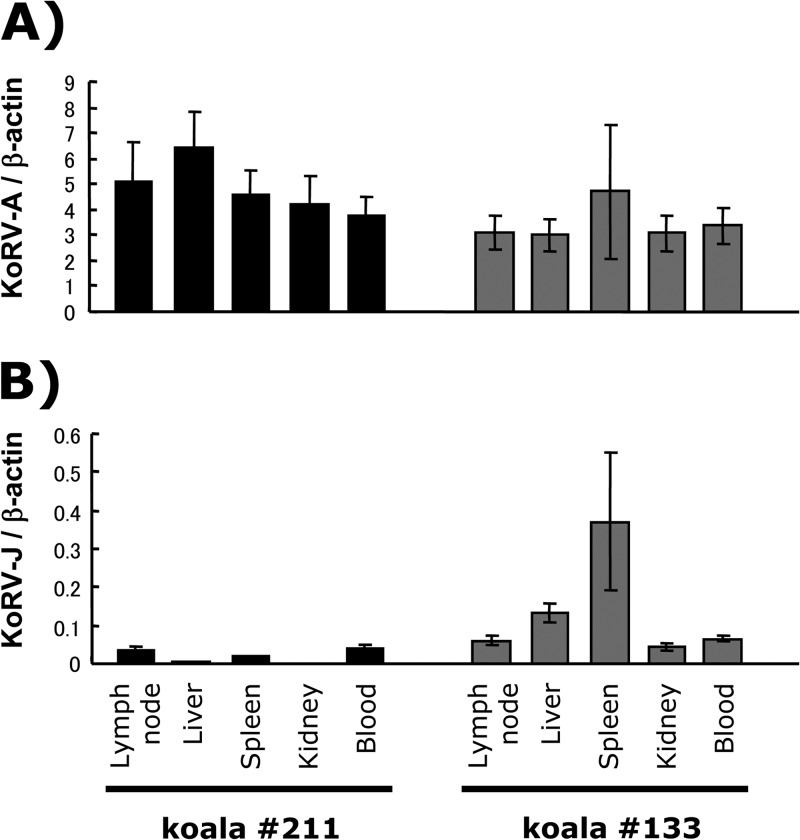
To determine whether KoRV-J is exogenous or endogenous, we determined the copy numbers of each subgroup in the genomes of different tissues in individual animals. Copy numbers of each subgroup in tissues of Queensland koalas numbers 211 and 133 (KoRV-A positive, KoRV-B positive) were measured by quantitative real-time PCR. Quantitative real-time PCR was performed using the SYBR green detection system (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Aliquots (100 ng) of each genomic DNA sample extracted from lymph node, liver, spleen, and whole blood of the koalas were used as PCR templates with primers specific to KoRV-A, KoRV-J, and the β-actin gene of koalas (Table 1). Amplification and signal detection were carried out using an ABI 7000 real-time PCR system (Applied Biosystems). The standard curves were generated by using serially diluted pFBKoRV-A, pFBKoRV-J, or a plasmid containing a partial sequence of the koala β-actin gene. Copy numbers of each subgroup per cell were normalized to the copy numbers of the β-actin gene. Approximately 3 to 6 copies of KoRV-A per cell were present in the tissues tested (Fig. 4A). In contrast to the relatively constant copy numbers of KoRV-A among tissues, the copy numbers of KoRV-J were less than 1 copy per cell and varied in tissues in both koalas (Fig. 4B). These data suggest that KoRV-J is not an ERV, at least not in these two koalas.
Fig 4.
Real-time PCR to detect KoRV-A (A) and KoRV-J (B) env genes. Genomic DNA samples from several tissues and blood were isolated from koalas numbers 211 and 133 and subjected to real-time PCR. Data were obtained from three independent experiments, and the values are expressed as the mean ± 1 standard deviation.
In this study, we identified a new subgroup of KoRV, named KoRV-J, which utilizes THTR1 as a receptor for entry into target cells. KoRV-A has become endogenous in northern koalas but not in most southern koalas in Australia (2, 8, 10). In koalas reared in Japanese zoos, KoRV-J was found only in northern koalas originating from Queensland and New South Wales (Table 1). Therefore, it is plausible that a KoRV-J-infected northern koala(s) was introduced into a Japanese zoo(s) rather than the virus being derived from other animals within the facilities, especially given that koalas are kept separately from other animals except humans.
The origin of KoRV-J is unknown at present. The low amino acid similarity on the surface of Env was not simply caused by nucleotide insertions and/or deletions (data not shown). Furthermore, it is unlikely that KoRV-J was generated from KoRV-A due to an accumulation of nucleotide mutations. KoRV-J could have been prevalent in an unknown host species in Australia that infected a population of northern koalas quite recently. It is also possible that KoRV-J may be the result of a recombination event between KoRV-A and another KoRV-related gammaretrovirus. Thus far, we have been unable to find any KoRV-J VRA-like sequences in the NCBI nr/nt database, indicating that further studies are needed to elucidate the origin of the virus.
Different receptor usage of KoRV subgroups may explain the wide range of diseases seen in koalas. In FeLV, different pathogenicities of FeLV subgroups have been identified (31). The commonly isolated FeLV-A viruses are generally of low to moderate pathogenicity, but FeLV-B viruses are more common in leukemic cats than in FeLV-A-infected healthy cats, and FeLV-B may contribute to the leukemogenic process. It is interesting that megaloblastic anemia has been linked with mutations in THTR1 in humans (32). KoRV-J may disrupt thiamine transport function and cause anemia, which is seen in some captive koalas in Japan. Because the number of koalas is limited in Japan, it is difficult to draw any conclusions regarding differences in the pathogenicities between KoRV-A and -J at this stage. Further long-term surveillance is needed.
Nucleotide sequence accession number.
The sequences of the KoRV env variants in this study were deposited in GenBank and assigned accession numbers AB822553 (clone 11-4), AB823238 (clone 11-5), AB828004 (clone 11-1), and AB828005 (clone 11-2).
ACKNOWLEDGMENTS
We are grateful to John Hunger (RSPCA, Brisbane, Queensland, Australia) for providing pcindy. We thank Hisashi Hashikawa and Masami Kurobe (koala specialists for the Japanese Association of Zoos and Aquariums) for helpful advice and the staffs at Japanese zoos for the collection of koala samples. We are grateful to Paul Young and Joanne Meers (University of Queensland, Brisbane, Queensland, Australia) for helpful discussions. We thank Peter Gee (Kyoto University) for his generous help in the preparation of the manuscript. We are grateful to Masakazu Hattori (Kyoto University, Kyoto, Japan) for providing recombinant human interleukin 2-producing Ltk− IL-2.23 cells.
This study was supported by grants from the Ministry of Education, Culture, Science and Sports of Japan and from the Bio-Oriented Technology Research Advancement Institution.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Gifford R, Tristem M. 2003. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315 [DOI] [PubMed] [Google Scholar]
- 2.Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442:79–81 [DOI] [PubMed] [Google Scholar]
- 3.Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. 2000. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J. Virol. 74:4264–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin J, Herniou E, Cook J, O'Neill RW, Tristem M. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canfield PJ, Sabine JM, Love DN. 1988. Virus particles associated with leukaemia in a koala. Aust. Vet. J. 65:327–328 [DOI] [PubMed] [Google Scholar]
- 6.Tarlinton R, Meers J, Hanger J, Young P. 2005. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J. Gen. Virol. 86:783–787 [DOI] [PubMed] [Google Scholar]
- 7.Tarlinton R, Meers J, Young P. 2008. Biology and evolution of the endogenous koala retrovirus. Cell. Mol. Life Sci. 65:3413–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown AS, Girjes AA, Lavin MF, Timms P, Woolcock JB. 1987. Chlamydial disease in koalas. Aust. Vet. J. 64:346–350 [DOI] [PubMed] [Google Scholar]
- 9.Simmons G, Young P, McKee J, Meers J. 2011. The epidemiology of koala retrovirus. J. Vet. Epidemiol. 15:1–9 [Google Scholar]
- 10.Simmons GS, Young PR, Hanger JJ, Jones K, Clarke D, McKee JJ, Meers J. 2012. Prevalence of koala retrovirus in geographically diverse populations in Australia. Aust. Vet. J. 90:404–409 [DOI] [PubMed] [Google Scholar]
- 11.Oliveira NM, Farrell KB, Eiden MV. 2006. In vitro characterization of a koala retrovirus. J. Virol. 80:3104–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi Y, Vile RG, Simpson G, O'Hara B, Collins MK, Weiss RA. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Hara B, Johann SV, Klinger HP, Blair DG, Rubinson H, Dunn KJ, Sass P, Vitek SM, Robins T. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119–127 [PubMed] [Google Scholar]
- 14.Miyazawa T, Shojima T, Yoshikawa R, Ohata T. 2011. Isolation of koala retroviruses from koalas in Japan. J. Vet. Med. Sci. 73:65–70 [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Cosset FL, Takeuchi Y, Battini JL, Weiss RA, Collins MK. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430–7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battini JL, Danos O, Heard JM. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. 2007. Changes in viral protein function that accompany retroviral endogenization. Proc. Natl. Acad. Sci. U. S. A. 104:17506–17511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bae Y, Kingsman SM, Kingsman AJ. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnett AL, Davey RA, Cunningham JM. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. U. S. A. 98:4113–4118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavillette D, Kabat D. 2004. Porcine endogenous retroviruses infect cells lacking cognate receptors by an alternative pathway: implications for retrovirus evolution and xenotransplantation. J. Virol. 78:8868–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette D, Ruggieri A, Russell SJ, Cosset FL. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 24.Shojima T, Hoshino S, Abe M, Yasuda J, Shogen H, Kobayashi T, Miyazawa T. 2013. Construction and characterization of an infectious molecular clone of koala retrovirus. J. Virol. 87:5081–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakata R, Miyazawa T, Shin YS, Watanabe R, Mikami T, Matsuura Y. 2003. Reevaluation of host ranges of feline leukemia virus subgroups. Microbes Infect. 5:947–950 [DOI] [PubMed] [Google Scholar]
- 26.Porter CD, Collins MK, Tailor CS, Parkar MH, Cosset FL, Weiss RA, Takeuchi Y. 1996. Comparison of efficiency of infection of human gene therapy target cells via four different retroviral receptors. Hum. Gene Ther. 7:913–919 [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986–9991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J. 2006. Transspecies transmission of the endogenous koala retrovirus. J. Virol. 80:5651–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendoza R, Anderson MM, Overbaugh J. 2006. A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J. Virol. 80:3378–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendoza R, Miller AD, Overbaugh J. 2013. Disruption of thiamine uptake and growth of cells by feline leukemia virus subgroup A. J. Virol. 87:2412–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarrett O. 1992. Pathogenicity of feline leukemia virus is commonly associated with variant viruses. Leukemia. 6(Suppl 3):153S–154S [PubMed] [Google Scholar]
- 32.Diaz GA, Banikazemi M, Oishi K, Desnick RJ, Gelb BD. 1999. Mutations in a new gene encoding a thiamine transporter cause thiamine-responsive megaloblastic anaemia syndrome. Nat. Genet. 22:309–312 [DOI] [PubMed] [Google Scholar]